Abstract
Skin is the largest organ in the human body, and the interplay between the environment factors and human skin leads to some skin diseases, such as acne, psoriasis, and atopic dermatitis. As the first line of human immune defense, skin plays significant roles in human health via preventing the invasion of pathogens that is heavily influenced by the skin microbiota. Despite being a challenging niche for microbes, human skin is colonized by diverse commensal microorganisms that shape the skin environment. The skin microbiota can affect human health, and its imbalance and dysbiosis contribute to the skin diseases. This review focuses on the advances in our understanding of skin microbiota and its interaction with human skin. Moreover, the potential roles of microbiota in skin health and diseases are described, and some key species are highlighted. The prevention, diagnosis and treatment strategies for microbe-related skin diseases, such as healthy diets, lifestyles, probiotics and prebiotics, are discussed. Strategies for modulation of skin microbiota using synthetic biology are discussed as an interesting venue for optimization of the skin-microbiota interactions. In summary, this review provides insights into human skin microbiota recovery, the interactions between human skin microbiota and diseases, and the strategies for engineering/rebuilding human skin microbiota.
Keywords: Skin, Microbiota, Commensal microbes, Synthetic biology, Omics technologies, Host-skin microbiota interaction, Cutaneous diseases, Acne
Introduction
Human skin, the body’s largest and most exposed organ, functions as a physical barrier; It is not only blocking the entry of pathogens from the environment, but also providing a large-scale ecological niche to an enormous variety of microbes [1]. The biochemical conditions of the skin are stringent, such as low pH, high salinity, exsiccosis and extensive exposure to the environmental factors. Nevertheless, many microbial species successfully colonize the skin, including bacteria, fungi, viruses (especially bacteriophages) [2]. The composition of the human skin microbiome is determined by the genetics, environmental factors, and the local microenvironment [3, 4]. Thus, the human skin microbiota varies from one body part to another, and can be very different among persons of different race, age, sex and state of health [5]. The skin microbiomes are ecosystems of diverse microorganisms that interact with the human body, including host epithelial and immune cells, as well as with other microorganisms sharing the same niche [6]. Usually, the interactions between skin microbiome and hosts are mutually reinforced. The hosts provide microbiome with ‘home’ and ‘food’, while the microbiome ‘guards’ the hosts against pathogen invasion and ‘educates’ the immune system. However, the human skin microbiome can be pathogenic, and this is closely associated with host homeostasis [7].
Some skin diseases are associated with pathogens, such as acne vulgaris [8–14], psoriasis [15–19], atopic dermatitis [20, 21], chronic wounds [22] (Table 1). Information about causal links between these diseases and key species of human microbiota is limited. With the rapid development of sequencing technologies, especially next-generation sequencing techniques [23] and long-read sequencing technologies [24], some pathogens from the human microbiota have been identified. In such cases, species-level information is not enough to identify pathogenic strains, and strain-level information is required. For example, some Staphylococcus epidermidis and Propionibacterium acnes (Current name is Cutibacterium acnes) strains contribute to acne and other skin diseases, while several other strains of the same species help to promote skin health by inhibiting the growth and invasion of pathogens [10]. Based on the identified microbes and their microbiological pathogenesis, insights into molecular and immunological mechanisms of microbiome-host interaction are essential.
Table 1.
Skin microbiome and associated diseases
| Disease type | Key Points | Major findings | References |
|---|---|---|---|
| Acne vulgaris | P. acnes | Although the relative abundances of P. acnes were similar, certain strains were highly associated with acne and healthy skin | Sorel Fitz-Gibbon et al. [8] |
| S. epidermidis | S. epidermidis mediates fermentation to inhibit the growth of P. acnes, which can be implications of probiotics in acne vulgaris | Yanhan Wang et al. [9] | |
| S. epidermidis & P. acnes | S. epidermidis and P. acnes are thought to contribute to the disease, but they are also known to promote health by inhibiting the growth and invasion of pathogens | Alan M. O’Neill et al. [10] | |
| Dysbiosis & Balance | The mere presence of disease-associated strains, as well as the balance between metagenomic elements shapes the overall virulence property of the skin microbiota. Dysbiosis is the process leading to a disturbed skin barrier and disequilibrium of the cutaneous microbiome | Emma Barnard et al. [11], B. Dreno [12], Chun-xi LI et al. [13] | |
| Androgen hormone activity | Increases sebum production inside the pilosebaceous follicle, adjusting the environment for P. acnes which triggers inflammation | M. A. Rocha et al. [14] | |
| Psoriasis | Diversity & Stability | Psoriasis induces physiological changes both at the lesion site and at the systemic lever, with increased diversity and reduced stability compared to the healthy skin microbiome | Alexander V Alekseyenko et al. [15], Daniel J. Lewis et al. [16] |
| Skin microbiome | Increased abundance of Corynebacterium, Staphylococcus, and Streptococcus, and decreased abundance of Malassezia, Propionibacterium, Cutibacterium genera versus controls | Di Yan et al. [17], Hsin-Wen Chang et al. [18] | |
| Gut microbiome | The gut microbiome composition in psoriasis patients has been altered markedly, and the ratio of Firmicutes and Bacteroidetes was perturbed in psoriatic individuals compared to healthy controls | Xinyue Zhang et al. [19], Di Yan et al. [17] | |
| Atopic dermatitis | S. aureus | AD has long been associated with S. aureus skin colonization or infection, and increases in Streptococcus, Propionibacterium, and Corynebacterium species were observed following therapy | Heidi H. Kong et al. [21], Tetsuro Kobayashi et al. [20] |
| Chronic wound | S. aureus & P. aeruginosa | S. aureus and Pseudomonas aeruginosa are the most common bacteria isolated from chronic wounds | Raffaele Serra et al. [22] |
| Skin and soft tissue infection | Cutibacterium acne | C. acnes has the potential to directly and indirectly cause inflammation and tissue damage | Laurice Floweis et al. [6] |
In the future, treatment of skin diseases should not be limited only to antibiotics, topical corticosteroids, laser therapy, or other traditional strategies. Modulation of human skin and gut microbiota with healthy diet and other strategies should be considered as well. Similar to fecal microbiota transplantation to regulate gut microbiota, combinations of probiotics, skin microbiota transplantation and other developing strategies are promising venues for skin disease treatment. Building synthetic microbiota with defined and controllable properties has been applied to evaluate the principle of skin microbial interactions and dynamics [25, 26]. Engineering and rebuilding of skin microbiota could become a powerful tool to help skin diseases treatment in the future.
Interactions between microbiota and human skin
Human skin tissue surface covers approximately 1.8 m2. Together with hair follicles, sebaceous glands and other associated appendages, human skin provides a habitat for > 1010 microbes, with 1 million microbes present per 1 cm2 [27, 28]. Based on the key conditions and the composition of skin microbiota, human skin can be divided into four types of environments: dry, moist, sebaceous (oil) and foot (Fig. 1) [1]. The diversity and abundance of skin microbiota in the state of health and disease are very different [6]. Most skin diseases are proved to be associated with the dysbiosis and imbalance of the skin microbiome [11, 15, 17]. Some strains and their key metabolites may be the biomarkers for diagnosis or therapeutic targets of skin diseases. Studying the interactions between the microbiota and the human skin (including the normal skin microbiota), the factors that disturb the skin microbiota, and the microbial roles in skin wound recovery, is of great interest.
Fig. 1.
Distribution of microorganisms in human skin. Human skin area can be divided into four microenvironments, including dry (forearm and palm), sebaceous (face, back and chest), moist (groin, bend of elbow, and inguinal canal) and foot (plantar heel and toe web space). The relative abundance of viral, bacterial, and fungal components of corresponding skin microbiota are indicated. Bar charts represent relative abundance of microorganism distributed in the human skin, and the white spare in the bar charts represents other bacterial or fungal categories except the described microbes in the bar chart. The front of the human body is shown
Colonization and dynamics of skin microbiota
Microorganisms that colonize the skin include bacteria, eukaryotes and viruses, and their distribution critically depends on the environmental conditions on the skin surface (Fig. 1). In general, people without skin diseases colonized with similar skin microbiota. At the kingdom level, bacteria are the most dominant. At the genus level, Propionibacterium, Corynebacterium and Staphylococcus represent the three most dominant microbes in the skin microbiota, each contributing positively to human health [29]. Since sebaceous glands secrete lipid-rich sebum, sebaceous sites are dominated by P. acnes and other lipophilic Propionibacterium species [30]. The Staphylococcus, Corynebacterium, and other humidity-loving species are abundant in moist areas [31, 32]. Among fungi [33], Malassezia species dominates the core-body and arm sites. Foot sites, the major sites of fungal infection, harbor complex fungal communities composed of Malassezia, Aspergillus, Cryptococcus, Rhodotorula, Epicoccum and other species.
Like the gut microbiome, the skin microbiome is a dynamic system. At the age of three, human gut microbiota typically converges toward an adult-like profile with dramatic transient shifts [34]. By contrast, the skin microbiota undergoes two dramatical changing stages. The first stage is mainly determined by the delivery mode. The skin microbiota of babies delivered vaginally matures earlier than that of babies delivered by cesarean section. Specifically, the alpha diversity of the skin microbiota of babies delivered with cesarean section is lower than that of babies that are vaginally delivered [35]. High levels of Propionibacterium and Streptococcus species were found in babies delivered with cesarean section, while high level of Lactobacillus species was found in babies that were vaginally delivered, presumably derived from the mother’s vagina. After birth, the skin environment undergoes dynamic structural and functional changes, including shifts in pH, water content, trans-epidermal water loss, and sebum production, all of which may influence the maturation of the skin microbiota [36]. The second dramatical change happens at adolescence stage. During puberty and sexual maturation, sebum secretion is exuberant, which supports extensive proliferation of lipophilic bacteria in the skin microbiota.
After these two phases, healthy adults maintain skin microbiota in a dynamic balance, despite the skin microbiota exposure to the environment and other individuals [3]. In fact, skin microbiota undergoes small changes during daily life due to the alteration of host biology and exposure to different environments [4, 37]. Personal care products induce highly individual skin microbiome responses, including alterations in steroid and pheromone levels, dynamics of bacterial and archaeal structure [38]. Many skin care products contain plant-derived extracts which have antimicrobial activities [39], and the antimicrobial extracts may provide a selection pressure, leading to enrichment of resistant strains [40].
The interactions between healthy skin and microbiota
Skin is composed of the surface, epidermis, and dermis (Fig. 2). Numerous microorganisms, resident or transient, colonize the surface, and they use cell debris, sebum, and mineral salts in sweat as nutrients [6]. Compared with the gut milieu, it is more difficult for the environmental microorganisms to colonize the skin. Moreover, some potential antibacterial molecules, such as natural antibiotics released by pioneering microorganisms, free fatty acids and antimicrobial peptides (AMPs) that symbiotic microorganisms secrete, can function as a protective skin barrier [29]. Staphylococcus strains are predominantly present at the skin surface, as they are tolerant of high salt concentration and may even utilize sweat components as nutrients. Some of them co-evolve to exchange mutual benefits with the host [41]. On the skin surface, hairs grow from the skin pores originating from hair follicles. Sebaceous glands are situated at the end of each hair follicle, providing lubrication for the hair follicle. Lipids secreted by sebaceous glands can serve as a source of nutrition for microbial growth. Hair follicle and sebaceous gland form a relatively anaerobic environment, which recruits anaerobic microbes. The microbiome of the deeper layers was described as the host indigenous microbiome, which commonly contains few microorganisms [42].
Fig. 2.
Skin structure and pathogenesis of acne. The skin structure consists of epidermis and dermis. On the skin surface, there are many hair pores, and numerous microorganisms attached. Deep in the dermis, the structure is complex, and it is composed of blood vessels, sweat glands, sebaceous glands, adipocytes, hair follicles, and immune cells. The antigen-presenting cells (APC) identify the abnormalities of attached microbes and secrete lipids, presenting a signal to the T lymphocytes (T cell), which leads to the secretion of inflammatory cytokines, such as INF γ. This leads to eradication of microorganisms by recruited neutrophil and monocytes, but contributes to the redness and formation of acne in the epidermis
The diverse microbial species in the skin environments promote immune tolerance [43], trigger pro-inflammatory responses, and help to maintain skin health. In parallel, the host provides nutrients to the microbes and thus shapes the composition of the microbiota. S. epidermidis provides a good example of a close association between microbiota and the host. S. epidermidis is common on healthy human skin, and it is believed to be a benign microbe. Certain S. epidermidis strains have protective effects, achieved by secreting specific chemicals. In addition, S. epidermis can promote wound repair, enhance skin immunity, and inhibit pathogen infection [44]. This microbiota-host interaction contributes to the stability of microbiota and skin integrity. Some other species, such as Roseomonas mucosa and Malasecia species, can modulate keratinocytes and host immune responses in an environment-dependent manner. P. acnes, an anaerobic bacterium, is one of the most common and essential symbiotic bacteria of human skin [8]. P. acnes metabolizes sebum secretions into fatty acid for survival [45] and helps to maintain the acidic pH of the skin, which provides suitable acidic environment for specific microorganisms [46]. Skin microbiota is a collection of microorganisms that are interacting heavily among themselves, and with our skin. Thus, understanding and maintaining the delicate balance between skin and microbiota are essential steps to give insight into the mechanisms responsible for maintaining healthy skin [47].
Roles of skin microbiota in acne
Acne vulgaris is a chronic inflammatory skin disease, and it is prevalent among teenagers. It affects approximately 85% of young persons, of which 15–20% of cases are severe [48, 49]. In addition to the formation of permanent scars, acne blocks and damages the hair follicles of adolescents. The acne is characterized by features, such as pimples, pustules and nodular cystic lesions, caused by bacteria that enter hair follicles [50, 51]. The occurrence of acne has a negative impact on the physical and psychological health of teenagers; it results in various inconveniences and feeling of inferiority [52–54]. Generally, the sebaceous gland, P. acnes and follicular keratinocytes are considered as the three key factors involved in the development of acne. Firstly, the excessive production of sebum from sebaceous glands blocks the hair follicles [55], leading to a relatively enclosed and anaerobic microenvironment with inflammation. This supports excessive colonization by P. acnes, where the increase of androgen and sebum provides a suitable environment for the growth of P. acnes inside hair follicles [14, 56]. Moreover, some Malassezia species have also been identified as the cause of refractory acne [57, 58].
S. epidermidis, sometimes been regarded as probiotic skin bacteria, can perform glycerol fermentation to produce short-chain fatty acids (SCFAs) which have antimicrobial activities to suppress the growth of P. acnes [9]. The incidence of acne is associated with disorder of the skin microbiota [11, 13]. Concretely, acne might be the result of an unbalanced state between P. acnes and S. epidermidis [59]. The microbiota of acne lesions is diverse at different zones of the body [60]. With the help of omics technologies, the incidence of acne was identified to be associated with some specific P. acnes and S. epidermidis strains in the skin cuticle [12, 13]. Therefore, a specific S. epidermidis strain and other strains in the skin microbiota of the acne can be potential biomarkers to predict the development of acne. Moreover, they can be the targets for accurate diagnosis and treatment of acne. The skin microbiome of patients with grade 1–3 acne was similar. However, patients with grade 4 acne showed a significantly different skin microbiome, including increased alpha diversity and overall increased presence of Gram-negative bacteria [13].
The knowledge of pathophysiology of skin disorder is limited. Host-microbiome interactions, which affect both innate and adaptive immune homeostasis, appear to be an essential factor in acne disease [10]. Besides, acne is mediated by immunity [61], as well as affected by genetics, diet and hygiene factors [54, 62–64]. At present, antibiotics were used to inhibit the growth of P. acnes [65]. As antibiotic resistance is becoming a growing concern in the clinical practice, it is critical to understand the skin microbiome associated with acne and find other strategies for acne treatment.
Roles of skin microbiota in psoriasis, atopic dermatitis, rosacea and other skin diseases
Psoriasis is a common cutaneous disease with multifactorial etiology including genetic and non-genetic factors like diet, drugs, smoking, infection, and mental stress [66]. The pathogenesis of psoriasis is thought to be driven by the interactions between innate immune cells, adaptive immune cells and keratinocytes, in a process mediated by cytokines (including interleukins IL-6, IL-17 and IL-22, interferon and tumor necrosis factor) and other signaling molecules [67]. Significant skin and gut dysbiosis among patients were found to be associated with psoriasis [68]. In psoriasis-affected skin, the alpha diversity of the microbiota is found to be decreased [69]. The microbiota of the psoriasis-affected and immediately adjacent skin was similar. Specially, psoriasis lesioned skin was linked to the increase of Firmicutes, Bacteroidetes and Streptococcus and decrease of Actinobacteria and Propionibacterium [17, 70]. Xanthomonadaceae, assigned to be Proteobacteria and known to be keratolytic, was associated with the clinical improvement after a 3-week balneotherapy treatment [71]. Orally administered probiotics have a positive influence on the course of psoriasis [72]. Therefore, it is possible to develop accurate molecular signatures for the diagnosis of psoriasis from skin microbiome data [73]. Strain-level analyses pointed to psoriatic niche-specific strain adaptation or selection, through revealing strain heterogeneity colonization and functional variability linked with psoriasis [74].
Several different but interdependent factors might lead to atopic dermatitis (AD). The impaired barrier function of AD patients’ skin is very different from healthy skin [75]. The dysbiosis of the skin microbiota was associated with increased colonization of pathogens and a decrease in numbers of beneficial commensals [76, 77]. While the role of dysbiosis in the pathogenesis of atopic dermatitis is unclear, AD patients generally have low-diversity skin microbiota, with predominance of S. aureus [20]. In some situations, overgrowth of S. aureus precedes the development of AD [78]. In another study, One-year old infants with AD were not colonized with S. aureus before developing AD symptoms [77], therefore, investigation of association between abundance of Staphylococci and AD symptoms is of great interest.
The involvement of skin microbiota in rosacea has not been extensively studied, as the main focus has remained on Demodex mites [79]. These ectoparasites are improbable to be the only agent involved in the progression of rosacea. Though antibiotics are effective in treating most rosacea patients [80], it has no effects on Demodex, thus, microbes may be an important pathogenic factor [81]. In many clinical cases, systemic antibiotics are extensively used to control the pustules and papules of rosacea. Comparison of the skin microbiota in rosacea before and after taking oral antibiotics showed that naturally occurring S. aureus and Corynebacterium bovis colonization engendered inflammation in eczematous dermatitis [20].
The host barrier and microbial infection
If the skin barrier is intact, it is normally difficult for the pathogens to invade. Chronic leg ulcers and many other chronic wounds affect 1–2% of the population, and cause increased morbidity and health costs [22]. Skin microbiota, especially specific pathogens, contribute to microbial infection of chronic wounds. S. aureus is often the cause of atopic dermatitis (AD) [82], although S. aureus is at other occasions just a commensal in the skin microbiome [83]. As S. aureus has antibiotic resistance genes, it can result in severe skin and soft tissue infection [84]. Moreover, S. aureus strains are highly diverse, and different patients may be colonized by different S. aureus strains. In a cohort study of diabetic foot ulcers, S. aureus was significantly enriched in non-healing wounds, and some S. aureus strains were associated with high morbidity [85]. R. mucosa depressed the growth of S. aureus in a murine model and in human skin via promoting production of IL-6 [86], showing the possibility to recover novel strains in the skin microbiome and use them to ameliorate dermatological disease.
S. aureus and P. aeruginosa are the most common bacteria isolated from chronic wounds [22]. They can express surface proteins and virulence factors to inhibit and decelerate wound healing. The co-infection of S. aureus and P. aeruginosa is more virulent than that of single infection [22]. The S. aureus strains can weaken skin barrier and activate deleterious host immune reactions [4]. It is known that dysbiosis of human skin microbiota could trigger immune dysregulation and subsequently an inflammatory response. Thus, human skin microbiota plays a key role in clinical manifestations [87].
Gut-brain-skin axis
The gut microbiome influences skin and other distant organ systems [88]. The relationships of gut microbiome with distant organs are conceptualized as the gut–brain axis, gut–lung axis, gut–skin axis and other gut-X axes [89, 90]. The skin and gut barrier are highly similar, and they share many features [91]. Both gut and skin surfaces are covered by epithelial cells (ECs), and they contact with the exogenous environment directly [92]. With high cellular turnover rate, the adherence and infection to the gut and skin by microbes are difficult [93]. The association between skin and gut is mediated by the host immune system [94].Normally, allergic food ingredients impair the intestinal barrier and lead to food allergies, including gut and skin symptoms [95].
There is bidirectional link between gut and skin dysbiosis, and the gut microbiota dysbiosis is the response to the pathophysiology of multiple inflammatory skin diseases [96]. The composition of gut microbiota in psoriasis patients was found to vary significantly, and the Firmicutes and Bacteroidetes ratios of persons with psoriasis differ from healthy ones [17, 19]. Skin exposure to ultraviolet B light (UVB) can enhance serum vitamin D levels indirectly, which can be correlated to an increase in alpha and beta diversities of Proteobacteria phylum in the gut microbiota [97]. Moreover, gastrointestinal disorders are associated with certain dermatoses, and 7–11% of patients with IBD suffer from psoriasis [98]. Augmenting or repairing a leaky gut barrier is often applied as adjuvant therapy in the treatment of inflammatory skin diseases, which helps to increase the efficacy of standard dermatotherapy.
In the future, the skin disease treatment strategy using gut microbiota might be realized by modulation of the gut microbiota using dietary agents or selected natural/synthetic microbiota [99]. The gut–skin axis was proposed to be an integral part of the gut–brain–skin axis [100]. For example, chronic wound conditions and depression share some common pathologic features, such as altered microbiome and dysregulated inflammation. It is believed that intestine and skin can be seen as one system, and the gut-skin axis should be applied in the treatment of skin diseases. Traditional Chinese medicine (TCM) has clinical experience on the modulation of the intestinal tract and skin diseases. Nowadays, the gut-skin axis evidence is based on limited cohort studies, and a multi-systematic study with a large cohort would provide better insights into the skin microbiota and their relationships with gut microbiota [101]. This approach of studying the gut-skin axis would highlight and reveal mechanisms of action of TCM.
Strategies for skin diseases treatment
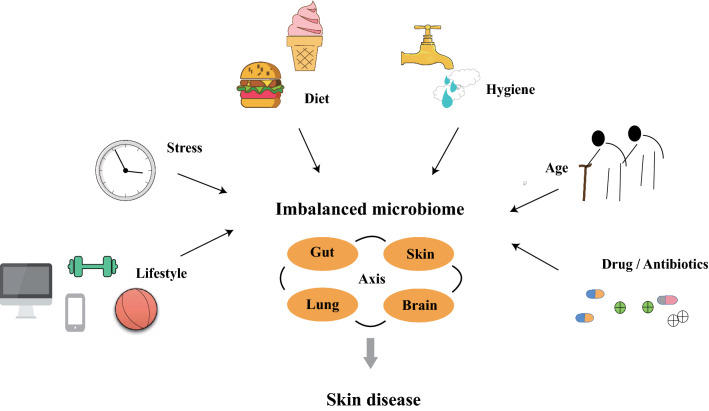
Unbalanced microbiota and specific strains can cause or contribute to skin diseases. Several lifestyles are associated with skin health (Fig. 3). Exercising is beneficial to maintaining healthy skin [102]. Although regular exercise can protect the skin from free radicals, training with extreme intensity or duration or lack of training may mediate oxidative stress and contribute to skin carcinogenesis [103]. Living in a polluted environment may reduce skin moisture, increase the rate of sebum excretion, and tends to exacerbate the symptoms of chronic inflammatory skin diseases [104]. The ‘‘hygiene hypothesis’’ illustrates that lack of exposure to a full range of microbiota during childhood may lead to failure in “training” the immune system [105]. This is proposed to decrease the resistance to microbial pathogens and increase vulnerability to infection and other ailments. AD and other skin atopic diseases might be related to the excessively clean (abiotic) environment [77].
Fig. 3.
Factors associated with skin diseases. Lifestyle, stress, diet, hygiene, age, and drug (antibiotics) application are closely related to skin diseases through the links between gut, skin, lung and brain axis
Traditional treatments for acne and other skin diseases
Treating inflammatory or mixed acne with topical antibiotics has been prevalent for over 40 years [106]. In addition to antimicrobial effects targeting P. acnes, oral antibiotics have anti-inflammatory effects via host immune responses [107]. Topical ofloxacin possess potent antimicrobial activity against the Propionibacterium and Staphylococci strains isolated from acne patients, making it an effective therapeutic drug for acne vulgaris [108]. Based on anti-inflammatory properties, topical retinoids are effective in the treatment of inflammatory lesions [109]. Several treatment guidelines and expert consensus documents suggest that macrolides, clindamycin and tetracyclines are recommended as the first-line therapy drugs in acute inflammatory phase of acne [110, 111].
Nowadays, nucleic acids (NAs) play significant roles in the treatment of several diseases. Topical NAs or NAs-based delivery of drugs have special advantages in skin disease treatment, due to the efficient NAs transfer and direct targeting to the skin disease sites [112]. Multitudes of NA-based therapeutics, including genes, siRNA, aptamers, antisense oligodeoxynucleotides (ODNs), and CpG oligonucleotides, have been applied for disease treatment [112]. Transdermal drug delivery (TDDS) has been highly explored, due to the advantages of special tissue targeting, improved drug release, avoiding the presystemic metabolism, high tolerance in patients, and less toxic to the patient’s liver [113]. Currently, a variety of nanoparticles and nanoemulsions are applied in TDDS to treat psoriasis, wound healing, melanoma and other skin diseases [114]. Low-level laser (light) therapy (LLLT) is a fast-developing technology for the treatment of diseases that require relief of pain and inflammation, function restoration, or healing stimulation. LLLT has positive effects on wrinkles, acne scars, hypertrophic scars, healing of burns, psoriasis and acne, and other inflammatory diseases [115]. LLLT can reduce UV damages both as a treatment strategy and as a prophylaxis. Its non-invasive characteristics and almost complete absence of side-effects warrant further exploration and application in dermatology.
Healthy diets and lifestyles affect skin microbiota
Despite strong lay beliefs that diet is not a dominant factor in acne, increased self-reported cases show that adolescent acne is associated with frequent consumption of milk and milk-containing foods (instant breakfast drink, cottage cheese, and cream cheese), and nonfat portion of milk seems to have a stronger association with acne than that of whole or low-fat milk [116, 117]. The skimmed milk contains hormones and bioactive molecules, and it may have an acne stimulating effect due to androgens, progesterone, and insulin growth factor-1 (IGF-1) [116]. Western diets, which are typically high-glycemic-load (HGL) diets, can elevate IGF-1 and blood insulin levels chronically or acutely, which leads to increased sebum production and even acne [118]. There are positive correlations between acne severity and high glycemic load foods [64]. As a result, dermatologists usually suggest that acne patients avoid high glycemic index foods [64].
Personal hygiene is closely related to acne, and too much washing may actually worsen the condition. Generally, washing twice daily appears to be the best recommendation [119]. Two independent groups of studies among young students suggested a correlation between acne severity and stress levels during examination periods [120, 121]. Regular exercise (4 h per week high-intensity aerobic exercise) reduced the thinning of the stratum corneum in patients compared with that in sedentary controls (1 h per week high-intensity aerobic exercise) [122]. The expression level of Pgc-1 (the master regulator of mitochondrial biogenesis) increased after exercise [123], which can decrease aging of skin cells. Additionally, vigorous exercise, especially calisthenics and aerobic exercise, was independently linked to reduced risk of psoriasis in US women [124]. Therefore, proper diet, cleaning, moisturizing and exercise should be applied together with other adopted skin disease therapeutic strategies [14].
Designed mixture of probiotics and prebiotics
The first generation of microbiota therapies consisted of probiotics and prebiotics. They are designed to maintain, restore and optimize the skin microbiota in different ways. Topical applications of probiotics are beneficial for cutaneous immune responses and eliminate pathogens by enhancing the skin natural defense barriers or producing antimicrobial peptides [125]. Prebiotics are non-digestible foods or metabolites degraded by the gut bacteria [126]. Prebiotics were applied in cosmetic formulations directly, in order to promote the growth and activity of beneficial skin microbiota [125]. Among the prebiotics, plants, especially traditional Chinese medical plants, can provide diverse natural products for skin microbiota. Insights into the traditional Chinese medical plants identified various bioactive components. As plant growth takes long time and extraction of the bioactive natural products is difficult, engineered yeasts were used for the efficient production of these bioactive natural products [127, 128]. Several traditional Chinese medical plant-derived natural products, such as ginsenosides, monoterpenoids, glycyrrhetinic acids, have been successfully synthesized using engineered yeasts [129, 130].
Based on the gut and skin microbiome, new probiotic and prebiotic products for the treatment of multitudinous skin conditions can be developed [131] (Fig. 4A). Probiotics derived from S. epidermidis can help restore the naturally balanced microbiota and regulate the host’s AMP mediators [59]. Coagulase-negative Staphylococci (CoNS) strains are commonly distributed on the skin surfaces of healthy persons, but are rare on that of the AD patients [132]. The antimicrobial activity of CoNS species, such as S. epidermidis and S. hominis, is based on AMPs they produce. Absence of CoNS strains may lead to excessive colonization of S. aureus on the skin of AD patients. The AMPs were found to be strain-specific and highly potent; They can selectively kill S. aureus and synergize with the human AMP LL-37 [132].
Fig. 4.
Microbiome strategies used to modulate skin microbiome in a balanced state. A Methods to discover/build new probiotics, prebiotics and synbiotics. Isolating and identifying bacteria from healthy gut or skin microbiome to discover new probiotics would lead to discover of functional probiotics. Probiotics with beneficial functions to skin health can be engineered or built using synthetic biology strategies. Combination of these probiotics and functional prebiotics form synbiotics. B Functional verification of synbiotics and healthy microbiome in vivo. Designed synbiotics (A) and balanced microbiome from the healthy mice to the mice with skin diseases can revert their imbalanced microbiome. C Microbiome transplantation to balance skin microbiota in clinical applications. Transplantation of healthy microbiome from healthy persons’ skin as a whole or specifically isolated probiotics and engineered probiotics, can revert the imbalance
Mixtures of different skin microorganisms in definite proportions can change the composition of the recipients’ skin microbiomes [133]. After sequential applications of a donor microbiome, the recipients’ microbiota becomes similar to the donors’ microbiota gradually, showing that using living bacteria to modulate skin microbiome composition is possible (Fig. 4B). Moreover, transferring probiotic solutions from facial skin microbes of healthy volunteers to the faces of acne patients can improve skin health (Fig. 4C). Application of natural bacteria onto the skin can improve moisture retention and decrease skin pH [134], and it opens the possibility to develop probiotic solutions which might help the human skin revert from disease microbiota state to a healthy state.
Manipulation of skin microbiota with increased abundance of beneficial species may reduce the presence of undesirable pathogens and promote skin health directly [135]. Some specific microorganisms in the skin microbiome can reduce the colonization and overgrowth of P. acnes by fermenting glycerol and creating inhibition zones [136]. Clinical isolates of CoNS species residing on normal skin microbiota produce autoinducing peptides to disturb the quorum sensing system of S. aureus, which decreases phenol-soluble modulin (PSM) expression and abolishes biofilm attachment and regrowth. A clinical isolate of S. hominis synthesizes an autoinducing peptide (SYNVCGGYF), which is a highly potent inhibitor of S. aureus Agr-mediated quorum sensing, and this could prevent S. aureus–mediated epithelial damage and inflammation on murine skin [137]. Oral probiotic interventions were investigated for clinical application in diverse diseases, but external skin commensals used to treat skin diseases are rarely reported [138]. Further discovery of probiotic functions in skin microbiota would be useful in future skin disease treatments. Synbiotics is the combination of prebiotics and probiotics. In the future, synbiotics will expand out possibilities to effectively treat skin diseases (Fig. 4).
Engineering and rebuilding of skin microbiota
Synthetic biology strategies applied on the microbiome can lead to characterization of underlying roles of skin microbiota in disease development, and develop novel diagnostic and therapeutic strategies for skin diseases. Different omics techniques: metagenome, metatranscriptome, metaproteome, and metabolome, can give systematic and global view of the skin microbiota. Biosensors, memory arrays, engineered bacteria, and other tools can be sued to rewire the microbiome [139]. Quick sensing of a stimulus in situ that can immediately trigger a precise therapeutic strategy would rebalance the dysbiosis of the skin microbiome and contribute to curing the skin diseases [139].
The knowledge gap in our understanding of the link between the skin microbiota and skin diseases is still substantial, and the application of probiotics in skin microbiota modulation has not been realized yet. It remains a challenge to reach a conclusion about the proper dose and formulation of probiotics [140]. Moreover, identification of skin probiotics is difficult, due to the genetic differences between strains [141]. Therefore, development of engineered strains with desired beneficial functions is necessary, and synthetic biology strategies could provide gene editing and other tools for strain reprograming (Fig. 4A). The engineered probiotics and the functional prebiotics they produce could be applied for the treatment of skin disease directly. The gene editing tool can be delivered with phages to remove designated pathogenic strains [142], showing the potential for future skin disease treatment based on targeted microbiota modulation. In summary, advanced synthetic biology enables new approaches to design and reprogram multispecies microbiota, which presents an exciting opportunity to rationally engineer or rebuild skin microbiota for skin disease treatment [26, 139].
Future perspectives and conclusion
Skin microbiota plays essential roles in skin disease occurrence and development. Therefore, modulation of skin microbiota is one of the best strategies used for skin disease treatment. At present, diverse microbiota modulation strategies are available, including prebiotics and probiotics. Nowadays, some skin care products contain herbal ingredients and other prebiotics, in order to maintain skin healthy. Though oral probiotics products are regularly used to improve intestinal flora via relieving indigestion and intestinal inflammation, few probiotics are applied in the modulation of skin microbiota. This is mainly due to the fact that the available information on skin microbiota is limited. The omics technologies can give insights into the interaction between skin microbiota and skin diseases and identify microbial markers for the diagnosis and treatment of skin diseases. Species-level and strain-level information of the microbiota related with diverse skin diseases need to be revealed. Clinical isolation of pathogenic strains and probiotics will help reveal the causal relationship between skin microbiota and skin diseases.
In the future, in situ monitoring of skin microbiota at the disease sites and identifying the pathogenic strains will provide crucial preliminary information for skin disease treatment. Further precise treatment strategies, such as addition of prebiotics and probiotics, engineering or rebuilding synthetic microbiota with desired characteristics, will be applied in clinics. The future treatment pipeline will comprise the following steps: recovering personal skin microbiota, isolating the key pathogenic bacteria, revealing pathogenic mechanisms, and developing effective prebiotics/probiotics agents to counter specific pathogens. The skin microbiota modulation strategy will not only relieve the occurrence and development of skin diseases, but also improve the appearance and maintain physical and mental health.
Author contributions
YW conceived the study. YY and YW drafted the manuscript, and YY prepared the figures. LQ and IM revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32111530179), Clinical Laboratories, Shenyou Bio, and Novo Nordisk Foundation (NNF10CC1016517) to IM.
Availability of data and materials
Not Applicable.
Declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that there are no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 2.Lunjani N, Hlela C, O'Mahony L. Microbiome and skin biology. Curr Opin Allergy Clin Immunol. 2019;19:328–333. doi: 10.1097/ACI.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 3.Oh J, et al. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemter AM, Nagler CR. Influences on allergic mechanisms through gut, lung, and skin microbiome exposures. J Clin Invest. 2019;129:1483–1492. doi: 10.1172/JCI124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langan EA, et al. The role of the microbiome in psoriasis: moving from disease description to treatment selection? Br J Dermatol. 2018;178:1020–1027. doi: 10.1111/bjd.16081. [DOI] [PubMed] [Google Scholar]
- 6.Flowers L, Grice EA. The skin microbiota: balancing risk and reward. Cell Host Microbe. 2020;28:190–200. doi: 10.1016/j.chom.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YE, Fischbach MA, Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553:427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitz-Gibbon S, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Investig Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. A precision microbiome approach using sucrose for selective augmentation of staphylococcus epidermidis fermentation against propionibacterium acnes. Int J Mol Sci. 2016 doi: 10.3390/ijms17111870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Neill AM, Gallo RL. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome. 2018;6:177. doi: 10.1186/s40168-018-0558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. 2016;6:39491. doi: 10.1038/srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreno B. What is new in the pathophysiology of acne, an overview. J Eur Acad Dermatol Venereol. 2017;31(Suppl 5):8–12. doi: 10.1111/jdv.14374. [DOI] [PubMed] [Google Scholar]
- 13.Li CX, You ZX, Lin YX, Liu HY, Su J. Skin microbiome differences relate to the grade of acne vulgaris. J Dermatol. 2019;46:787–790. doi: 10.1111/1346-8138.14952. [DOI] [PubMed] [Google Scholar]
- 14.Rocha MA, Bagatin E. Skin barrier and microbiome in acne. Arch Dermatol Res. 2018;310:181–185. doi: 10.1007/s00403-017-1795-3. [DOI] [PubMed] [Google Scholar]
- 15.Alekseyenko AV, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis DJ, Chan WH, Hinojosa T, Hsu S, Feldman SR. Mechanisms of microbial pathogenesis and the role of the skin microbiome in psoriasis: a review. Clin Dermatol. 2019;37:160–166. doi: 10.1016/j.clindermatol.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Yan D, et al. The role of the skin and gut microbiome in psoriatic disease. Curr Dermatol Rep. 2017;6:94–103. doi: 10.1007/s13671-017-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang HW, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. 2018;6:154. doi: 10.1186/s40168-018-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Shi L, Sun T, Guo K, Geng S. Dysbiosis of gut microbiota and its correlation with dysregulation of cytokines in psoriasis patients. BMC Microbiol. 2021;21:78. doi: 10.1186/s12866-021-02125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42:756–766. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serra R, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. 2015;13:605–613. doi: 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 23.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 24.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji B, Herrgård MJ, Nielsen J. Microbial community dynamics revisited. Nature Comput Sci. 2021;1:640–641. doi: 10.1038/s43588-021-00144-6. [DOI] [PubMed] [Google Scholar]
- 26.Johns NI, Blazejewski T, Gomes AL, Wang HH. Principles for designing synthetic microbial communities. Curr Opin Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grice EA, Segre JA. Erratum: the skin microbiome. Nat Rev Microbiol. 2011;9:626–626. doi: 10.1038/nrmicro2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scharschmidt TC, Fischbach MA. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov Today Dis Mech. 2013 doi: 10.1016/j.ddmec.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 31.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu DM, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Younge NE, Araujo-Perez F, Brandon D, Seed PC. Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome. 2018;6:98. doi: 10.1186/s40168-018-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkins D, Leung MH, Lee PK. Microbiota fingerprints lose individually identifying features over time. Microbiome. 2017;5:1. doi: 10.1186/s40168-016-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouslimani A, et al. The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 2019;17:47. doi: 10.1186/s12915-019-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kareru PG, et al. Antimicrobial activities of skincare preparations from plant extracts. Afr J Tradit Complement Altern Med. 2010;7:214–218. doi: 10.4314/ajtcam.v7i3.54777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat Commun. 2020;11:1427. doi: 10.1038/s41467-020-15222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeeuwen PL, et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012;13:R101. doi: 10.1186/gb-2012-13-11-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25:370–377. doi: 10.1016/j.smim.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, et al. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown SK, Shalita AR. Acne vulgaris. Lancet. 1998;351:1871–1876. doi: 10.1016/s0140-6736(98)01046-0. [DOI] [PubMed] [Google Scholar]
- 46.Youn SH, Choi CW, Choi JW, Youn SW. The skin surface pH and its different influence on the development of acne lesion according to gender and age. Skin Res Technol. 2013;19:131–136. doi: 10.1111/srt.12023. [DOI] [PubMed] [Google Scholar]
- 47.Sfriso R, Egert M, Gempeler M, Voegeli R, Campiche R. Revealing the secret life of skin—with the microbiome you never walk alone. Int J Cosmet Sci. 2020;42:116–126. doi: 10.1111/ics.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhate K, Williams HC. Epidemiology of acne vulgaris. Br J Dermatol. 2013;168:474–485. doi: 10.1111/bjd.12149. [DOI] [PubMed] [Google Scholar]
- 49.Ghodsi SZ, Orawa H, Zouboulis CC. Prevalence, severity, and severity risk factors of acne in high school pupils: a community-based study. J Invest Dermatol. 2009;129:2136–2141. doi: 10.1038/jid.2009.47. [DOI] [PubMed] [Google Scholar]
- 50.Kurokawa I, et al. New developments in our understanding of acne pathogenesis and treatment. Exp Dermatol. 2009;18:821–832. doi: 10.1111/j.1600-0625.2009.00890.x. [DOI] [PubMed] [Google Scholar]
- 51.Zaenglein AL, et al. Guidelines of care for the management of acne vulgaris. J Am Acad Dermatol. 2016;74:945–973 e933. doi: 10.1016/j.jaad.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 52.Duru P, Orsal O. The effect of acne on quality of life, social appearance anxiety, and use of conventional, complementary, and alternative treatments. Complement Ther Med. 2021;56:102614. doi: 10.1016/j.ctim.2020.102614. [DOI] [PubMed] [Google Scholar]
- 53.Clark AK, Haas KN, Sivamani RK. Edible plants and their influence on the gut microbiome and acne. Int J Mol Sci. 2017 doi: 10.3390/ijms18051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, et al. Daily intake of soft drinks and moderate-to-severe acne vulgaris in chinese adolescents. J Pediatr. 2019;204:256–262 e253. doi: 10.1016/j.jpeds.2018.08.034. [DOI] [PubMed] [Google Scholar]
- 55.Tom WL, Barrio VR. New insights into adolescent acne. Curr Opin Pediatr. 2008;20:436–440. doi: 10.1097/MOP.0b013e328305e273. [DOI] [PubMed] [Google Scholar]
- 56.Kanwar IL, et al. Models for acne: a comprehensive study. Drug Discov Ther. 2018;12:329–340. doi: 10.5582/ddt.2018.01079. [DOI] [PubMed] [Google Scholar]
- 57.Akaza N, et al. Malassezia globosa tends to grow actively in summer conditions more than other cutaneous Malassezia species. J Dermatol. 2012;39:613–616. doi: 10.1111/j.1346-8138.2011.01477.x. [DOI] [PubMed] [Google Scholar]
- 58.Numata S, et al. Analysis of facial skin-resident microbiota in Japanese acne patients. Dermatology. 2014;228:86–92. doi: 10.1159/000356777. [DOI] [PubMed] [Google Scholar]
- 59.Claudel JP, et al. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology. 2019;235:287–294. doi: 10.1159/000499858. [DOI] [PubMed] [Google Scholar]
- 60.Dreno B, et al. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp Dermatol. 2017;26:798–803. doi: 10.1111/exd.13296. [DOI] [PubMed] [Google Scholar]
- 61.Dréno B, et al. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32:5–14. doi: 10.1111/jdv.15043. [DOI] [PubMed] [Google Scholar]
- 62.Melnik B. Acne vulgaris. Role of diet. Hautarzt. 2010;61:115–125. doi: 10.1007/s00105-009-1831-0. [DOI] [PubMed] [Google Scholar]
- 63.Jung JY, et al. Effect of dietary supplementation with omega-3 fatty acid and gamma-linolenic acid on acne vulgaris: a randomised, double-blind, controlled trial. Acta Derm Venereol. 2014;94:521–525. doi: 10.2340/00015555-1802. [DOI] [PubMed] [Google Scholar]
- 64.Mahmood SN, Bowe WP. Diet and acne update: carbohydrates emerge as the main culprit. J Drugs Dermatol. 2014;13:428–435. [PubMed] [Google Scholar]
- 65.Xu H, Li H. Acne, the skin microbiome, and antibiotic treatment. Am J Clin Dermatol. 2019;20:335–344. doi: 10.1007/s40257-018-00417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng J, Luo S, Huang Y, Lu Q. Critical role of environmental factors in the pathogenesis of psoriasis. J Dermatol. 2017;44:863–872. doi: 10.1111/1346-8138.13806. [DOI] [PubMed] [Google Scholar]
- 67.Martins AM, Ascenso A, Ribeiro HM, Marto J. The brain-skin connection and the pathogenesis of psoriasis: a review with a focus on the serotonergic system. Cells. 2020 doi: 10.3390/cells9040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sikora M, et al. Gut microbiome in psoriasis: an updated review. Pathogens. 2020 doi: 10.3390/pathogens9060463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hidalgo-Cantabrana C, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br J Dermatol. 2019;181:1287–1295. doi: 10.1111/bjd.17931. [DOI] [PubMed] [Google Scholar]
- 70.Langan EA, et al. Combined culture and metagenomic analyses reveal significant shifts in the composition of the cutaneous microbiome in psoriasis. Br J Dermatol. 2019;181:1254–1264. doi: 10.1111/bjd.17989. [DOI] [PubMed] [Google Scholar]
- 71.Martin R, Henley JB, Sarrazin P, Seite S. Skin microbiome in patients with psoriasis before and after balneotherapy at the Thermal Care Center of La Roche-Posay. J Drugs Dermatol. 2015;14:1400–1405. [PubMed] [Google Scholar]
- 72.Olejniczak-Staruch I, et al. Alterations of the skin and gut microbiome in psoriasis and psoriatic arthritis. Int J Mol Sci. 2021 doi: 10.3390/ijms22083998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Statnikov A, et al. Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci Rep. 2013;3:2620. doi: 10.1038/srep02620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tett A, et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes. 2017;3:14. doi: 10.1038/s41522-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387:1109–1122. doi: 10.1016/s0140-6736(15)00149-x. [DOI] [PubMed] [Google Scholar]
- 76.Myles IA, et al. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight. 2016 doi: 10.1172/jci.insight.86955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kennedy EA, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139:166–172. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meylan P, et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol. 2017;137:2497–2504. doi: 10.1016/j.jid.2017.07.834. [DOI] [PubMed] [Google Scholar]
- 79.Chang YS, Huang YC. Role of Demodex mite infestation in rosacea: a systematic review and meta-analysis. J Am Acad Dermatol. 2017;77:441–447 e446. doi: 10.1016/j.jaad.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 80.van Zuuren EJ, Kramer SF, Carter BR, Graber MA, Fedorowicz Z. Effective and evidence-based management strategies for rosacea: summary of a Cochrane systematic review. Br J Dermatol. 2011;165:760–781. doi: 10.1111/j.1365-2133.2011.10473.x. [DOI] [PubMed] [Google Scholar]
- 81.Jarmuda S, et al. Potential role of Demodex mites and bacteria in the induction of rosacea. J Med Microbiol. 2012;61:1504–1510. doi: 10.1099/jmm.0.048090-0. [DOI] [PubMed] [Google Scholar]
- 82.von Eiff C, Proctor RA, Peters G. Coagulase-negative Staphylococci—pathogens have major role in nosocomial infections. Postgrad Med. 2015;110:63–76. doi: 10.3810/pgm.2001.10.1046. [DOI] [PubMed] [Google Scholar]
- 83.Krismer B, Weidenmaier C, Zipperer A, Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol. 2017;15:675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- 84.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalan LR, et al. Strain- and species-level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe. 2019;25:641–655 e645. doi: 10.1016/j.chom.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romano-Bertrand S, et al. Skin microbiota is the main reservoir of Roseomonas mucosa, an emerging opportunistic pathogen so far assumed to be environmental. Clin Microbiol Infect. 2016;22(737):e731–737.e737. doi: 10.1016/j.cmi.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 87.Fercek I, et al. Features of the skin microbiota in common inflammatory skin diseases. Life (Basel) 2021 doi: 10.3390/life11090962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salem I, Ramser A, Isham N, Ghannoum MA. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front Microbiol. 2018;9:1459. doi: 10.3389/fmicb.2018.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arck P, et al. Is there a 'gut-brain-skin axis'? Exp Dermatol. 2010;19:401–405. doi: 10.1111/j.1600-0625.2009.01060.x. [DOI] [PubMed] [Google Scholar]
- 90.Wei Y, Li X, Ji B, Qu L. Recent advances on the recovery, modulation and synthetic biology of gut microbiota and hosts. Scientia Sinica Vitae. 2022;52:249. doi: 10.1360/SSV-2021-0088. [DOI] [Google Scholar]
- 91.De Pessemier B, et al. Gut-skin axis: current knowledge of the interrelationship between microbial dysbiosis and skin conditions. Microorganisms. 2021 doi: 10.3390/microorganisms9020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaykhiev R, Bals R. Interactions between epithelial cells and leukocytes in immunity and tissue homeostasis. J Leukoc Biol. 2007;82:1–15. doi: 10.1189/jlb.0207096. [DOI] [PubMed] [Google Scholar]
- 93.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 94.Salem I, Ramser A, Isham N, Ghannoum MA. The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol. 2018;9:1459. doi: 10.3389/fmicb.2018.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y, et al. Advances in the relationships between cow’s milk protein allergy and gut microbiota in infants. Front Microbiol. 2021 doi: 10.3389/fmicb.2021.716667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shah KR, Boland CR, Patel M, Thrash B, Menter A. Cutaneous manifestations of gastrointestinal disease. J Am Acad Dermatol. 2013;68(189):e181–189.e121. doi: 10.1016/j.jaad.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 97.Bosman ES, Albert AY, Lui H, Dutz JP, Vallance BA. Skin exposure to narrow band ultraviolet (UVB) light modulates the human intestinal microbiome. Front Microbiol. 2019;10:2410. doi: 10.3389/fmicb.2019.02410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang BL, Chandra S, Shih DQ. Skin manifestations of inflammatory bowel disease. Front Physiol. 2012;3:13. doi: 10.3389/fphys.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Neill CA, Monteleone G, McLaughlin JT, Paus R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. BioEssays. 2016;38:1167–1176. doi: 10.1002/bies.201600008. [DOI] [PubMed] [Google Scholar]
- 100.Bowe WP, Logan AC. Acne vulgaris, probiotics and the gut-brain-skin axis—back to the future? Gut Pathog. 2011;3:1. doi: 10.1186/1757-4749-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hadian Y, et al. Microbiome-skin-brain axis: a novel paradigm for cutaneous wounds. Wound Repair Regen. 2020;28:282–292. doi: 10.1111/wrr.12800. [DOI] [PubMed] [Google Scholar]
- 102.Yeh C, Flatley E, Elkattawy O, Berger L, Rao B. Exercise in dermatology: exercise's influence on skin aging, skin cancer, psoriasis, venous ulcers, and androgenetic alopecia. J Am Acad Dermatol. 2021 doi: 10.1016/j.jaad.2021.07.023. [DOI] [PubMed] [Google Scholar]
- 103.Kruk J, Duchnik E. Oxidative stress and skin diseases: possible role of physical activity. Asian Pac J Cancer Prev. 2014;15:561–568. doi: 10.7314/apjcp.2014.15.2.561. [DOI] [PubMed] [Google Scholar]
- 104.Fuks KB, Woodby B, Valacchi G. Skin damage by tropospheric ozone. Hautarzt. 2019 doi: 10.1007/s00105-018-4319-y. [DOI] [PubMed] [Google Scholar]
- 105.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Titus S, Hodge J. Diagnosis and treatment of acne. Am Fam Physician. 2012;86:734–740. [PubMed] [Google Scholar]
- 107.Dreno B, et al. Antibiotic stewardship in dermatology: limiting antibiotic use in acne. Eur J Dermatol. 2014;24:330–334. doi: 10.1684/ejd.2014.2309. [DOI] [PubMed] [Google Scholar]
- 108.Nakajima A, et al. Antimicrobial activities of ozenoxacin against isolates of propionibacteria and staphylococci from Japanese patients with acne vulgaris. J Med Microbiol. 2016;65:745–750. doi: 10.1099/jmm.0.000293. [DOI] [PubMed] [Google Scholar]
- 109.Thielitz A, Abdel-Naser MB, Fluhr JW, Zouboulis CC, Gollnick H. Topical retinoids in acne - an evidence-based overview. J Dtsch Dermatol Ges. 2008;6:1023–1031. doi: 10.1111/j.1610-0387.2008.06741.x. [DOI] [PubMed] [Google Scholar]
- 110.Hayashi N, et al. Japanese dermatological association guidelines: guidelines for the treatment of acne vulgaris 2017. J Dermatol. 2018;45:898–935. doi: 10.1111/1346-8138.14355. [DOI] [PubMed] [Google Scholar]
- 111.Thiboutot DM, et al. Practical management of acne for clinicians an international consensus from the Global Alliance to Improve Outcomes in Acne. J Am Acad Dermatol. 2018;78(23):S1–S23 e21. doi: 10.1016/j.jaad.2017.09.078. [DOI] [PubMed] [Google Scholar]
- 112.Zakrewsky M, Kumar S, Mitragotri S. Nucleic acid delivery into skin for the treatment of skin disease: proofs-of-concept, potential impact, and remaining challenges. J Control Release. 2015;219:445–456. doi: 10.1016/j.jconrel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Palmer BC, DeLouise LA. Nanoparticle-enabled transdermal drug delivery systems for enhanced dose control and tissue targeting. Molecules. 2016 doi: 10.3390/molecules21121719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carter P, Narasimhan B, Wang Q. Biocompatible nanoparticles and vesicular systems in transdermal drug delivery for various skin diseases. Int J Pharm. 2019;555:49–62. doi: 10.1016/j.ijpharm.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 115.Avci P, et al. Low-level laser (light) therapy (LLLT) in skin: stimulating, healing, restoring. Semin Cutan Med Surg. 2013;32:41–52. [PMC free article] [PubMed] [Google Scholar]
- 116.Adebamowo CA, et al. High school dietary dairy intake and teenage acne. J Am Acad Dermatol. 2005;52:207–214. doi: 10.1016/j.jaad.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 117.Aghasi M, et al. Dairy intake and acne development: a meta-analysis of observational studies. Clin Nutr. 2019;38:1067–1075. doi: 10.1016/j.clnu.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 118.Smith RN, Mann NJ, Braue A, Makelainen H, Varigos GA. A low-glycemic-load diet improves symptoms in acne vulgaris patients: a randomized controlled trial. Am J Clin Nutr. 2007;86:107–115. doi: 10.1093/ajcn/86.1.107. [DOI] [PubMed] [Google Scholar]
- 119.Choi JM, Lew VK, Kimball AB. A single-blinded, randomized, controlled clinical trial evaluating the effect of face washing on acne vulgaris. Pediatr Dermatol. 2006;23:421–427. doi: 10.1111/j.1525-1470.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 120.Yosipovitch G, et al. Study of psychological stress, sebum production and acne vulgaris in adolescents. Acta Derm Venereol. 2007;87:135–139. doi: 10.2340/00015555-0231. [DOI] [PubMed] [Google Scholar]
- 121.Chiu A, Chon SY, Kimball AB. The response of skin disease to stress—changes in the severity of acne vulgaris as affected by examination stress. Arch Dermatol. 2003;139:897–900. doi: 10.1001/archderm.139.7.897. [DOI] [PubMed] [Google Scholar]
- 122.Crane JD, et al. Exercise-stimulated interleukin-15 is controlled by AMPK and regulates skin metabolism and aging. Aging Cell. 2015;14:625–634. doi: 10.1111/acel.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baar K, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 124.Frankel HC, Han J, Li T, Qureshi AA. The association between physical activity and the risk of incident psoriasis. Arch Dermatol. 2012;148:918–924. doi: 10.1001/archdermatol.2012.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Al-Ghazzewi FH, Tester RF. Impact of prebiotics and probiotics on skin health. Benef Microbes. 2014;5:99–107. doi: 10.3920/BM2013.0040. [DOI] [PubMed] [Google Scholar]
- 126.Delbes AS, et al. Prebiotics supplementation impact on the reinforcing and motivational aspect of feeding. Front Endocrinol (Lausanne). 2018;9:273. doi: 10.3389/fendo.2018.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wei Y, Ji B, Ledesma-Amaro R, Chen T, Ji X-J. Editorial: engineering yeast to produce plant natural products. Front Bioengineering Biotechnology. 2021 doi: 10.3389/fbioe.2021.798097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wei Y. Synthetic biology of yeasts: tools and applications. Berlin: Springer International Publishing; 2022. pp. 181–205. [Google Scholar]
- 129.Gao Q, Wang L, Zhang M, Wei Y, Lin W. Recent advances on feasible strategies for monoterpenoid production in Saccharomyces cerevisiae. Front Bioengineering Biotechnology. 2020;8:1372. doi: 10.3389/fbioe.2020.609800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guan R, et al. Metabolic engineering for glycyrrhetinic acid production in Saccharomyces cerevisiae. Front Bioengineering Biotechnology. 2020;8:1318. doi: 10.3389/fbioe.2020.588255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maguire M, Maguire G. The role of microbiota, and probiotics and prebiotics in skin health. Arch Dermatol Res. 2017;309:411–421. doi: 10.1007/s00403-017-1750-3. [DOI] [PubMed] [Google Scholar]
- 132.Nakatsuji T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paetzold B, et al. Skin microbiome modulation induced by probiotic solutions. Microbiome. 2019;7:95. doi: 10.1186/s40168-019-0709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nodake Y, et al. Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe–a blinded randomized clinical trial. J Dermatol Sci. 2015;79:119–126. doi: 10.1016/j.jdermsci.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 135.Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21:660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. 2013;98:411–424. doi: 10.1007/s00253-013-5394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Williams, M. R. et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med. 1126/scitranslmed.aat8329 (2019).10.1126/scitranslmed.aat8329 (2019). [DOI] [PMC free article] [PubMed]
- 138.Yu Y, Dunaway S, Champer J, Kim J, Alikhan A. Changing our microbiome: probiotics in dermatology. Br J Dermatol. 2020;182:39–46. doi: 10.1111/bjd.18088. [DOI] [PubMed] [Google Scholar]
- 139.Ezzamouri B, Shoaie S, Ledesma-Amaro R. Synergies of systems biology and synthetic biology in human microbiome studies. Front Microbiol. 2021;12:681982. doi: 10.3389/fmicb.2021.681982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brussow H. Probiotics and prebiotics in clinical tests: an update. F1000Res. 2019 doi: 10.12688/f1000research.19043.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cresci GAM, Lampe JW, Gibson G. Targeted approaches for in situ gut microbiome manipulation. JPEN J Parenter Enteral Nutr. 2020;44:581–588. doi: 10.1002/jpen.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hamilton TA, et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10:4544. doi: 10.1038/s41467-019-12448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not Applicable.