Abstract
Background and Objective
Postexercise heart rate recovery (HRR) is an important indicator of cardiac autonomic function and abnormal HRR is associated with adverse outcomes. We hypothesized that deep learning on resting electrocardiogram (ECG) tracings may identify individuals with impaired HRR.
Methods
We trained a deep learning model (convolutional neural network) to infer HRR based on resting ECG waveforms (HRRpred) among UK Biobank participants who had undergone exercise testing. We examined the association of HRRpred with incident cardiovascular disease using Cox models, and investigated the genetic architecture of HRRpred in genome-wide association analysis.
Results
Among 56,793 individuals (mean age 57 years, 51% women), the HRRpred model was moderately correlated with actual HRR (r = 0.48, 95% confidence interval [CI] 0.47–0.48). Over a median follow-up of 10 years, we observed 2060 incident diabetes mellitus (DM) events, 862 heart failure events, and 2065 deaths. Higher HRRpred was associated with lower risk of DM (hazard ratio [HR] 0.79 per 1 standard deviation change, 95% CI 0.76–0.83), heart failure (HR 0.89, 95% CI 0.83–0.95), and death (HR 0.83, 95% CI 0.79–0.86). After accounting for resting heart rate, the association of HRRpred with incident DM and all-cause mortality were similar. Genetic determinants of HRRpred included known heart rate, cardiac conduction system, cardiomyopathy, and metabolic trait loci.
Conclusion
Deep learning–derived estimates of HRR using resting ECG independently associated with future clinical outcomes, including new-onset DM and all-cause mortality. Inferring postexercise heart rate response from a resting ECG may have potential clinical implications and impact on preventive strategies warrants future study.
Keywords: Electrocardiogram, Risk factor, Diabetes mellitus, Heart failure, Machine learning
Key Findings.
-
•
A deep learning model of resting electrocardiogram (ECG) tracings can estimate heart rate recovery (HRR), an important measure of cardiac autonomic dysfunction that usually requires exercise provocation testing to ascertain.
-
•
Deep learning–derived estimates of HRR using resting ECG independently associated with future clinical outcomes, including new-onset diabetes mellitus and all-cause mortality. Specifically, individuals with impaired HRRpred had a more than 80% greater risk of future diabetes and 27% greater risk of death even after accounting for clinical risk factors including resting heart rate.
-
•
Artificial intelligence–enhanced interpretation of a resting ECG may serve as a proxy for HRR and lend insight into the association of cardiac autonomic dysfunction with clinical outcomes. The ability to infer postexercise heart rate response using widely scalable methods from a resting ECG harbors potential clinical implications, although utility with respect to potential screening, identification of at-risk individuals, and impact on preventive strategies will require further study.
Introduction
Cardiac autonomic dysfunction including sympathetic overactivation and parasympathetic withdrawal has important clinical consequences, and can occur in the setting of diabetes mellitus with primary autonomic failure or as a result of cardiovascular disease (CVD), including heart failure and ischemic heart disease. The ascertainment of autonomic dysfunction is challenging, and traditional metrics have required either 24-hour Holter electrocardiogram (ECG) monitoring, tilt table testing, or exercise provocation that may shed light on specific aspects of cardiac autonomic control.1 One such metric that is ascertained with clinical exercise testing is heart rate recovery (HRR), which is the dynamic decline in heart rate after an acute bout of exercise and reflects both parasympathetic reactivation and sympathetic withdrawal in the recovery period.2 Cardiac autonomic dysfunction as measured by abnormal HRR has been independently associated with all-cause mortality across healthy individuals, as well as patients with diabetes mellitus and heart failure.3, 4, 5, 6, 7 Importantly, the clinical implications of abnormal HRR appear independent of resting heart rate, clinical risk factors, and beta-blocker use. Further, initial studies have shown that HRR is modifiable with exercise training.8,9
Recent studies have demonstrated the potential clinical applicability of artificial intelligence–enhanced interpretation of ECGs, including deep learning–based ECG phenotyping that may enhance detection of CVD beyond clinical ECG interpretation.10 Because HRR is a clinically relevant measure of autonomic dysfunction but requires exercise testing that may not be immediately available at scale or feasible in all individuals, we investigated whether deep learning on widely available resting ECG tracings may identify individuals with impaired HRR. Further, we sought to examine whether predicted HRR from a resting ECG would be associated with clinical outcomes including incident diabetes mellitus, CVD, and all-cause mortality. Additionally, we sought to examine the genetic architecture of predicted HRR based on our resting ECG deep learning model, since prior studies have indicated that HRR is heritable.11 We used a unique resource of genome-wide genotyping to perform a genome-wide association study of predicted HRR in order to identify potential genetic and biological pathways regulating cardiac autonomic function.
Methods
Study sample
The UK Biobank is a cohort study of 503,325 adults aged 40–69 years recruited between 2006 and 2010.12 Of this sample, a total of 96,567 eligible participants underwent exercise testing with ECG monitoring at the baseline examination and 75,766 had available full-disclosure ECG data for this analysis, including pretest resting ECG tracings. We excluded individuals who withdrew consent (n = 6), those without exercise (n = 2306), those missing full-length pretest/rest phase data (n=10,163), those with nonphysiologic HRR (n = 615), those with ECG artifact (n = 5637), and those missing key clinical covariates (n = 246), leaving 56,793 participants for analysis (Supplemental Figure 1). For diabetes-related analyses, an additional 8693 individuals missing fasting glucose values were excluded. All UK Biobank participants provided electronic signed consent at recruitment, and the study protocol was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). Use of data (under UKB applications 7089 and 28807) for the current study was approved by the Mass General Brigham Institutional Review Board.
Exercise testing
Exercise testing was performed using an upright cycle ergometer (eBike, Firmware v1.7, GE Healthcare, Amersham, United Kingdom) among 76,146 participants.13 In brief, participants underwent assessment of risk factors and an exercise protocol was selected based on predicted absolute maximum workload. Each protocol consisted of 2 minutes of loaded exercise at constant power, followed by incremental ramp over 4 minutes to peak power. Participants were instructed to pedal at 60 revolutions per minute with total exercise duration of 6 minutes. ECG leads were applied over bilateral antecubital fossa and wrists prior to the start of exercise with continuous acquisition 4-lead ECG monitoring starting 15 seconds before exercise until 1 minute into recovery (CAM-USB 6.5, Cardiosoft v6.51, GE Healthcare, Amersham, United Kingdom, and Activwave Cardio device, CamNTech, Papworth, United Kingdom). Continuous ECG data were recorded at 500 Hz and stored in XML files for analysis.
ECG waveform processing
Three-lead ECG tracings obtained at the time of exercise testing were downloaded in XML format from the UK Biobank. In order to calculate the resting, peak, and recovery heart rate, 3 segments were extracted from each tracing (Supplemental Figure 2): (1) pretest: the first 15 seconds of the ECG before exercise; (2) peak: starting 5 seconds before the beginning of the recovery phase and ending 5 seconds after the start of the recovery phase; and (3) recovery: starting 50 seconds after the beginning of the peak trace and ending 50 seconds after the end of the peak trace.
The UK Biobank provided irregularly sampled heart rate measurements throughout the waveform. In order to get heart rate measurements at specific times, a Python library, BioSPPy, was used to calculate pretest, peak, and recovery heart from raw ECG tracings by taking the median heart rate among the 3 leads.14 Using BioSPPy also enabled comparison of heart measurements across the leads as a quality control measure. HRR was defined as the peak minus the recovery heart rate.
Deep learning model to predict heart rate recovery
To test whether HRR could be predicted from a resting ECG tracing, we used convolutional neural networks (CNN), a type of neural network architecture specialized for learning from spatially or temporally structured data (eg, images and time series). CNNs were trained to regress HRR on traces from the pretest resting phase of the exercise tests. To obtain model predictions on the entire cohort, the cohort was split into 5 folds with nonoverlapping test sets with 20% of the ECGs. The remaining 80% of each fold was then split into 70% for training and 10% for validation. A 1-dimensional CNN based on the DenseNet architecture was trained on each training split.15 DenseNet is a type of CNN with many skip connections, which have been shown to facilitate model training using typical backpropagation-based optimizers.16 The input to the CNNs were 10-second segments of lead I of the 15 seconds of pretest trace. Each time a trace was input into the CNN during training, a random contiguous 10 seconds of the 15-second trace was selected, with the goal of aiding the model to learn invariance to the phase of the ECG signal. The CNNs were trained to predict HRR standardized by the split’s training sample mean and standard deviation. The loss function was the log-cosh. Each CNN was trained with the Adam optimizer with learning rate 0.001 decayed by a factor of 10 2 times when the validation loss failed to improve for 20 epochs of the training data until early stopping.17 The CNNs used the swish activation and were trained with a spatial dropout rate of 10% on the convolutional layers and a dropout rate of 50% on the final fully connected layer.18, 19, 20 The optimizer settings, architecture, and the swish activation were picked using performance on each fold’s validation set (details in Supplemental Table 1).
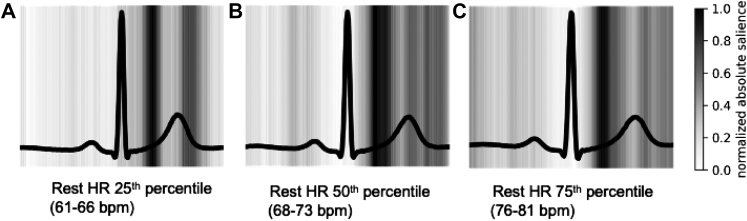
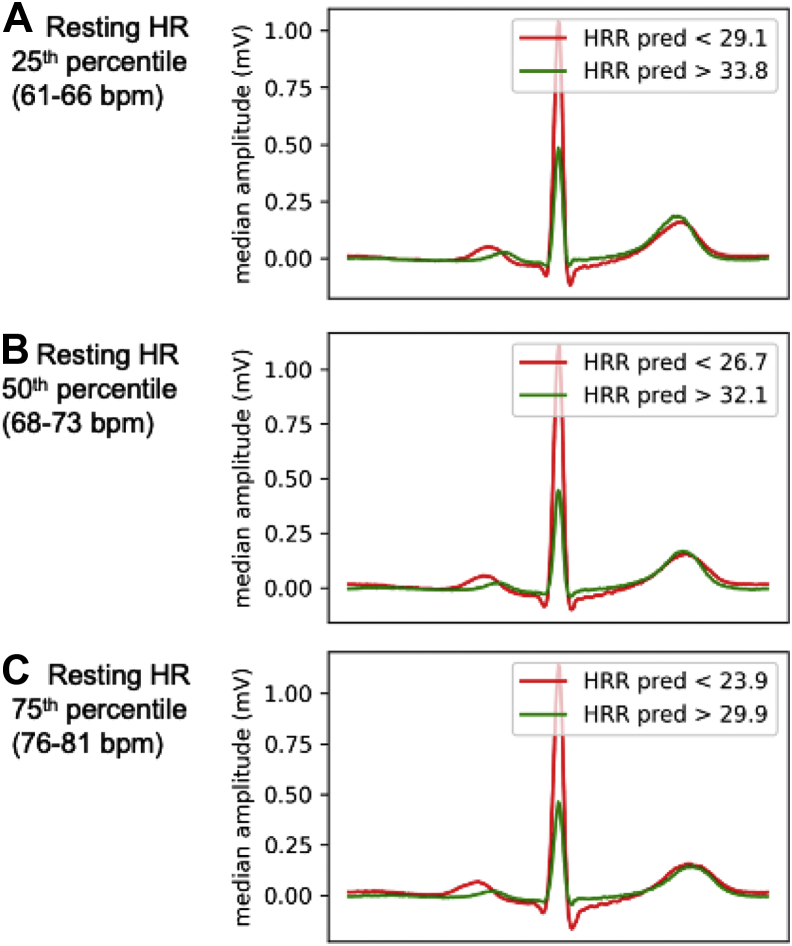
The features learned by the CNNs were visualized using 2 methods. First, we examined saliency maps to demarcate areas of the ECG waveform with greatest influence on the CNN HRR predictions. Second, we graphed median ECG waveforms for participants with high predicted HRR (HRRpred) and with low HRRpred. In order to eliminate potential effects of resting heart rate, ECG visualizations were stratified by resting heart rate (within 5 beats per minute [bpm] range of the 25th, 50th, and 75th percentiles of resting heart rate). Within each group, we visualized the median ECG waveform of participants in the 5th and 95th percentile of HRRpred to examine what ECG features were most relevant to the deep learning model.
Clinical endpoints
Participants were followed longitudinally for the occurrence of clinical outcomes of interest, including incident diabetes mellitus, congestive heart failure, CVD (defined as myocardial infarction, heart failure, or stroke), and all-cause mortality. Outcomes were defined using combinations of self-report, inpatient International Classification of Diseases, 9th and 10th revision, and death registry information as previously outlined.21 Start of follow-up was defined as the date of exercise testing, and was censored at the time of last available linked hospital data. Prevalent disease was excluded for incident event-specific analyses.
Genome-wide association study
To examine genetic correlates of HRR predicted from resting ECG, we performed a genome-wide association study (GWAS) among 43,722 individuals of European ancestry with genetic information available. As described previously, UK Biobank samples underwent genotyping using either the UK BiLEVE or UK Biobank Axiom arrays, with imputation using the Haplotype Reference Consortium panel and UK10K+1000 Genomes panel.22 We used BOLT-REML v2.3.4 to assess heritability of HRRpred and BOLT-LMM23 to examine the association of single nucleotide polymorphisms (SNPs) with HRRpred in analyses adjusted for age, sex, array, and the first 5 principal components of genetic ancestry. Results were deemed genome-wide significant at P = 5 × 10-8.
To determine independently significant SNPs, we performed pruning and thresholding using Plink v1.90 (–clump-kb 5000 –clump-r2 0.01 –clump-p1 5e-8) with linkage disequilibrium based on the hard-called variants in our study sample (n = 43,722). We constructed a polygenic risk score from the 8 identified independent SNPs meeting genome-wide significance in the HRRpred GWAS with the score being the sum of effect size × count of effect alleles. We examined the association with incident disease outcomes among 21,865 individuals after excluding individuals with available HRR data in order to specify a discrete “validation” sample. Lastly, we constructed polygenic risk scores of HRR from previously published studies to examine associations with HRRpred.24,25
Statistical analysis
Clinical characteristics of our sample were summarized by HRRpred tertile. In order to assess HRRpred model fit, we plotted actual vs predicted HRR and examined Pearson correlation coefficients, mean error, and R2 in the out-of-sample test set. To quantify how much of the HRR variance could be explained by HRRpred and other clinical covariates such as resting heart rate, we used a linear regression model. To compare the CNN model to prediction of HRR using resting heart rate, we constructed a random forest model and compared R2 coefficients via 5-fold cross-validation. In secondary analyses, we defined impaired HRR as the lowest tertile of HRR in the test set (recognizing there is no clinically accepted cut point to define impaired HRR) and examined HRRpred as a predictor of impaired HRR using multivariable logistic regression, with adjustment for age and sex. We calculated the % variance explained by HRRpred and assessed model discrimination by calculating the c-statistic. To examine clinical correlates of HRRpred (dependent variable) we used a linear regression model with clinical covariates including age, sex, systolic blood pressure, use of antihypertensive medications, body mass index, smoking status, diabetes mellitus, and prevalent CVD. To examine contributions of clinical covariates to HRRpred we took a stepwise approach with P for entry <.1 and P for retention <.05.
We then examined the association of HRRpred with incident disease outcomes including diabetes mellitus, heart failure, CVD, and all-cause mortality using multivariable Cox models. Models were first adjusted for age, sex, systolic blood pressure, use of antihypertensive medications, body mass index, smoking status, diabetes mellitus (with exception of diabetes analyses), and prevalent CVD (with exception of CVD models). Models for incident diabetes were additionally adjusted for fasting glucose. In secondary analyses, we accounted for resting heart rate in the multivariable models. We quantified the degree of information gained with the addition of HRRpred to the multivariable model by examining the model area under the curve with and without HRRpred.26 Analyses were performed using the lifelines Python package for survival analyses. For outcome associations, a 2-sided P value <.05 was deemed significant.
Results
We studied 56,793 individuals with mean age 57 ± 8 years and 51% women. Few participants had prevalent CVD, with 1% of individuals that had a prior history of atrial fibrillation and 0.2% prior heart failure. Actual and predicted HRR were 27 bpm (interquartile range (IQR) 21, 34) and 28 bpm (IQR 25, 31), respectively. Individuals in the lowest vs highest HRRpred tertile were more likely to be men (58% vs 41%), with higher body mass index (28.6 vs 25.2 kg/m2) and greater prevalence of comorbid conditions, including treated hypertension (29% vs 12%) and diabetes mellitus (4% vs 2%, Table 1).
Table 1.
Baseline clinical characteristics of UK Biobank sample by predicted heart rate recovery tertile
| Clinical characteristic | HRRpred tertile |
|||
|---|---|---|---|---|
| Total sample |
Tertile 1 |
Tertile 2 |
Tertile 3 |
|
| N = 56,793 | N = 18,931 | N = 18,931 | N = 18,931 | |
| Age, years | 57.1 (8.1) | 57.8 (8.1) | 57.5 (8.1) | 56.1 (8.2) |
| Men, n (%) | 27,578 (49%) | 10,947 (58%) | 8834 (47%) | 7797 (41%) |
| Race, n (%) | ||||
| White | 51,977 (91%) | 17,382 (91%) | 17,389 (91%) | 17,206 (91%) |
| Black | 1539 (3%) | 507 (3%) | 477 (3%) | 555 (3%) |
| Other | 3277 (6%) | 1042 (6%) | 1065 (6%) | 1170 (6%) |
| Body mass index, kg/m2 | 27.4 (4.4) | 29.2 (4.7) | 27.3 (4.1) | 25.7 (3.6) |
| Systolic blood pressure, mm Hg | 137 (17) | 141 (16) | 138 (17) | 134 (17) |
| Hypertension treatment, n (%) | 11,131 (20%) | 5395 (29%) | 3477 (18%) | 2259 (12%) |
| Diabetes mellitus, n (%) | 1225 (2%) | 722 (4%) | 348 (2%) | 155 (1%) |
| Current smoker, n (%) | 4708 (8%) | 1750 (9%) | 1517 (8%) | 1441 (8%) |
| Total cholesterol, mmol/L | 5.7 (4.9, 6.4) | 5.7 (4.9, 6.5) | 5.7 (5.0, 6.5) | 5.7 (5.0, 6.4) |
| HDL cholesterol, mmol/L | 1.4 (1.2, 1.7) | 1.3 (1.1, 1.6) | 1.4 (1.2, 1.7) | 1.5 (1.3, 1.8) |
| Lipid-lowering therapy | 10,141 (18%) | 4634 (25%) | 3243 (17%) | 2264 (12%) |
| Prevalent CVD, n (%) | 1521 (3%) | 543 (3%) | 546 (3%) | 432 (2 %) |
| Prevalent heart failure, n (%) | 136 (0.2%) | 71 (0.4 %) | 43 (0.2%) | 22 (0.1%) |
| Prevalent atrial fibrillation, n (%) | 757 (1%) | 291 (2%) | 262 (1%) | 204 (1%) |
| Resting heart rate, bpm | 71 (63, 79) | 81 (75, 88) | 70 (65, 75) | 62 (57, 67) |
| Observed HRR at 50 seconds, bpm | 27 (21, 34) | 22 (16, 28) | 28 (22, 33) | 32 (26, 38) |
| Predicted HRR at 50 seconds, bpm | 28 (25, 31) | 23 ( 21, 25) | 28 (27, 29) | 33 (31, 34) |
Data are expressed as n (%), mean (standard deviation), or median (25th, 75th percentile) as appropriate.
CVD = cardiovascular disease; HDL = high-density lipoprotein; HRR = heart rate recovery; HRRpred = predicted HRR from resting electrocardiogram convolutional neural network model.
Deep learning to estimate HRR using resting ECG
Deep-learned HRRpred was moderately correlated with measured HRR (Pearson r = 0.48 [95% confidence interval (CI) 0.47–0.48], mean error = 6.78, 95% CI 6.73–6.82, R2 = 0.23). HRRpred generally made conservative HRR predictions; Supplemental Figure 3 shows distributions of HRR vs HRRpred. By comparison, a random forest model of resting heart rate as a predictor of HRR explained only 14.6% of the variance in HRR (Pearson r = 0.38, 95% CI 0.36–0.40, mean error = 7.17, 95% CI 7.01–7.31). To examine which portions of the resting ECG waveform were particularly relevant for estimating HRRpred, we examined saliency maps to illustrate areas of the ECG waveform with greatest influence on HRRpred (Figure 1). We also examined ECG waveforms for those predicted to have high vs low HRRpred. Median waveforms were aggregated for individuals at 25th, 50th, and 75th percentile of resting heart rate (Figure 2). For example, for individuals with median heart rate, those in the 5th percentile of HRRpred had longer PR intervals, greater QRS voltage, and greater QRS width compared with those in the 95th percentile of HRRpred.
Figure 1.
Representations of deep learning electrocardiogram (ECG) model behavior, part 1. Saliency maps of the ECG convolutional neural network model with areas on the ECG waveform of greatest influence on heart rate recovery predictions shown in darker gray and black. Saliency was averaged over 200 individuals and grouped based on resting heart rate (within 5 beats/min of the 25th, 50th, and 75th percentile of resting heart rate). bpm = beats per minute; Rest HR = resting heart rate.
Figure 2.
Representations of deep learning electrocardiogram (ECG) model behavior, part 2. Median ECG waveforms for a random sample of 100 individuals, each with high (green) vs low (red) predicted heart rate recovery (HRRpred) (90th and 10th percentile), grouped based on resting heart rate (HR). A: Within 5 beats per minute (bpm) of the 25th percentile of resting HR. B: Within 5 bpm of the 50th percentile of resting HR. C: Within 5 bpm of the 75th percentile of resting HR).
When defining impaired HRR using the lowest tertile, we found that HRRpred correctly classified 70.5% of individuals as having impaired vs normal HRR and incorrectly classified 29.5% of individuals. We next examined HRRpred as a predictor of impaired HRR using multivariable logistic regression (Supplemental Figure 4). A univariable model with HRRpred showed a c-statistic of 0.735. When added to a model with age and sex, HRRpred resulted in improved discrimination as ascertained by the c-statistic (0.657 vs 0.770, P < .0001).
Association of HRRpred with clinical correlates and cardiovascular outcomes
In cross-sectional multivariable analyses, we found that clinical correlates of lower HRRpred included older age, male sex, higher body mass index, higher systolic blood pressure, diabetes mellitus, and current smoking status (Supplemental Table 2). The strongest predictor of lower HRRpred was higher resting heart rate. Specifically, a 1 standard deviation (SD) higher resting heart rate was associated with a 3.6 bpm (standard error 0.01) lower HRRpred. Taken together, clinical covariates explained 72.4% of the variance of HRRpred.
Over a median follow-up time of 9.9 (IQR 9.8, 10.1) years, we observed 2361 incident diabetes, 862 incident heart failure, 1591 incident CVD events, and 2065 deaths. We found that higher HRRpred was associated with lower risk of incident diabetes mellitus, heart failure, CVD, and death in multivariable-adjusted Cox models (Table 2). Specifically, a 1 SD higher HRRpred was associated with lower risk of new-onset diabetes (hazard ratio [HR] 0.79, 95% CI 0.76–0.83, P < .0001), a 11% incident heart failure (HR 0.89, 95% CI 0.83–0.95, P = .0009), and death (HR 0.83, 95% CI 0.79–0.86, P < .0001). These findings mirrored associations of observed HRR with outcomes (Table 2).
Table 2.
Association of predicted heart rate recovery and heart rate recovery with longitudinal outcomes
| Outcome | Predictor | Multivariable model† |
Multivariable + resting HR |
||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Incident DM | HRRpred | 0.79 (0.76, 0.83) | <.0001 | 0.77 (0.71, 0.83) | <.0001 |
| N = 2060 | HRR | 0.87 (0.83, 0.92) | <.0001 | 0.94 (0.89, 0.99) | .02 |
| Incident HF | HRRpred | 0.89 (0.83, 0.95) | .0009 | 0.95 (0.84, 1.08) | .43 |
| N = 862 | HRR | 0.86 (0.79, 0.93) | .0002 | 0.89 (0.82, 0.97) | .009 |
| Incident CVD | HRRpred | 0.89 (0.85, 0.94) | <.0001 | 0.95 (0.87, 1.04) | .27 |
| N = 1591 | HRR | 0.87 (0.83, 0.93) | <.0001 | 0.90 (0.85, 0.96) | .002 |
| All-cause death | HRRpred | 0.83 (0.79, 0.86) | <.0001 | 0.87 (0.81, 0.95) | .0009 |
| N = 2065 | HRR | 0.84 (0.80, 0.88) | <.0001 | 0.89 (0.84, 0.94) | <.0001 |
CVD = cardiovascular disease; DM = diabetes mellitus; HF = heart failure; HR = hazard ratio; HRR = heart rate recovery; HRRpred = predicted HRR.
Multivariable model adjusted for age, sex, systolic blood pressure, hypertension treatment, body-mass index, smoking status, diabetes mellitus (prevalent DM excluded from incident DM analyses), and prevalent cardiovascular disease (prevalent CVD excluded from incident CVD analyses). DM models additionally adjusted for fasting glucose. Hazard ratios are expressed per 1 standard deviation change in predictor variable (SD for HRRpred was 4.7 beats/min, and for HRR was 9.8 beats/min).
After addition of resting heart rate to the multivariable model, the association of HRRpred with incident diabetes mellitus and all-cause mortality were similar (diabetes: HR 0.77, 95% CI 0.71–0.83; mortality: HR 0.87, 95% CI 0.81–0.95, P ≤ .001 for both). By contrast, associations of HRRpred with incident heart failure and incident CVD were attenuated (P > .2 for both).
When added to a clinical model predicting incident diabetes mellitus, HRRpred improved model discrimination (c-statistic 0.805 [95% CI 0.798–0.809] vs 0.808 [95% CI 0.802–0.816], P < .0001 for difference, Supplemental Table 3). Interestingly, a multivariable clinical model containing HRRpred had better model discrimination with respect to future risk of diabetes (c-statistic 0.808 [95% CI 0.802–0.816]) compared with a model containing resting HR (c-statistic 0.807 [95% CI 0.802–0.814]) as well as observed HRR (c-statistic 0.806 [95% CI 0.800–0.812]).
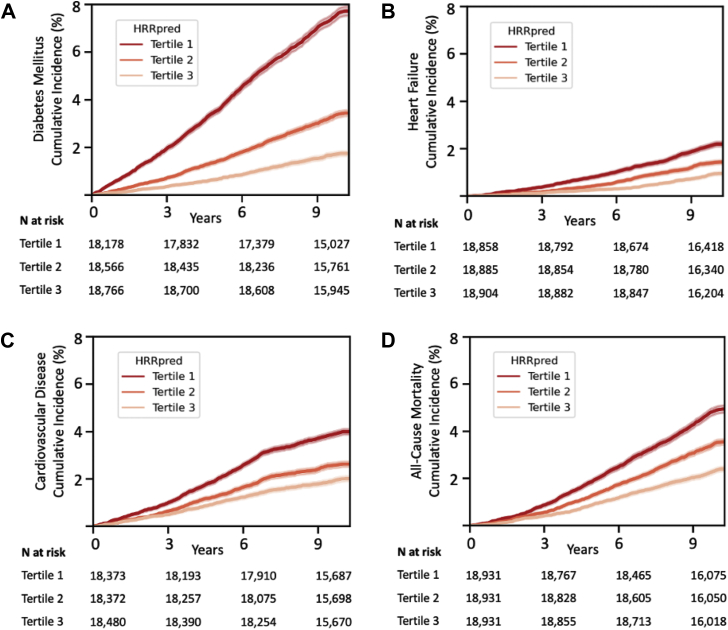
We next examined the association of impaired HRRpred as defined by the lowest HRRpred tertile (compared with highest tertile) with incident outcomes (Figure 3). Participants with impaired HRRpred had an 83% higher risk of future diabetes (multivariable-adjusted HR 1.83, 95% CI 1.55–2.16) and 27% higher risk of all-cause death (HR 1.27, 95% CI 1.09–1.49) even after accounting for resting heart rate (Supplemental Table 4).
Figure 3.
Overall cumulative incidence of cardiovascular events by predicted heart rate recovery (HRRpred) tertile. Panels show plots for future risk of A: diabetes mellitus, B: heart failure, C: cardiovascular disease, and D: all-cause mortality across HRRpred tertiles, with tertile 1 representing most impaired HRRpred. Predicted HRR ranges in tertile 1: 8.9–26.2 beats per minute (bpm); tertile 2: 26.2–30.3 bpm; tertile 3: 30.3–44.2 bpm. Numbers of individuals at risk in each tertile are shown at the bottom of each panel.
Genetic determinants of HRRpred
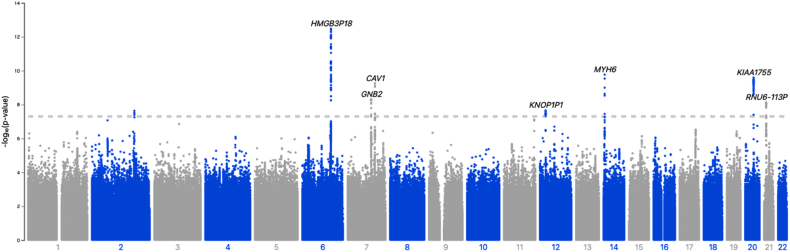
The estimated age- and sex-adjusted heritability of HRRpred was h2 = 0.209 (standard error 0.014). The genetic correlation between actual HRR and HRRpred was 0.541. In a GWAS of HRRpred (genomic inflation factor lambda 1.048) we found 8 genome-wide significant loci associated with HRRpred (Table 3, Figure 4) with the following genes in closest proximity: CCDC141, GJA1, GNB2, CAV1, BCAG1, MYH6, KIAA1755, and C21orf37. Among these loci, variants near CCC141, BCAT1, and KIAA1755 have been previously related to heart rate or related traits in other genetic studies,27,28 and variants near GJA1, GNB1, and MYH6 have been associated with conduction system abnormalities and other structural heart disease (Table 3).29, 30, 31 Lastly, CAV1 has previously been associated with insulin resistance and metabolic syndrome,32,33 in addition to HRR and other ECG-related traits.24,34 In comparing SNPs associated with HRRpred against published GWAS on type 2 diabetes mellitus, we found that rs6127466 in KIAA1755 was also associated with diabetes (P = 1.1 × 10-4).35
Table 3.
Genetic loci associated with predicted heart rate recovery
| Chr | SNP | Effect allele / Referent allele | EAF | Gene | Location | Beta | s.e. | P | Selected prior trait associations |
|---|---|---|---|---|---|---|---|---|---|
| 2 | rs142556838 | C/T | 0.91 | CCDC141 | Intron | 0.306 | 0.055 | 2.30E-08 | Heart rate27 |
| 6 | 6:122113614 | CT/C | 0.90 | GJA1 | 0.378 | 0.052 | 3.30E-13 | Cardiac conduction29 | |
| 7 | rs221789 | C/T | 0.15 | GNB2 | 5' utr | -0.246 | 0.043 | 4.80E-09 | Familial sinus node and atrioventricular conduction dysfunction30,33 |
| 7 | rs1997571 | A/G | 0.41 | CAV1 | Intron | -0.197 | 0.032 | 5.60E-10 | Insulin resistance, metabolic syndrome32 |
| 12 | rs4963772 | A/G | 0.85 | BCAT1 | Intergenic | -0.244 | 0.044 | 2.20E-08 | Heart rate variability28 |
| 14 | rs422068 | T/C | 0.64 | MYH6 | Intron | 0.205 | 0.033 | 1.70E-10 | Familial cardiomyopathy, rare variant also associated with sick sinus syndrome31,42 |
| 20 | rs6127466 | G/A | 0.53 | KIAA1755 | Intron | -0.197 | 0.031 | 2.50E-10 | Heart rate27 |
| 21 | rs2846867 | C/T | 0.98 | C21orf37 | Intergenic | 0.181 | 0.031 | 7.40E-09 |
BCAT1 = branched chain amino acid transaminase 1; CAV1 = caveolin-1; CCDC141 = coiled-coil domain containing 141; Chr = chromosome; EAF = estimated allele frequency; GJA1 = gap junction protein alpha 1; GNB2 = G protein subunit beta 2; MYH6 = myosin heavy chain 6; s.e. = standard error; SNP = single nucleotide polymorphism.
Figure 4.
Manhattan plot of genome-wide association study (GWAS) of predicted heart rate recovery. Chromosomes are represented across the x-axis, and -log10(P value) on the y-axis. The dashed line indicates genome-wide significant P value threshold of 5 × 10-8. Most significant genetic loci are annotated on the plot. Sample size for GWAS was n = 43,722.
We next created 2 polygenic risk scores of HRR from prior published GWAS to examine its association with HRRpred. After adjustment for age, sex, array, and 5 principal components, we found that HRR polygenic risk scores were associated with HRRpred (P = 1.6 × 10-15 for SNPs identified in Ramirez and colleagues25; P = 3.2 × 10-31 for 22 SNPs in Verweij and colleagues24, Supplemental Figure 5). Further, we constructed a polygenic risk score for HRRpred, and found associations with incident diabetes (HR per 1 SD increase in polygenic risk score 0.97, 95% CI 0.96–0.99, P = 4.1 × 10-5) but not heart failure (P = .43), mirroring previous trait analyses.
Discussion
Our study demonstrates that a deep learning model of resting ECG tracings can estimate HRR, an important measure of cardiac autonomic dysfunction that usually requires exercise provocation testing to ascertain. While HRRpred was only moderately correlated with actual HRR, it was independently associated with incident clinical outcomes, including new-onset diabetes and all-cause mortality. Specifically, individuals with impaired HRRpred had a more than 80% greater risk of future diabetes and 27% greater risk of death even after accounting for clinical risk factors including resting heart rate. Lastly, the genetic correlates of HRRpred include known heart rate, cardiac conduction system, cardiomyopathy, and metabolic trait loci. Taken together, these findings support the idea that artificial intelligence–enhanced interpretation of a resting ECG may serve as a proxy for HRR and lend insights into the association of cardiac autonomic dysfunction with clinical outcomes. The ability to infer postexercise heart rate response using widely scalable methods from a resting ECG harbors potential clinical implications, although utility with respect to potential screening, identification of at-risk individuals, and impact on preventive strategies will require further study.
Recent studies have demonstrated the potential value of deep learning methods including CNNs applied to the ECG, which have enabled detection of silent or future atrial fibrillation, asymptomatic left ventricular systolic dysfunction, and overall survival, among other health conditions.10,36, 37, 38 CNNs in particular enable the application of a sequence of learned spatial filters (convolutions) to data and have proven effective on time series data including ECG tracings. By extracting complex and subtle information contained within the ECG, these methods have the potential to enhance human ECG interpretation in a meaningful way. We now show that a deep learning model can infer postexercise HRR from a resting ECG, and that predicted HRR is strongly and independently associated with clinical outcomes. We note that resting ECGs are scalable and widely available across health systems, and that continued advances in wearable ECG monitors may expand potential applicability even further. Whether deployment of deep learning inferences on HRR as an important measure of cardiac autonomic dysfunction may be used as a screening tool remains unknown, though this concept is well supported by a recent pragmatic, randomized clinical trial that demonstrated that an artificial intelligence–enabled ECG algorithm was feasibly deployed within the primary care setting and enabled early diagnosis of patients with a reduced ejection fraction.37
The clinical importance of HRR has long been recognized, with known associations with all-cause mortality and sudden cardiac death among relatively healthy adults.3,4 More importantly, these associations appear independent of exercise workload, change in heart rate, or use of beta-blockers. Prognostic value has also been demonstrated across individuals with heart failure and left ventricular systolic dysfunction.5,6 Although HRRpred is related to resting heart rate, we found that associations with clinical outcomes appear to be robust even after accounting for resting heart rate in our study, underlining the potential additive prognostic information contained within HRRpred. In addition to all-cause mortality, we find that HRRpred is strongly and independently associated with future risk of diabetes mellitus. This mirrors prior clinical studies showing that heart rate–related measures of autonomic dysfunction were associated longitudinally with incident diabetes.39,40 Interestingly, measures of autonomic dysfunction including HRR improved among individuals randomized to the lifestyle modification group in the Diabetes Prevention Program, and were associated with lower risk of diabetes development independent of weight change.41 These findings highlight the clinical importance of HRR as an early harbinger of cardiometabolic disease and the future potential of HRRpred in this context as a widely available screening tool.
We studied genetic determinants of HRRpred and found 8 genome-wide significant loci, which demonstrate overlap with prior heart rate–related traits including resting heart rate, cardiac conduction dysfunction, and heart rate variability. Further, genetic loci associated with HRRpred were also associated with structural heart disease, as well as cardiometabolic disease including insulin resistance and metabolic syndrome. For example, rs6127466 in KIAA1755 has previously been associated with resting heart rate and diabetes mellitus27,35; the C allele was associated with genetically lower HRRpred and higher risk of diabetes, mirroring clinical trait associations. We also found that genetic associations of HRR identified in prior GWAS are also associated with HRRpred. Taken together, these studies confirm plausible biologic genetic associations with HRRpred that indicate both cardiovascular and metabolic underpinnings and further substantiate our observed associations of HRRpred with incident diabetes and cardiovascular outcomes.
Several limitations deserve mention. We recognize that the correlation of HRRpred with observed HRR was modest, and potential misclassification of individuals with and without impaired HRR could be possible. However, even with modest performance, the potential of HRRpred as a screening tool to enrich for high-risk individuals with cardiac autonomic dysfunction may be useful in the context of clinical trials or other settings. We note that HRRpred was based on 3-lead resting ECG tracings prior to exercise testing, and future refinements using 12-lead ECG may be able to improve upon our current model. Further, the sample included in this study was subject to potential selection bias, given that exercise protocols among UK Biobank participants were based on clinical risk factor assessment, and individuals deemed at high risk did not undergo exercise testing.13 In addition, exercise testing protocols in the UK Biobank were not symptom-limited but rather time-limited, which could have affected peak heart rate achieved during the test. In the context of an observational study, we acknowledge that causal inferences cannot be drawn, and while we accounted for traditional cardiovascular risk factors, residual confounding may be present. Lastly, future studies across the disease spectrum may address generalizability to other samples.
Conclusion
In sum, we show that a deep learning model of resting ECG tracings can estimate HRR, an important measure of cardiac autonomic dysfunction. We find that HRRpred is independently associated with new-onset diabetes and all-cause mortality in a large population-based cohort. Genetic determinants of HRRpred substantiate associations with heart rate, cardiac conduction system, cardiomyopathy, and metabolic trait loci. Taken together, our study demonstrates that the resting ECG may serve as a proxy for HRR and lend insights into the association of cardiac autonomic dysfunction with clinical outcomes. The utility of HRRpred as a widely scalable tool to infer postexercise heart rate response, and subsequent screening and therapeutic implications, are important areas of future study.
Acknowledgments
Funding Sources
Dr Weng is supported by AHA 18SFRN34110082. Dr Lau is supported by AHA Career Development Award 853922. Dr Khurshid is supported by NIH T32HL007208. Dr Ellinor is supported by NIH 1R01HL092577, K24HL105780, AHA 18SFRN34110082, and MAESTRIA (965286). Dr Anderson is supported by NIH R01NS103924, U01NS069673, AHA 18SFRN34250007, and AHA-Bugher 21SFRN812095. Dr Lubitz is supported by NIH 1R01HL139731 and AHA 18SFRN34250007. Dr Ho is supported by NIH R01HL134893, R01HL140224, and K24HL153669. This work was sponsored by Bayer AG.
Disclosures
Dr Friedman receives sponsored research support from Bayer AG and IBM. Dr Diedrich, Dr Mielke, Dr Eilken, Dr Derix, and Ms Ghadessi are employees of Bayer AG. Dr Ellinor receives sponsored research support from Bayer AG and IBM Research and has consulted for Bayer AG, Novartis, and MyoKardia. Dr Anderson receives sponsored research support from Bayer AG and has consulted for ApoPharma and Invitae. Dr Philippakis receives sponsored research support from Bayer AG, IBM, Intel, and Verily. He has also received consulted fees from Novartis and Rakuten. He is a Venture Partner at GV and is compensated for this work. Dr Batra receives sponsored research support from Bayer AG and IBM, and consults for Novartis. Dr Lubitz receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM, and has consulted for Bristol Myers Squibb / Pfizer, Bayer AG, and Blackstone Life Sciences. Dr Ho has received sponsored research support from Bayer AG and research supplies from EcoNugenics.
Author Contributions
All authors attest they meet the current ICMJE criteria for authorship. Conception and design of the work (N.D., P.D.A., P.B., S.A.L., J.E.H.); acquisition, analysis, and interpretation of data (N.D., P.D.A., L.-C.W., M.G., P.B., S.A.L., J.E.H.); drafted the work or substantively revised it and approved the submitted version and be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature (all authors).
Patient Consent
All patients provided written informed consent.
Ethics Statement
The authors designed the study and gathered and analyzed the data according to the Helsinki Declaration guidelines on human research. The research protocol used in this study was reviewed and approved by the institutional review board.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2022.06.001.
Appendix. Supplementary data
References
- 1.Goldberger J.J., Arora R., Buckley U., Shivkumar K. Autonomic nervous system dysfunction: JACC Focus Seminar. J Am Coll Cardiol. 2019;73:1189–1206. doi: 10.1016/j.jacc.2018.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai K., Sato H., Hori M., et al. Vagally mediated heart rate recovery after exercise is accelerated in athletes but blunted in patients with chronic heart failure. J Am Coll Cardiol. 1994;24:1529–1535. doi: 10.1016/0735-1097(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 3.Cole C.R., Blackstone E.H., Pashkow F.J., Snader C.E., Lauer M.S. Heart-rate recovery immediately after exercise as a predictor of mortality. N Engl J Med. 1999;341:1351–1357. doi: 10.1056/NEJM199910283411804. [DOI] [PubMed] [Google Scholar]
- 4.Jouven X., Empana J.P., Schwartz P.J., Desnos M., Courbon D., Ducimetiere P. Heart-rate profile during exercise as a predictor of sudden death. N Engl J Med. 2005;352:1951–1958. doi: 10.1056/NEJMoa043012. [DOI] [PubMed] [Google Scholar]
- 5.Kubrychtova V., Olson T.P., Bailey K.R., Thapa P., Allison T.G., Johnson B.D. Heart rate recovery and prognosis in heart failure patients. Eur J Appl Physiol. 2009;105:37–45. doi: 10.1007/s00421-008-0870-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipinski M.J., Vetrovec G.W., Gorelik D., Froelicher V.F. The importance of heart rate recovery in patients with heart failure or left ventricular systolic dysfunction. J Card Fail. 2005;11:624–630. doi: 10.1016/j.cardfail.2005.06.429. [DOI] [PubMed] [Google Scholar]
- 7.Tang Y.D., Dewland T.A., Wencker D., Katz S.D. Post-exercise heart rate recovery independently predicts mortality risk in patients with chronic heart failure. J Card Fail. 2009;15:850–855. doi: 10.1016/j.cardfail.2009.06.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara J., Murakami H., Maeda S., Kuno S., Matsuda M. Change in post-exercise vagal reactivation with exercise training and detraining in young men. Eur J Appl Physiol. 2001;85:259–263. doi: 10.1007/s004210100443. [DOI] [PubMed] [Google Scholar]
- 9.Hao S.C., Chai A., Kligfield P. Heart rate recovery response to symptom-limited treadmill exercise after cardiac rehabilitation in patients with coronary artery disease with and without recent events. Am J Cardiol. 2002;90:763–765. doi: 10.1016/s0002-9149(02)02607-3. [DOI] [PubMed] [Google Scholar]
- 10.Siontis K.C., Noseworthy P.A., Attia Z.I., Friedman P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat Rev Cardiol. 2021;18:465–478. doi: 10.1038/s41569-020-00503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Vegte Y.J., Tegegne B.S., Verweij N., Snieder H., van der Harst P. Genetics and the heart rate response to exercise. Cell Mol Life Sci. 2019;76:2391–2409. doi: 10.1007/s00018-019-03079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UKBB Biobank Cardio Assessment (version 1.0). Accessed May 25, 2021. https://biobank.ndph.ox.ac.uk/ukb/ukb/docs/Cardio.pdf.
- 14.Carreiras C., Alves A.P., Lourenço A., et al. BioSPPy - Biosignal Processing in Python. 2015. https://github.com/PIA-Group/BioSPPy/ Accessed November 4, 2021.
- 15.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2016:arXiv:1608.06993.
- 16.Khurshid S., Friedman S., Reeder C., et al. ECG-based deep learning and clinical risk factors to predict atrial fibrillation. Circulation. 2022;145:122–133. doi: 10.1161/CIRCULATIONAHA.121.057480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014:arXiv:1412.6980.
- 18.Ramachandran P, Zoph B, Le QV. Searching for activation functions. 2017:arXiv:1710.05941.
- 19.Tompson J, Goroshin R, Jain A, LeCun Y, Bregler C. Efficient object localization using convolutional networks. 2014:arXiv:1411.4280.
- 20.Srivastava N., Hinton G., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Machine Learning Res. 2014;15:1929–1958. [Google Scholar]
- 21.Khurshid S., Friedman S., Pirruccello J.P., et al. Deep learning to predict cardiac magnetic resonance-derived left ventricular mass and hypertrophy from 12-lead ECGs. Circ Cardiovasc Imaging. 2021;14 doi: 10.1161/CIRCIMAGING.120.012281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh P.R., Kichaev G., Gazal S., Schoech A.P., Price A.L. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verweij N., van de Vegte Y.J., van der Harst P. Genetic study links components of the autonomous nervous system to heart-rate profile during exercise. Nat Commun. 2018;9:898. doi: 10.1038/s41467-018-03395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez J., Duijvenboden S.V., Ntalla I., et al. Thirty loci identified for heart rate response to exercise and recovery implicate autonomic nervous system. Nat Commun. 2018;9:1947. doi: 10.1038/s41467-018-04148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pencina M.J., D'Agostino R.B. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 27.den Hoed M., Eijgelsheim M., Esko T., et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerr K.F., Avery C.L., Lin H.J., et al. Genome-wide association study of heart rate and its variability in Hispanic/Latino cohorts. Heart Rhythm. 2017;14:1675–1684. doi: 10.1016/j.hrthm.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao S., Shimura D., Baum R., et al. Auxiliary trafficking subunit GJA1-20k protects connexin-43 from degradation and limits ventricular arrhythmias. J Clin Invest. 2020;130:4858–4870. doi: 10.1172/JCI134682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stallmeyer B., Kuss J., Kotthoff S., et al. A Mutation in the G-protein gene GNB2 causes familial sinus node and atrioventricular conduction dysfunction. Circ Res. 2017;120:e33–e44. doi: 10.1161/CIRCRESAHA.116.310112. [DOI] [PubMed] [Google Scholar]
- 31.Holm H., Gudbjartsson D.F., Sulem P., et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abaj F., Saeedy S.A.G., Mirzaei K. Are caveolin-1 minor alleles more likely to be risk alleles in insulin resistance mechanisms in metabolic diseases? BMC Res Notes. 2021;14:185. doi: 10.1186/s13104-021-05597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Souza G.M., de Albuquerque Borborema M.E., de Lucena T.M.C., et al. Caveolin-1 (CAV-1) up regulation in metabolic syndrome: all roads leading to the same end. Mol Biol Rep. 2020;47:9245–9250. doi: 10.1007/s11033-020-05945-y. [DOI] [PubMed] [Google Scholar]
- 34.Ntalla I., Weng L.C., Cartwright J.H., et al. Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction. Nat Commun. 2020;11:2542. doi: 10.1038/s41467-020-15706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan A., Taliun D., Thurner M., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Attia Z.I., Noseworthy P.A., Lopez-Jimenez F., et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–867. doi: 10.1016/S0140-6736(19)31721-0. [DOI] [PubMed] [Google Scholar]
- 37.Yao X., Rushlow D.R., Inselman J.W., et al. Artificial intelligence-enabled electrocardiograms for identification of patients with low ejection fraction: a pragmatic, randomized clinical trial. Nat Med. 2021;27:815–819. doi: 10.1038/s41591-021-01335-4. [DOI] [PubMed] [Google Scholar]
- 38.Raghunath S., Ulloa Cerna A.E., Jing L., et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat Med. 2020;26:886–891. doi: 10.1038/s41591-020-0870-z. [DOI] [PubMed] [Google Scholar]
- 39.Carnethon M.R., Golden S.H., Folsom A.R., Haskell W., Liao D. Prospective investigation of autonomic nervous system function and the development of type 2 diabetes: the Atherosclerosis Risk In Communities study, 1987-1998. Circulation. 2003;107:2190–2195. doi: 10.1161/01.CIR.0000066324.74807.95. [DOI] [PubMed] [Google Scholar]
- 40.Qiu S.H., Xue C., Sun Z.L., Steinacker J.M., Zugel M., Schumann U. Attenuated heart rate recovery predicts risk of incident diabetes: insights from a meta-analysis. Diabet Med. 2017;34:1676–1683. doi: 10.1111/dme.13517. [DOI] [PubMed] [Google Scholar]
- 41.Carnethon M.R., Prineas R.J., Temprosa M., et al. The association among autonomic nervous system function, incident diabetes, and intervention arm in the Diabetes Prevention Program. Diabetes Care. 2006;29:914–919. doi: 10.2337/diacare.29.04.06.dc05-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carniel E., Taylor M.R., Sinagra G., et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.