SUMMARY
RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT2C serotonin receptor, which undergoes 32 distinct RNA-editing events leading to 24 protein isoforms, is a notable example of this process. These 5HT2C isoforms display differences in constitutive activity, agonist/inverse agonist potencies, and efficacies. To elucidate the molecular mechanisms responsible for these effects of RNA editing, we present four active-state 5HT2C-transducer-coupled structures of three representative isoforms (INI, VGV, and VSV) with the selective drug lorcaserin (Belviq) and the classic psychedelic psilocin. We also provide a comprehensive analysis of agonist activation and constitutive activity across all 24 protein isoforms. Collectively, these findings reveal a unique hydrogen-bonding network located on intracellular loop 2 that is subject to RNA editing, which differentially affects GPCR constitutive and agonist signaling activities.
Graphical Abstract
In brief
It is established that the 5HT2C receptor undergoes RNA editing leading to 24 isoforms. Several isoforms exhibit changes in basal activity and are linked to pathologies. Gumpper et al. have done a systematic structure-function characterization of all the isoforms revealing the underlying mechanisms that govern basal activity of the 5HT2C receptor.
INTRODUCTION
RNA editing occurs throughout all taxa of life and is responsible for regulating a myriad of biological processes, including neurodegenerative diseases, brain development, and viral immune defenses (Bass and Weintraub, 1988; Benne et al., 1986; Ekdahl et al., 2012; Hood and Emeson, 2012; Krestel and Meier, 2018; Nishikura, 2016; Simpson and Shaw, 1989; Sommer et al., 1991; Ward et al., 2011; Yu et al., 2003). RNA-editing machinery has evolved to include base substitutions, insertions, deletions, and various capping mechanisms (Nishikura, 2016). One such well-studied substitution is the deamination of adenine (A) to inosine (I) (A-to-I) (Bass and Weintraub, 1988; Nishikura, 2016). In vertebrates, this reaction is catalyzed by several adenosine deaminases acting on RNA (ADARs) (Bass and Weintraub, 1988; Nishikura, 2016). ADARs selectively deaminate adenosines located on double-stranded RNAs and are most often located within inverted Alu repetitive elements (Bass and Weintraub, 1988; Nishikura, 2016). A-to-I editing can also occur at a specific coding locus to inform the translational machinery to read the I as a guanosine (G), thereby recruiting a different tRNA, creating a mutant phenotype of the desired protein. A-to-I editing can also lead to ribosomal stalling (Licht et al., 2019a; 2019b). These RNA editing events are widely used to expand the transcriptome by allowing for a diversification of function at a single gene locus.
It has been established that serotonin 2C (5HT2C) receptor transcripts undergo A-to-I editing at five different sites encompassing amino acids 156–160 (Figure 1A) (Niswender et al., 1999, 2001; Zhu et al., 2012). This extensive post-transcriptional editing leads to 32 edited isoforms comprising 24 distinct protein modifications at positions 156, 158, and 160 located on intracellular loop 2 (ICL2) (Abbas et al., 2010; Niswender et al., 1999). The wild-type isoform contains an I, N, and I at each respective position, while each successive editing event changes the amino acid content of ICL2 (Figure 1A). The first editing position can be edited to a V or an M, while the second position is changed to a D, G, or S. Finally, the third position is edited to a V (Figure 1A). Consequently, the constitutive activity, pharmacology, and signaling of the 5HT2C receptor are altered (De Deurwaerdère et al., 2004; Niswender et al., 1999, 2001; Rauser et al., 2001). Altering 5HT2C RNA editing in vivo leads to diverse neuropathological outcomes reminiscent of Prader-Willi syndrome (Morabito et al., 2010) and profound metabolic alterations (Kawahara et al., 2008; Xie et al., 2021). Dysregulated 5HT2C RNA editing has also been implicated in several neuropsychiatric disorders, including depression, substance abuse, and schizophrenia (O’Neil and Emeson, 2012; Warhaftig et al., 2021; Zhu et al., 2012). Previous reports have suggested that highly edited isoforms VSV and VNV are found in particularly high levels in rat, mouse, and human brains (Niswender et al., 1999; Zhu et al., 2012), while both the unedited (INI) and the less edited INV isoforms have been identified in the choroid plexus (Niswender et al., 1999, 2001). This gives further indication that altered RNA editing could lead to disease states in a tissue-dependent manner.
Figure 1. RNA editing of the 5HT2C receptor leads to isoforms with differing constitutive activity and potencies.
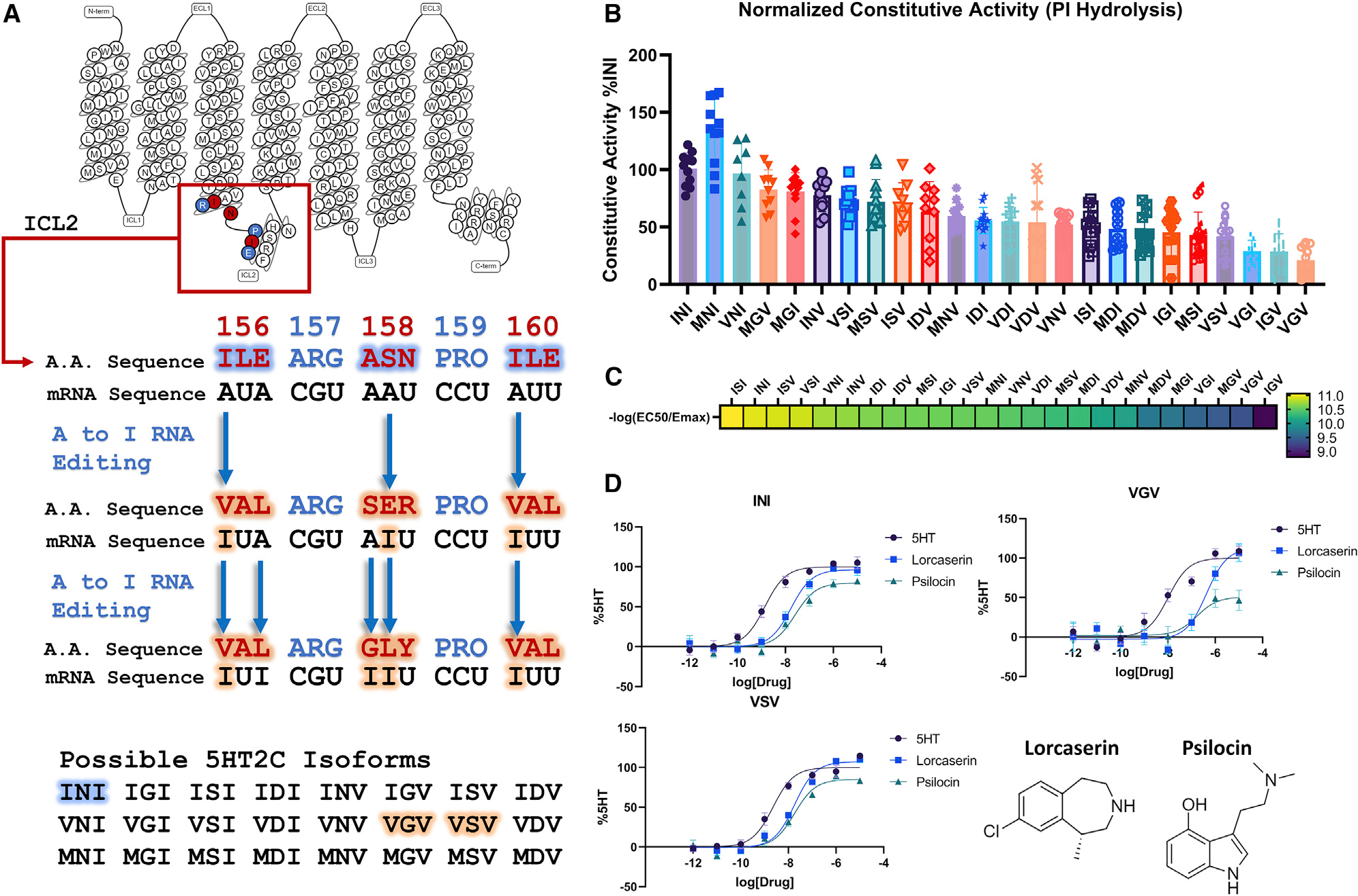
(A) Schematic of the RNA editing that 5HT2C undergoes to produce the 24 possible isoforms taken from GPCRdB (Pándy-Szekeres et al., 2018).
(B) PI-hydrolysis assay done in HEK293 cells to test the basal activity of the 24 different isoforms. Data represent mean values ± SEM of n = 3 biological replicates, with each technical replicate being plotted (n = 9–12) and from a single point on the titration curve (30 ng of plasmid DNA).
(C) Heatmap of different isoforms displaying the transduction coefficient (−log(EC50/Emax)) from n = 3 biological replicates. Complete data are shown in Figure S1 and Table S1.
(D) TRUPATH assays comparing the unedited INI isoform and VGV/VSV isoforms normalized to the %5HT response for each isoform. As one notices, these curves exhibit different potencies for 5HT, lorcaserin (studied in this work), and psilocin (studied in this work) across the different isoforms. Data represent means ± SEM of n = 3 biological replicates.
One of the most profound consequences of 5HT2C RNA editing is the selective modulation of constitutive (a.k.a. basal or ligand independent) GPCR signaling (Niswender et al., 1999, 2001), which has significant functional and pathological consequences (Kawahara et al., 2008; Morabito et al., 2010; O’Neil and Emeson, 2012; Rauser et al., 2001; Warhaftig et al., 2021; Xie et al., 2021; Zhu et al., 2012). The most common 5HT2C edited isoforms—VGV and VSV—both exhibit a loss of constitutive activity (Figure 1B) and reduced potency for 5HT (Berg et al., 2005; Niswender et al., 1999, 2001; Rauser et al., 2001; Zhu et al., 2012). Mice expressing fully edited 5HT2C receptors suffer from a host of pathological phenotypes, including hyperphagia and increased energy expenditure (Kawahara et al., 2008; Morabito et al., 2010). It has also been shown that pancreatic cells overexpressed with various 5HT2C isoforms exhibit reduced glucose-stimulated insulin secretion (Xie et al., 2021).
To elucidate the molecular mechanisms by which RNA editing alters 5HT2C constitutive activity, we solved the INI (wild type [WT]), VGV, and VSV active-state receptors complexed with Gαq and β/γ subunits by single-particle cryoelectron microscopy (cryo-EM), utilizing both the selective 5HT2C agonist lorcaserin (Belviq) and the classic psychedelic psilocin. The structures revealed a hydrogen-bonding network including E16134.52, N1583.56, R1573.55, and Y1533.51 (Y of the DRY motif) that is disrupted in the VGV and VSV isoforms, while N1583.56 preserves this network in the WT unedited isoform. This conformation maintains the loop positioning to expose I16034.51 to multiple hydrophobic contacts on the C terminus of Gαq. Bolstered by a comprehensive set of biochemical and computational investigations, our studies reveal structural mechanisms for 5HT2C contributing to the RNA-editing-induced modulation of a prototypical GPCR.
RESULTS
Isoform differences modulate Gq coupling and constitutive activity
Initially, we investigated the constitutive activity of all the 24 protein-coding isoforms of the 5HT2C receptor. Previously, the VGV isoform was found to exhibit a significant reduction in constitutive activity, while the INI isoform displayed the highest basal phosphoinositide (PI) hydrolysis activity (Niswender et al., 1999, 2001). Confirming these prior studies, we similarly found that the INI isoform showed some of the highest basal activity, while VGV and VSV showed significantly reduced basal activity (Figure 1B). To rule out differences in constitutive activity related to expression levels, we measured the cell-surface expression of each isoform and found no statistically significant differences in expression levels (Figure S2). Interestingly, exploring the constitutive activity across all possible 5HT2C isoforms, we found that the MNI, INI, and VNI isoforms exhibited the highest basal activity. These isoforms have NI in positions 158 and 160, respectively. In contrast, we noticed that the placement of a G or S at position 158 tended to decrease basal PI hydrolysis activity, particularly with I or V in flanking positions (positions 156 and 160).
We next evaluated the ability of each isoform to activate the Gαq heterotrimer utilizing our recently described TRUPATH biosensor platform (Olsen et al., 2020). Using this approach, which is relatively insensitive to receptor reserve and downstream amplification, we found that VGV showed reduced Emax and/or potency for 5HT, while VSV’s reduced potency was not statistically significant (Figures 1C and S1; Table S1) (Niswender et al., 1999, 2001). Also, to minimize possible kinetic effects of G-protein coupling to the receptor, the ligand was incubated with the receptor for 30 min prior to reading. Interestingly, the ISI isoform was the most efficient transducer, while the IGV isoform was the least (Figures 1C and S1; Table S1). Combining these parameters (i.e., constitutive activity, potency, and transduction coefficient estimates), we were able to cluster the 24 isoforms into six major clades (Figure S1). The INI, MNI, and VNI isoforms belong to a single cluster, which exhibits higher basal signaling, while the cluster containing VGV consists of isoforms (three of the four isoforms in this cluster) that have a G in position 158 (Figure S1) and exhibit the lowest basal signaling. Based on these findings, we hypothesized that “NI” in positions 158 and 160 is important for higher constitutive activity, while a G in position 158 is associated with lower constitutive activity. The other possible choices, while they are as efficient as an N in position 158, allow for quantifiable levels of basal PI hydrolysis activity.
To investigate whether the NI motif exhibited structural differences between the INI, the VGV, and the VSV isoforms on Gαq coupling, we focused on the 5HT2C-preferring agonist lorcaserin (Belviq) (Aschenbrenner, 2020). Lorcaserin (Belviq) is a potent and selective 5HT2C agonist that was approved for weight loss but was recently removed from the market after long-term clinical trials showed that patients were diagnosed with cancer at significantly higher rates (Aschenbrenner, 2020). Recent reports suggest that activation of the 5HT2C receptor with lorcaserin could be used to treat addiction for nicotine dependence and/or psychostimulant abuse (Gerak et al., 2019; Higgins et al., 2012; Tabbara et al., 2021). Utilizing our previously developed TRUPATH system, we explored the ability of lorcaserin to activate heterotrimer G-protein dissociation with the INI, VGV, and VSV isoforms (Olsen et al., 2020) (Figure 1D). Similar to 5HT, we found that lorcaserin displays a reduction in potency for the VGV isoform compared with INI (logEC50 INI, −7.77; VGV, −6.37; p < 0.01), but not for the VSV isoform (logEC50 INI, −7.77; VSV, −7.74; p > 0.05; Table S2). We chose lorcaserin for structural studies because of its chemical stability, full agonist activity at each tested isoform, and ability to stabilize 5HT2C receptor-G protein complexes. To investigate the role a common partial agonist could play in Gαq coupling, we chose the classic hallucinogen psilocin to solve the INI structure (Sard et al., 2005). Psilocin has also been known to be a potent partial agonist at 5HT2C, but has not been extensively studied at the different isoforms. We noticed a similar pattern with psilocin, although not statistically significant, with a right shift in potency only in the VGV isoform across INI, VGV, VSV (logEC50 −7.59, −6.77, and −7.74; p > 0.05; Table S2). With this in mind, we also structurally characterized the INI isoform bound with psilocin to understand the differences between a full and a partial agonist.
Structures of the 5HT2C receptor in complex with mini-GαqiN heterotrimer
To improve 5HT2C expression and stability, both the N and the C termini were minimally truncated to Δ23N and Δ414C; these alterations did not affect G-protein recruitment (Figure S1). To assemble the Gαq receptor complex, we utilized our previously published mini-GαqiN-Gβ1-Gγ2 heterotrimer coexpression system, which has also been recently used by others (Kim et al., 2020; Mobbs et al., 2021; Xia et al., 2021). The receptor (INI, VGV, or VSV), G-protein complex (mini-GαqiN-Gβ1-Gγ2), and stabilizing single-chain antibody scFv16 (Koehl et al., 2018) were expressed separately in Spodoptera frugiperda (Sf9) insect cells. The complexes were then assembled in the presence of lorcaserin or psilocin and then subjected to single-particle cryo-EM analysis (Figures 2, S3, and S4; Table S3). The INI (lorcaserin), INI (psilocin), VGV (lorcaserin), and VSV (lorcaserin) Gαq-coupled complexes were solved to global resolutions of 2.8, 3.6, 3.2, and 3.4 Å, respectively (Figures 2, S3, and S4; Table S3). This resolution allowed for complete modeling of the ligand in all three isoforms as well as the active-state conformation of ICL2. The overall receptor-G protein interfaces were consistent with our previous work on the 5HT2A Gαq ternary complex (Kim et al., 2020) and other GPCR-Gq complexes (Mobbs et al., 2021; Xia et al., 2021). One notable difference, however, was a cholesterol molecule bound between transmembrane domain 3 (TM3) and TM4 over ICL2 (Figure 2), which is not modeled in our 5HT2A Gαq ternary complex structure (Kim et al., 2020) or other recent GPCR-Gq structures (Mobbs et al., 2021; Xia et al., 2021).
Figure 2. Structure of 5HT2C-mini-GαqiN isoforms.
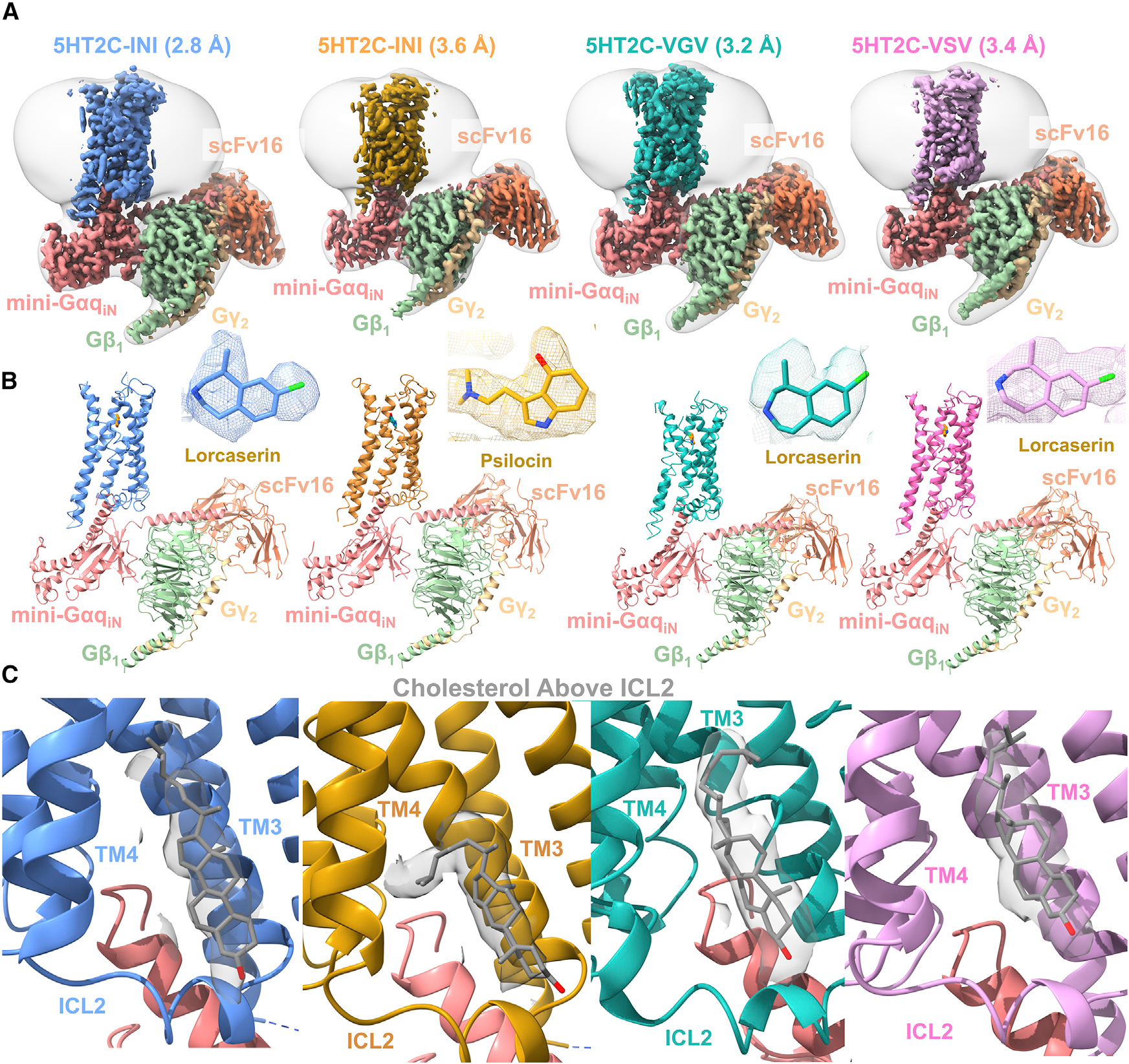
(A) Colored maps are DEEPemHancer-sharpened maps of the different isoforms of 5HT2C-miniGαqiN structures. From left to right, INI-lorcaserin shown in blue, INI-psilocin shown in orange, VGV-lorcaserin shown in cyan, and VSV-lorcaserin shown in magenta. Structures are colored by chain, and a low-pass filtered map is shown in transparent white.
(B) Models built into the maps with their respective colors and labeled as in (A). Ligand density for each model is shown in the upper right of each image.
(C) Density for a cholesterol molecule present between TM3 and TM4 just above ICL2 in each structure (gray).
Lorcaserin/psilocin binding pocket and potential for 5HT2C selectivity
To understand if the modulation of constitutive activity by RNA editing could be explained by changes within the orthosteric pocket, we examined the differences in the lorcaserin binding pose. Given the resolution, the lorcaserin molecules can be placed in the structure, although the precise molecular pose is estimated. The molecules were modeled in such a way that satisfies the coulombic density as well as molecular contacts. These contacts were then verified via functional assays and the reported distances below are based on our models. In addition, the ligand positions were further validated using the GlideEM pipeline (Figure S5), with each structure exhibiting a significant amount of overlap (~1 Å root-mean-square deviation [RMSD]) to the best scored docking pose (Robertson et al., 2020). Across the three isoforms, lorcaserin shows similar binding poses, with the INI and VSV poses showing significant overlap (0.96 Å difference in Cl-to-Cl distance and 0.83 Å difference in N-to-N). The lorcaserin pose in the VGV structure is slightly shifted by 1.81 Å (Cl-to-Cl) and 2.18 Å (N-to-N) compared with the INI pose. However, even with these modest shifts we do not observe any differences in the ligand contacts with the receptor between the three isoforms. All the ligands make H-bonding contacts with the canonical D1343.32 residue and π-π stacking/hydrophobic interactions with F3286.52. Interestingly, most of the binding pockets show significant structural conservation between the three isoforms except for W1303.28. In the VGV isoform, the six-membered ring of W1303.28 is rotated compared with the INI and VSV isoforms (Figure 3A). This rotation places the indole N of W1303.28 closer to D134 (3.93 Å), possibly explaining the slight shift in the ligand binding pose. To functionally verify these binding pocket residues, we tested the D134A, F328A, and W130A mutants. Across all three isoforms D134A abolished both lorcaserin and 5HT Gq activity (Figures 3A, 3Dii, and S6; Table S1). Disrupting the π-stacking interactions, the F328A mutation also significantly reduces the potency of lorcaserin for Gq activation in the INI and VSV isoforms, right shifting the logEC50 by +1.69 (p < 0.0001) and +1.39 (t(4) = 13.07, p < 0.0001), respectively, while completely abolishing activity at the VGV isoform (Figures 3Dii and S5). The W130A mutation reduced the activity of both 5HT (Figure 3Di and S6) and lorcaserin in the VGV isoform, shifting the logEC50 for 5HT by +1.36 (p < 0.05). It is noted that the right shift in the curve for the VGV-W130A mutation in lorcaserin is so extreme that a complete dose-response curve could not be obtained. For the INI isoform, 5HT and lorcaserin logEC50 were changed by +0.50 (p < 0.05) and +0.64 (p < 0.05), respectively, while the VSV isoforms were shifted by +0.88 (p < 0.05) and +1.0 (p < 0.05), respectively. This would indicate that W1303.28 could play an important role in stabilizing D1343.32 across the various isoforms.
Figure 3. The orthosteric binding pocket reveals ligand interactions.
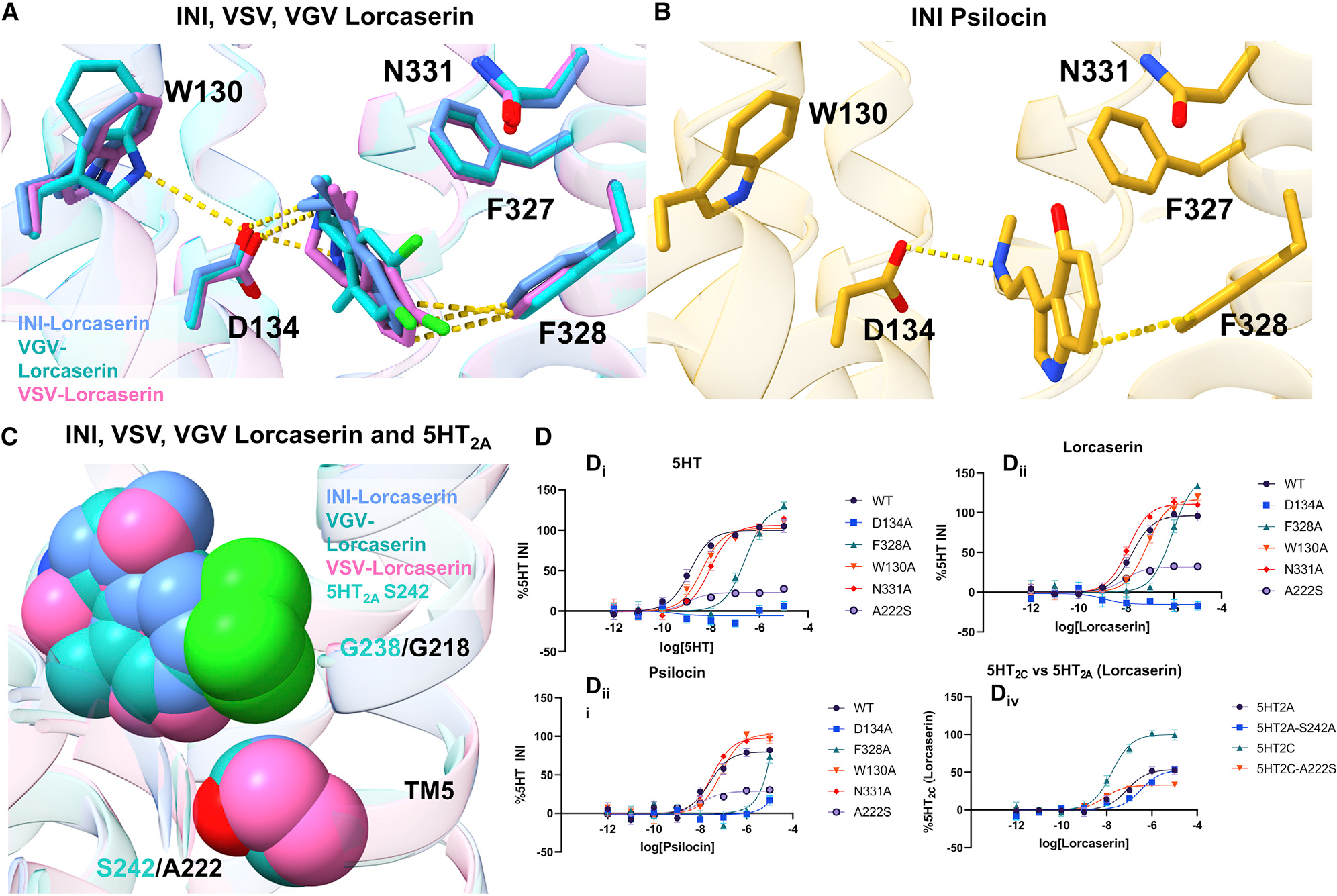
(A) Lorcaserin bound to the INI (cornflower blue), VSV (magenta), and VGV (dark cyan). Yellow lines are indicative of either hydrogen-bonding interactions or π-stacking interactions.
(B) Psilocin bound to the INI (orange) isoform. The yellow lines are indicative of either hydrogen-bonding interactions or π-stacking interactions.
(C) Lorcaserin shown as VDW spheres and A222/S242 from the 5HT2C/5HT2A receptor (PDB: 6WHA is used for the 5HT2A receptor). It is shown that the bulky nature of the chlorine sits in a hydrophobic pocket close to TM5. The presence of a serine in position 5.46 effects the selectivity of lorcaserin for the 5HT2C receptor.
(D) (i–iii) Mutation of the binding pocket for the INI isoform (other isoforms are shown in Figure S4A and Table S2), as well as ICL2 for both lorcaserin and psilocin. All binding pocket mutations affect ligand potency and/or Emax. (iv) Mutations that reflect the slight changes in potencies when mutating A222S (5HT2C) and S242A (5HT2A). Data represent means ± SEM and n = 3 biological replicates.
The selectivity of lorcaserin for the 5HT2 family of receptors (Figure 3C) was previously proposed to be due to the position of the chlorine atom on the benzene ring of the benzazepine and interactions with G2185.42 (Peng et al., 2018). The 5HT2A receptor has a serine (S242) in position 5.46, while 5HT2C has an alanine (A222) at this position. These differing residues change the binding pocket nearing TM5, which could theoretically shift the position of the Cl interaction. This structural hypothesis was confirmed by our functional assays, and the non-synonymous 5HT2A receptor mutation can be made to the 5HT2C receptor (A222S5.46) to test this hypothesis. This mutation modestly increases the potency of lorcaserin for 5HT2C but is not statistically significant (logEC50 increase of +0.336, p > 0.05), while Emax is dramatically decreased (96% versus 32%, p < 0.0001) (Figure 3Div; Table S2). Mutating residue S242A5.46 to an alanine in the 5HT2A receptor slightly decreases the potency but is not statistically significant (right shifting logEC50 by +0.62, p > 0.05) for lorcaserin, while retaining the same Emax as the WT (~52% WT and 51% S242A5.46, p > 0.05) (Figure 3Div; Table S1). This finding reinforces the idea that the selectivity of lorcaserin for the 5HT2C receptor is driven by the hydrophobic nature of A2225.46 and is reflected in the statistically significant drop in Emax of G-protein activation. However, the absence of a change when making a synonymous mutation in 5HT2A would indicate that there are other factors influencing lorcaserin’s activation of the 5HT2A receptor.
Examining the INI-psilocin structure, we noticed that psilocin makes similar contacts in the orthosteric pocket compared with lorcaserin. D1343.32 makes a salt-bridge interaction with the psilocin amino tail, while the tryptamine ring participates in π-stacking interactions with F3286.52 (Figure 3A). Alanine mutations of these important residues completely abolish Gαq activation at the INI isoform (Figure 3Diii; Table S1). Furthermore, based on the coulombic density for the ligand, we can clearly place the 4-OH of psilocin making a clear contact with N3316.55 (Figures 2 and 3B). Interestingly, when mutating this residue to an alanine, we see a slight right shift, but not statistically significant, in potency (a change of 0.12 pEC50, p > 0.05) and a slight increase in Emax (+18%, p < 0.01). This would indicate that the interaction between the 4-OH and N3316.55 is not important for receptor stimulation by psilocin, while other interactions (D1343.32 and F3286.52) drive Gαq activation.
Even though it has been shown that the TRUPATH platform is relatively insensitive to receptor reserve and downstream amplification (Cao et al., 2021; Olsen et al., 2020), to ensure that the observed differences in potency and Emax across all the mutations were accurate, we measured the cell-surface expression of each construct (Figure S2). While there are small observed differences in expression, they did not reach a level of statistical significance. These findings predict that, for the functional data presented here, differences in expression do not affect estimates for either Emax or potency.
Comparison of 5HT2C inactive/active ICL2 conformations
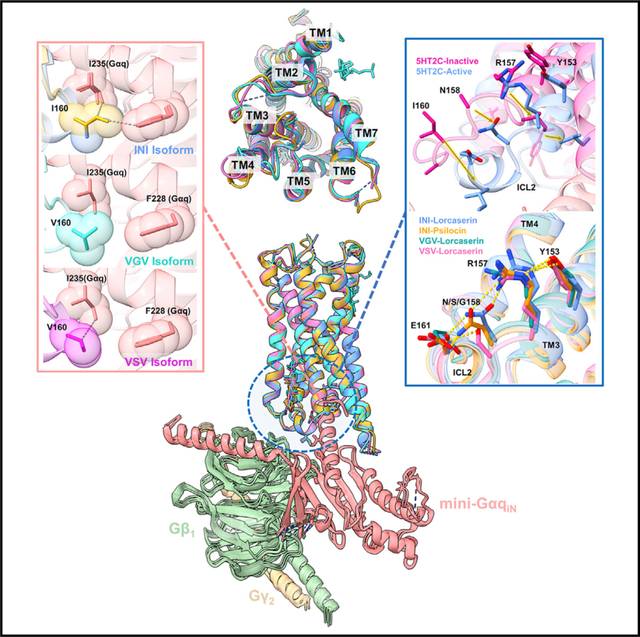
Examining the prior 5HT2C X-ray crystal structures, we noticed that our new G-protein-coupled cryo-EM structures were reminiscent of the previously published “active-state” crystal structure (PDB: 6BQG). Significantly, in Peng et al. (2018), ICL2 in the previous “active-state” crystal structure (PDB: 6BQG) was unresolved, presumably due to the absence of the heterotrimer. This is clear in our previous 5HT2A-heterotrimer structure, which makes several critical interactions between ICL2 and the α5 helix of Gαq, which stabilizes ICL2 in the active-state conformation (Kim et al., 2020). In our new structures, all the isoforms exhibit sufficient coulombic density to resolve most of the side chains and the backbone of ICL2 (Figures S3 and S4). Furthermore, the INI, VSV, and VGV isoforms show remarkable similarity in ICL2 conformation and positioning as well as global structural alignment (Figure 4).
Figure 4. Structural comparison of the 5HT2C isoforms.
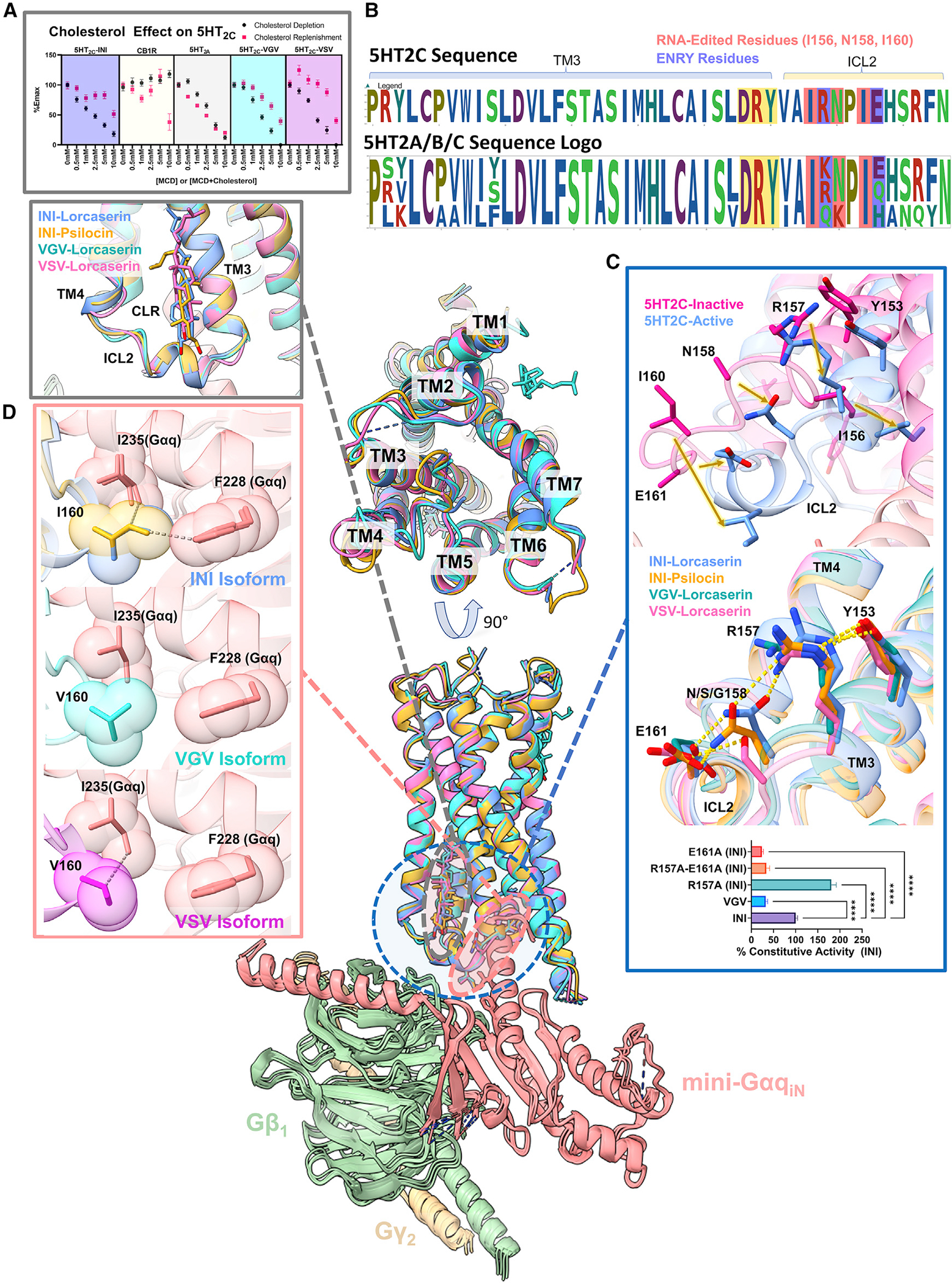
The middle shows the receptor alignment of all the isoforms, showing the global similarity between all of the structures. INI-lorcaserin is shown in cornflower blue, INI-psilocin in goldenrod orange, VGV-lorcaserin in cyan, and VSV-lorcaserin in magenta.
(A) (Top) Plot of Emax for the various 5HT2Cs, 5HT2A, and CB1R (control) as a function of methyl-β-cyclodextrin (MCD) and MCD + cholesterol complexes. Full dose-response curves can be seen in Figure S4. All data are representative of the mean values ± SEM and n = 3 biological replicates. (Bottom) The cholesterol present in all of the isoforms above ICL2 and sandwiched in between TM3/4.
(B) Sequence logos of the 5HT2A, 5HT2B, and 5HT2C receptors of TM3 and the ICL2 loop. Residues shaded in red get RNA edited to form 5HT2C isoforms, residues shaded in blue partake in the ENRY H-bond network, and residues highlighted in gold represent the canonical DRY motif. This is to note that R157 and E161 are non-conserved residues across the 5HT2 family.
(C) (Top) The inactive-to-active state transition of ICL2. The pink is PDB: 6BQH and the cornflower blue is the INI-lorcaserin structure from this work. (Middle) The ENRY H-bond network stretching from Y153 (Y of the DRY motif) to E161 on ICL2 for the INI isoform structures. Also present is the disruption of the H-bond network for the VSV and VGV isoforms. (Bottom) PI hydrolysis for basal activity of the ICL2 mutants performed the same as in Figure 1B. Data are representative of mean values ± SEM and n = 3 biological replicates. For statistical significance ANOVA was done on the constitutive activity and Dunnett’s multiple comparison test was done to compare each mutant with the WT (****p < 0.0001).
(D) Hydrophobic interactions (shown as dashed lines) of I/V160 with F228 (Gαq) and I235 (Gαq) shown in the INI-lorcaserin, INI-psilocin, VGV-lorcaserin, and VSV-lorcaserin structures. These interactions are important for modulating potency and constitutive activity.
Using the inactive state structure for comparison (PDB: 6BQH) (Peng et al., 2018), 5HT2C bound with the inverse agonist ritanserin, we were able to refine the ICL2 transition, which occurs upon G-protein interaction with the active receptor. Upon activation, ICL2 undergoes a significant structural rearrangement (Figure 4C, top), transitioning from an unstructured loop to a helix, positioning key residues for interactions with Gαq. I16034.51 is translated by 7.3 Å and rotated by 18.8° (inactive I16034.51 Cα: active C1463.44 Cα: active I16034.51 Cα), E161 is translated by 4.3 Å and rotated by 9.8° (inactive E16134.52 Cα: active C1463.44 Cα: active E16134.52 Cα), and N1583.56 is translated by 6.0 Å and rotated by 18.5° (inactive N1583.56 Cα: active C1463.44 Cα: active N1583.56 Cα).
We also noted the cholesterol above ICL2 (Figures 2C and 4), which is not present in either of the previously published inactive- or active state-crystal structures. To test if cholesterol differentially acts as a modulator between INI, VSV, and VGV isoforms, we examined Gαq activation upon depletion of cholesterol by methyl-β-cyclodextrin (MCD) and subsequent replenishment. As a control, we utilized CB1R, which was previously shown to be negatively modulated by cholesterol, and utilized a similar cholesterol depletion/addition assay via MCD (Bari et al., 2005a, 2005b). As can be seen in Figures 4A and S6B, CB1R activation is insensitive to cholesterol depletion as a function of increasing MCD concentrations (Bari et al., 2005a). In addition, activity can be depleted through cholesterol replenishment at all concentrations of MCD + cholesterol complexes (Figures 4A and S6B) (Bari et al., 2005b), recapitulating the notion that cholesterol negatively modulates CB1 activity. An inverse phenomenon is present across the 5HT2C isoforms; upon cholesterol depletion Gαq activity decreases, but Gαq activation during cholesterol replenishment is minimally rescued (Figures 4A and S6B). Since this process appears to be only partially reversible, this would indicate that bulk cholesterol plays an important role in the functional stability of the 5HT2C receptor. Moreover, we do not observe rescue of the activity upon cholesterol replenishment in the 5HT2A receptor (Figures 4A and S6B). However, after close examination of the deposited map (EMD-21669), we noticed that there is an unmodeled density between TM3/4 that resembles a cholesterol molecule (Figure S6) (PDB: 6WHA) (Kim et al., 2020). While the density is in a slightly different place compared with our 5HT2C structures, this is also indicative of bulk cholesterol playing a stabilizing structural role for 5HT2A and correlates with our functional data. This would lead us to believe that annular shell cholesterols possibly play a small role in stabilizing the active state of 5HT2C/5HT2A. Furthermore, since this phenomenon is observable across the three different isoforms, where basal activity is significantly different, this would indicate that cholesterol is not involved in modulating constitutive activity. It is also unclear whether this cholesterol could play a role in coupling to other transducers (i.e., arrestin or other Gq family members). In addition, there are several in silico studies for both the 5HT2C and the 5HT2A receptors that indicate that cholesterol could be important for structural stabilization or dimerization (Massaccesi et al., 2020; Ramírez-Anguita et al., 2018).
ICL2-Gαq interactions mediate 5HT2C constitutive activity
After ruling out differences in the orthosteric site upon ligand activation and changes to the global structure of 5HT2C, we next examined the interactions between the RNA-edited residues and Gαq. RNA editing results in mutated residues located at the end of TM3 and beginning of ICL2 (I1563.54, N1583.56, and I6034.51) (Figures 1A and 4C, middle). Closely examining both the structure and the sequence homology of the 5HT2 family in this region, we noticed that two non-conserved residues, E16134.52 and R1573.55, flank the RNA-edited residues (Figures 4A and 4B). The INI structure shows that a hydrogen-bonding network forms from E16134.52 all the way to Y1533.51 of the DRY motif (Figure 4C). These interactions place I16034.51 in the optimal position to make hydrophobic contacts with I285 (Gαq)/F228 (Gαq) (Figure 4D). The different isoforms modulate these interactions through the creation or disruption of this hydrogen-bonding network. In this case, the INI isoform has been shown to have a high level of constitutive activity (Figure 1B), whereas VSV and VGV show reduced levels of basal activity and do not have this network (Figure 4D). In these cases, V16034.51 in VSV makes only one hydrophobic contact with the α5 helix of Gαq, while VGV makes none (Figure 4D).
Based on the structural evidence, the stabilization of the active conformation in ICL2 by the E16134.52-N1583.56-R1573.55-Y1533.51 (ENRY) network allows for constitutive activation of Gq and is further stabilized by the multiple hydrophobic interactions between I16034.51 and Gαq (Figure 4D). With the disruption of these interactions, constitutive activity is significantly reduced while maintaining other key interactions between ICL2 and Gαq for agonist-promoted G-protein activation (i.e., VGV). To further probe the importance of the ENRY network, we created E161A, R157A, and E161A/R157A (double mutation) mutants for the INI isoform. The rationale was that if we could modulate the constitutive activity through disruption of this network on an isoform with high basal activity, this would support our hypothesis on the importance of the network in stabilizing a constitutively active conformation of ICL2. Interestingly, we found that deletion of E16134.52 from the ENRY network reduced the levels of basal activity to that of VGV, while deletion of R1573.55 significantly increased constitutive activity compared with INI (Figure 4C, bottom). In fact, the double mutation R157A/E161A, much like E161A, also showed attenuated constitutive activity comparable to VGV. This indicates that E16134.52 is essential for the placement of ICL2 in a favorable position for ligand-independent activation of the G-protein heterotrimer. Disruption of this network (i.e., G at position 158) through RNA editing allows for modulation of basal activity through optimal positioning of I16034.51 to interact with the α5 helix of Gαq and potentially regulate the loop dynamics of ICL2 from the inactive to the active state.
ICL2 flexibility correlates with constitutive activity
To further test whether disruption of the ENRY network via RNA editing changes the loop dynamics, we examined the conformational space of the ICL2 loop using Rosetta Backrub (Friedland et al., 2008; Smith and Kortemme, 2008). It has been previously shown that Rosetta Backrub can recapitulate the natural conformational variability of protein loops, often covering structural space not accessible to molecular dynamics (MD) simulations except at exceptionally long time points and/or high temperatures (Smith and Kortemme, 2008). To understand how the loop dynamics change between the various isoforms (INI, VSV, and VGV), we ran five simulations for each of the isoforms with 2,000,000 Monte Carlo steps (Figures 5A and S6D). These simulations revealed that INI isoforms exhibit a more stable loop conformation with an average RMSD of 0.22 Å, while VSV and VGV exhibited average RMSDs of 0.43 and 0.45 Å (INI to VSV, p < 0.01, and INI to VGV, p < 0.0001), respectively (Figure S6D). These results predict that the ENRY network is important for stabilizing the active-state loop conformation for constitutive activity. We also calculated the RMSF of the residues across the simulation and noticed that the end of TM3 and beginning of ICL2 varied between the isoforms, with INI showing the least flexibility and VSV/VGV displaying increased dynamics in this region (Figure 5A). To examine this phenomenon further, we examined the local resolution of the INI, VSV, and VGV lorcaserin structures (for the sake of continuity) (Figure 5C). The trend observed in the RMSF is also exhibited in the local resolution, with INI exhibiting a higher relative resolution at the end of TM3 and beginning of ICL2, indicating higher homogeneity or less flexibility, where VSV/VGV have a lower relative resolution across the TM3-ICL2-TM4 interface, signifying less homogeneity or more flexibility (Figure 5C). This would corroborate with our hypothesis that the ENRY network stabilizes this region of ICL2 so that important hydrophobic contacts can be made with the α5 helix of Gq.
Figure 5. Rosetta Backrub simulations show that ICL2 stability is isoform dependent and R157 is important for transitioning to the active state.
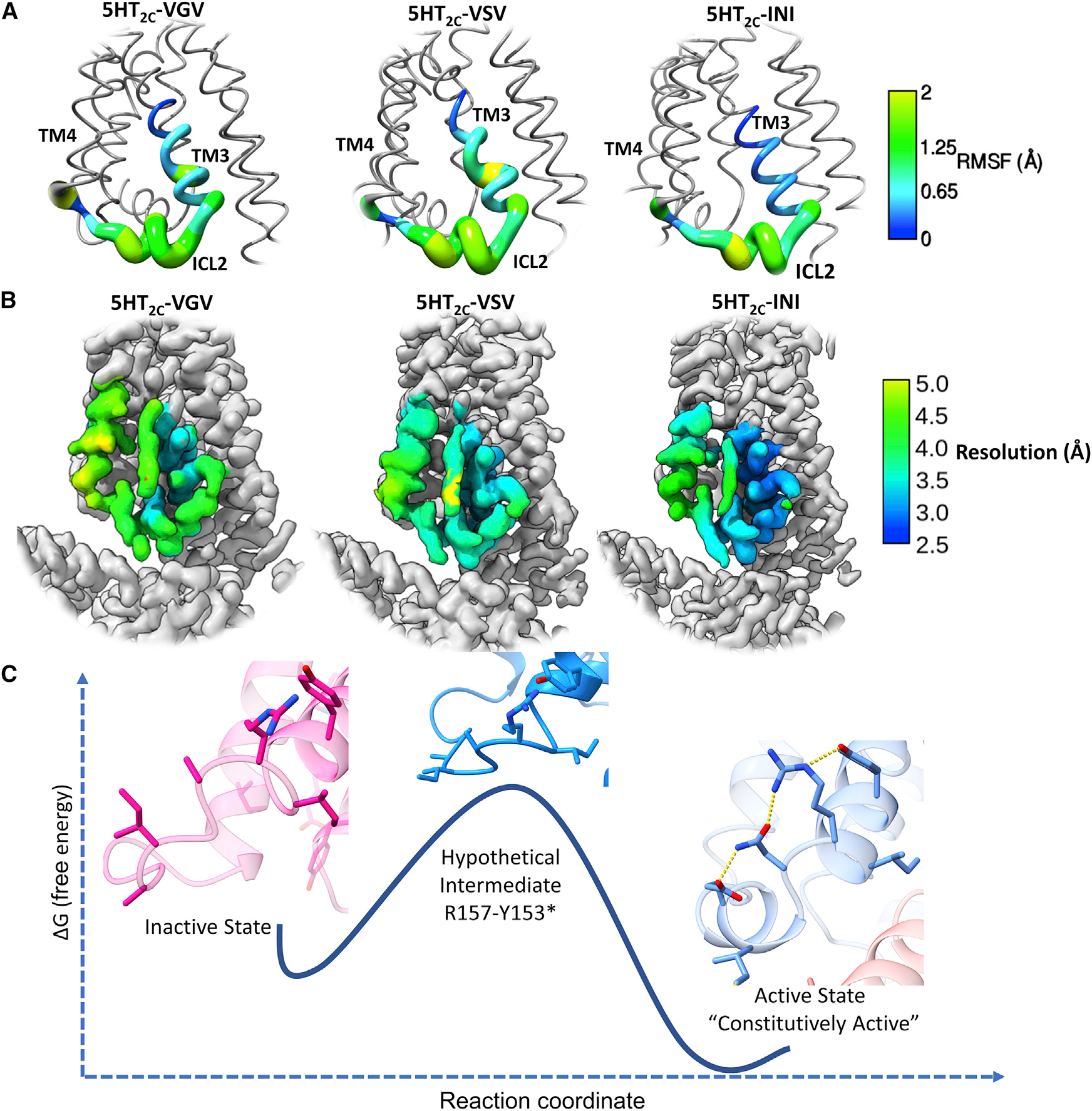
(A) Worm structures of the RMSF values from the Backrub simulations across the three different isoforms.
(B) Local resolution of the cryo-EM structures across the INI, VSV, and VGV isoforms bound with lorcaserin for continuity. The local resolution ranges for the displayed region are 3.2–4.5 Å for VGV, 3.1–4.6 Å for VSV, and 2.5–3.8 Å for INI.
(C) Proposed model of the inactive (PDB: 6BQH)-to-active state (INI-lorcaserin) transition. Significant movement of ICL2 must occur for the receptor to become constitutively active. Based on the active-state structure, R157 is most likely to make H-bonding contacts with Y153 of the DRY motif. This would switch on the active state of ICL2, and the rest of the hydrogen-bonding network would fall into place waiting for Gαq.
Interestingly, we noticed that the MXX isoforms did not follow the predicted pattern for constitutive activity (i.e., MGV, MGI) (Figure 1B). To better understand this pattern, and as an additional orthogonal method to test our hypothesis, we employed a protocol utilizing untemplated AlphaFold predictions to examine predicted conformational states of receptors and transporters (Del Alamo et al., 2022). Using this protocol, we generated 50 structures for INI, VGV, MGV, and MGI isoforms, respectively (Figure S7). The INI exhibited little variation on ICL2 and more closely resembled the active state (Figure S7), whereas the VGV, MGV, and MGI showed a much more dispersed pattern compared with the active/inactive state structures. These results further support the importance of ENRY stabilization of ICL2 on Gαq coupling, as both MGV and MGI exhibit significantly reduced potency (Table S1) (p < 0.0001), but this does not describe the elevated levels of constitutive activity. However, upon examination of the structures, we noticed that the M in the MGV/MGI isoforms points into the receptor pocket (Figure S7). More specifically, the M pushes outward on TM5 and TM6, potentially opening the receptor pocket. With the increased flexibility of ICL2 in these isoforms, this would improve the probability of Gαq interacting with the receptor in an open/active state, serving as a model for explaining the elevated levels of constitutive activity.
DISCUSSION
Here we provide the first molecular details that provide insight into how RNA editing modulates GPCR signaling. It has long been known that the 5HT2C receptor has various isoforms that arise from post-transcriptional RNA editing (Niswender et al., 1999, 2001; Zhu et al., 2012). RNA editing effectively mutates residues 156, 158, and 160 located in ICL2 to M/V at the first position, D/G/S at the second position, and V in the final position. Here we present four active-state structures of the 5HT2C receptor and a model for constitutive activity. These structures are the first that characterize the different isoforms that are formed by RNA editing and provide an explanatory model for the constitutive activity of the 5HT2C receptor centered around the formation of the ENRY network.
We also provide a model based on functional and structural data for spontaneous activation of the receptor, which leads to basal activity (Figure 5C). When examining the transition from inactive state to active state (Figure 5C), we noticed that R157 would have to make a hydrogen-bonding contact with Y153 and stabilize the DRY motif into the active-state position. Due to the positioning of these residues, they are only 4.3 Å apart in the inactive state, and natural movement of R157 would most likely create a hydrogen bond between the two. Mutation of R1573.55 to A significantly increases the constitutive activity of the 5HT2C-INI receptor, allowing for further spontaneous activation of the ICL2 loop through H-bonding between E16134.52 and N1583.56. These interactions act in concert to stabilize an active-state ICL2 conformation, optimally positioning I16034.51 to interact with the α5 helix of Gαq, forming a stabilizing network of interactions allowing for the correct position to couple with G protein without any ligand-induced conformational changes. The E161A removes these stabilizing interactions, or raises the energy barrier, and disrupts the ENRY network, drastically reducing basal activity. This is recapitulated through disruption of this network due to RNA editing, as well as changing receptor/Gαq interactions, as seen in the various isoforms, and explains the differences in basal activity. While it is still unclear what has the greatest effect on constitutive activity, whether it is the receptor isomerization constant (stabilization by the ENRY network) or changes in G-protein affinity (interactions with Gαq), it is probable that both mechanisms play a significant role in modulating basal activity.
There is significant evidence that the 5HT2C receptor and its isoforms are responsible for the expression of various pathological phenotypes, which can be linked to differences in basal activity (Kawahara et al., 2008; Morabito et al., 2010; Warhaftig et al., 2021). Since the orthosteric site remains unchanged across the isoforms, and our structural findings suggest that the differences in Gαq activation are due to the changes in interactions between the receptor and G protein mediated by ICL2, one could imagine that an isoform-selective allosteric modulator could be designed. We have also found a potential cholesterol binding site between TM3/TM4 above ICL2, which could be exploited by small-molecule allosteric modulators. The functional ramifications of altering basal activity in an isoform-dependent manner remain to be explored. Finally, differences in G-protein selectivity/arrestin among the differing isoforms, both ligand-dependent and independent activation, remain to be explored in 5HT2C pharmacology.
Limitations of study
There are several potential limitations of this study, which could be adequately addressed with follow-up experiments. First, the mechanistic underpinnings of constitutive activity and isoform-specific G-protein coupling could be further revealed by additional active-state structures with isoforms that contain an M or D. Also, transducer-free active-state structures could be obtained to further validate the importance of the ENRY H-bonding motif. These structures could be further supplemented with a kinetic (coupling to G proteins) and protein dynamics study across the different isoforms. This would reveal whether the constitutive activity is due to the kinetic effect of G-protein coupling or conformational selectivity between the isoforms (or if a certain isoform favors the active versus the inactive state). In this study, these important molecular underpinnings remain opaque. Finally, it is unclear how these isoforms affect the constitutive activity as well as transducer coupling across other Gαs and β-arrestins and how an array of ligands/allosteric modulators affects the receptor in an isoform-dependent manner. Future studies will be needed to address these possibilities.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
All inquiries for further information regarding requests for reagents or resources should be directed to the lead contact: Bryan L. Roth (bryan_roth@med.unc.edu).
Materials Availability
All plasmids and/or resources generated by this study will be available through the lead contact.
Data and code availability
All the analyzed and/or generated data is available in this manuscript/Supplementary information. All structural data has been deposited in the Electron Microscopy Databank (EMDB) (cryo-EM maps) and Protein Databank (PDB) (cryo-EM maps and models). They have been deposited under the PDB: 8DPF/EMD-27633 for INI-lorcaserin, PDB: 8DPG/EMD-27634 for INI-psilocin, 8DPH/EMD-27635 for VGV-lorcaserin, and PDB: 8DPI/EMD-27636 for VSV-lorcaserin.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For protein expression Spodoptera fugiperda (SF9, Expression Systems) cells were grown in cell suspension using ESF-921 medium at 120 rpm and 27°C. For the in vitro pharmacology assays HEK293T cells were obtained from the American Type Culture Collection (ATCC, CRL-11268). Cells were maintained at 37°C incubator with 5% CO2 and DMEM medium (VWR, #45000) supplemented with 10% (v/v) fetal bovine serum (FBS, VWR, #89510-186) and 100 I.U./mL penicillin and 100 mg/mL streptomycin. For all assays the media was replaced with DMEM supplemented with 1% (v/v) dialyzed FBS prior to plating.
METHODS DETAILS
Bioluminescence resonance energy transfer assays (TRUPATH BRET2)
In order to measure G-protein activation by the receptor, we employed our previously published TRUPATH system and all assays were carried out as previously described (Olsen et al., 2020). In short, HEK293T cells were plated in 10cm dishes 20–24 hours prior to transfection at ~2 million cells per dish. Two hours prior to transfection, the media was replaced with DMEM containing 1% dialyzed FBS to prevent receptor internalization. Cells were then transfected in a ratio of 1:1:1:1 of receptor:Gα:Gβ:Gγ with 1μg of each plasmid using Transit 2020 (Mirus Biosciences) (using the manufacturer’s recommendation) at a concentration of 3μLs per μg of plasmid and a volume of 10 ng/μL of Opti-MEM. 24 hours post transfection, cells were then plated into poly-l-lysine coated 96-well plates at a concentration of 40–50k cells per well in DMEM containing 1% dialyzed FBS. The next day, the media was aspirated and cells were immediately covered with 60μLs of buffer (1 × HBSS, 20 mM HEPES pH 7.4) and allowed to incubate at 37°C for 10 minutes. 30μLs of a 3× drug binding buffer (1 × HBSS, 20 mM HEPES pH 7.4, 0.3% (w/v) BSA, 0.03% (w/v) Ascorbic Acid, plus concentration of ligand to be tested) were added and allowed to incubate for additional 10 minutes at RT. Finally, 10μLs of coelenterazine 400a was added to each well and allowed to incubate for an additional 10 minutes prior to reading the BRET ratio (395nm/510nm) on a PHERAstar FSX. Each plate was read a total of 5 times for a total of 1 second per well. The fifth read was taken for each plate for a total of 30 minutes of incubation time with the appropriate agonist and data was normalized to a %WT using. All 24 isoforms were generated via site-directed mutagenesis using the 5HT2C-INI as the backbone and PrimeSTAR Max DNA Polymerase according to the manufacturer’s protocols.
Constitutive activity assay (PI-hydrolysis)
PI-hydrolysis at several different plasmid concentrations was done to determine the constitutive activity of the different isoforms/mutations. Transfections were carried out similarly to the TRUPATH assays, except in 12-well plates. Concentrations of 10ng, 30ng, 100ng, and 300ng of receptor were transfected for each isoform and pcDNA3.1 was added to keep the transfection efficiency (total DNA added 300 ng) the same across all receptors in DMEM containing 1% dialyzed FBS. The next day cells were plated in poly-l-lysine coated 96-well plates at a concentration of 50k cells per well in myo-inositol deficient DMEM containing 1% dialyzed FBS. Tritiated inositol was added at 0.1uci per well and allowed to incubate overnight. The next day the media was removed and 50μLs of fresh assay buffer (1xHBSS, 24mM NaHCO3, 11mM Glucose, 15mM LiCl) was added per well and allowed to incubate for 60 minutes at 37°C. The assay buffer was then carefully removed and 50μLs of 50mM formic acid was added to each well and was allowed to incubate for 30 minutes at 4°C. Following the cell lysis, the formic acid solution was removed and added to a 96-well sample plate (MicroBeta Sample Plates) and 75μLs of RNA binding beads (1.2 mg/mL) were added to each well and allowed to incubate for 30 minutes at 4°C. The plates were then counted using a MicroBeta reader as a Counts per Minute (CPM). For quantification of constitutive activity, the 30ng point was used and all counts were normalized to the INI isoform of 100% and pcDNA3.1 control as 0% using GraphPad Prism software v9.0.
Cell surface expression
The cell surface expression of each isoform and mutant presented in the work was measured by ELISA chemiluminescence. In 6 well plates 200 ng of the respective construct, (mock was pcDNA3.1) was transfected like the BRET based assays. 24hrs post transfection cells were plated in white 384 well plates. 48hrs post-transfection the cells were fixed using 20 μLs of 4% paraformaldehyde for 10 minutes. The wells were then washed twice with 40 μLs of PBS and subsequently incubated with 20 μLs of 5% BSA (in PBS) for one hour. The cells were then incubated with 20 μL of an anti-Flag-horseradish peroxidase conjugated antibody (1:10,000 dilution in 5% BSA in PBS) for an additional hour. The wells were then washed four times with 100 μL of PBS and then 20 μL of Super Signal Enzyme-Linked Immunosorbent Assay Pico Substrate was added per well. Luminesence was measured using a PHERAstar FSX and the data was analyzed using GraphPad Prism v9. ANOVA and Dunnett’s multiple comparison test was then carried out for statistical significance. Data is available in Figure S2.
G-protein activation upon cholesterol depletion and replenishment
G-protein activation was measured using the TRUPATH platform in the same manner as described above, but with a cholesterol depletion or replenishment step as previously described (Bari et al., 2005a; 2005b). To achieve cholesterol depletion, the day after plating cells in 96-well plates a solution of DMEM+β-methyl-cyclodextrin (MCD) (either 0.5mM, 1mM, 2.5mM, 5mM, or 10 mM MCD) were allowed to incubate for 1hr. After 1hr, the process of drug incubation and plate reading was then carried out as normal. To achieve cholesterol replenishment, after the 1hr of MCD incubation, the cells were treated with varying concentration of MCD + cholesterol complexes (either 0.5mM, 1mM, 2.5mM, 5mM, or 10mM MCD + cholesterol) and allowed to incubate for 1hr.
5HT2C receptor expression and purification
Using the Bac-to-Bac Baculovirus expression system (Invitrogen) to generate recombinant baculovirus, a Δ23N-terminal truncation and Δ414C-terminal truncation construct was made. Viral titer was then determined by flow cytometry using the gp64-PE to ascertain the % of cells stained. Sf9 cells were then infected at a concentration of 2 million cells/mL and an MOI of 3. The cells were then harvested after 48hrs by centrifugation at 3000 g for 15 minutes. After the initial cell pellet the cells were then washed and centrifuged again with buffer (20 mM Tris pH 7.4, 100 mM NaCl) and either purified immediately or stored at −80°C for future use.
The 5HT2C receptor was purified using the same method as previously (Kim et al., 2020). In short, the cell membranes were disrupted using a hypotonic buffer (10 mM HEPES pH 7.5, 10 mM MgCl2, 20mM KCl, protease inhibitors (500 μM AEBSF, 1 μM E−64, 1 μM Leupeptin, and 0.15 μM Aprotinin), 20 μM Ligand (Lorcaserin/Psilocin)) with douncing. Soluble proteins were then removed with a high-osmotic wash (10 mM HEPES pH 7.5, 1000 mM NaCl, 10 mM MgCl2, 20mM KCl, protease inhibitors, and 20 μM Ligand) for 2 hours at 4°C. 2.0 mg/mL of iodoaceamide (Sigma) was then added for 30 minutes prior to addition of of 5mM β-mercaptoethanol. The membranes were then solubilized in 50 mM HEPES pH 7.5, 1% (w/v) n-dodecyl-beta-D-maltopyranoside (DDM, Anatrace), 0.2% (w/v) cholesterol hemisuccinate (CHS, Sigma), 150 mM NaCl, and 20μM of ligand at 4°C for 2 hrs. The solubilized receptor was then isolated by ultra-centrifugation at 60,000 rpm at 4°C for 1 hr. The supernatant was then collected and TALON IMAC resin, final concentration of NaCl 500 mM, a final concentration of 20 mM Imidazole, and 20μM of ligand was added and allowed to incubate overnight at 4°C with gentle agitation. The next day, the resin was then washed with 10 column volumes (cv) of 50 mM HEPES pH 7.5, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 800 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole, and 20μM of desired ligand. A subsequent was was then done with 10 cv of 50 mM HEPES pH 7.5, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 500 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole, and 20μM of desired ligand. The protein was then eluted in 3 cv of 50 mM HEPES pH 7.5, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 500 mM NaCl, 10% (v/v) glycerol, 250 mM imidazole, and 20μM of desired ligand. Immediately following elution, the protein is then buffer exchanged into 20 mM HEPES pH 7.5, 100 mM NaCl, 0.5% (w/v) LMNG, 0.05% CHS, 0.00025% (w/v) GDN, 100 μM TCEP, and 20 μM of desired ligand.
scFv16 and mini-GαqiN expression
ScFv16 was expressed and purified as previously reported (Koehl et al., 2018). Purified scFv16 from media was concentrated and subjected to size-exclusion chromatography on a S200 10/300 column equilibrated with 20 mM HEPES pH 7.5, 0.001% (w/v) LMNG, 0.0001% CHS, 0.00025% GDN, 100 μM TCEP. Fraction containing pure protein were collected and stored either at 4°C or at −80°C for complex formation. The mini-GαqiN was expressed and purified as was previously reported and was also subjected to size exclusion on a S200 10/300 column equilibrated with 20 mM HEPES pH 7.5, 0.001% (w/v) LMNG, 0.0001% CHS, 0.00025% GDN, 100 μM TCEP (Kim et al., 2020).
Generation of the 5HT2C, mini-GαqiN heterotrimer, and scFv16 complex
Purified receptor, mini-GαqiN heterotrimer, and scFv16 were mixed in a 2:1 molar ratio of mini-GαqiN-heterotrimer:receptor and scFv16:receptor. Apyrase (to catalyze the hydrolysis of unbound GDP), PNGase F (to cleave any glycosylation on the receptor), and HRV3C (to cleave all the His-tags on the proteins) were added to the mixture and allowed to incubate overnight 4°C. The complex was then subjected to a final round of size exclusion chromatography on a Superdex 200 10/300 column equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl, 0.001% (w/v) LMNG, 0.0001% (w/v) CHS, 0.000025% GDN, 100 μM TCEP, and 20 μM of the desired ligand. The peak fractions were then collected and concentrated to ~2mg/mL for cryo-EM studies.
Single particle cryo-EM data collection and processing
Using a Vitrobot mark IV (FEI) glow discharged Quantifoil R 1.2/1.3 Au 300 holey carbon film grids were individually frozen in an ethane/propane mixture (60/40). The Vitrobot was set to 95% humidity at 4°C with a blot time ranging from 2.5–5 seconds. Images were collected on a 200keV G3 Talos Arctica equipped with a Gatan K3 direct electron detector at a physical pixel size of 0.91 Å. Images were recorded for ~2.7 seconds yielding 60 subframes to give a total exposure of ~50 electrons/Å2. The movies were recorded automatically using SerialEM and a multi-shot array (Mastronarde, 2005). After manual curation and inspection, the micrographs were processed using cryoSPARC v3.1 (Punjani et al., 2017, 2020). An example processing tree can be found in Figure S2 and S3. In brief, a subset of micrographs were used as a training set in the TOPAZ particle picking software (Bepler et al., 2019, 2020). After removal of duplicate particle picks, multiple rounds of reference free 2D classification and multi-reference 3D refinement led to a final stack of particles that was then subjected to Non-Uniform 3D refinement (Punjani et al., 2020). Resolution was determined by using the gold-standard Fourier shell correlation (GFSC) 0.143Å cut-off (Rosenthal and Henderson, 2003; Scheres and Chen, 2012). Map and model validations were performed using the half-maps and post-processed B-factor sharpened maps using Mtriage branch of the Phenix Software package (Afonine et al., 2018; Punjani et al., 2017). Alternative map sharpening was performed on the half-maps using deepEMhancer (Sanchez-Garcia et al., 2020) to assist in visualization and model building.
Model building and refinement
For the initial model, the receptor portion of 6BQG was aligned with the coordinates of 6WHA (5HT2A active state complex). The BRIL domain was then removed from the structure and the position of the receptor was used to place it on the mini-GαqiN heterotrimer/scFv16 complex. Initial placement of the model was done using fit to map in ChimeraX and further automated docking was done using “Dock in map” in Phenix. Once the model was docked to the map, the structure was then subjected to Phenix Real-space refinement with Rigid Body, Morphing, and Simulated Annealing turned on for a single round (Liebschner et al., 2019). Further modelling and structure validation was done using COOT and restraints were generated for both lorcaserin and psilocin using Phenix Elbow (Emsley et al., 2010; Liebschner et al., 2019). Final model validation used Phenix Mtriage to assess map-to-model quality and model statistics were validated using MolProbity (Williams et al., 2018). The final ligand poses were further validated by GlideEM following the previously published protocols (Robertson et al., 2020) (Figure S5). All structural figures were prepared either in PyMol or ChimeraX (DeLano, 2002; Pettersen et al., 2021).
Sequence alignment and visualization
The 5HT2 family (5HT2A, 5HT2B, 5HT2C) were aligned for TM3 and ICL2 using GPCRdb (Kooistra et al., 2020; Pándy-Szekeres et al., 2018). The aligned sequence was downloaded, and the sequence logo was made using the python library Logomaker (Tareen and Kinney, 2020).
Rosetta Backrub simulations
The final receptor models generated from the INI, VSV, and VGV lorcaserin structures were used in the Rosetta Backrub simulations. As previously mentioned, Rosetta backrub can be used to explore conformational space which may be inaccessible to MD simulations except at long time points or high temperatures (Smith and Kortemme, 2008). The Rosetta backrub implementation was used from Rosetta v3.1.2 and the receptor was placed in a membrane using RosettaMP. All atom Backrub simulations were kicked off on receptor residues 147–168 which includes part of TM3 and all ICL2, to explore the entire conformational space of ICL2. Each simulation was run for 2,000,000 monte-carlo steps with a temperature of 0.3 and a trajectory was written every 200 steps. The Backrub trajectories were then analysed with the MDAnalysis package to calculate the Cα RMSD of the loop per step, the average RMSD, and the all atom RMSF (Gowers et al., 2016). The average RMSD per simulation was then plotted in the GraphPad Prism v9.0. For the worm plots the average RMSF was calculated over the 5 runs with MDAanalysis and was written as the B-factor in the starting PDB. The graphics were then rendered by UCSF Chimera v1.14 (Pettersen et al., 2004).
Generation of AlphaFold structures
The code and protocol used was run in the Google Colab notebook found in https://github.com/delalamo/af2_conformations and described in (Del Alamo et al., 2022). In short, this protocol uses different MSA depths as input for the AlphaFold predictions. At the request of the authors, we sampled various sequence depths (128, 64, 32, 16, and 8). Levels 32, 64, and 128 yields a majority of conformationally uniform structures. However, at depth 8, it led to many misfolded proteins. Thus, we used an MSA depth of 16 which created a structurally diverse set of models while keeping proper folding. We created 50 structures for the isoforms INI, VGV, MGV, and MGI. To focus the structural alignment on the receptor core, the N-terminus, ICL3, and C-terminus were removed due to the unstructured nature of these regions. Each structure was then aligned to either the active (our INI lorcaserin structure) or the inactive state (PDB 6BQH) and the Cα RMSD was calculated of ICL2 (residues 150–177) using the MDAnalysis package.
QUANTIFICATION AND STATISTICAL ANALYSIS
In all figures/tables the data is reported as the mean ± standard error of the mean (SEM). The biological replicates and technical replicates are available in the figure/table legends, where “N” is the number of biological replicates. For the BRET based TRUPATH assays, the net-BRET ratio was plotted as a function of drug concentration and subsequently normalized to %5HT or reference agonist. The grouped data was then fit using nonlinear regression and the three-parameter “log-agonist vs response” function in GraphPad Prism v9. Subsequent EC50, Emax, and Transduction Coefficients were analyzed individually by analysis of variance (ANOVA) and significance was reported by using Dunnett’s multiple comparison test. p values are reported as ****p < 0.0001, ***p < 0.001, **p < 0.01,*p < 0.05, and ns (not significant or p > 0.05).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| gp64-PE antibody | Expression Systems | Cat#97-201 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Bac-to-Bac Baculovirus Expression System | Invitrogen | Cat#A11100 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| AEBSF | GoldBio | Cat#A-540-5 |
| Leupeptin | Sigma | Cat#L2884 |
| n-dodecyl-beta-D-Maltopyranoside (DDM) | Anatrace | Cat#D310 |
| Lauryl maltose neopentyl glycol (LMNG) | Anatrace | Cat#NG310 |
| Glyco-Diosgenin (GDN) | Anatrace | Cat#GDN101 |
| Cholestryl Hemisuccinate tris salt | Anatrace | Cat#CH210-25GM |
| His-tagged Precission protease | GenScript | Cat#Z0392-500 |
| Tris HCl | VWR | Cat#0497 |
| HEPES | Fisher Scientific | Cat#BP310 |
| Glycerol | Fisher Scientific | Cat#BP229 |
| Imidazole | Sigma | Cat#I0250 |
| Sodium Chloride | Fisher | Cat#BP358 |
| Potassium Chloride | Sigma | Cat#793590 |
| Magnesium Chloride | Sigma | Cat#M0250 |
| Talon IMAC resin | Clontech | Cat#635507 |
| Coelenterazine | Promega | Cat#S2001 |
| Poly-L-lysine | Sigma | Cat#P2636 |
| Penicillin/Streptomycin | Invitrogen | Cat#15140-122 |
| Sf-900 II SFM | Invitrogen | Cat#10902096 |
| ESF921 | Expression Systems | Cat#96-001-01 |
| DMEM | Sigma | Cat#D6429-6X50 |
| USDA FBS | Omega Scientific | Cat#3463 FB-01 |
| Dialyzed FBS | Omega Scientific | Cat#2459 FB-03 |
| 10×HBSS | Invitrogen | Cat#14065-056 |
| BSA, Fatty Acid Free | Akron | Cat#AK8909 |
| Ascorbic Acid | Sigma | Cat#A5960 |
| Cellfectin II | Invitrogen | Cat#10363-100 |
| TransIT-2020 | Mirus | Cat#MIR5400 |
| Serotonin (5HT) hydrochloride | Tocris | Cat#3547 |
| Psilocin Analytical Standard | Sigma | Cat#P-098-1ML |
| Lorcaserin (hydrochloride) | Caymen | Cat#1521 |
| 3H-inositol | Perkin Elmer | Cat#NET115005MC |
| RNA binding YSi SPA beads | Perkin Elmer | Cat#RPNQ0013 |
| Monoclonal ANTI-FLAG M2-peroxidase antibody | Sigma | Cat#A8592-0.2MG |
| Paraformaldehyde Powder | Sigma | Cat#158127 |
| SuperSignal Elisa Pico Chemiluminesence | ThermoFisher | Cat#37069 |
| Methyl-β-cyclodextrin | Sigma | Cat#C4555-10G |
|
| ||
| Deposited data | ||
|
| ||
| 5HT2C-(INI)-Lorcaserin | This paper | PDB: 8DPF |
| 5HT2C-(INI)-Psilocin | This paper | PDB: 8DPG |
| 5HT2C-(VGV)-Lorcaserin | This paper | PDB: 8DPH |
| 5HT2C-(VSV)-Lorcaserin | This paper | PDB: 8DPI |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Spodoptera frugiperda Sf9 cells | Expression Systems | Cat#94-001S |
| HEK293T | ATCC | Cat#CRL-11268 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pcDNA 3.1 | ThermoFisher | Cat#V79020 |
| Human-5HT2C gene | IDT | N/A |
| Mini-GαqiN chimera | IDT | N/A |
| scFv16 | IDT | N/A |
| TRUPATH (G-alpa, G-beta, G-gamma) | Addgene | Kit#1000000163 |
| pFastBac dual expression vector | Invitrogen | Cat#10712024 |
| pFastBac 1 expression vector | Invitrogen | Cat#10359016 |
|
| ||
| Software and algorithms | ||
|
| ||
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/ |
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Pettersen et al., 2021 | https://www.rbvi.ucsf.edu/chimerax/ |
| Phenix | Liebschner et al., 2019 | https://www.phenix-online.org |
| PyMOL | Schrodinger | www.pymol.org |
| Prism v9.0 | GraphPad Software Inc. | N/A |
| Python v3.9.5 | Python Software Foundation | www.python.org |
|
| ||
| Other | ||
|
| ||
| 100kDa Concentrator (20) Vivaspin | Sartorius Stedim | Cat#VS2042 |
| 100kDA Concentrator (500) Vivaspin | Sartorius Stedim | Cat#VS0142 |
| Superdex 200 Increase 10/300 column | GE Healthcare | Cat#289909944 |
| 96-well black plates | Greiner Bio-one GmbH | Cat#655090 |
| 96-well white plates | Greiner Bio-one GmbH | Cat#655098 |
| 384-well white plates | Greiner Bio-one GmbH | Cat#781073 |
| Amicon Ultra-4 10K filter Unit | Merk Millipore | Cat#UFC801024 |
| Amicon Ultra-4 30K filter Unit | Merk Millipore | Cat#UFC803024 |
| Quantifoil, Au300-R1.2/1.3 | Electron Microscopy Sciences | Cat#Q350AR13A |
Highlights.
5HT2C receptor RNA editing modulates constitutive activity through interactions on ICL2
Structural evidence reveals that RNA editing modulates a hydrogen-bonding network on ICL2
Evidence of ICL2 isomerization and changes in G-protein affinity that alter basal activity
ACKNOWLEDGMENTS
This work was supported by NIH grants RO1MH112205 and R37DA045657 and DARPA. The Titan X Pascal used for this research was kindly donated to J.F.F. by the NVIDIA Corporation. We would like to acknowledge Dr. Kuglae Kim and Dr. Yongfeng Liu for initial guidance on protein purification and PI-hydrolysis assays, respectively. Also, we would like to thank the UNC cryo-EM facility, specifically, Jared Peck and Dr. Joshua Strauss, for technical assistance using the Talos Arctica.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111211.
REFERENCES
- Abbas AI, Urban DJ, Jensen NH, Farrell MS, Kroeze WK, Mieczkowski P, Wang Z, and Roth BL (2010). Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method. Nucleic Acids Res. 38, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, and Urzhumtsev A (2018). New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner DS (2020). FDA requests withdrawal of weight-loss drug from market. Am. J. Nurs. 120, 22. [DOI] [PubMed] [Google Scholar]
- Bari M, Battista N, Fezza F, Finazzi-Agrò A, and Maccarrone M (2005a). Lipid rafts control signaling of type-1 cannabinoid receptors in neuronal cells. Implications for anandamide-induced apoptosis. J. Biol. Chem. 280, 12212–12220. [DOI] [PubMed] [Google Scholar]
- Bari M, Paradisi A, Pasquariello N, and Maccarrone M (2005b). Cholesterol-dependent modulation of type 1 cannabinoid receptors in nerve cells. J. Neurosci. Res. 81, 275–283. [DOI] [PubMed] [Google Scholar]
- Bass BL, and Weintraub H (1988). An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, and Tromp MC (1986). Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826. [DOI] [PubMed] [Google Scholar]
- Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, and Berger B (2019). Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T, Kelley K, Noble AJ, and Berger B (2020). Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun. 11, 5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KA, Harvey JA, Spampinato U, and Clarke WP (2005). Physiological relevance of constitutive activity of 5-HT2A and 5-HT2C receptors. Trends Pharmacol. Sci. 26, 625–630. [DOI] [PubMed] [Google Scholar]
- Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, et al. (2021). Structure, function and pharmacology of human itch GPCRs. Nature 600, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Alamo D, Sala D, Mchaourab HS, and Meiler J (2022). Sampling alternative conformational states of transporters and receptors with AlphaFold2. ELife 11, e75751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92. [Google Scholar]
- De Deurwaerdère P, Navailles S, Berg KA, Clarke WP, and Spampinato U (2004). Constitutive activity of the serotonin2C receptor inhibits in vivo dopamine release in the rat striatum and nucleus accumbens. J. Neurosci. 24, 3235–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl Y, Farahani HS, Behm M, Lagergren J, and Öhman M (2012). A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 22, 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland GD, Linares AJ, Smith CA, and Kortemme T (2008). A simple model of backbone flexibility improves modeling of side-chain conformational variability. J. Mol. Biol. 380, 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerak LR, Collins GT, Maguire DR, and France CP (2019). Effects of lorcaserin on reinstatement of responding previously maintained by cocaine or remifentanil in rhesus monkeys. Exp. Clin. Psychopharmacol. 27, 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers R, Linke M, Barnoud J, Reddy T, Melo M, Seyler S, Domański J, Dotson D, Buchoux S, Kenney I, et al. (2016). Mdanalysis: a python package for the rapid analysis of molecular dynamics simulations. In Proceedings of the 15th Python in Science Conference (SciPy), pp. 98–105. [Google Scholar]
- Higgins GA, Silenieks LB, Rossmann A, Rizos Z, Noble K, Soko AD, and Fletcher PJ (2012). The 5-HT2C receptor agonist lorcaserin reduces nicotine self-administration, discrimination, and reinstatement: relationship to feeding behavior and impulse control. Neuropsychopharmacology 37, 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, and Emeson RB (2012). Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 353, 61–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, and Nishikura K (2008). Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 28, 12834–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Che T, Panova O, DiBerto JF, Lyu J, Krumm BE, Wacker D, Robertson MJ, Seven AB, Nichols DE, et al. (2020). Structure of a hallucinogen-activated gq-coupled 5-ht2a serotonin receptor. Cell 182, 1574–1588.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehl A, Hu H, Maeda S, Zhang Y, Qu Q, Paggi JM, Latorraca NR, Hilger D, Dawson R, Matile H, et al. (2018). Structure of the μ-opioid receptor-Gi protein complex. Nature 558, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, Keserű GM, and Gloriam DE (2020). GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krestel H, and Meier JC (2018). RNA editing and retrotransposons in neurology. Front. Mol. Neurosci. 11, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K, Kapoor U, Amman F, Picardi E, Martin D, Bajad P, and Jantsch MF (2019a). A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 29, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K, Hartl M, Amman F, Anrather D, Janisiw MP, and Jantsch MF (2019b). Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 47, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaccesi L, Laudadio E, Mobbili G, Minnelli C, and Galeazzi R (2020). Cholesterol-mediated oligomerization pathways of serotonin G-coupled receptor 5-HT2C. Int. J. Biol. Macromol. 160, 1090–1100. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mobbs J, Belousoff MJ, Harikumar KG, Piper SJ, Xu X, Furness SGB, Venugopal H, Christopoulos A, Danev R, Wootten D, et al. (2021). Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins: insight into mechanisms of G protein selectivity. Preprint at bioRxiv. 10.1371/journal.pbio.3001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, and Emeson RB (2010). Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol. Dis. 39, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, and Sanders-Bush E (1999). RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 274, 9472–9478. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, and Sanders-Bush E (2001). RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology 24, 478–491. [DOI] [PubMed] [Google Scholar]
- O’Neil RT, and Emeson RB (2012). Quantitative analysis of 5HT(2C) receptor RNA editing patterns in psychiatric disorders. Neurobiol. Dis. 45, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RHJ, DiBerto JF, English JG, Glaudin AM, Krumm BE, Slocum ST, Che T, Gavin AC, McCorvy JD, Roth BL, and Strachan RT (2020). TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat. Chem. Biol. 16, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, Bojarski AJ, and Gloriam DE (2018). GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, McCorvy JD, Harpsøe K, Lansu K, Yuan S, Popov P, Qu L, Pu M, Che T, Nikolajsen LF, et al. (2018). 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. Cell 172, 719–730.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221. [DOI] [PubMed] [Google Scholar]
- Ramírez-Anguita JM, Rodríguez-Espigares I, Guixà-González R, Bruno A, Torrens-Fontanals M, Varela-Rial A, and Selent J (2018). Membrane cholesterol effect on the 5-HT2A receptor: insights into the lipid-induced modulation of an antipsychotic drug target. Biotechnol. Appl. Biochem. 65, 29–37. [DOI] [PubMed] [Google Scholar]
- Rauser L, Savage JE, Meltzer HY, and Roth BL (2001). Inverse agonist actions of typical and atypical antipsychotic drugs at the human 5-hydroxytryptamine(2C) receptor. J. Pharmacol. Exp. Ther. 299, 83–89. [PubMed] [Google Scholar]
- Robertson MJ, van Zundert GCP, Borrelli K, and Skiniotis G (2020). GemSpot: a pipeline for robust modeling of ligands into cryo-EM maps. Structure 28, 707–716.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia R, Gomez-Blanco J, and Cuervo A (2020). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sard H, Kumaran G, Morency C, Roth BL, Toth BA, He P, and Shuster L (2005). SAR of psilocybin analogs: discovery of a selective 5-HT 2C agonist. Bioorg. Med. Chem. Lett. 15, 4555–4559. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, and Chen S (2012). Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, and Shaw J (1989). RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell 57, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, and Kortemme T (2008). Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J. Mol. Biol. 380, 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, and Seeburg PH (1991). RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19. [DOI] [PubMed] [Google Scholar]
- Tabbara RI, Li Z, Fletcher PJ, and Lê AD (2021). The serotonin 2C receptor agonist lorcaserin, alone and in combination with the opioid receptor antagonist naltrexone, attenuates binge-like ethanol drinking. Addict. Biol. 26, e13040. [DOI] [PubMed] [Google Scholar]
- Tareen A, and Kinney JB (2020). Logomaker: beautiful sequence logos in Python. Bioinformatics 36, 2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SV, George CX, Welch MJ, Liou L-Y, Hahm B, Lewicki H, de la Torre JC, Samuel CE, and Oldstone MB (2011). RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. USA 108, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warhaftig G, Sokolik CM, Khermesh K, Lichtenstein Y, Barak M, Bareli T, Levanon EY, and Yadid G (2021). RNA editing of the 5-HT2C receptor in the central nucleus of the amygdala is involved in resilience behavior. Transl. Psychiatry 11, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Wang N, Xu Z, Lu Y, Song J, Zhang A, Guo C, and He Y (2021). Cryo-EM structure of the human histamine H1 receptor/Gq complex. Nat. Commun. 12, 2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K-Y, Chien S-J, Tan BC-M, and Chen Y-W (2021). RNA editing of 5-HT2C R impairs insulin secretion of pancreatic beta cells via altered store-operated calcium entry. FASEB J. 35, e21929. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, and Yu X-F (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- Zhu H, Urban DJ, Blashka J, McPheeters MT, Kroeze WK, Mieczkowski P, Overholser JC, Jurjus GJ, Dieter L, Mahajan GJ, et al. (2012). Quantitative analysis of focused a-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS One 7, e43227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the analyzed and/or generated data is available in this manuscript/Supplementary information. All structural data has been deposited in the Electron Microscopy Databank (EMDB) (cryo-EM maps) and Protein Databank (PDB) (cryo-EM maps and models). They have been deposited under the PDB: 8DPF/EMD-27633 for INI-lorcaserin, PDB: 8DPG/EMD-27634 for INI-psilocin, 8DPH/EMD-27635 for VGV-lorcaserin, and PDB: 8DPI/EMD-27636 for VSV-lorcaserin.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


