Abstract
Responding to the fast-spreading SARS-CoV-2 Omicron variant, to improve screening efficiency, rapid antigen tests (RATs) were first added as a supplementary detection method in China in mid-March, 2022. What and how big a role RATs should play need to be supported by clinical data. Here, RAT performance and relevant factors in comparison with nucleic acid amplification tests (NAATs) were assessed in Omicron-infected inpatients. From the NAAT results, nasopharyngeal swabs (NPs) performed better than oropharyngeal swabs (OPs). RATs tested on NAAT positive NPs performed better than those with OP-positive samples. The RAT positivity rate was strongly associated with high levels of N and OFR1ab genes, especially in NPs where patients also had significantly longer hospital stays and shorter days from symptom onset to RAT testing. Self-performed RATs had a detection accuracy that was comparable to professionally performed RATs when the subjects were well guided. The antigen negative rate of the studied patients was 100% at discharge. These findings suggest that, in addition to a supplementary detection role, RATs can be an important strategy for evaluating the disease progression of Omicron-infected inpatients. This study provides important clinical data to support better rules regarding RATs under China's COVID-19 prevention and control policy.
Keywords: SARS-CoV-2, Omicron variant, Rapid antigen tests (RATs), Dynamic zero-COVID policy, COVID-19 progression evaluation
Highlights
-
•
Disease progression time and gene level in nasopharynx were relevant factors on RAT.
-
•
Accuracy rate of self-performed RATs was comparable to professionally-performed RAT.
-
•
RATs may be an important strategy for COVID-19 progression evaluation.
1. Introduction
The global transmission of SARS-CoV-2 poses a serious threat to worldwide public health. By August 5, 2022, more than 578 million cases and more than 6.40 million deaths from SARS-CoV-2 have been reported, with a global case fatality rate of up to 1.11% (WHO, 2022).
The new SARS-CoV-2 Omicron variant, which has become the primary strain worldwide, has been shown to be more transmissible than the Delta variant and has a worse prognosis in older adults, thereby presenting new challenges for many countries (Cai et al., 2022; Viana and Moyo, 2022; Zhang et al., 2022). China is one of these nations, as Chinese mainland has a large population base with a rising proportion of elderly (≥ 60 years) citizens, reaching 18.7% in 2020 (China National Bureau of Statistics, 2021). The Omicron variant hit Shanghai, the biggest city in China, causing the largest SARS-CoV-2 infection wave, with 572,329 asymptomatic cases, 57,114 confirmed cases and 571 deaths from February 22, 2022 to May 14, 2022 (Shanghai Municipal Health Commission, 2022).
To respond to SARS-CoV-2 variants, China adopted and maintained a dynamic zero-COVID strategy. One model-based study predicted that the Omicron variant would cause a serious healthcare burden in China without implementation of this strategy (may cause approximately 1.55 million deaths) (Cai et al., 2022), making its appropriate execution essential. The zero-COVID strategy employed nucleic acid amplification test (NAAT) technology to comprehensively screen SARS-CoV-2 infected persons. While NAAT has been shown to be an effective screening tool, it can only be performed in specialized laboratories by professionals and usually requires 4–24 h to produce results. Self point-of-care testing (POCT) was therefore urgently in need in Chinese mainland, particularly in the setting of large outbreaks in super cities.
In mid-March of 2022, the National Health Commission of the People's Republic of China revised the eighth edition of the Diagnosis and Treatment Protocols for COVID-19 (National Health Commission of the People's Republic of China, 2021) into the ninth trial edition (National Health Commission of the People's Republic of China, 2022). Changes included the addition of rapid antigen testing (RAT) as a supplementary detection method to improve screening performance.
Several works outside of China have evaluated the performance of RATs in real-world situations (Erman Daloğlu et al., 2022; Mane et al., 2022; Parikh et al., 2022; Schrom et al., 2022). However, what and how big a role RATs should play in China as part of its disease prevention policy are still unclear. To provide clinical evidence for these problems and to help to make science-driven decisions, this study enrolled SARS-CoV-2 Omicron variant-infected patients admitted to Taizhou Hospital of Zhejiang Province affiliated to Wenzhou Medical University and evaluated the diagnostic performance of RATs, identified factors that contributed to RAT efficacy and monitored dynamic changes in RAT results over disease progression.
2. Materials and methods
2.1. Sample collection and preservation
After admission to the hospital, respiratory tract samples were collected for SARS-CoV-2 detection. Nasal swab samples were used for RATs. Samples were collected by the patients themselves or by nurses according to the antigen detection kit instruction manual. After noses were blown, swabs were inserted 1–1.5 cm into both nasal passages, rotated at least four times and maintained for longer than 15 s. Because prescriptive sampling is important to detection results, the patients were self-trained by learning videos and written materials or trained face-to-face by professionals. The nasal swab samples were immediately put into extracting solution for RATs.
Nasopharyngeal (NP) and oropharyngeal (OP) swab samples were used for NAATs. Samples were collected by nurses, stored in virus preservation solution and immediately sent to the clinical laboratory for NAATs at −4 °C according to the Technical Guidelines for SARS-CoV-2 Sample Collection and Detection in the Diagnosis and Treatment Protocols for COVID-19 (the eighth trial edition) released by the National Health Commissions of the People's Republic of China (National Health Commission of the People's Republic of China, 2021).
2.2. RATs
RATs were performed using the SARS-CoV-2 Antigen Detection Kit (Colloidal gold immunochromatography) (Labnovation, China) according to the manufacturer's instructions. Patients enrolled in the self-performed RATs were guided by videos or face-to-face instruction by professionals. In brief, the extracting solution was mixed for more than 30 s and swabs were squeezed at least five times. Three drops of the extracted solution were dripped onto the detection card and results were read in 15–20 min. Results were positive if a band was observed in the testing area, and negative when no band was observed. RAT results were valid only when a control band was observed.
2.3. NAATs
NAATs were performed using the Nucleic Acid Extraction Reagent (Magnetic Beads) (Wuhan Easydiagnosis Biomedicine, China) and the Automatic Nucleic Acid Extraction and Purification Instrument (Wuhan Easydiagnosis Biomedicine, China) according to the instruction manuals. To measure the SARS-CoV-2 nucleic acid level, reverse-transcriptive real-time PCR (RT-PCR) was performed using the COVID-19 (SARS-CoV-2) Nucleic Acid Test Kit (Wuhan Easydiagnosis Biomedicine, China) and a 7500 Real-Time PCR System (Applied Biosystems, USA) according to the instruction manuals. Results were interpreted by professionals. Ct values ≥ 38 were considered negative.
2.4. Data analysis
Mean age, mean days from symptom onset to hospitalization, mean length of hospitalization and mean days from symptom onset to RATs and their corresponding standard deviations were calculated. Gender ratios and the percentage of patients with clinical symptoms were also calculated. Mean Ct values were used to analyze gene levels. Negative results without Ct value (not amplified) were assigned Ct values of 40. Statistical analysis comparing RAT positive and negative patients was performed and P values were calculated using unpaired t-tests.
3. Results
3.1. Study population
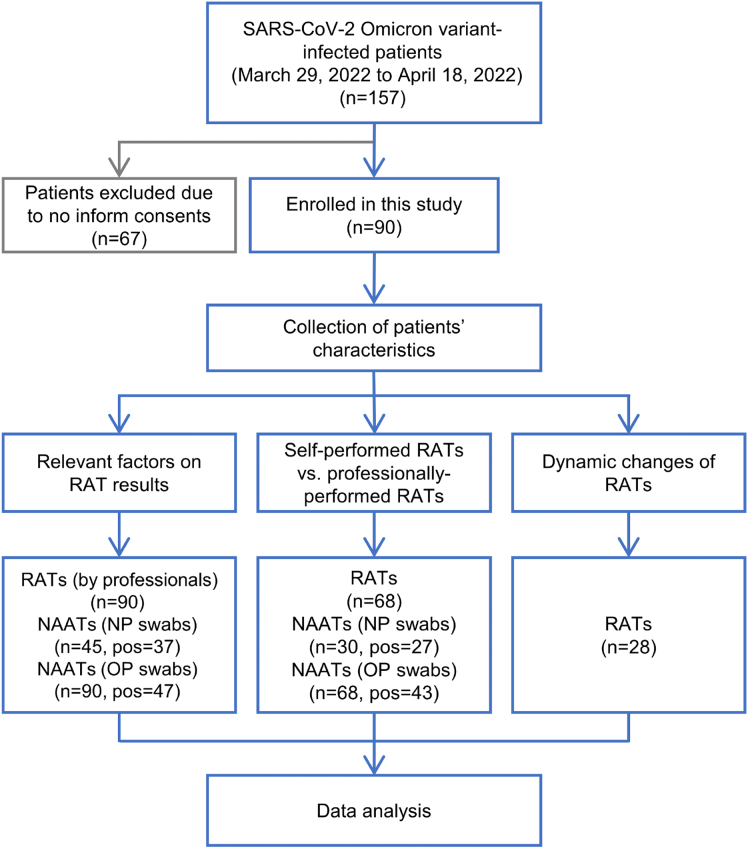
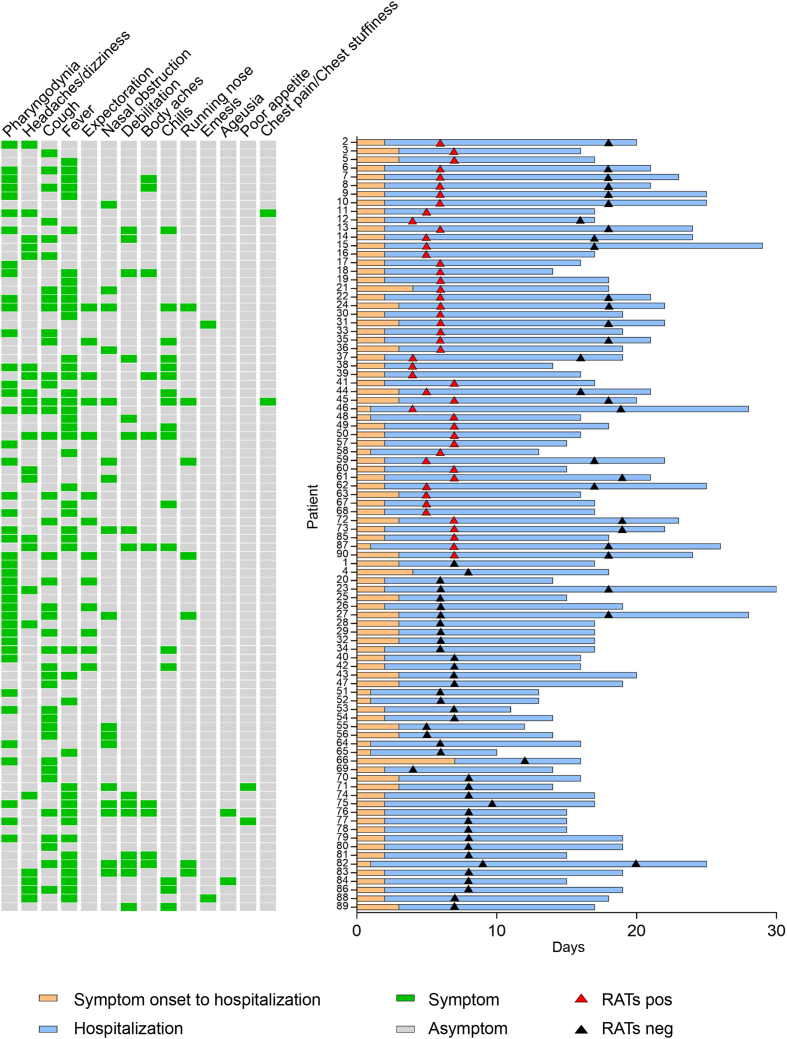
From March 30, 2022 to April 18, 2022, 157 patients infected with the SARS-CoV-2 Omicron variant were admitted to Taizhou Hospital of Zhejiang Province affiliated to Wenzhou Medical University. Of these, 90 signed informed consent forms and were enrolled into this study (Group 1). Sixty-eight of these patients were used to study the impact of testers (patients vs. professionals) on RAT results (Group 2), while 28 were used to evaluate dynamic changes in RAT results (Group 3) (Fig. 1). The mean ages of Groups 1 to 3 were 35.03 ± 10.15, 35.93 ± 11.04 and 31.89 ± 10.82 years and included 28.89%, 36.76% and 25.00% females, respectively. The average days from symptom onset to hospitalization and length of hospitalization were 2.26 ± 0.82 and 15.98 ± 4.03 for Group 1, 2.32 ± 0.87 and 15.87 ± 4.16 for Group 2 and 2.18 ± 0.55 and 20.71 ± 2.94 for Group 3, respectively. The major clinical manifestations of all three groups were pharyngodynia, headaches/dizziness, cough and fever (Table 1 and Fig. 2).
Fig. 1.
Study flowchart. RAT, rapid antigen test; NAAT, nucleic acid amplification test; NP, nasopharyngeal; OP, oropharyngeal; pos, positive.
Table 1.
Characteristics of subjects.
| Group 1 | Group 2 | Group 3 | |
|---|---|---|---|
| N | 90 | 68 | 28 |
| Age (y), mean ± SD | 35.03 ± 10.15 | 35.93 ± 11.04 | 31.89 ± 10.82 |
| Gender, n (%) | |||
| Male | 64 (71.11) | 43 (63.24) | 21 (75.00) |
| Female | 26 (28.89) | 25 (36.76) | 7 (25.00) |
| Symptom onset to hospitalization (d), mean ± SD | 2.26 ± 0.82 | 2.32 ± 0.87 | 2.18 ± 0.55 |
| Length of hospitalization (d), mean ± SD | 15.98 ± 4.03 | 15.87 ± 4.16 | 20.71 ± 2.94 |
| Symptoms, n (%) | |||
| Pharyngodynia | 42 (46.67) | 36 (52.94) | 15 (53.57) |
| Headaches/dizziness | 22 (24.44) | 14 (20.59) | 9 (32.14) |
| Cough | 39 (43.33) | 31 (45.59) | 13 (46.43) |
| Fever | 44 (48.89) | 27 (39.71) | 15 (53.57) |
| Expectoration | 13 (14.44) | 10 (14.71) | 5 (17.86) |
| Nasal obstruction | 17 (18.89) | 11 (16.18) | 9 (32.14) |
| Debilitation | 15 (16.67) | 4 (5.89) | 6 (21.43) |
| Body aches | 10 (11.11) | 4 (5.89) | 4 (14.29) |
| Chills | 17 (18.89) | 12 (17.65) | 8 (28.57) |
| Running nose | 7 (7.78) | 4 (5.89) | 6 (21.43) |
| Emesis | 2 (2.22) | 1 (1.47) | 1 (3.57) |
| Ageusia | 2 (2.22) | 0 (0) | 0 (0) |
| Poor appetite | 2 (2.22) | 0 (0) | 0 (0) |
| Chest pain/chest stuffiness | 2 (2.22) | 2 (2.94) | 1 (3.57) |
Group 1: all enrolled RATs; Group 2: RATs (professional-performed vs. self-performed); Group 3: dynamic RATs.
SD, standard deviation; RAT, rapid antigen test.
Fig. 2.
Clinical symptoms, COVID-19 progression and RAT results. RAT, rapid antigen test; pos, positive; neg, negative.
3.2. Relevant factors influencing RAT results
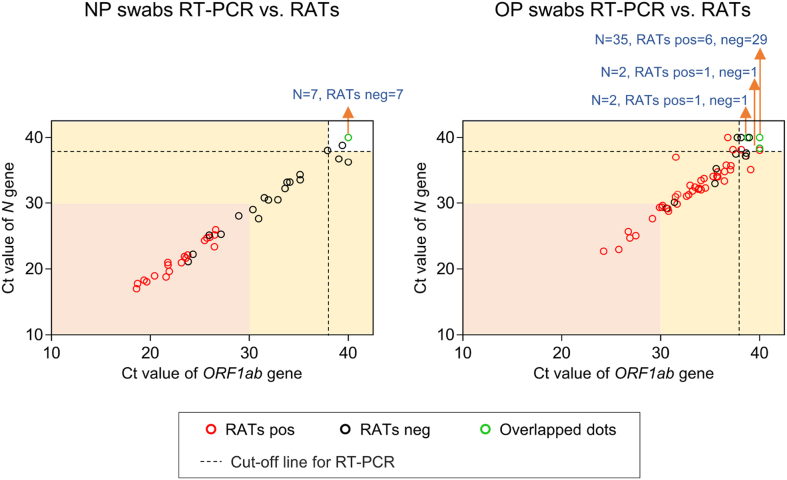
All 90 patients in Group 1 received at least one RAT after admission. To measure the nucleic acid level of the Omicron variant, all enrolled patients received NP and/or OP swab NAATs on the day of their first RAT. The clinical symptoms, days from symptom onset to hospitalization, length of hospitalization, days of RATs performed and RAT results for Group 1 are shown in Fig. 2. On the day of the first RAT, 45 patients received NP swab NAATs and 37 were positive. Of the NP swab NAAT positive patients, 19 were RAT positive and 18 were RAT negative. All of the patients who were NP swab NAAT negative were RAT negative. All RAT positive patients had a high nucleic acid level in their nasopharynx (Ct value < 30) (Fig. 3). Factors such as gender, age, nucleic acid level, disease progression time and clinical symptoms were analyzed by comparing the two sub-groups. Data showed that the ORF1ab and N gene levels of RAT-positive patients were significantly higher than those of RAT-negative patients, their length of hospitalizations were significantly longer, and their days from symptom onset to RAT testing were significantly shorter. All other factors had no significant effect (Table 2 and Fig. 3).
Fig. 3.
ORF1ab and N gene Ct values and professionally-performed RAT results in 90 patients. RAT, rapid antigen test; NP, nasopharyngeal; OP, oropharyngeal; pos, positive; neg, negative.
Table 2.
Characteristics of RATs-positive and RATs-negative subjects.
| NP swabs RT-PCR pos |
OP swabs RT-PCR pos |
|||||
|---|---|---|---|---|---|---|
| RATs pos | RATs neg | P value | RATs pos | RATs neg | P value | |
| N | 19 | 18 | 39 | 8 | ||
| Age (y), mean ± SD | 34.79 ± 12.85 | 35.83 ± 9.29 | 0.7789 | 34.62 ± 10.53 | 32.88 ± 7.53 | 0.6598 |
| Gender ratio (M: F) | 16:3 | 17:1 | 0.3299 | 28:11 | 5:3 | 0.6098 |
| Ct value (ORF1ab), mean ± SD | 22.87 ± 2.75 | 32.03 ± 4.72 | < 0.0001 | 32.98 ± 3.64 | 35.71 ± 3.15 | 0.0550 |
| Ct value (N), mean ± SD | 21.31 ± 2.79 | 30.42 ± 4.84 | < 0.0001 | 31.81 ± 4.00 | 34.97 ± 3.87 | 0.0469 |
| Symptom onset to hospitalization (d), mean ± SD | 2.11 ± 0.46 | 2.11 ± 0.76 | 0.9774 | 2.21 ± 0.57 | 2.13 ± 0.83 | 0.7402 |
| Length of Hospitalization (d), mean ± SD | 17.42 ± 4.19 | 14.39 ± 3.33 | 0.0205 | 17.79 ± 4.08 | 14.75 ± 4.83 | 0.0686 |
| Symptom onset to RATs (d), mean ± SD | 5.95 ± 1.18 | 7.22 ± 1.17 | 0.0022 | 5.85 ± 1.01 | 6.38 ± 0.74 | 0.17 |
| Symptoms, n (%) | ||||||
| Pharyngodynia | 8 (42.11) | 7 (38.89) | 0.8474 | 18 (46.15) | 4 (50.00) | 0.8467 |
| Headaches/dizziness | 7 (36.84) | 3 (16.67) | 0.1765 | 13 (33.33) | 1 (12.50) | 0.2499 |
| Cough | 5 (26.32) | 10 (55.56) | 0.0736 | 15 (38.46) | 5 (62.50) | 0.2190 |
| Fever | 7 (36.84) | 8 (44.44) | 0.6489 | 19 (48.72) | 3 (37.50) | 0.5723 |
| Expectoration | 2 (10.53) | 0 (0) | 0.1658 | 6 (15.38) | 3 (37.50) | 0.1541 |
| Nasal obstruction | 3 (15.79) | 6 (33.33) | 0.2250 | 7 (17.95) | 1 (12.50) | 0.7160 |
| Debilitation | 3 (15.79) | 5 (27.78) | 0.3900 | 4 (10.26) | 0 (0) | 0.3545 |
| Body aches | 0 (0) | 3 (16.67) | 0.0662 | 4 (10.26) | 0 (0) | 0.3545 |
| Chills | 2 (10.53) | 1 (5.56) | 0.5921 | 10 (25.64) | 0 (0) | 0.1111 |
| Running nose | 1 (5.26) | 2 (11.11) | 0.5282 | 4 (10.26) | 1 (12.50) | 0.8552 |
| Emesis | 0 (0) | 0 (0) | / | 1 (2.56) | 1 (12.50) | 0.2132 |
| Ageusia | 0 (0) | 1 (5.56) | 0.3109 | 0 (0) | 0 (0) | / |
| Poor appetite | 0 (0) | 0 (0) | / | 0 (0) | 0 (0) | / |
| Chest pain/chest stuffiness | 1 (5.26) | 0 (0) | 0.3374 | 2 (5.13) | 0 (0) | 0.5232 |
NP, nasopharyngeal; OP, oropharyngeal; RAT, rapid antigen test; pos, positive; neg, negative; M, male; F, female; SD, standard deviation.
Statistical analysis was performed by unpaired t-tests. Bold represents statistically significant difference.
All Group 1 patients received OP swab NAATs, which had 47 positive results and 43 negative results. Thirty-nine of the 47 NAAT positive results were also RAT positive, while eight were negative. The N gene level in the oropharynx was significantly higher in RAT positive patients. However, age, gender, ORF1ab gene level in the oropharynx, disease progression time and symptoms were all equivalent between the two groups (Table 2 and Fig. 3). Ten OP swab NAAT negative patients were RAT positive that seemed to be false positive, and this will be further analyzed in 3.3 sub-section to determine whether they were true positives or false positives (Fig. 3).
Compared with NP NAAT, RAT performance (performed by professionals) with nasal swabs (n = 45) collected on 6.8 ± 1.53 days after symptom onset was evaluated. When Ct value of NAAT <30 defines positive RT-PCR (Schrom et al., 2022), specificity and sensitivity of RATs are 100% and 79.17%, respectively. When Ct value < 38 defines positive (referred to the instruction manual of NAAT kit), specificity and sensitivity are 100% and 51.35%, respectively (Supplementary Table S1).
3.3. Comparison of self-performed and professionally-performed RATs
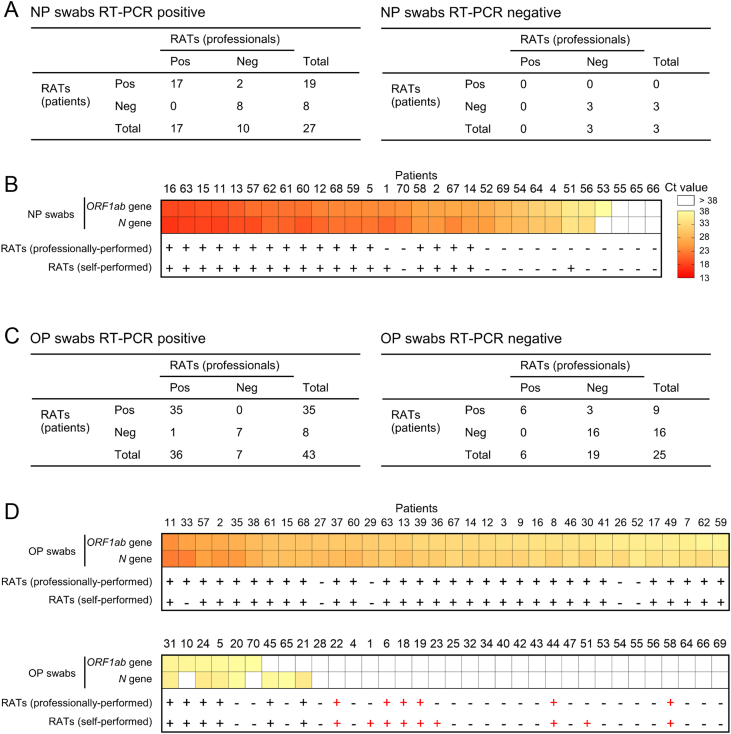
Thirty patients in Group 2 received NP swab NAATs, of which 27 were positive and three were negative. All negative NAATs had negative RATs as well. Seventeen of the 27 positive NAATs were positive in self- and professionally-obtained RATs, eight were negative after both strategies, and two samples had positive self-performed RATs but negative professionally-performed RATs (Fig. 4A and B).
Fig. 4.
Comparison of professionally-performed and self-performed RATs. A RAT results of NP swab NAAT-positive patients (left) and NP swab NAAT-negative patients (right). BORF1ab and N gene Ct values from NP NAAT samples and RAT results. C RAT results in OP swab NAAT-positive patients (left) and OP swab NAAT-negative patients (right). DORF1ab and N gene Ct values from OP NAAT samples and RAT results. NP, nasopharyngeal; OP, oropharyngeal; RAT, rapid antigen test; NAAT, nucleic acid amplification test.
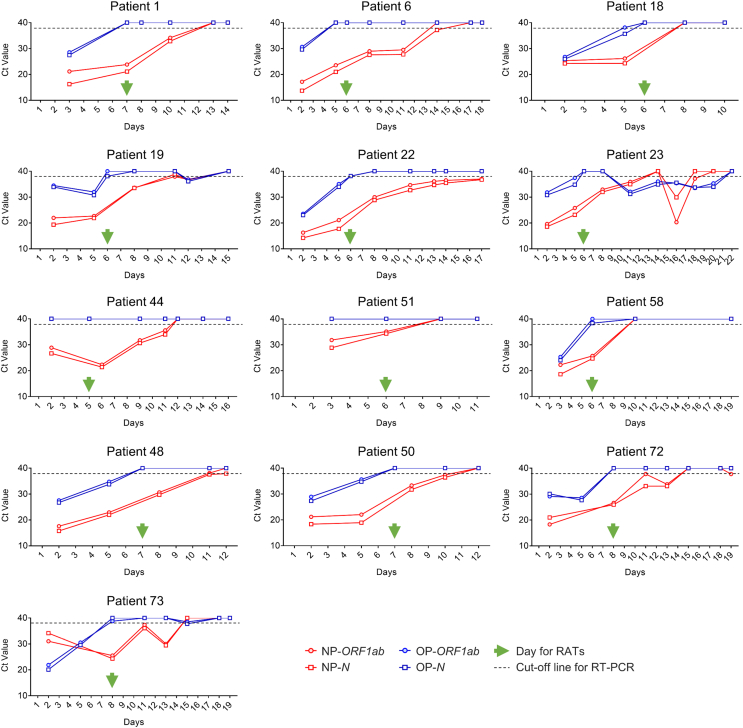
Sixty-eight patients received OP swab NAATs, 43 of whom were positive. Of the 25 negative samples, 16 self- and professionally-obtained RATs were negative, 6 were positive from both RAT types (Patients 6, 18, 19, 22, 44 and 58), and three self-performed RATs were positive but professionally negative (Patients 1, 23 and 51) (Fig. 4C and D). To make it clear whether “false positive” RATs were actually false, we analyzed variation in the ORF1ab and N genes in the nasopharynx and oropharynx. Patients 6, 19, 22, 23, 44 did not have a NP NAAT performed on the study day but had persistently positive NP samples before and after, with nucleic acid negative conversion observed at least 3 days later. Patients 1, 51 and 58 had positive NP NAATs on the day for RATs (Fig. 5). These results indicate that the RAT results were true-positive. Of the OP swab positive patients, 35 patient and professional RATs were positive, 7 were dual negative and 1 was professionally positive but patient-obtained negative. The other four RAT-positive (only performed by professionals)/NAAT-negative (OP swab) had the same results (Patients 48, 50, 72 and 73) (Fig. 4C and D, Fig. 5), thus proved to be true positive results with RAT.
Fig. 5.
Variation in the RT-PCR Ct values of 13 RAT-positive/OP swab NAAT-negative subjects (symptom onset is defined as day 1). RAT, rapid antigen test; NAAT, nucleic acid amplification test; NP, nasopharyngeal; OP, oropharyngeal.
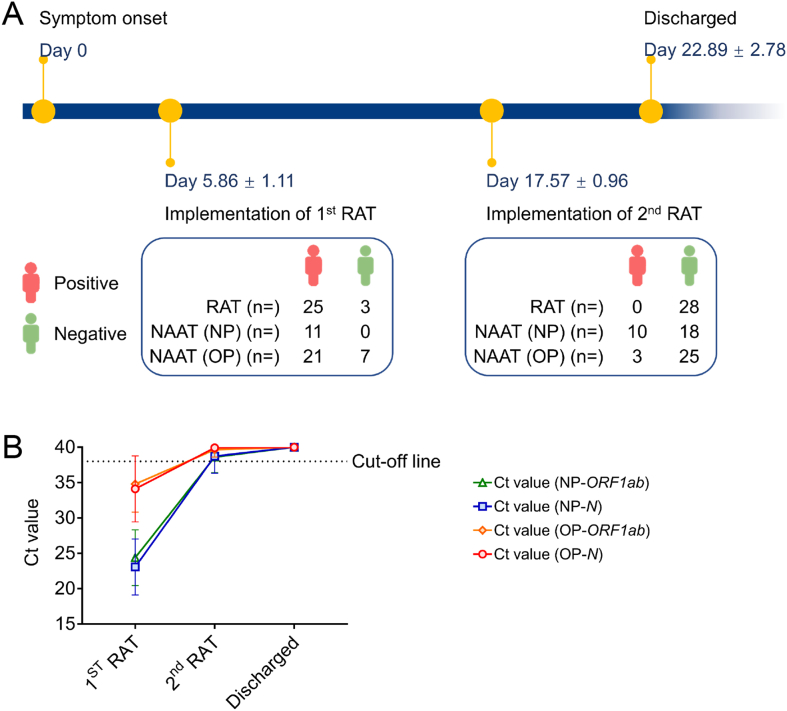
3.4. Dynamic monitoring of RAT results with COVID-19 progression
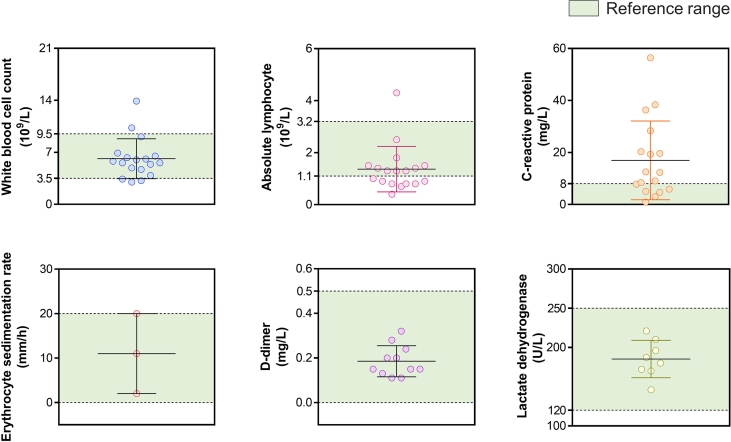
Twenty-eight patients (Group 3) received a second RAT 17.57 ± 0.96 days after symptom onset (5.32 ± 2.83 days before being discharged). The antigen tests of 25 patients turned negative and 3 patients remained negative (performed by professionals). NAATs performed on the day of or after the second RAT had low gene levels or negative results: 18 patients had negative nasopharynx and oropharynx samples, 7 patients had negative oropharynx samples and the mean Ct values were 38.61 ± 2.20 (NP swab ORF1ab gene), 38.75 ± 2.42 (NP swab N gene), 39.65 ± 1.04 (OP swab ORF1ab gene) and 39.92 ± 0.40 (OP swab N gene). All 28 patients had relatively mild symptoms. Of these, 18 patients underwent the serological tests and 12 patients received a chest CT scan on admission. White blood cell count was slightly reduced in 3/18 patients while 2/18 had increased counts. The absolute lymphocyte count of 8/18 patients was low while 1/18 was high. Increased C-reactive protein level was seen in 11/17 patients. The level of D-dimers of all of the tested patients (11/11) was normal, so were the erythrocyte sedimentation rate and the level of lactate dehydrogenase (Supplementary Fig. S1).
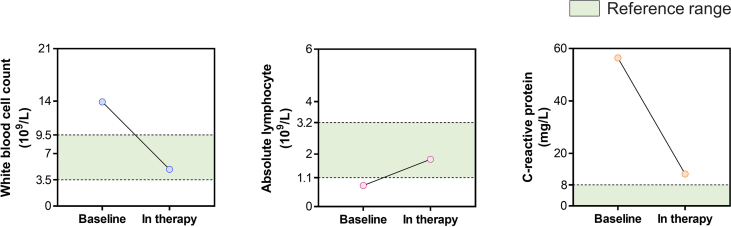
Chest CT scans showed that 4/12 patients presented with SARS-CoV-2-associated pneumonia. Treatment and auxiliary inspections were guided by the ninth trial edition of the Diagnosis and Treatment Protocols for COVID-19 (National Health Commission of the People's Republic of China, 2021). Except for Patient 45, all patients improved to the point that they no longer met standard for serological monitoring or chest imaging. Patient 45 received one chest CT scan test and one serological test, which demonstrated an improvement (Supplementary Fig. S2). The patient's C-reactive protein level dropped to a nearly normal level, the white blood cell count and absolute lymphocyte count normalized and lung inflammation resolved. By the day of the second RAT, all patients had made good progress according to their clinicians' judgement. On their day of discharge, all patients had clinically recovered and the ORF1ab gene and N gene levels in both of their nasopharynx and oropharynx presented the negative results (Fig. 6).
Fig. 6.
Dynamic monitoring of RAT and NAAT results over COVID-19 disease progression. A Timeline of the dynamic monitoring of RAT and NAAT results in 28 Omicron-infected inpatients. B Average Ct values of the ORF1ab and N gene of the Omicron variant in NP and OP swab samples on the day of the 1st RAT, on the day or the next day after the 2nd RAT and the day of discharge. RAT, rapid antigen test; NAAT, nucleic acid amplification test; NP, nasopharyngeal; OP, oropharyngeal.
4. Discussion
As China has a large population, COVID-19 prevention and control has been a considerable challenge. To respond to the Omicron variant while maintaining dynamic zero-COVID policies, China added RATs as a supplementary detection method in mid-March of 2022. In the early days following the implementation of this trial policy, clinical feedback of RATs performed on Omicron infected cases can contribute to policy optimization and scientific adjustment.
Most published studies evaluated RAT performance with different SARS-CoV-2 variants (including Alpha, Beta, Gamma, Delta and Omicron) using different specimens from different countries (Galliez et al., 2022; Kyritsi et al., 2022; Hardick et al., 2022; Weishampel et al., 2022). Sporadic worldwide studies compared NAATs and RATs during the Omicron surge (Schrom et al., 2022; Lin et al., 2022). However, RATs may perform differently and play a unique role in the setting of different prevention policies. Chinese mainland adopted a rigorous zero-COVID policy that is entirely different from others, so few relevant studies are available. Our study helps to fill the gaps. Rapid antibody tests were also recommended for inclusion in the COVID-19 diagnostic algorithms proposed by prior works (Yıldırım et al., 2021). However, after the performance of such tests on SARS-CoV-2 infections was tested, there remains disagreement on their diagnostic value (Kızıloglu et al., 2021; Yıldırım et al., 2021). It takes 7–14 days from symptom onset for an antibody response to the virus to form, and antibodies produced by a natural infection or following vaccination are hard to distinguish (Peeling et al., 2022). Rapid antibody tests were therefore not adopted as a detection or supplementary detection modality for diagnosing SARS-CoV-2 infections.
This study evaluated the performance of RATs in Omicron variant-infected patients and analyzed factors predictive of that performance. Both NP and OP swabs can be used for SARS-CoV-2 NAATs according to the Diagnosis and Treatment Protocols for COVID-19 (National Health Commission of the People's Republic of China, 2022). This study sampled one or both of them based on clinical demand. When NP swab NAATs were considered the gold standard, 20.83% of patients with high viral gene levels (Ct < 30) were RAT negative and all low-gene-level patients were negative. This may be because the time from symptom onset to RAT testing was longer than 7 days, allowing antigen levels to drop below the detection limit (Peeling et al., 2022). Our data showed that the Ct values of both viral genes and disease timing were relevant predictors of RAT performance. Gene levels were higher and length of hospitalizations was longer for RAT positive patients, while days from symptom onset to positive RATs were shorter. This suggests that RAT results are associated disease progression. However, the agreement between OP swab NAAT and RAT results was poor (Table 2, Figs. 2, Fig. 3).
Because NAATs can only be performed by professionals in specialized biosafety laboratories, the shipment, pretreatment and detection of positive specimens may pose infectious risks to staff. Encouragingly, RATs can be self-performed at the bedside. There is therefore an urgent need for clinical data evaluating the performance of self-performed RATs. We evaluated self-performed vs. professionally-performed RATs of 68 hospitalized patients infected with the Omicron variant. Inspiringly, the accuracy rate of self-performed RATs was exactly comparable to that of professionally-performed RATs (Figs. 4 and 5), although the results of self-performed RATs were not 100% consistent with those of professionally-performed RATs. Because no true false-positives occurred in either self- or professionally-performed tests, the reason for this inconsistency was likely operation, caused by sampling technique (in-nose retention time of the swab and sampling force) and elution efficiency of the virus. It was also worth noting that these performances cannot be separated with well-instructed subjects. This study therefore provides us with great confidence that self-administered RATs can be performed in Chinese mainland adequately when subjects are provided with good guidance.
False positive tests are a major challenge with SARS-CoV-2 RATs (Gans et al., 2022; Yang et al., 2022) and may lead to unnecessary additional burden on the healthcare system. This study analyzed true/false positive RATs in detail. Thirteen NAAT (OP swab)-negative/RAT-positive patients (14.44%) were identified from a total of 90 patients, and were considered likely false-positive results. However, analysis of variations of NAAT Ct values confirmed that these were true-positive results. In these 13 patients, the measurement of Omicron variant nucleic acid levels in the oropharynx turning negative was earlier than the RAT measurement time, compared to levels in the nasopharynx that were positive and remained as such (Fig. 5). Dynamic monitoring of RATs showed that 89.29% of patients turned antigen negative and the remainder stayed negative until the day of discharge. We can draw two conclusions here: first, RATs can be an important strategy for evaluating COVID-19 progression under China's dynamic zero-COVID policy; second, RATs can be an important supplementary test for OP swab NAATs.
This study is limited by its small sample size. In the near future, conclusions drawn from this work should be further validated with multicenter studies and large-scale samples.
5. Conclusions
Our study provided new insights for defining the role that RATs should play as part of China's dynamic zero-COVID policy. Our findings suggest that RATs can optimize COVID-19 prevention policies and improve our respond to the Omicron and future variants.
Data availability
All data generated from the present study are included in this manuscript.
Ethics statement
This study was ethically approved by the Ethics Committee of Taizhou Hospital of Zhejiang Province affiliated to Wenzhou Medical University (No. K20220133). Informed consent was obtained from each subject. All participants were notified of the study purpose and research procedure at the time of enrollment.
Author contributions
Mengyuan Chen: investigation, visualization, writing-original draft preparation. Jiaqin Xu: data curation, methodology. Lingjun Ying: investigation. Miaoguo Cai: investigation. Tao-Hsin Tung: supervision. Kai Zhou: data curation. Yufen Zheng: funding acquisition, resources. Xiaojie Bi: formal analysis, resources. Jing Wang: methodology. Xi Tu: formal analysis, funding acquisition. Dongqing Lv: conceptualization, project administration. Bo Shen: conceptualization, project administration.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This study was funded by the Medical Science and Technology Project of Zhejiang Province (Grant Numbers 2021KY394 and 2018KY912). We would like to thank the nurses at Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University, for their kind help with sample collection, and thank the patients for being enrolled in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.08.008.
Contributor Information
Bo Shen, Email: shenb@enzemed.com.
Dongqing Lv, Email: lvdq@enzemed.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
Supplementary Fig. S2.
References
- Cai J., Deng X., Yang J. Modeling transmission of sars-cov-2 omicron in China. Nat. Med. 2022;28:1468–1475. doi: 10.1038/s41591-022-01855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China National Bureau of Statistics Interpretation for the 7th census of China. 2021. http://www.stats.gov.cn/xxgk/jd/sjjd2020/202105/t20210512_1817342.html
- Erman Daloğlu A., Er H., Sepin Özen N., Çekin Y. Evaluation of the rapid antigen detection kit with the polymerase chain reaction for detection of sars-cov-2 in respiratory samples. Mikrobiyoloji Bulteni. 2022;56:263–273. doi: 10.5578/mb.20229806. [DOI] [PubMed] [Google Scholar]
- Galliez R.M., Bomfim L., Mariani D., Leitão I.C., Castiñeiras A.C.P., Gonçalves C.C.A., Ortiz da Silva B., Cardoso P.H., Arruda M.B., Alvarez P., Brindeiro R., Ota V.A., Rodrigues D.G.M., da Costa L.J., Ferreira O.D.C., Jr., Castiñeiras T.M.P.P., Faffe D.S., Tanuri A. Evaluation of the panbio COVID-19 antigen rapid diagnostic test in subjects infected with omicron using different specimens. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01250-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans J.S., Goldfarb A., Agrawal A.K., Sennik S., Stein J., Rosella L. False-positive results in rapid antigen tests for sars-cov-2. JAMA. 2022;327:485–486. doi: 10.1001/jama.2021.24355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardick J., Gallagher N., Sachithanandham J., Fall A., Siddiqui Z., Pekosz A., Manabe Y.C., Mostafa H.H. Evaluation of four point of care (POC) antigen assays for the detection of the SARS-CoV-2 variant omicron. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01025-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kızıloglu I., Sener A., Siliv N. Comparison of rapid antibody test and thorax computed tomography results in patients who underwent RT-PCR with the pre-diagnosis of COVID-19. Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyritsi M.A., Speletas M., Mouchtouri V., Vachtsioli E., Babalis D., Kouliou O., Tsispara A., Tseroni M., Hadjichristodoulou C. Performance evaluation of a rapid antigen test (RAT) during omicron pandemic wave in Greece, conducted by different personnel, and comparison with performance in previous wave (Alpha variant) period. Diagnostics. 2022;12:1048. doi: 10.3390/diagnostics12051048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Frediani J.K., Damhorst G.L., Sullivan J.A., Westbrook A., McLendon K., Baugh T.J., O’Sick W.H., Roback J.D., Piantadosi A.L., Waggoner J.J., Bassit L., Rao A., Greenleaf M., O’Neal J.W., Swanson S., Pollock N.R., Martin G.S., Lam W.A., Levy J.M. Where is omicron? Comparison of SARS-CoV-2 RT-PCR and antigen test sensitivity at commonly sampled anatomic sites over the course of disease. medRxiv. 2022;9 2022.02.08.22270685. [Google Scholar]
- Mane A., Jain S., Jain A., Pereira M., Sirsat A., Pathak G., Bhoi V., Bhavsar S., Panda S. Diagnostic performance of oral swab specimen for sars-cov-2 detection with rapid point-of-care lateral flow antigen test. Sci. Rep. 2022;12:7355. doi: 10.1038/s41598-022-11284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health Commission of the People’s Republic of China Diagnosis and treatment protocols for covid-19 (the ninth trial edition) 2022. http://www.nhc.gov.cn/yzygj/s7653p/202203/b74ade1ba4494583805a3d2e40093d88.shtml
- National Health Commission of the People’s Republic of China Diagnosis and treatment protocols for covid-19 (the eighth trial edition) 2021. http://www.nhc.gov.cn/jkj/s3577/202105/6f1e8ec6c4a540d99fafef52fc86d0f8.shtml
- Parikh A., Cooper L., Frogel D., Le Benger K., Cooper C.K., Parvu V. Large-scale sars-cov-2 antigen testing with real-world specimens. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.836328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Heymann D.L., Teo Y.Y., Garcia P.J. Diagnostics for covid-19: moving from pandemic response to control. Lancet. 2022;399:757–768. doi: 10.1016/S0140-6736(21)02346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrom J., Marquez C., Pilarowski G. Comparison of sars-cov-2 reverse transcriptase polymerase chain reaction and binaxnow rapid antigen tests at a community site during an omicron surge : a cross-sectional study. Ann. Intern. Med. 2022;175:682–690. doi: 10.7326/M22-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanghai Municipal Health Commission News release. 2022. https://wsjkw.sh.gov.cn/xwfb/index.html
- Viana R., Moyo S. Rapid epidemic expansion of the sars-cov-2 omicron variant in southern africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weishampel Z.A., Young J., Fischl M., Fischer R.J., Donkor I.O., Riopelle J.C., Schulz J.E., Port J.R., Saturday T.A., van Doremalen N., Berry J.D., Munster V.J., Yinda C.K. OraSure InteliSwab™ rapid antigen test performance with the SARS-CoV-2 variants of concern-alpha, Beta, Gamma, Delta, and omicron. Viruses. 2022;14:543. doi: 10.3390/v14030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Who coronavirus (covid-19) dashboard. 2022. https://covid19.who.int/
- Yang Y.P., Huang L.L., Pan S.J., Xu D., Jiesisibieke Z.L., Tung T.H. False-positivity results in rapid antigen tests for sars-cov-2: an umbrella review of meta-analyses and systematic reviews. Expert Rev. Anti Infect. Ther. 2022;4:1–9. doi: 10.1080/14787210.2022.2070152. [DOI] [PubMed] [Google Scholar]
- Yıldırım F., Gulhan P.Y., Diken Ö.E., Capraz A., Simsek M., Yildirim B.B., Taysi M.R., Ozturk S.Y., Demirtas N., Ergil J., Dirican A., Uzar T., Karaman I., Ozkaya S. Role of serological rapid antibody test in the management of possible COVID-19 cases. World J. Exp. Med. 2021;11:44–54. doi: 10.5493/wjem.v11.i4.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang W., Chen S. Shanghai’s life-saving efforts against the current omicron wave of the covid-19 pandemic. Lancet. 2022;399:2011–2012. doi: 10.1016/S0140-6736(22)00838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated from the present study are included in this manuscript.