Abstract
Intraepithelial T cells (IETs) are in close contact with intestinal epithelial cells and the underlying basement membrane, and they detect invasive pathogens. How intestinal epithelial cells and basement membrane influence IETs survival and function, at steady state or following infection, is unclear. The herpes virus entry mediator (HVEM), a member of the TNF receptor superfamily, is constitutively expressed by intestinal epithelial cells and is important for protection from pathogenic bacteria. Here, we showed that at steady-state LIGHT, an HVEM ligand, binding to epithelial HVEM promoted the survival of small intestine IETs. RNA-seq and addition of HVEM ligands to epithelial organoids indicated that HVEM increased epithelial synthesis of basement membrane proteins, including collagen IV, which bound to β1 integrins expressed by IETs. Therefore, we proposed IETs survival depended on β1 integrin binding to collagen IV, and showed β1 integrin-collagen IV interactions supported IETs survival in vitro. Moreover, the absence of β1 integrin expression by T lymphocytes decreased TCR αβ+ IETs in vivo. Intravital microscopy showed that the patrolling movement of IETs was reduced without epithelial HVEM. As likely consequences of decreased number and movement, protective responses to Salmonella enterica were reduced in mice lacking either epithelial HVEM, HVEM ligands, or β1 integrins. Therefore, tissue-resident IETs, at steady state and following infection, depended on HVEM expressed by epithelial cells for the synthesis of collagen IV by epithelial cells. Collagen IV engaged β1 integrins on IETs that were important for their maintenance and ultimately for the protective function of IETs in mucosal immunity.
One sentence summary:
Epithelial HVEM maintains intraepithelial T cell survival and function and supports host defense.
INTRODUCTION
Intestinal intraepithelial lymphocytes (IELs) are one of the largest population of lymphocytes in the body. They are found above the basement membrane within the intestinal epithelium, and they interact extensively with intestinal epithelial cells (IEC) by actively patrolling the basement membrane and by migration into the lateral intercellular space (1–4). IEL are believed to be crucial for maintenance of the intestinal barrier integrity, wound repair, and protection from pathogens (5). They include innate lymphoid cells (ILCs)(6) but are mostly T lymphocytes, referred to as intraepithelial T cells (IETs).
Based on development and antigen-recognition, IETs are divided into two groups termed “induced” IETs (also known as type a IETs) and “natural” IETs (also known as type b IETs) (5, 7, 8). Induced IETs express αβ TCRs and one of the TCR co-receptors that promotes TCR signaling, CD4 or CD8αβ. Induced IETs are tissue resident memory cells derived from antigen-primed conventional or mainstream CD4+ or CD8αβ+ T cells (9, 10). By contrast, natural or type b IETs express either αβ TCRs or γδ TCRs, but lack the expression of the TCR co-receptors. Natural IETs in mice are approximately 50% of the total population and they can be either CD4−, CD8α− double negative (DN), but often express CD8αα homodimers, which lack TCR co-receptor function (11). TCRαβ+ precursors of natural IETs undergo an alternative, self-antigen-based or agonist thymic selection and maturation process (12–14) and they have a distinct set of specificities compared to TCRαβ+ CD8αβ lymphocytes (15–17). The functions of natural IETs have not been fully characterized, however, TCRγδ+ IETs are implicated in host defense and repair of damaged epithelium by various mechanisms (18–22).
The factors important for the maintenance of IETs are not completely defined, but the aryl hydrocarbon receptor (AHR), T-bet and MyD88 are implicated for both TCRαβ+ and TCRγδ+ IET populations (23–25). G protein-coupled receptor 18 (GPR18) positively regulates TCRγδ+ IETs (3) while GPR55 is a negative regulator of these γδ cells (4). IL-15 contributes to maintenance of IETs, providing an example of an epithelial cell-derived influence (26–28). Epithelial expression of the thymus leukemia antigen, a nonclassical class I molecule, shapes the population of induced TCRαβ+ CD8αβ+ IETs via induction of co-expression of CD8αα homodimers, which interact with the thymus leukemia antigen (29) and promote the survival of these cells (30). The homing and maintenance of intestinal IETs in the epithelium depends on expression of β7 integrins, including α4β7, which binds MAdCAM-1 and the αEβ7 integrin, which interacts with E-cadherin expressed by IEC (31–33).
HVEM is a member of the tumor necrosis factor receptor superfamily (TNFRSF14) expressed by intestinal epithelial cells, lymphocytes and other cell types. It is important for the regulation of barrier or mucosal immunity in mice in the context of infection or inflammation in a variety of contexts (34–41). HVEM binds to several types of proteins. One of these is LIGHT (homologous to lymphotoxin, exhibits inducible expression and competes with HSV glycoprotein D for binding to herpesvirus entry mediator, a receptor expressed on T lymphocytes) or TNFSF14 (42, 43), which engages HVEM in a trimeric form (44). It also binds immunoglobulin superfamily (IgSF) proteins including B and T lymphocyte attenuator (BTLA or CD272), CD160 and SALM5(45). In binding these proteins, HVEM engages in bidirectional signaling, also serving as a ligand for IgSF receptor signaling (45).
Here, we showed that HVEM expressed by small intestine (SI) epithelial cells was involved in the homeostasis of natural IETs, the patrolling function of IETs at steady-state, and the response to pathogenic bacteria. Ligand binding to epithelial HVEM at steady state stimulated the synthesis of extracellular membrane proteins, a key one being collagen IV. Collagen IV affected cell survival by binding to β1 integrins expressed by the IETs. Analysis of gene deficient mice showed that this homeostatic circuit, including epithelial HVEM, HVEM ligands and β1 integrins, was involved in protective responses to a bacterial pathogen. Therefore, the data reveal how epithelial responses influencing the basement membrane, a structural element of tissue, are integrated to regulate tissue resident T cells in the intestine at steady-state and following infection.
RESULTS
HVEM is involved in maintaining IETs
Because of the engagement of HVEM to CD160 expressed on IETs is involved in the protection of mice from oral infection with Citrobacter rodentium(38), we determined if HVEM also is involved in the regulation of IETs in the intestine at steady state. We used flow cytometry to examine the total CD45+ cell number from mice with a germline deletion of the Hvem (Tnfrsf14) gene. Total CD45+ cells were significantly reduced throughout the SI, but not in the cecum and colon (Fig. 1A). We also observed a decrease in CD45+ cells throughout the SI epithelium in Hvem−/− mice with confocal microscopy (Figs. 1B, 1C). Most of the CD45+ cells in the epithelium were T lymphocytes, including TCRγδ+ cells and TCRαβ+ cells, with a smaller population TCR negative cells (Fig. 1E) that are mostly ILC1 (6). We gated on the IETs subsets as shown in Fig. S1A. Considering the different subsets, TCRαβ+CD8αα+ IETs exhibited the most pronounced reduction in cell frequency (Fig. 1D) and cell number (Fig. 1E). Natural γδ+CD8αα+ and induced TCRαβ+CD8αβ+ were also decreased (Fig. 1E). Furthermore, induced TCRαβ+CD4+ IETs, including cells that co-express CD8αα+ and which contain CD4+ T cells that acquire the function of cytotoxic T lymphocytes (CTLs) (46, 47), were also decreased (Fig. 1E). These data indicated that HVEM was involved in the accumulation of the three most numerically prevalent IET populations in the intestine at steady state.
Fig. 1. HVEM is important for maintaining IET.
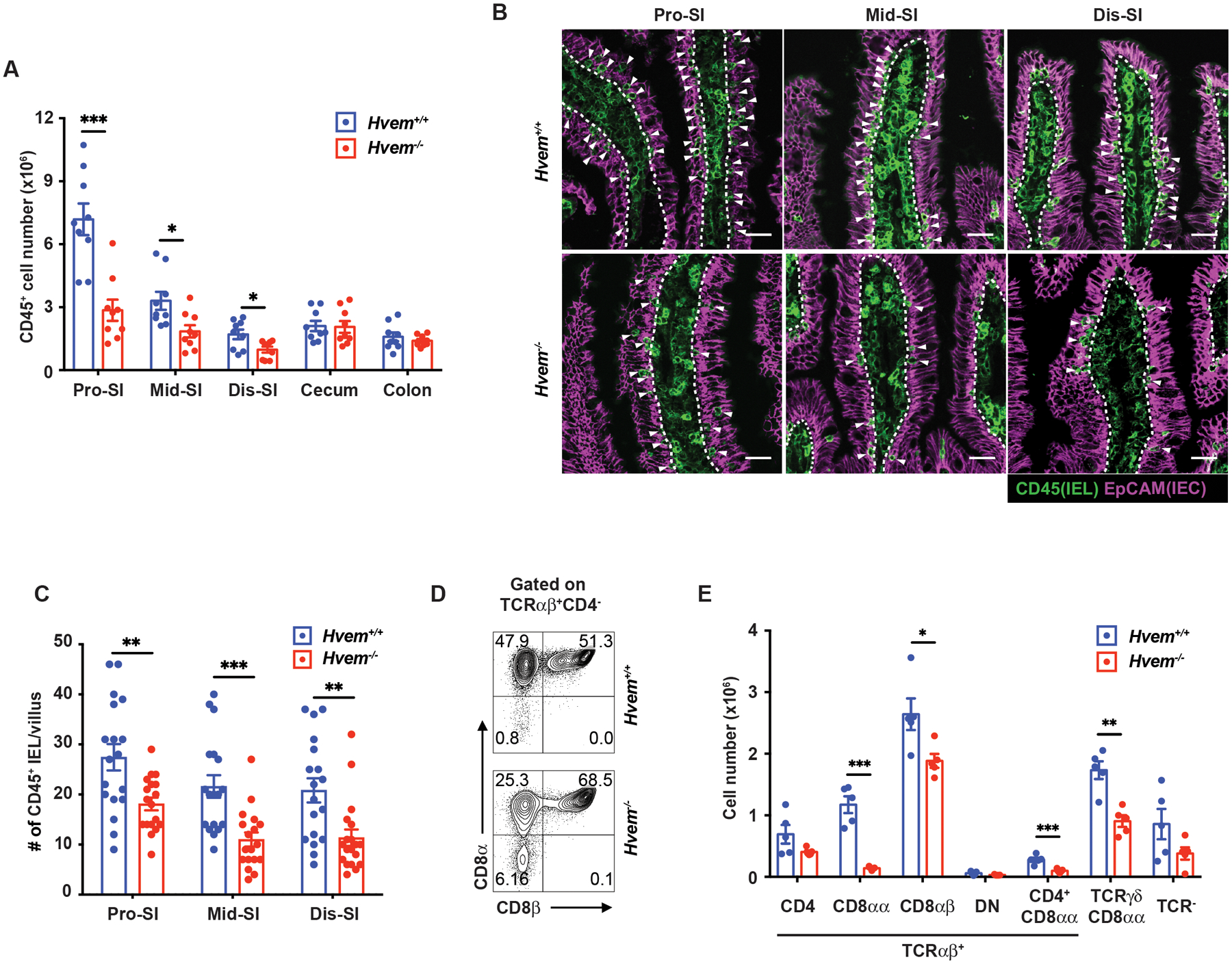
(A) Total IEL numbers by flow cytometry in proximal SI (Pro-SI), middle SI (Mid-SI), distal SI (Dis-SI), cecum and colon from Hvem+/+ (n=9) and Hvem−/− (n=9) mice. (B) Representative immunofluorescence staining of CD45+ cells from the SI in Hvem+/+ and Hvem−/− mice. White arrowheads indicate CD45+ intraepithelial cells (IEL) in the epithelium. Dashed white lines indicate the interface between the epithelium and lamina propria. Scale bars, 25μm. (C) Quantification of CD45+ IEL in villi from Hvem+/+ (n=4) and Hvem−/− (n=4) mice. (D) Representative plots of TCRαβ+CD8αα+ and TCRαβ+CD8αβ+ IET in TCRαβ+ IET from proximal SI in Hvem+/+ and Hvem−/− mice. (E) Absolute numbers of the indicated subsets in total IEL from proximal SI in Hvem+/+ (n=5) and Hvem−/− (n=5) mice. Statistical analysis was performed using an unpaired t-test (A, C, E). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data in A, C, and E show means ± SEM. In A and E, each symbol represents a measurement from a single mouse. In C, each symbol represents cell numbers of CD45+ IEL per a boundary approximating a villus. Data represent pooled results from at least two independent experiments with at least four mice per group in each experiment (A), compiled from four independent experiments (B, C) or representative results from one of at least two independent experiments with at least four mice in each experimental group (E). Groups of co-housed mice were analyzed.
Epithelial HVEM maintains IETs
Considering the importance of HVEM for TCRαβ+ CD8αβ+ memory and resident memory T cell formation (34, 36), we tested for a T cell intrinsic effect of HVEM expression. Because of the striking reduction in TCRαβ+CD8αα+ IETs, initially we focused on this population and analyzed the number of CD4−, CD8α− double-negative (DN) TCRαβ+ (DN TCRαβ+) thymocytes, which are the precursors of TCRαβ+ CD8αα+ IETs (5, 16, 48). We found no difference between wild type and Hvem−/− mice (Fig. S1B). We transferred these precursor cells, NK1.1−TCRγδ−TCRαβ+ thymocytes, into Rag1−/− mice. A 1:1 co-transfer of donor thymocytes from Hvem−/− mice (CD45.2) and congenic, wildtype CD45.1+ C57BL/6 mice was performed (Fig. S1C). There was no defect in the ability of the Hvem−/− donor thymocytes to give rise to TCRαβ+CD8αα+ IETs. In fact, the Hvem−/− donor cells were more highly represented at four weeks after transfer but the populations were equivalent at six weeks (Fig. S1C). We also analyzed mice with a T cell-specific deletion of the Hvem gene driven by CD4-Cre. These Cd4-cre × Hvemfl/fl (HvemΔCD4) mice showed no difference in the numbers of the most prevalent IET subsets in the proximal SI (Fig. S1D), although the TCRαβ+ CD4+ IETs, whether CD8αα+ or not, were reduced. Considering these results, an intrinsic or T cell role for HVEM expression was not the major influence in determining the size of the total IET population.
As IECs have direct contact with resident IETs, we wanted to know the role of HVEM at steady-state in IECs. HVEM was predominantly on the basolateral surface of epithelial cells (Fig. S2A), an optimal position for contact with immune cells, and it was expressed throughout the intestine epithelium, although at a higher amount in IEC from the proximal SI compared to the middle portion (Fig. S2B). Interestingly, HVEM expression by IEC was not increased during infection (38) nor was it diminished in IEC from germ-free mice (Fig. S2C). Also, HVEM was expressed in proximal small intestinal organoid cultures (Fig. S2D, S2E). Therefore, constitutive HVEM expression by IEC was unaffected by acute infection or the absence of either the microbiota or IEL.
To test if there is a role for epithelial cell-expressed HVEM, we crossed Villin-cre mice with Hvemfl/fl mice to generate mice with HVEM expression ablated only in epithelial cells (HvemΔIEC mice, Fig. S2F, S2G). Flow cytometric analyses indicated that HvemΔIEC mice had significantly decreased CD45+ cell numbers in the proximal and middle small intestine compared to Hvemfl/fl controls (Fig. 2A). The proximal SI has the greatest number of IETs, and when this segment was analyzed in more detail, TCRαβ+CD8αα+ were most decreased, similar to Hvem−/− mice, but the TCRγδ+CD8αα+ IETs and TCRαβ+CD4+ IETs also were reduced (Fig. 2B, 2C). These trends were partially replicated in the middle SI but less so in the distal SI (Fig. S2H). The effect on TCRαβ+CD4+CD8α+ CTL suggests there could have been HVEM contributions to this population that are both T cell-intrinsic and epithelial cell dependent. TCRαβ+ CD8αβ+ IETs were not reduced in HvemΔIEC mice although they were reduced in Hvem−/− mice. The complexities regarding induced IET subsets may reflect their history, which unlike natural IETs, require priming in lymph nodes and many of the cell types they encounter during priming (5, 10) and while circulating express HVEM and HVEM ligands. Regardless, the effects were specific to the SI, as no IET populations were diminished in the colon of HvemΔIEC mice (Fig. S2H).
Fig. 2. Epithelial HVEM maintains IETs.
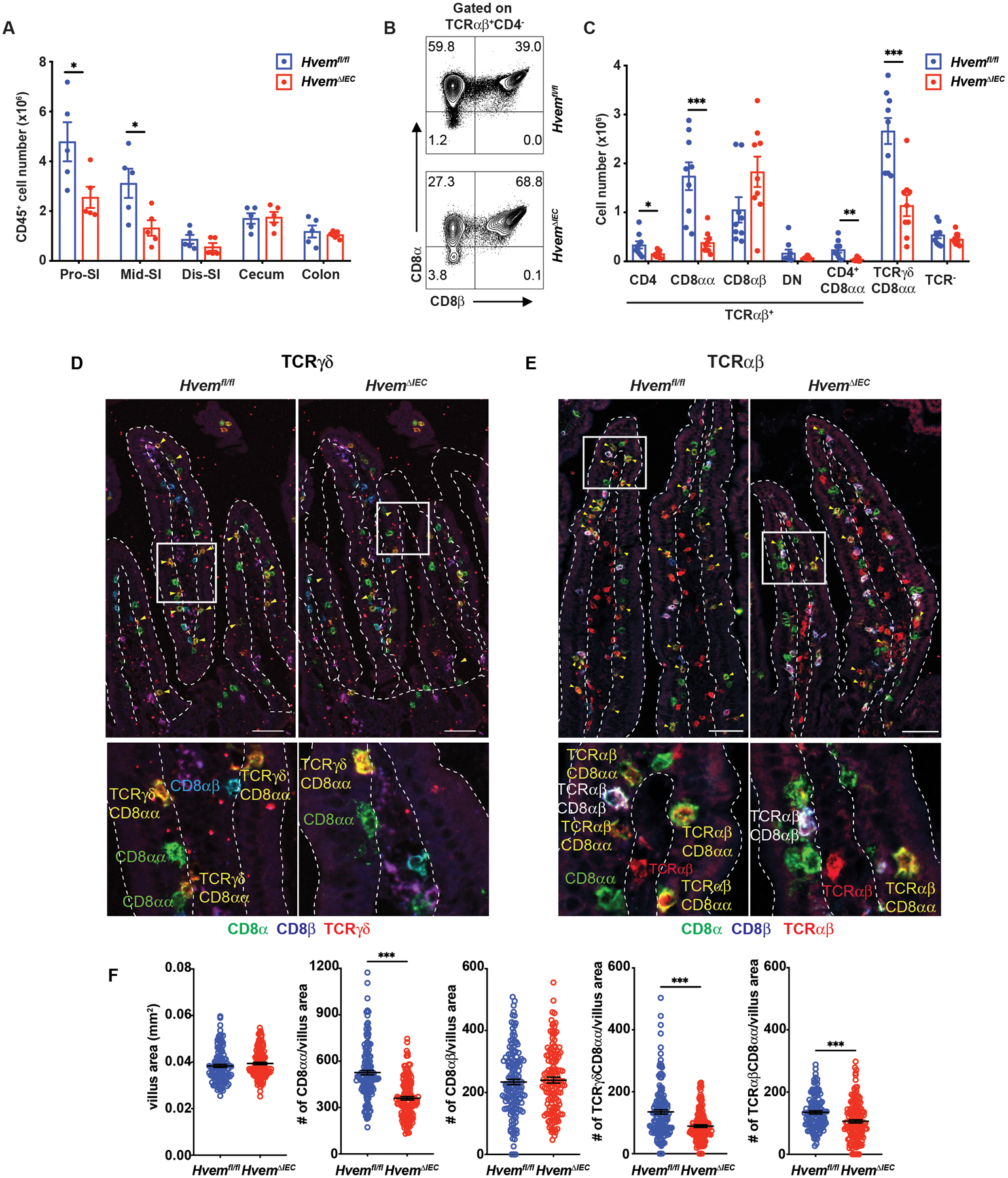
(A) Total IEL numbers in portions of the intestine from Hvemfl/fl (n=5) and HvemΔIEC (n=5) mice. (B) Representative plots of TCRαβ+CD8αα+ and TCRαβ+CD8αβ+ in TCRαβ IET from proximal SI in Hvemfl/fl and HvemΔIEC mice. (C) Absolute numbers of indicated IET subsets in total IEL from proximal SI in Hvemfl/fl (n=9) and HvemΔIEC (n=9) mice. (D-E) Representative immunofluorescence staining of TCRγδ IET (D) and TCRαβ IET (E) from proximal SI in Hvemfl/fl and HvemΔIEC mice. Composite images in which the three channels were merged. Composite images depict expression of CD8α (green), CD8β (blue), and TCRδ (red, D) or TCRβ (red, E). Yellow arrowheads highlight TCRγδ+CD8αα+ (D) and TCRαβ+CD8αβ+ IET (E). Single-channel images are in Figure S3. Dashed white lines indicate the boundaries of the epithelium, approximately one villus in each case, used for the quantitation. Scale bars, 50μm. Two independent experiments were carried out yielding similar results. (F) Quantification of TCRγδ+CD8αα+ and TCRαβ+CD8αβ+ per mm2 from proximal SI in Hvemfl/fl (n=6) and HvemΔIEC (n=7) mice. Statistical analysis was performed using unpaired t-test (A, C, F). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data in A, C, and F show means ± SEM. In A and C, each symbol represents a measurement from a single mouse. In F, each symbol represents a calculation to give the number per mm2. Data are representative results from one of at least two independent experiments with at least four mice in each experimental group (A) or pooled results from at least two independent experiments with at least four mice per group in each experiment (C, F). Groups of co-housed littermates were analyzed.
To confirm that the homeostasis of IET populations in the small intestine was affected by epithelial HVEM expression, we performed in situ immunofluorescence (IF) multi-color staining in frozen sections from the proximal small intestine of HvemΔIEC and control mice. To identify TCRγδ+CD8αα+ IETs, we used fluorochrome-coupled antibodies to TCRδ, CD8α, CD8β and to detect TCRγδ+CD8αα+ IETs (Fig. S3A). Similarly, to identify TCRαβ+CD8αα+ IETs, we used fluorochrome-coupled antibodies to TCRβ, CD8α, CD8β (Fig. S3B). Although this method did not distinguish TCRαβ+CD8αα+ IETs from TCRαβ+CD4+CD8α+ CTL, the CD4+ CTL were less frequent (Figs. 1E, 2C, S2H). After imaging by confocal microscopy, the IET subsets were quantified (Figs. 2D–2F). The total villus area in the proximal SI was comparable between Hvemfl/fl and HvemΔIEC mice (Fig. 2F). Similar to the flow cytometry results, HvemΔIEC mice had a decrease of total CD8αα+ IETs, including individual populations of TCRαβ+CD8αα+ plus TCRαβ+CD4+CD8α+ CTL and TCRγδ+CD8αα+ IETs, but no decrease in CD8β+ cells in the proximal small intestine (Figs. 2D–2F). In summary, loss of epithelial HVEM expression led to decreases in natural IET subsets.
Epithelial HVEM is required for IETs survival
Among the possible mechanism(s) for the decreased number of IETs in HvemΔIEC mice, we tested for components involved in homing to the intestine or the maintenance of the cells in the epithelial site, for IETs proliferation and for IETs survival. Flow cytometric analysis indicated that the expression of the integrin β7 chain and the integrin αE were not affected in IETs from HvemΔIEC mice (figs. S4A, S4B). The expression of mRNA for the Cdh1 gene encoding E-cadherin in HvemΔIEC IEC was normal as well (fig. S4C).
To address if IETs proliferation was responsible, Ki-67 staining and EdU incorporation assays were performed. Although the trend suggested increased Ki-67, the IET from HvemΔIEC mice were not significantly different from the controls (Fig. S4D). Similarly, after six days of continuous EdU labeling, the percent of labeled IETs was not different in HvemΔIEC mice (Fig. 3A). When the EdU label was chased or removed for 14 days, this led to a decreased percentage in EdU+ in the two main natural IET subsets compared to control mice (Fig. 3A). These data suggested there was a survival defect in the TCRαβ+CD8αα+ and TCRγδ+CD8αα+ IETs in HvemΔIEC mice. Compared with control mice, cells from HvemΔIEC mice exhibited higher Annexin V+ labeling in both the TCRαβ+CD8αα+ and TCRγδ+CD8αα+ IET subsets when analyzed ex vivo (Fig. 3B, Fig S4E). They also expressed greater amounts of the pro-apoptotic Bcl2 family proteins Bim and Bax, while even the TCRαβ+ CD8αβ+ IETs expressed more Bax (Figs. 3C, 3D), suggesting there could be a small effect on these cells. To confirm that the absence of epithelial HVEM affected IETs survival, we analyzed mice with a TCRαβ cell-specific overexpression of the pro-survival Bcl-2 family gene Bclxl (Bcl2l1) driven by the Lck proximal promoter (Lckpr-Bcl-xLTg) crossed to either Hvemfl/fl or HvemΔIEC mice. HvemΔIEC mice expressing the Bclxl transgene had a normal number of TCRαβ+CD8αα+ IETs. The increase in TCRγδ+CD8αα+ IETs was not significant (Fig. 3E), but the proximal Lck promoter does not work efficiently in TCRγδ+ cells (49). To gain further insight into the effects of epithelial HVEM on TCRαβ+CD8αα+ IETs, we performed RNA-seq analysis on the most affected population, TCRαβ+CD8αα+ IETs sorted from SI of HvemΔIEC and co-housed littermate Hvemfl/fl mice. The top 50 differentially expressed genes included 19 genes that have been reported to affect cell cycle, cell survival or cell growth (Figs. 3F, 3G). Among those genes reduced in IETs from HvemΔIEC mice were autophagy and beclin 1 regulator 1 (Ambr1), which promotes cell survival under stress conditions by inducing autophagy (50), cullin associated and neddylation dissociated gene 1 (Cand1), with reduced expression associated with apoptosis in cancer cells (51) and also, there was decreased expression of some genes in the NF-κB pathway. Overall, the data were consistent with the hypothesis that epithelial HVEM affected the survival of the natural IETs, including TCRαβ+CD8αα+ and TCRγδ+CD8αα+ IETs.
Fig. 3. Epithelial HVEM affects CD8αα+ IET survival.
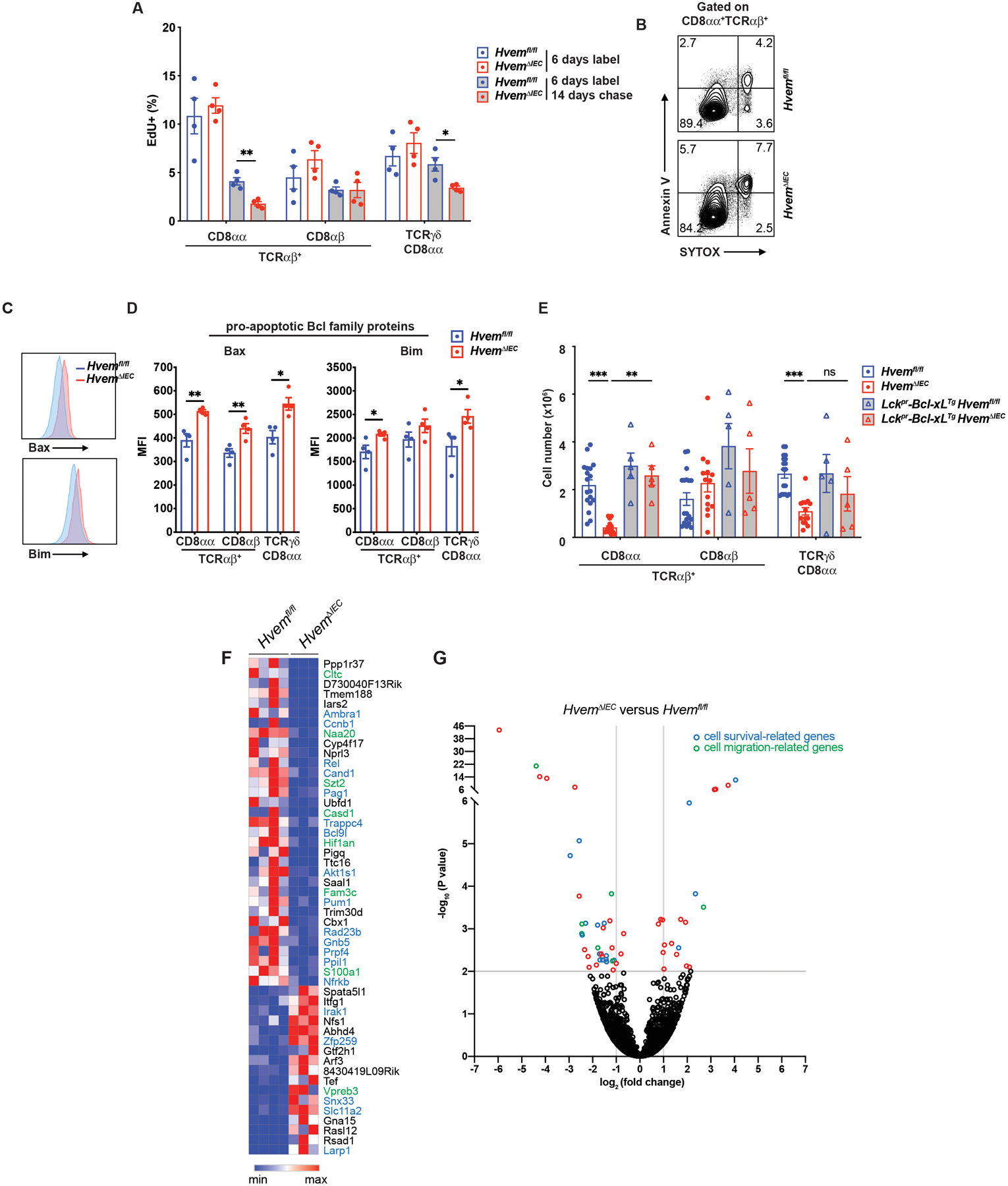
(A) Frequencies of EdU + cells from proximal SI IET of Hvemfl/fl (n=4) and HvemΔIEC (n=4) mice. Mice were administered EdU once per day for 6 days. Cells were isolated from proximal SI and EdU+ cells were measured at either day 6 or day 20. (B) Representative flow cytometry images detecting apoptotic cells in TCRαβ+CD8αα+ IET from proximal SI in Hvemfl/fl and HvemΔIEC mice.
(C, D) Representative flow cytometry showing expression of pro-apoptotic Bcl family proteins (Bax and Bim) on TCRαβ+CD8αα+ IET from proximal SI of Hvemfl/fl (n=4) and HvemΔIEC (n=4) mice (C) and compiled MFI from multiple individual mice (D). (E) Absolute cell number of TCRαβ+CD8αα+, TCRαβ+CD8αβ+, and TCRγδ+CD8αα+ IET subsets from Hvemfl/fl (n=18), HvemΔIEC (n=14), Lckpr-Bcl-xLTgHvemfl/fl (n=5) and Lckpr-Bcl-xLTgHvemΔIEC (n=5) mice. Statistical analysis was performed using an unpaired t-test (A, D) or 2 way ANOVA with Bonferroni’s multiple comparison test (E). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data in A and C show means ± SEM. In A, D and E, each symbol represents a measurement from a single mouse. Data are representative results of one of at least two independent experiments with at least four mice in each experimental group. (F) Sorted TCRαβ+CD8αα+ IET from SI of Hvemfl/fl and HvemΔIEC mice were analyzed by RNA-seq. The top 50 most differentially expressed genes with respect to P-value. (G) Volcano plots showing mean log2-transformed fold change (x axis) and significance (−log10 (adjusted P value)) of differentially expressed genes between the TCRαβ+CD8αα+ IET from the SI of Hvemfl/fl and HvemΔIEC mice. In F and G, genes in blue font or blue symbol are associated with cell survival or proliferation and those designated with green are associated with cell migration. Groups of co-housed littermates (A-D) or cohoused mice (E) were analyzed.
HVEM signaling increases basement membrane synthesis
To interpret how epithelial HVEM affects CD8αα+ IETs, especially IETs survival, we performed RNA-seq analysis on IEC sorted from proximal SI of HvemΔIEC and co-housed littermate Hvemfl/fl mice. By gene set enrichment analysis, we observed a significant down-regulation of a number of genes associated with the extracellular matrix in HvemΔIEC mice (fig. S5A). Down-regulated genes in IEC from HvemΔIEC mice revealed significant enrichment in genes associated with the proteinaceous extracellular matrix according to gene set enrichment analysis (Fig. 4A). In agreement with this, analysis of differentially expressed genes also identified extracellular matrix, basement membrane and collagen, all categories decreased in the absence of epithelial HVEM expression (figs. S5B, S5C). These results suggested a possible role for epithelial HVEM in supporting IETs survival and migration in the epithelium through stimulation of production of extracellular matrix proteins.
Fig. 4. HVEM signaling contributes to induction of basement membrane synthesis.
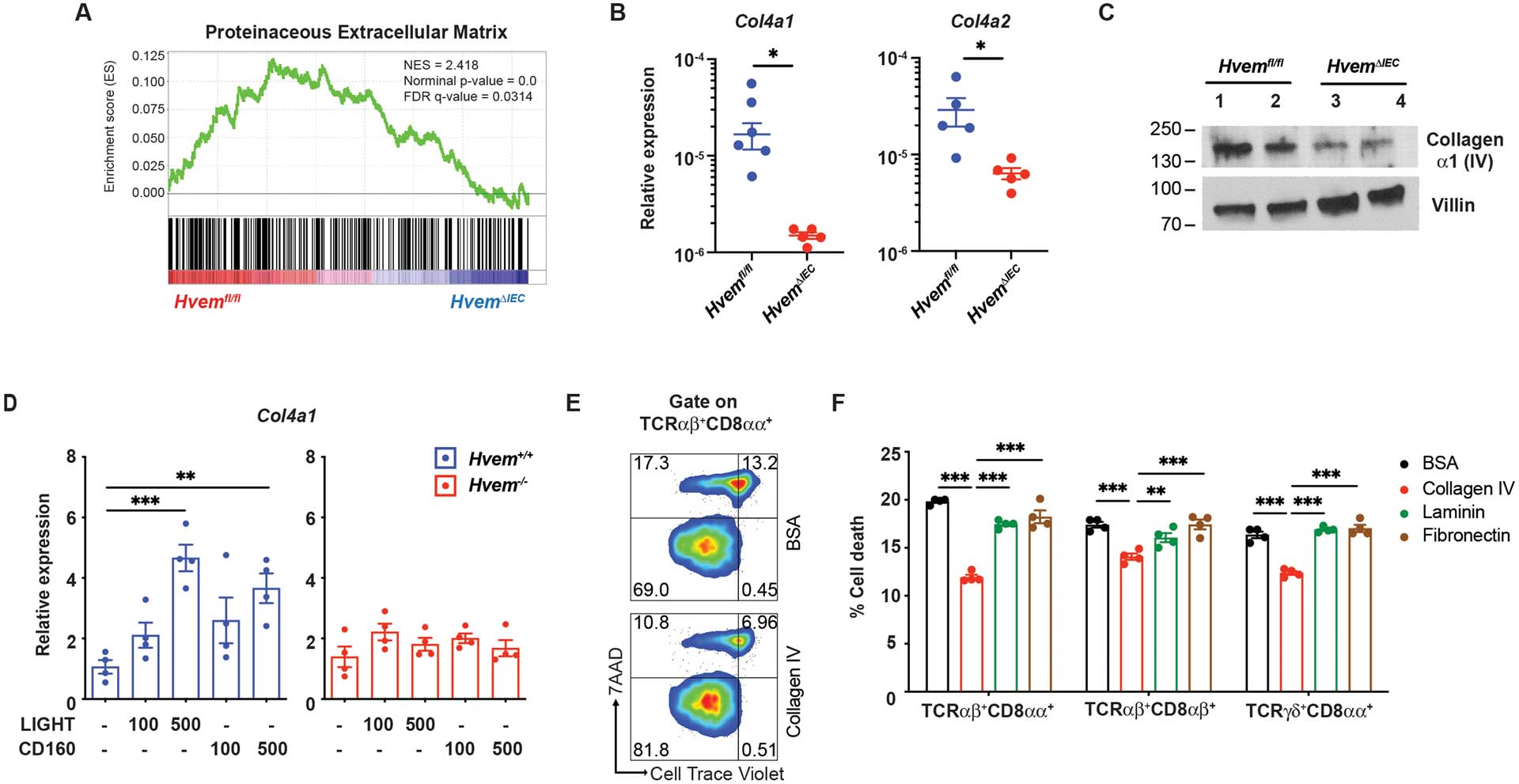
(A) Gene set enrichment analysis (GSEA) of transcripts from isolated IEC (CD31−CD45−EpCAM+) showing downregulation of GO Proteinaceous Extracellular Matrix genes in HvemΔIEC mice. (B) Gene expression of Col4a1 and Col4a2 mRNA in IEC from Hvemfl/fl (n=5) and HvemΔIEC mice (n=5) by q-PCR. Data are normalized to the Actb housekeeping gene. (C) Expression of collagen α1 (IV) in sorted IEC from Hvemfl/fl and HvemΔIEC mice by Western blot. The expression level of villin is used as a control for the amount of protein lysates loaded. (D) Gene expression of Col4a1 mRNA determined by q-PCR in cells from intestinal organoid cultures from proximal SI of Hvemfl/fl (n=5) and HvemΔIEC mice (n=5). Isolated crypts from Hvem+/+ and Hvem−/− mice were cultured with growth factors in the presence or absence of HVEM ligands, indicated concentrations in ng/ml, for 7 days. Data are normalized to Rpl32 as the housekeeping gene. (E-F) Isolated SI IET subsets (CD4− from wild type mice were labeled with CellTrace™ Violet (CTV) and cultured on plates coated with the indicated mouse basement proteins or BSA control (n=4 per group). After 3 days, cells were stained with antibodies and 7AAD for monitoring cell death by flow cytometry. Representative plots (E) and calculated of % cell death (F) in IET subsets. Statistical analysis was performed using an unpaired t-test (B) or 2 way ANOVA with Bonferroni’s multiple comparison test (D, F). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data in B, D, and F show means ± SEM. Each symbol represents a measurement from a single mouse (B), single organoid culture (D) or an individual well (E, F). Data are representative results from one of at least two independent experiments. Groups of co-housed littermates were analyzed.
Collagen IV is a major component of basement membranes and there is evidence that IETs bind to collagen (52, 53). Collagen IV composed of the α1 and α2 chains is present in nearly all basement membranes (54, 55). To further test if epithelial HVEM regulated the production of collagen IV in vivo, we carried out quantitative real-time PCR (qPCR) and showed reduced expression of Col4a1 and Col4a2, mRNA, encoding the collagen α1 (IV) and α2 (IV) chains, respectively, by IEC from HvemΔIEC mice (Fig. 4B). HvemΔIEC IEC also exhibited reduced collagen α1 (IV) protein expression by Western blot (Fig. 4C).
To address if HVEM signaling induced Col4a1 mRNA expression in IEC, intestinal organoid cultures from HvemΔIEC and control mice were stimulated in vitro with HVEM ligands, LIGHT or CD160. Col4a1 mRNA expression was increased by either HVEM binding partner in the wild type organoid culture, but not in HvemΔIEC organoids (Fig. 4D). Together, these data indicated that epithelial HVEM signaled to stimulate increased synthesis of mRNA for collagen IV.
To investigate the role of collagen IV and other basement membrane proteins, such as laminin and fibronectin, in promoting IETs survival, we carried out in vitro cultures. Sorted IET populations were expanded briefly in IL-15 and cultured on plates coated with basement membrane proteins, and cell death was monitored by flow cytometry. We gated on the two natural IET populations, TCRαβ+CD8αα+ and TCRγδ+CD8αα+ IETs, as well as induced TCRαβ+CD8αβ+ IETs. When cultured on collagen-coated wells, each of these three cell subsets had decreased cell death compared to IETs cultured on either laminin, fibronectin or BSA (Figs. 4E, 4F).
Collagen-binding integrins increase IET survival
Integrins containing the β1 chain are known to bind collagen IV, especially α1β1 integrin, with α2β1 integrin binding to a lesser extent (52). The three prevalent IET populations, TCRαβ+CD8αα+, TCRγδ+CD8αα+ and TCRαβ+CD8αβ+ IETs expressed relatively high amounts of integrin α1 and β1, in agreement with a previous report (56), although expression was lower by TCRαβ+CD4+ IETs (Fig. 5A). To investigate the role of integrins in IETs survival in vitro, we blocked α1β1 integrin binding to collagen IV with a β1-specific antibody. This led to increased death of all three of the most prevalent IET populations (Fig. 5B). Cell death was also increased using the peptide disintegrin (inhibitor), Obtustatin (Fig. 5B). These data suggested that collagen IV might have influenced IETs survival in part through binding to α1β1 and perhaps other collagen-binding β1 integrins.
Fig. 5. Collagen-binding integrins influence IET survival and host defense.
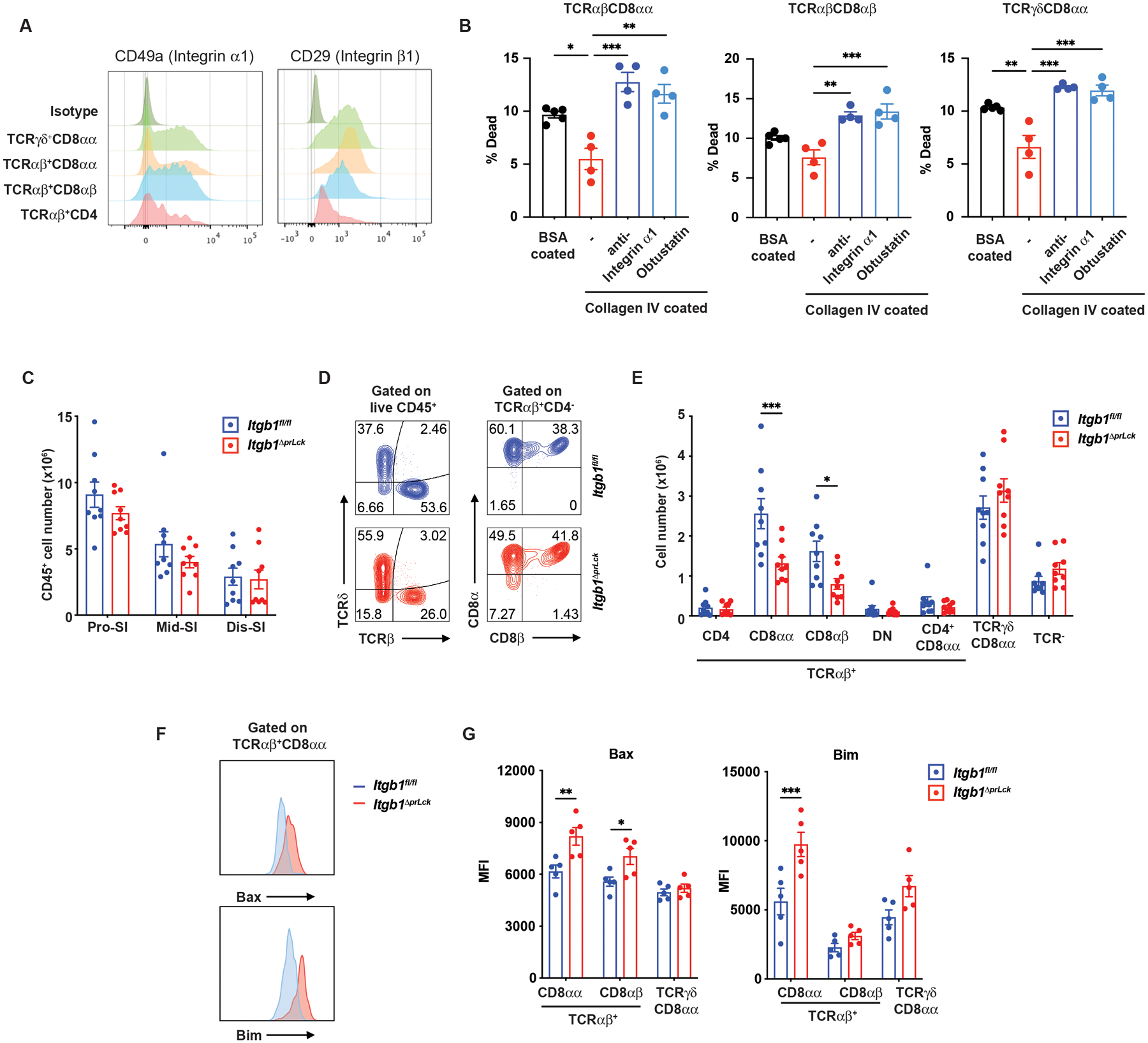
(A) Expression of integrin α1 and β1 subunits by IET populations from proximal SI of wild type mice by flow cytometry. (B) Isolated IET subsets from SI of wild type mice labeled with CellTrace™ Violet (CTV) and cultured on plates coated with BSA control (n=5) or collagen IV in the presence or absence of anti-integrin α1 mAb or inhibitor Obtusatin (n=4 per group). After 3 days, cells were monitored for viability by flow cytometry as described in Materials and Methods. (C) Total IEL numbers in segments of SI from Itgb1fl/fl (n=9) and Itgb1ΔprLck (n=9) mice. (D) Representative plots of TCRαβ+ and TCRγδ+ among CD45+ IEL (left) and TCRαβ+CD8αα+ and TCRαβ+CD8αβ+ among TCRαβ IET (right) from proximal SI in Itgb1fl/fl and Itgb1ΔprLck mice. (E) Absolute numbers of indicated subsets in total IEL from proximal SI in Itgb1fl/fl (n=9) and Itgb1ΔprLck (n=9) mice. (F, G) Representative flow cytometry shows expression of pro-apoptotic Bcl family proteins (Bax and Bim) on TCRαβ+CD8αα+ IET from proximal SI of Itgb1fl/fl (n=5) and Itgb1ΔprLck (n=5) mice (F). The MFI on CD8α+ IET from multiple individual mice was measured by flow cytometry (G). Statistical analysis was performed using 2 way ANOVA with Bonferroni’s multiple comparison test (B) or unpaired t-test (C, E, G). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data show means ± SEM. Each symbol represents a measurement from an individual well (B) or a single mouse (C, E, G). Data represent representative results from one of at least two independent experiments (B, G) or pooled results from at least two independent experiments (C, E). Groups of co-housed littermates were analyzed.
To determine if there was a role for T cell-expressed integrins in CD8αα+ IETs survival in vivo, we generated Itgb1ΔprLck mice by crossing Lckpr-Cre mice with Integrin b1fl/fl (Itgb fl/fl) mice. Flow cytometric analyses indicated that Itgb1ΔprLck mice had no difference in total CD45+ cell numbers throughout the SI compared to Itgb1fl/fl controls (Fig. 5C). However, the TCRαβ+CD8αα+ and TCRαβ+CD8αβ+ IETs in the proximal SI of Itgb1ΔprLck mice were decreased (Figs. 5D–5E), in agreement with the greater activity of the Lckpr-Cre in TCRαβ cells (49). TCRαβ+CD4+ IETs were not affected, consistent with their reduced expression of β1 integrins. In line with the in vitro survival results, TCRαβ+CD8αα+ IETs expressed greater amounts of the pro-apoptotic Bcl2 family proteins Bim and Bax, while TCRαβ+ CD8αβ+ expressed higher Bax in Itgb1ΔprLck mice (Figs. 5F–5G). Therefore, the selective effect in the Itgb1ΔprLck mice was consistent with the hypothesis that collagen IV promoted IETs survival, in part through α1β1 integrin binding.
LIGHT expression is required for IETs
We tested for the effect of the loss of HVEM binding partners expressed by IETs on their survival. CD160, important for anti-bacterial responses in the intestine (38, 57, 58), was expressed most prominently in the distal small intestine (Fig. S6A). In contrast, BTLA protein could not be detected on any IET subset besides intraepithelial ILC (Fig. S6B). We could not examine protein expression of LIGHT in IETs due to the absence of a reliable antibody for detecting mouse LIGHT. TCRαβ+CD8αα+ IETs expressed RNA encoding LIGHT and CD160, but not BTLA (Fig. S6C). Whole body deletion of Cd160 had no effect on IETs number (Fig. S6D–S6E), in agreement with a previous report (57) and the expression of the pro-apoptotic Bcl2 family proteins Bim and Bax was not increased (Fig. S6F). In contrast, by flow cytometric analysis, whole body deletion of the gene encoding LIGHT (Tnfsf14) led to significantly decreased total IETs in the proximal and middle SI (Fig. 6A). In the proximal SI, the mice had significant reductions in the two natural IET populations. The percentage of induced TCRαβ+CD8αβ+ IETs actually was increased (Fig. 6B), because these cells were less affected by LIGHT deficiency, but the number of TCRαβ+CD8αβ+ IETs also was reduced (Fig. 6C). These findings were corroborated by fluorescent microscopy-based in situ analysis (Fig. 6D–6F). The three predominant populations of IETs expressed higher Bax in mice lacking LIGHT protein (LIGHT−/−) (Fig. 6G), consistent with the possibility that decreased survival contributed to the decreased IET subpopulations. Therefore, although HVEM has multiple binding partners, LIGHT is important for survival of IETs at steady-state.
Fig 6. LIGHT is required for IET homeostasis.
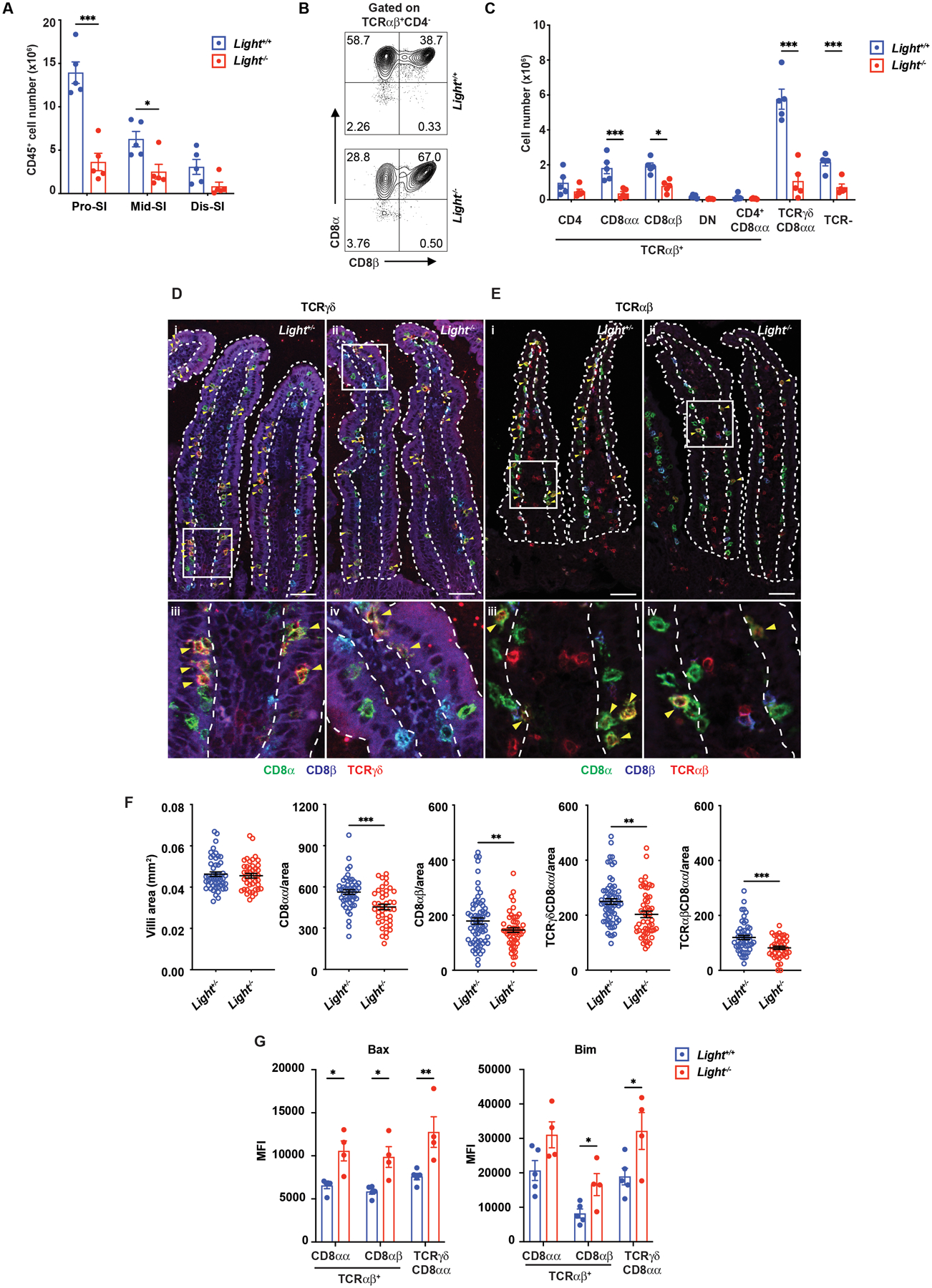
(A) Total IEL numbers in portions of the intestine from Light+/+ (n=5) and Light−/− (n=5) mice. (B) Representative plots of TCRαβ+CD8αα+ and TCRαβ+CD8αβ+ in TCRαβ IET from proximal SI in Light+/+ and Light−/− mice. (C) Absolute numbers of indicated IET subsets in total IEL from proximal SI in Light+/+ (n=5) and Light−/− (n=5) mice. (D-E) Representative immunofluorescence staining of TCRγδ IET (D) and TCRαβ IET (E) from proximal SI in Light+/+ and Light−/− mice. Composite images in which the three channels were merged. Composite images depict expression of CD8α (green), CD8β (blue), and TCRδ (red, D) or TCRβ (red, E). Yellow arrowheads highlight TCRγδ+CD8αα+ (D) and TCRαβ+CD8αβ+ IET (E). Dashed white lines indicate the boundaries of the epithelium, approximately one villus in each case, used for the quantitation. Scale bars, 50μm. Two independent experiments were carried out yielding similar results. (F) Quantification of TCRγδ+CD8αα+ and TCRαβ+CD8αβ+ per mm2 from proximal SI in Light+/+ (n=5) and Light−/− (n=4) mice. (G) The MFI of Bax and Bim expression on CD8α+ IET from Light−/− (n=5) and Light+/+ (n=4) mice. Statistical analysis was performed using unpaired t-test (A, C, F, G). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data show means ± SEM. In A, C and G, each symbol represents a measurement from a single mouse. In F, each symbol represents a calculation to give the number per mm2. Data are representative results from one of at least two independent experiments with at least four mice in each experimental group (A, C, G) or pooled results from at least two independent experiments with at least four mice per group in each experiment (F). (A-G) Groups of co-housed littermates were analyzed.
HVEM affects IET patrolling
The top 50 differentially expressed genes in IETs from wild type and HvemΔIEC mice also contained eight genes that influence cell migration (Figs. 3F, 3G). Included in these genes with decreased expression in HvemΔIEC mice were clathrin heavy chain (Cltc), which among its different functions influences lymphocyte migration by inducing actin accumulation at the cellular leading edge (59), N-α-acetyltransferase 20 (Naa20), which acetylates some terminal methionine amino acids and affects the actomyosin fibers needed for cell migration (60), and CAS1 domain containing protein 1 (Casd1), which catalyzes the 9-O-acetylation of sialic acids and which is required for cell migration in the nervous system (61).
To further examine if epithelial HVEM deficiency affected IETs migration, we used intravital imaging to track lymphocytes labeled with a CD8α-specific mAb in the SI epithelium of live mice (1, 3). We used confocal reflection microscopy to provide tissue context for the CD8α-labeled cells. With this modality, we were able to observe the epithelial layer, underlying blood capillaries, and lamina propria extending below (Fig. S7). In some cases, we used an EpCAM-specific antibody to label the epithelial cells. Our analysis was constrained to the cells in the epithelial layer. The average coverage of the villus area by migrating CD8α labelled cells was greatly reduced in HvemΔIEC mice (Figs. 7A–7B). This decrease in coverage reflected not only the decreased cell number described above, but also decreased average movement. The mean speed, track length, and track displacement length of CD8α mAb-labeled IETs were all reduced in HvemΔIEC mice (Figs. 7C–7F, movies S1 and S2). Most IETs, including CD8αα+, CD8αβ+ and CD4+CD8α+ IETs, express CD8α (10). Therefore, the intravital microscopy data suggested that epithelial HVEM expression was required for the patrolling behavior of most IETs, including natural and induced subsets.
Fig. 7. Epithelial HVEM influences patrolling by CD8α+ IET.
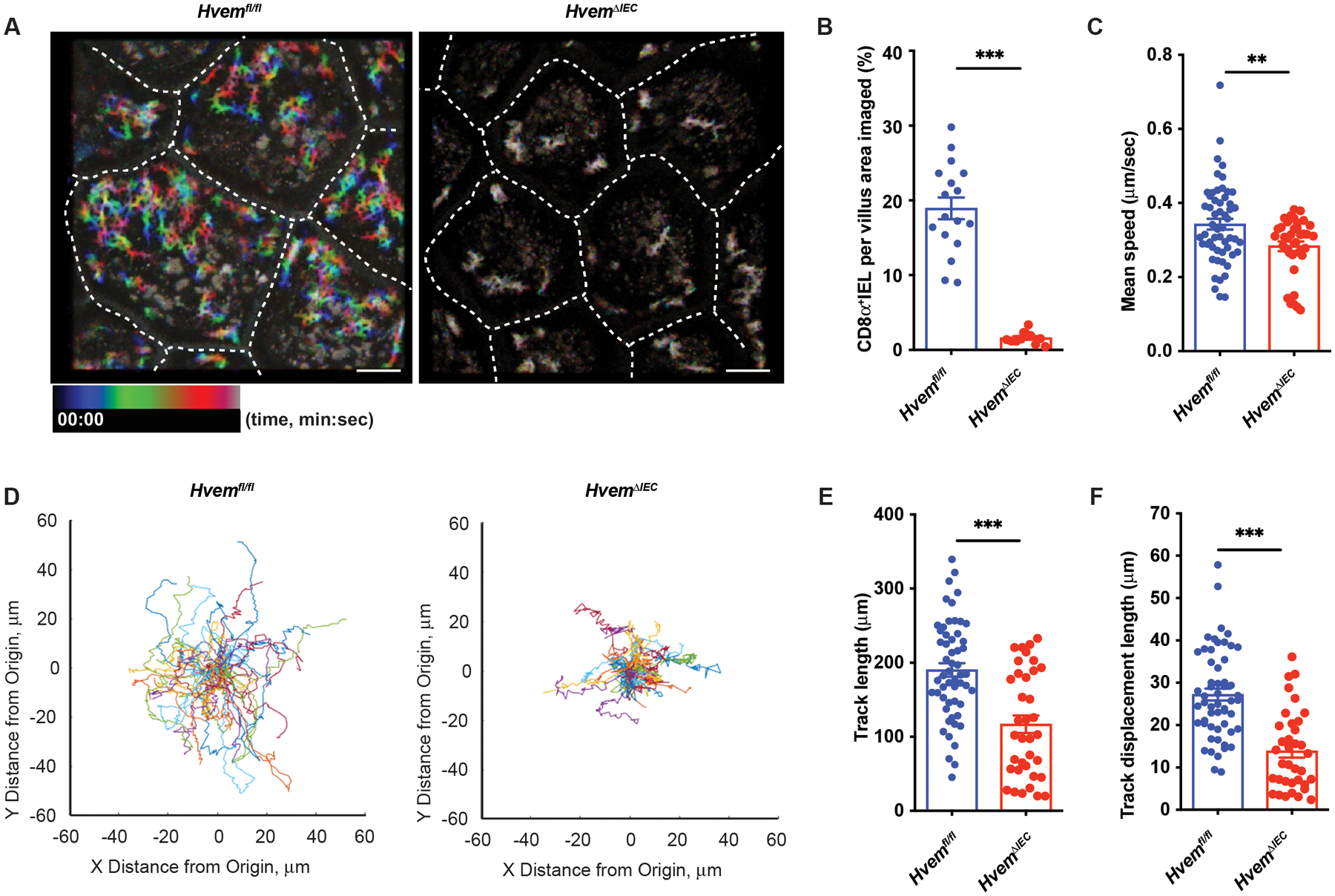
Mice were injected with anti-CD8α before imaging of the epithelium by taking Z-stacks encompassing the epithelium and the upper layers of the lamina propria at a 2.8 μm step size every 10 sec. Each XY plane spans 365 × 365 μm2. Fiji was used to calculate the coverage area and generate representative images. Movement metrics (mean speed, track length, track displacement length, and paths) for each CD8α+ lymphocyte were calculated with Imaris. (A) Rainbow plot time projection reveals the area covered by CD8α+ IET during 10 min, starting from time 0 (00:00) to 10 mins (10:00). Rainbow color shows the tracks of moving CD8α+ IET. Blue denotes starting position at the beginning of the recording it goes to green and then red, denotes ending position, as the time elapses. If the cell does not move, the superposition of colors blends to give a color closer to white. Scale bars, 50μm. (B-C) Quantification of the fraction of villus area covered by CD8α+ cells (B), mean speed (C) in a 10 min recording (Hvemfl/fl, n=3; HvemΔIEC, n=3). (D) Spider plots of tracks show the migratory path of each CD8+ IEL in the 10 min recording. Each cell path was translated to begin at (0,0) and randomly colored. (E-F) Quantification of track length (E), and the displacement between beginning and ending positions (F) in a 10 min recording (Hvemfl/fl, n=3; HvemΔIEC, n=3). Statistical analysis was performed using unpaired t-test (B-C, E-F). Statistical significance is indicated by *, p < 0.05; ***, p < 0.001. In B-C and E-F, bars show the mean and each symbol represents a measurement from a single cell. Dashed white lines (A) indicate the boundaries of each quantified villus. Data represent combined results of two independent experiments with at least three mice in each experimental group. Groups of co-housed littermates were analyzed.
Epithelial HVEM is involved in host defense against bacteria
We have thus far established a role for epithelial HVEM in regulating IETs homeostasis and migration at steady state, and therefore we interrogated if there were correlated effects on host defense. TCRγδ+ IETs play a critical role in preventing invasion of Salmonella enterica serovar typhimurium (S. typhimurium), which causes gastroenteritis in humans and other mammals, including mice(62). IETs migration among epithelial cells in contact with S. typhimurium is essential for T cell surveillance and immediate host defense (2, 63). To determine if epithelial HVEM expression contributed to host protection, mice were orally infected with S. typhimurium and analyzed. HvemΔIEC mice had a decreased survival rate (Fig. 8A), rapid weight loss (Fig. 8B), and increased bacterial burden in the spleen, liver, small intestine, and colon (Fig. 8C) after oral S. typhimurium administration. After S. typhimurium infection, Cd160−/− mice had rapid weight loss (Fig. S8A) and increased bacterial colonies in the spleen and liver (Fig. S8B). LIGHT-deficient Tnfsf14−/− mice also had rapid weight loss (Fig. S8C) and a trend toward increased colonies in the spleen and liver that did not reach statistical significance (Fig. S8D). Btla−/− mice, however, exhibited no differences in body weight, and bacterial burden in the spleen and liver (Fig. S8E–S8F). Therefore, the data suggested that epithelial HVEM expression, probably with interactions with both CD160 and LIGHT, contributed to host defense against S. typhimurium infection.
Fig. 8. Epithelial HVEM is required for host defense against S. typhimurium infection.
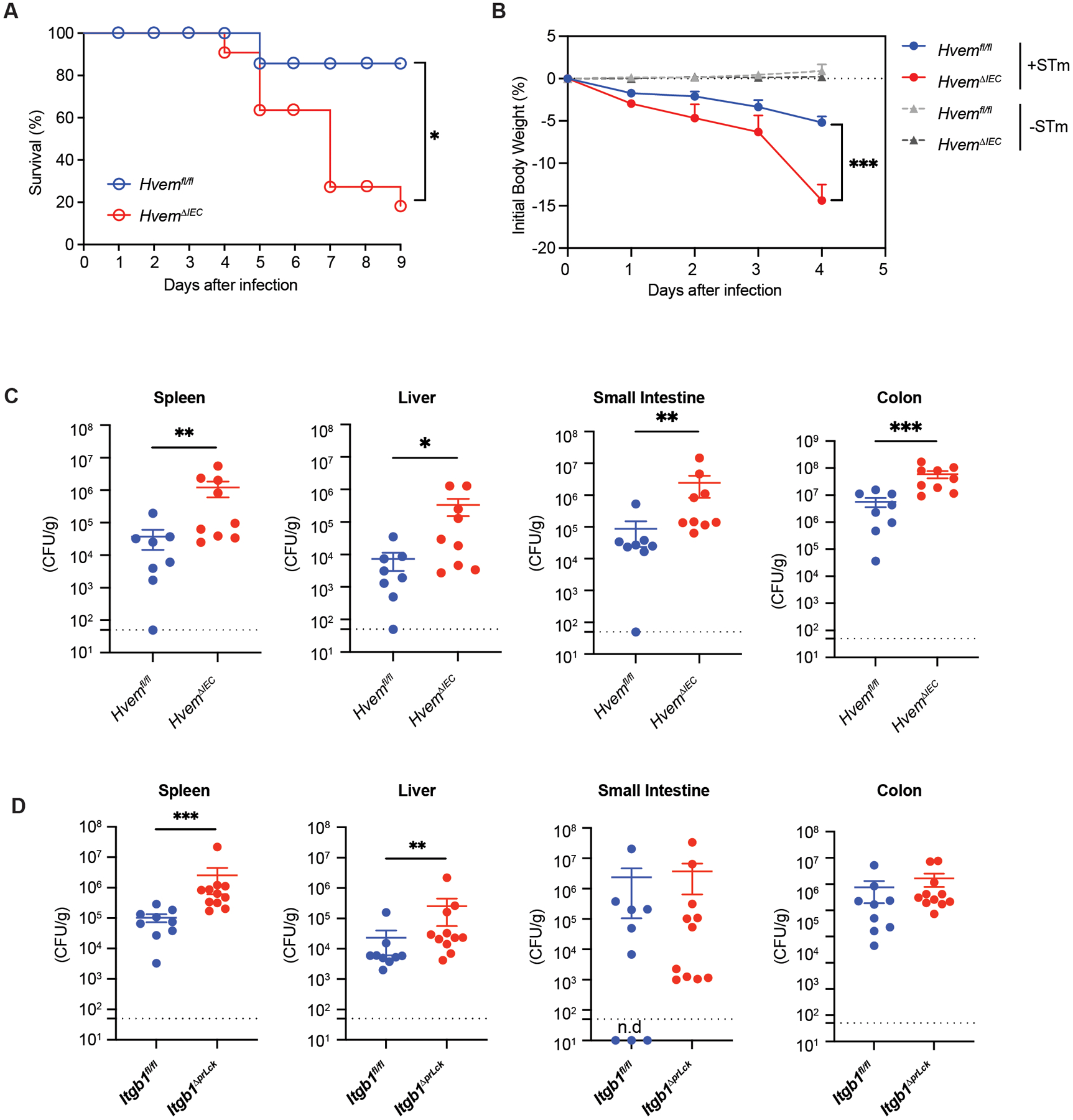
HvemΔIEC mice and control Hvemfl/fl mice were infected orally with 1 × 107 S. typhimurium colony-forming units (CFU)/mouse (A-C). (A) Survival curves (Hvemfl/fl, n=8; HvemΔIEC, n=11). (B) Changes in body weight (% change from baseline) (infected Hvemfl/fl, n=8; infected HvemΔIEC, n=9; uninfected Hvemfl/fl, n=11; uninfected HvemΔIEC, n=8). (C) Bacterial burdens at day 4 p.i. (Hvemfl/fl, n=8; HvemΔIEC, n=9). (D) Itgb1fl/fl (n=9) and Itgb1ΔprLck (n=11) mice were infected orally with S. typhimurium and bacterial burdens measured at days 3–4 p.i. Dashed line represents the limit of detection (C, D). Statistical analysis was performed using Log-rank test (A), 2 way ANOVA with Bonferroni’s multiple comparison test (B), or Mann-Whitney test (C, D). Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001. Data shown are means ± SEM (A). Bars show the mean, symbols represent individual mice. Data represent pooled results from at least two independent experiments having at least three mice per group in each experiment. Groups of co-housed littermates were analyzed.
The proposed mechanism for HVEM affecting the survival of natural IETs is indirect, through stimulation of synthesis of β1 integrin ligand. To investigate if the same mechanism might be relevant for the anti-bacterial response, we infected Itgb1ΔprLck mice with S. typhimurium. Although weight loss was not affected, the deficiency of β1 integrin expression only in TCRαβ+ cells led to an increased bacterial burden in the spleen and liver (Fig, 8D). These data suggest that the interaction between β1 integrins expressed by IETs and the basement membrane, a mechanism that contributed to the maintenance of natural IETs, might also contribute to protection from mucosal infection.
DISCUSSION
The mechanisms by which IEC regulate IET homeostasis in the intestine have not been fully elucidated, although they include IL-15 (26–28) and epithelial expression of the thymus leukemia antigen (30). Here, we provided evidence that HVEM, a TNFR superfamily member that is constitutively expressed by IEC, was highly involved in regulating different aspects of IETs behavior, including the survival of natural IETs, the migration of most small intestine IETs, and in the protective response to an intestinal bacterial infection that requires IET responses. We propose that some of these HVEM-mediated influences at steady state, especially natural IET subsets survival, were due to LIGHT-mediated stimulation of HVEM, that caused the increased synthesis of basement membrane proteins that interacted with β1 integrins expressed by IETs. In contrast, we propose that the interaction of HVEM with CD160 might have contributed by triggering the protective response of IETs in the context of an infection.
While present throughout the SI, the effects of epithelial HVEM deficiency on IETs survival were most prominent in the proximal segment, where the majority of these cells are found. They were not uniform across different IET populations, with TCRαβ+CD8αα+ IETs most severely reduced, although TCRγδ+ IETs, also were consistently diminished. HVEM effects on CD4+ IETs, which were less thoroughly analyzed, involved expression by both T cells and epithelial cells. In contrast, TCRαβ+ CD8αβ+ IETs, were not decreased in HvemΔIEC mice, although they had increased pro-apoptotic Bax protein expression in HvemΔIEC mice, they expressed β1 integrins, and β1 integrin-collagen IV interactions affected the survival of this subset in vitro. Additionally, germ line deficiency of the gene encoding LIGHT caused decreased TCRαβ+ CD8αβ+ IETs. A reduction in TCRαβ+ CD8αβ+ IETs also was observed in mice with deficiency for β1 integrin subunit in TCRαβ+ T cells, and TCRαβ+ CD8αβ+ IETs also had increased Bax expression in this strain. There could have been effects of the LIGHT or β1 integrin gene knockouts outside the epithelium, however, consistent with the decreased TCRαβ+ CD8αβ+ IETs in whole body knockouts of HVEM, that were not observed in HvemΔIEC mice. Despite this caveat, collectively the in vivo and in vitro data suggested that the HVEM-dependent mechanism for IETs homeostasis was compensated for or overridden by other survival mechanisms in TCRαβ+ CD8αβ+ IETs. There are precedents for signals that selectively influence IET subsets (23, 26–28, 64, 65), and therefore, variation of the effects in HvemΔIEC mice on the survival of different IET populations was not surprising.
Our data showed there was decreased extracellular matrix protein synthesis by epithelial cells from HvemΔIEC mice at steady-state. Furthermore, signaling by HVEM ligands in vitro increased Col4a1 and Col4a2 mRNA synthesis in intestinal organoid cultures, providing direct evidence that HVEM signaling in IEC could increase basement membrane protein synthesis. There are previous reports that collagen IV promotes survival and proliferation of different types of cancer cells (66, 67). Human and mouse IETs bind collagen (52, 56), and we showed IETs cultured on collagen IV coated microwells had increased survival. Therefore, the findings here provide a link between HVEM signals, extracellular matrix synthesis and IETs survival, although there could have been other effects of epithelial HVEM expression that promoted the accumulation of natural IET populations.
Various integrins containing a β1 subunit bind to extracellular membrane components, including different types of collagen and fibronectin (68). For example, integrin α1β1 preferentially binds collagen type IV, while α2β1 preferentially binds to collagen type I (52, 68, 69). Human IETs bind collagen VI through integrin α1β1 (52), and we confirmed that mouse IETs express collagen-binding integrins. The positive survival effect of IETs culture on collagen IV coated microwells could be reversed by blocking integrin binding. Moreover, mice with a deficiency for Itgb1 in TCR αβ+ cells had reduced TCRαβ+ IETs. We note that previous in vivo analyses of the role of α1β1 for IETs accumulation provides conflicting results. In agreement with our findings, mice with a full body deletion of the gene encoding the integrin α1 have reduced total IET (53). In contrast to the findings here, in bone marrow chimeras in which the donor cells have a poly I:C induced deletion of Itgb1, normal IET numbers are present (56). The gene deletion is not complete in these mice, however, with approximately 50% of the IET retaining integrin β1 expression. Also consistent with the data here, Very Late Activation Antigen 1 (VLA-1), or α1β1 integrin, is important for the retention of memory CD8+ T cells in the lung after influenza virus infection (70). Integrins activate downstream signaling pathways that affect survival and other processes, and although the signaling mechanism operating downstream of α1β1 engagement of collagen in mouse IETs has not been defined, in human peripheral blood T cells α2β1 interaction with collagen increased resistance to apoptosis through the Erk pathway (71).
IETs are migratory cells located above the basement membrane, which they patrol extensively (1). Movement and survival of IETs may be linked, as both were negatively regulated in TCRγδ+ IETs by GPR55 (4). Furthermore, collagen IV promotes migration of tumor cells and endothelial cells (72, 73). RNA-Seq analyses confirmed that the expression of genes related to migration and survival were reduced in TCRαβ+CD8αα+ IETs in the absence of epithelial HVEM. Consistent with this, in HvemΔIEC mice IETs speed, track length and displacement were decreased. Combined with fewer cells, this led to a greatly decreased area of the villi covered. Because these effects were evident when CD8α+ IETs were labeled, this indicated epithelial HVEM deficiency had a broad effect on migration of all or most IET subsets. Therefore, a plausible hypothesis is that epithelial HVEM deficiency affected not only the survival of natural IETs but also patrolling of IETs through altered basement membrane synthesis and the action of β1 integrins, although other HVEM-dependent effects on IETs migration have not been excluded.
IETs movement and surveillance likely are important for protecting the epithelium from injury or infection. TCRγδ+ IETs movement in the small intestine is dynamically regulated by MyD88-dependent signaling by IEC (2). Furthermore, exposure to invasive Salmonella bacteria enhanced the movement of TCRγδ+ IETs between the IEC in a “flossing” movement, and movement of γδ IEL to epithelial cells in contact with pathogen is important for immediate host defense from Salmonella infection (62). Epithelial deficiency of HVEM led to increased susceptibility to S. typhimurium infection, which may be related to reduced IETs migration as well as decreased cell number. Although for survival in the proximal small intestine LIGHT was the key HVEM ligand, following infection CD160 deficiency had an equal or even greater influence than LIGHT deficiency, consistent with the importance of CD160 during other mucosal infections(38, 57, 58). The role of CD160 during infection might be due in part to the signaling capacity of CD160, which might trigger the protective response of the IETs, as it has been reported to do for NK cells (74).
According to previous results (62), TCRγδ+ IETs could have been responding to Salmonella, but the TCRαβ+ IETs might have contributed as well, consistent with the increased bacterial colonies in Itgb1ΔprLck mice, in which TCRαβ+ cells were affected. We don’t know which population(s) were relevant, but CD4+ T cells are important for host defense against this microbe (75).
HVEM is a multi-functional protein that influences inflammation and the response to acute infections in the intestinal mucosae (34, 37, 38, 41). Here, our results showed that the interaction of LIGHT with epithelial HVEM influences the survival of natural IET subsets at steady-state, in part by promoting synthesis of β1 integrins ligands, whereas HVEM interaction with CD160 might exert an influence in enhancing the protective capacity of the responding IETs. Therefore, our data indicate that overall HVEM expression by IEC has a role in regulating mucosal immunity at steady state and following oral infection.
MATERIALS AND METHODS
Study design
HVEM had previously been shown to affect mucosal immune responses, and the goal of this study was to determine how it affected intraepithelial T cells (IETs) in the small intestine at steady state and after infection. We determined that although HVEM is expressed by many cell types, epithelial HVEM was critical for IETs. Using mice with intestine epithelial cell- specific deletion of Hvem, flow cytometry and in vitro cultures, we showed that epithelial HVEM influenced IETs survival, migration in the epithelium, and the host response to Salmonella typhimurium infection. RNA-seq of HVEM deficient and wildtype epithelial cells implicated the synthesis of basement membrane proteins as a key function reduced when HVEM was missing, and we confirmed this in organoid cultures. We showed with in vitro cultures that collagen IV, a basement membrane component dependent on HVEM expression, affected IETs survival. β1 integrins expressed by IETs bind collagen IV and these integrins mediated a survival benefit in vitro by binding collagen IV and in vivo. Mice deficient for epithelial HVEM, HVEM ligands or β1 integrins, were more susceptible to S. typhimurium, indicating the mechanisms governing steady state survival and the response to acute infection were similar. Experiments were done at least two times with at least four mice in each experimental group. Observational studies, such as intravital microscopy analysis, immunofluorescence analysis, or bacterial infection, were carried by an individual blinded as to the experimental conditions or a third party not involved in the study.
Mice
Hvem−/−, which is Hvemflox-neo/flox-neo (Hvemfn/fn) mice, Hvemfl/fl mice, Btla−/−, which is Btlaflox-neo/flox-neo (Btlafn/fn) mice, Btlafl/fl, which is a control for Btla−/− and LIGHT gene deficient (Tnfsf14−/−, listed as LIGHT−/− in the figures) mice were bred and described previously (37). These mice were generated by us except Tnfsf14−/− mice were a gift from Dr. Klaus Pfeffer (Heinrich Heine University, Dusseldorf, Germany). Cd160−/− mice were provided by Dr. Yang-Xin Fu (UT Southwestern, TX) (74). Cd4-Cre (STOCK Tg(Cd4-cre)1Cwi/BfluJ; Stock No: 017336), Villin-Cre (B6.Cg-Tg(Vil1-cre)997Gum/J, Stock No: 004586), Lckpr-Cre (B6.Cg-Tg(Lck-cre)548Jxm/J, Stock No:003802), Itgb1fl/fl (B6;129-Itgb1tm1Efu/J, Stock No: 004605) and Lckpr-Bcl-xLTg mice (B6.Cg-Tg(LCKprBCL2L1)12Sjk/J; Stock No: 013738) were all purchased from The Jackson Laboratory. Lckpr-Bcl-xLTg mice express the pro-survival Bcl2 family gene Bclxl (Bcl2l1) under the control of the mouse Lck proximal promoter (Lckpr Cre). Hvemfl/fl mice were bred to Villin-cre mice to generate conditional knockout mice, HvemΔIEC. Integrin b1fl/fl mice were bred to Lckpr Cre mice to generate conditional knockout mice, Itgb1ΔpLck mice. All Cre mouse strains were maintained on the C57BL/6 background or were backcrossed to C57BL/6 for at least for 6 generations. 15 to 17-week-old male mice on the C57BL/6 genetic background were used in this study. In most experiments, groups of co-housed control and gene knockout mice were analyzed to minimize the effect of housing conditions on experimental variation. In a few experiments, as indicated in the figure legends, co-housed mice that were not littermates were analyzed. For tissue or cell analyses, tissues were collected and used for immunofluorescence analysis and intraepithelial lymphocyte preparation. Mice were bred and housed under specific pathogen-free conditions in the vivarium of the La Jolla Institute for Immunology (LJI). All procedures were approved by the LJI Animal Care and Use Committee.
Germ-free mouse husbandry
Germ-free mice were bred and housed in the animal facility of POSTECH Biotech Center. Germ-free C57BL/6 (B6) mice were kindly provided originally by Drs. Andrew Macpherson (Bern Univ., Switzerland) and David Artis (Univ. Pennsylvania, USA) and maintained in sterile flexible film isolators (Class Biological Clean Ltd., USA). The sterility of germ-free mice was checked regularly by the absence of bacterial colonies in the culture experiment using their fecal pellets. Germ free C57BL/6 mice maintained in the animal facility of POSTECH Biotech Center in accordance with institutional ethical guideline and the protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the POSTECH.
Isolation of IEL
Intestines were collected from mice, and were divided into five parts, including proximal SI, middle SI, and distal SI, approximately one-third each, as well as cecum and colon. Peyer’s patches were carefully removed, and tissues were cut open longitudinally, briefly washed, and cut into 1.5 cm pieces. The tissue pieces were incubated in 20 mL of RPMI (5% FBS, 25 mM HEPES and 1 mM DTT) in a shaker at 200 rpm, 37°C, for 20 min, followed by incubated in 20 mL HBSS (25 mM HEPES and 20 mM EDTA) in a shaker at 200 rpm, 37°C, for 30 min. After each incubation, the cell suspension was filtered through a metal mesh and the supernatant was saved for IEL preparation. The flow-through cell suspension was spun down. The cell pellets were then re-suspended in 40% Percoll solution and overlaid above 80% Percoll solution carefully, followed by centrifugation at 2000 rpm, 25°C for 20 min without the brake. IEL were collected from the interface, washed once and re-suspended in the complete RPMI-1640 medium. These purified cells constituted the epithelial cell fraction enriched for IEL. The cells were used immediately for cell counting and staining.
Flow cytometry
Flow cytometry analysis was performed on an LSRII instrument (BD Biosciences). The data were analyzed by using FlowJo software (Tree Star). Absolute cell counts were obtained by using CountBright™ Absolute Counting Beads (Life Technologies). The following mAbs were used: TCRδ (eBioscience, eBioGL3), TCRβ (BioLegend, H57–597), CD4 (eBioscience, RM4–5), CD8α (BD Biosciences, 53–6.7), CD8β (eBioscience, eBioH35–17.2), CD45 (BioLegend, 30-F11), CD45.1 (BioLegend, A20), CD45.2 (Thermo Fisher Scientific, 104), HVEM (BioLegend, HMHV-1B18), integrin β7 (BD Biosciences, M293), integrin αE (Thermo Fisher Scientific, 2E7), Bax (Thermo Fisher Scientific, 6A7), Bim (Bio-Rad AbD Serotec, AHP933), CD49a (BD Biosciences, Ha31/8), CD29 (BioLegend, HMβ1–1), Ki-67 (Thermo Fisher Scientific, SP6), EdU (Thermo Fisher Scientific), Annexin V (BD Biosciences), SYTOX™ AADvanced™ Dead Cell Stain Kit (Thermo Fisher Scientific). All representative FACS gating not included in the main or supplemental figures is present in supplemental data file S2. Staining methods are further shown in data file S2.
Cryosection immunofluorescence
SI tissue was collected, opened longitudinally and the luminal contents washed in 20 mL RPMI in a 50 mL tube by inverting 20 times. The tissues were placed on foil with O.C.T. for the Swiss-roll technique. Once the entire intestine length was rolled up, the intestine Swiss roll was transferred into a tissue mold and was frozen on a thermo-conductive platform (ThermalTray, Biocision) pre-cooled with dry ice, and then placed into −80°C freezer. Frozen sections were cut to a 10 μm thickness. After drying at RT for 1 hr, frozen sections were fixed in pre-cooled ethanol at −20 °C for 10 min followed by pre-cooled acetone at −20 °C for 10 min. The fixed sections were used for immunofluorescence (IF) staining or kept in −80°C freezer for later use. Primary antibodies were diluted in PBST (PBS, 0.5% BSA, 0.1% Tween-20). Antibodies used were: CD45 (BioLegend, clone 30-F11), EpCAM (BioLegend, clone G8.8), TCRδ (BioLegend, clone GL3), TCRβ (BioLegend, clone H57–597), CD8α (BD, clone 53–6.7), CD8β (BD, clone H35–17.2). In Figs.1B–1C, fluorescence imaging was performed on an FV10i confocal microscope (Olympus). Areas from Pro-SI, Mid-SI or Dis-DI were outlined and CD45+ IEL per villus were counted manually. In Figs. 2D–2F, fluorescence imaging was performed on Zeiss LSM 780 or LSM 880 confocal microscopes (Zeiss). Data analysis was performed using ZEN software (Zeiss); areas of each villus from Pro-SI was outlined and measured and IET subsets of each villus were counted manually. The number of IET subsets was normalized to the area (mm2) in each villus. Fiji software (National Institutes of Health) were used for data for representative images.
Intravital microscopy
To track CD8α+ IEL in the epithelium, mice were injected with 15–20 μg of anti-CD8α-AF488 (eBioscience, clone 53–6.7) or anti- CD8α-AF647 (BD Biosciences, clone 53–6.7) with/without EpCAM-AF647 (BioLegend, clone G8.8) by retro-orbital injection 4 hr before imaging. After anesthetization, mice were positioned on a WPI ATC2000 heating pad ventral side up and kept at 37°C. The distal duodenum was exposed and opened along the antimesenteric border. The mucosal surface was hydrated with PBS and placed against a coverslip fitted onto a suction ring (76). All intravital imaging was done using 25× 0.95NA water immersion objective on the Leica SP5 upright confocal microscope with a resonant scanner and acquired using Leica Application Suite software. The sample was excited with 488 nm and 633 nm lasers using 488/543/633 triple dichroic mirror. The internal detectors were set to collect AF488 fluorescence at 494–563 nm, reflection image at 619–651 nm, and AF647 fluorescence at 651–722 nm. Images were acquired by taking Z-stacks encompassing the epithelium and the upper layers of the lamina propria at 2.8 um step size every 10 sec. Each XY plane spans 365 × 365 μm2. To calculate cell coverage, the data was median filtered in Fiji (77). Then, the sections from the top of the villi encompassing nearly exclusively epithelium were Z projected. A maximum intensity time projection was performed and coverage was calculated as the area with intensity values over a threshold, demonstrating it was visited by a cell. Additionally, a rainbow time projection was generated to visualize cell movement. Individual cells in Z stacks were tracked in Imaris (Bitplane) semi-automatically using the spots function. Cell movement metrics (mean speed, track length, track displacement length) for each CD8α+ lymphocyte cell were calculated with Imaris and plotted using Prism (version 8, GraphPad). Spider graphs of individual cell paths, each beginning at the origin, were plotted using Matlab (Mathworks, 2021A) using the positions output from Imaris.
Small intestinal organoid culture
SI organoids were derived from Hvem/− or littermate Hvem+/+ mice. The proximal SI was cut into 5-mm segments and incubated in 4°C 2 mM EDTA in PBS for 5 min and washed by pipetting. The segments were incubated in 2 mM EDTA in PBS for 30 min at 4°C, and crypts were isolated by pipetting with cold HBSS. Dissociated crypts were passed through a 70-μm cell strainer and pelleted by centrifuge at 600 rpm for 3 min at 4°C. The crypts were resuspended in Advanced DMEM/F12 medium (Thermo Fisher Scientific); the number of crypts was counted, and they were resuspended in Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning). The crypts were plated in a 24-well plate with organoid growth medium supplemented with 100 μg/ml penicillin, 100 U/ml streptomycin, 2 mM Glutamax, 1×N-2 supplement, 1×B27 supplement, 10 mM HEPES (Thermo Fisher Scientific), 1 mM N-acetylcysteine (Sigma-Aldrich), 100 ng/ml recombinant mouse Noggin (Peprotech), 50 ng/ml recombinant mouse EGF (BioLegend), 500 ng/ml recombinant human R-spondin 1 (Peprotech). Media were changed every 2 days.
Quantitative Real-time PCR
Total RNA extraction from IECs was performed with RNeasy Kit (Qiagen), according to the manufacturer’s instructions. cDNA synthesis was performed by using iScript Advanced cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR reactions were performed with SYBR Green I Master Kit and LightCycler 480 system (Roche). mRNA levels of Col4a1 were normalized to the housekeeping gene Actb or Rpl32. The primers were synthesized by the Integrated DNA Technologies. Primers used were:
Col4a1-forward 5’- TCCGGGAGAGATTGGTTTCC-3’
Col4a1-reverse 5’- CTGGCCTATAAGCCCTGGT-3’
Actb- reverse 5’- GATCTGGCACCACACCTTCT-3’
Actb - reverse 5’- GGGGTGTTGAAGGTCTCAAA-3’
Rpl32- reverse 5’- TTCCTGGTCCACAATGTCAA-3’
Rpl32 - reverse 5’- GGCTTTTCGGTTCTTAGAGGA-3’
RNA-seq library preparation
RNA-seq libraries were prepared from sorted proximal SI IEC or from sorted SI TCRαβ+CD8αα+ IET by the Sequencing Core at La Jolla Institute for Immunology using the SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (TaKaRa)(78). Libraries were sequenced on an Illumina HiSeq 2500, generating 50 bp single-end reads.
RNA-seq analysis
The single-end reads that passed Illumina filters were filtered for reads aligning to tRNA, rRNA, adapter sequences, and spike-in controls. The reads were then aligned to mm10 reference genome using TopHat (v 1.4.1) (79). DUST scores were calculated with PRINSEQ Lite (v 0.20.3) (80) and low-complexity reads (DUST > 4) were removed from the BAM files. The alignment results were parsed via the SAMtools (81) to generate SAM files. Read counts to each genomic feature were obtained with the htseq-count program (v 0.7.1) (82) using the “union” option. After removing absent features (zero counts in all samples), the raw counts were then imported to R/Bioconductor package DESeq2 (v 1.6.3) (83) to identify differentially expressed genes among samples. P-values for differential expression were calculated using the Wald test for differences between the base means of two conditions. These P-values were then adjusted for multiple test correction using the Benjamini-Hochberg algorithm (84). Principal Component Analysis (PCA) was performed using the ‘prcomp’ function in R. The sequences used in this article have been submitted to the Gene Expression Omnibus under accession number GSE 206183. Gene set enrichment analysis was done using the “GseaPreranked” method with “classic” scoring scheme. All the GO gene sets were downloaded from MSigDB in GMT format. Rank files for each DE comparison of interest were generated by assigning a rank of −log10(pValue) to genes with log2FoldChange greater than zero and a rank of log10(pValue) to genes with log2FoldChange less than zero. GO analysis of DEGs was performed with ToppGene (http://toppgene.cchmc.org) (85). FDR (Benjamini-Hochberg) < 0.05 was considered statistically significant. Heatmap was performed with Morpheus (https://software.broadinstitute.org/morpheus/). For classical pathway, Ingenuity Pathway Analysis (Qiagen) was used.
Western-blot analysis
Harvested IECs were lysed in triton lysis buffer (137 mM NaCl, 20 mM Tris base at pH 7.4, 10% glycerol and 1% Triton X-100) supplemented with a protease and phosphatase inhibitor mixture (Roche) for 20min on ice, and centrifuged at 15,000 rpm for 10 min at 4°C. 10 μg of denatured proteins were loaded onto a Mini-PROTEAN Precast Gel (Bio-Rad) and transferred onto a PVDF membrane (Thermo Fisher Scientific).The membranes were blocked by 5% skim milk in TBST (Tris-buffered saline /0.1% Tween 20, Bio-Rad) and then incubated with antibodies to collagen a1 (IV) (Origene) and Villin (Abcam). Immunoreactive bands were detected by chemiluminescence (ECL solution, Santa Cruz).
CD8α+ IEL culture
For the isolation of IEL, CD8α+ cells were enriched from the SI IEL preparation by negative and positive selection with the iMag cell separation system (BD Biosciences) according to the manufacturer’s instructions. Briefly, single-cell IEL suspension was incubated with staining buffer (PBS containing 2% FBS with 2 mM EDTA) containing a mixture of biotin-conjugated mAbs against CD4 (RM4–5; BioLegend), CD19 (6D5; BioLegend), CD326 (G8.8; BioLegend), and TER-119 (TER-119; BioLegend) in a 5-ml round tube (BD Biosciences) for 30 min at 4°C. Cells were washed with staining buffer and incubated with Streptavidin Particle Plus-DM (BD Biosciences) for 30 min at 4°C. The tube was placed on the Cell Separation Magnet at room temperature for 8 min. The supernatant was carefully aspirated off as a negative fraction. The negative fraction was incubated with biotin-conjugated anti-CD8α (53–6.7; BioLegend) in staining buffer for 30 min at 4°C, washed with staining buffer, and incubated with Streptavidin Particle Plus-DM in a 5-ml round tube for 30 min at 4°C. The tube was placed on the Cell Separation Magnet at room temperature for 8 min. The supernatant was carefully removed and CD8α+ positive fraction was washed and resuspended with complete medium. CD8α+IEL were expanded in complete medium containing 50 ng/ml recombinant mouse IL-15/IL-15R complex (Thermo Fisher Scientific) on a tissue culture plate at the concentration of 5×105 cells/ml for 3 days. CD8α+IEL were then cultured on a 96-well protein high binding plate (Corning) coated with 100 μg/ml bovine serum albumin (BSA, Thermo Fisher Scientific), type IV collagen (Corning), Laminin (Corning) or Fibronectin (Abcam). For monitoring cell death, isolated SI CD8α+ IET (EpCAM−CD4−CD19−TER119− CD8α+) from wild type mice were labeled with CellTrace™ Violet (CTV, Thermo Fisher Scientific), and then labeled cells were incubated in the presence of recombinant IL-15/15Rα (50ng/mL) in the presence of absence of anti-integrin α1 (10μg/mL, BD Biosciences) or Integrin α1β1 inhibitor Obtusatin (10nM, R&D Systems) in the coated culture plate. After 3 days of culture, cells were stained with antibodies and 7-aminoactinomycin D (7AAD, BD Biosciences) for viability assessment of the different CD8α+ IET subsets when analyzed by flow cytometry. % cell death in proliferating cells is calculated as: % cell death in proliferating cells = (CTV−7AAD+) / (CTV−7AAD−) + (CTV−7AAD+) × 100.
Bacterial infection
Salmonella enterica serovar Typhimurium strain IR715 (S. typhimurium), which is a naturally nalidixic acid resistant derivative of ATCC 14028, was obtained from Dr. M. Raffatellu (UCSD) (86) and used in the infection studies. Bacteria were aerobically grown overnight at 37°C in Luria-Bertani broth, then diluted 1:20 and subcultured for 4 hr in fresh medium. Bacteria were washed twice in ice-cold phosphate-buffered saline (PBS) and then suspended in cold PBS (1 × 107 colony forming units [CFU]/100 μl). Mice were pretreated with 20 mg streptomycin 24 h prior to infection with 1 × 107 CFU S. typhimurium by oral gavage in a total volume of 100μl PBS (87). Recipients were analyzed for weight loss until day 3 or 4 p.i. At the terminal endpoint, tissues were collected for CFU assays and histopathology analysis. For CFU assays, several tissues including spleen, liver, small intestine and colon were weighed, homogenized in sterile PBS, serially diluted and plated on LB agar plates supplemented with 200μg/mL nalidixic acid. Histopathology analysis of liver samples was performed on zinc formalin (Medical Chemical Corporation)-fixed tissue after hematoxylin and eosin (H&E) stain.
Statistical analysis
Details concerning the statistical analysis methods are provided in each figure legend. Briefly, all data were analyzed using GraphPad Prism 8 software and were shown as mean and the standard error of the mean (SEM). Statistical significance was determined by unpaired t-test for cell numbers, % of cell proliferation, cell survival and cell death and Mann-Whitney test for intravital microscopy and infection experiments, Log-rank test for survival curves, 2 way ANOVA with Bonferroni’s multiple comparison test for cell death. Statistical significance is indicated by *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Supplementary Material
Acknowledgments:
We thank the staff of the Microscopy & Histology Core, Flow Cytometry Core, Sequencing Core, Bioinformatics Core and the Department of Laboratory Animal Care (DLAC) at La Jolla Institute for Immunology for excellent technical assistance.
Funding:
National Institutes of Health grants P01 DK46763, R01 AI61516 and MIST U01 AI125955 (MK); MIST U01 AI125957 (HC); S10RR027366 (LJI Flow Cytometry Core Facility); S10OD021831 (LJI Microscopy Facility Core); and Crohn’s and Colitis Foundation of America grant CCFA-254582 (JWS); Uehrara Foundation grant (DT); Chan-Zuckerberg Initiative Imaging Scientist Grant (SM).
Footnotes
Competing interests: MK and TFC have filed a patent on uses of HVEM with selective binding to ligands.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the main text or the Supplementary Materials. All reasonable requests for materials including, mouse strains, will be available upon request to Mitchell Kronenberg (mitch@lji.org). Mouse strain # 030862 with a floxed allele of the gene encoding HVEM (B6;SJL-Tnfrsf14tm1.1Kro/J) has been deposited at the Jackson lab. RNA-seq data are available from GEO under accession code GSE206183.
References and notes
- 1.Edelblum KL, Shen L, Weber CR, Marchiando AM, Clay BS, Wang Y, Prinz I, Malissen B, Sperling AI, Turner JR, Dynamic migration of gammadelta intraepithelial lymphocytes requires occludin. Proceedings of the National Academy of Sciences of the United States of America 109, 7097–7102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D, Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell 171, 783–794 e713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Sumida H, Cyster JG, GPR18 is required for a normal CD8alphaalpha intestinal intraepithelial lymphocyte compartment. The Journal of experimental medicine 211, 2351–2359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumida H, Lu E, Chen H, Yang Q, Mackie K, Cyster JG, GPR55 regulates intraepithelial lymphocyte migration dynamics and susceptibility to intestinal damage. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheroutre H, Lambolez F, Mucida D, The light and dark sides of intestinal intraepithelial lymphocytes. Nature reviews. Immunology 11, 445–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M, Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 38, 769–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayday A, Theodoridis E, Ramsburg E, Shires J, Intraepithelial lymphocytes: exploring the Third Way in immunology. Nature immunology 2, 997–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Olivares-Villagomez D, Van Kaer L, Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends Immunol 39, 264–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masopust D, Vezys V, Marzo AL, Lefrancois L, Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Lefrancois L, Masopust D, T cell immunity in lymphoid and non-lymphoid tissues. Curr Opin Immunol 14, 503–508 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Cheroutre H, Lambolez F, Doubting the TCR coreceptor function of CD8alphaalpha. Immunity 28, 149–159 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H, Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity 16, 355–364 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Oh-Hora M, Komatsu N, Pishyareh M, Feske S, Hori S, Taniguchi M, Rao A, Takayanagi H, Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity 38, 881–895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamagata T, Mathis D, Benoist C, Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nature immunology 5, 597–605 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Mayans S, Stepniak D, Palida S, Larange A, Dreux J, Arlian B, Shinnakasu R, Kronenberg M, Cheroutre H, Lambolez F, alphabetaT cell receptors expressed by CD4(−)CD8alphabeta(−) intraepithelial T cells drive their fate into a unique lineage with unusual MHC reactivities. Immunity 41, 207–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald BD, Jabri B, Bendelac A, Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nature reviews. Immunology 18, 514–525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald BD, Bunker JJ, Ishizuka IE, Jabri B, Bendelac A, Elevated T cell receptor signaling identifies a thymic precursor to the TCRalphabeta(+)CD4(−)CD8beta(−) intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen MM, Witherden DA, Havran WL, gammadelta T cells in homeostasis and host defence of epithelial barrier tissues. Nature reviews. Immunology 17, 733–745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R, Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proceedings of the National Academy of Sciences of the United States of America 99, 14338–14343 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boismenu R, Havran WL, Modulation of epithelial cell growth by intraepithelial gamma delta T cells. Science 266, 1253–1255 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Komano H, Fujiura Y, Kawaguchi M, Matsumoto S, Hashimoto Y, Obana S, Mombaerts P, Tonegawa S, Yamamoto H, Itohara S, et al. , Homeostatic regulation of intestinal epithelia by intraepithelial gamma delta T cells. Proceedings of the National Academy of Sciences of the United States of America 92, 6147–6151 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, Ruhn KA, Hou B, DeFranco AL, Yarovinsky F, Hooper LV, Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proceedings of the National Academy of Sciences of the United States of America 108, 8743–8748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M, Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Klose CS, Blatz K, d’Hargues Y, Hernandez PP, Kofoed-Nielsen M, Ripka JF, Ebert K, Arnold SJ, Diefenbach A, Palmer E, Tanriver Y, The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8alphaalpha(+) intraepithelial lymphocyte development. Immunity 41, 230–243 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Yu Q, Tang C, Xun S, Yajima T, Takeda K, Yoshikai Y, MyD88-dependent signaling for IL-15 production plays an important role in maintenance of CD8 alpha alpha TCR alpha beta and TCR gamma delta intestinal intraepithelial lymphocytes. Journal of immunology 176, 6180–6185 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A, IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9, 669–676 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Hu MD, Ethridge AD, Lipstein R, Kumar S, Wang Y, Jabri B, Turner JR, Edelblum KL, Epithelial IL-15 Is a Critical Regulator of gammadelta Intraepithelial Lymphocyte Motility within the Intestinal Mucosa. Journal of immunology 201, 747–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma LJ, Acero LF, Zal T, Schluns KS, Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. Journal of immunology 183, 1044–1054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H, T cell responses modulated through interaction between CD8alphaalpha and the nonclassical MHC class I molecule, TL. Science 294, 1936–1939 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Park Y, Wang-Zhu Y, Larange A, Arens R, Bernardo I, Olivares-Villagomez D, Herndler-Brandstetter D, Abraham N, Grubeck-Loebenstein B, Schoenberger SP, Van Kaer L, Kronenberg M, Teitell MA, Cheroutre H, Mucosal memory CD8(+) T cells are selected in the periphery by an MHC class I molecule. Nature immunology 12, 1086–1095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins JM, Mandlebrot DA, Shaw SK, Russell GJ, Murphy EA, Chen YT, Nelson WJ, Parker CM, Brenner MB, Direct and regulated interaction of integrin alphaEbeta7 with E-cadherin. J Cell Biol 140, 197–210 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB, Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature 372, 190–193 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Schon MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR, Olson S, Lefrancois L, Brenner MB, Grusby MJ, Parker CM, Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. Journal of immunology 162, 6641–6649 (1999). [PubMed] [Google Scholar]
- 34.Steinberg MW, Huang Y, Wang-Zhu Y, Ware CF, Cheroutre H, Kronenberg M, BTLA interaction with HVEM expressed on CD8(+) T cells promotes survival and memory generation in response to a bacterial infection. PloS one 8, e77992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desai P, Tahiliani V, Hutchinson TE, Dastmalchi F, Stanfield J, Abboud G, Thomas PG, Ware CF, Song J, Croft M, Salek-Ardakani S, The TNF Superfamily Molecule LIGHT Promotes the Generation of Circulating and Lung-Resident Memory CD8 T Cells following an Acute Respiratory Virus Infection. Journal of immunology 200, 2894–2904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn R, Hutchinson T, Murphy KM, Ware CF, Croft M, Salek-Ardakani S, CD8 T cell memory to a viral pathogen requires trans cosignaling between HVEM and BTLA. PloS one 8, e77991 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo GY, Shui JW, Takahashi D, Song C, Wang Q, Kim K, Mikulski Z, Chandra S, Giles DA, Zahner S, Kim PH, Cheroutre H, Colonna M, Kronenberg M, LIGHT-HVEM Signaling in Innate Lymphoid Cell Subsets Protects Against Enteric Bacterial Infection. Cell Host Microbe 24, 249–260 e244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shui JW, Larange A, Kim G, Vela JL, Zahner S, Cheroutre H, Kronenberg M, HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature 488, 222–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herro R, Shui JW, Zahner S, Sidler D, Kawakami Y, Kawakami T, Tamada K, Kronenberg M, Croft M, LIGHT-HVEM signaling in keratinocytes controls development of dermatitis. The Journal of experimental medicine 215, 415–422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg MW, Turovskaya O, Shaikh RB, Kim G, McCole DF, Pfeffer K, Murphy KM, Ware CF, Kronenberg M, A crucial role for HVEM and BTLA in preventing intestinal inflammation. The Journal of experimental medicine 205, 1463–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breloer M, Hartmann W, Blankenhaus B, Eschbach ML, Pfeffer K, Jacobs T, Cutting Edge: the BTLA-HVEM regulatory pathway interferes with protective immunity to intestinal Helminth infection. Journal of immunology 194, 1413–1416 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Harrop JA, McDonnell PC, Brigham-Burke M, Lyn SD, Minton J, Tan KB, Dede K, Spampanato J, Silverman C, Hensley P, DiPrinzio R, Emery JG, Deen K, Eichman C, Chabot-Fletcher M, Truneh A, Young PR, Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. The Journal of biological chemistry 273, 27548–27556 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Harrop JA, Reddy M, Dede K, Brigham-Burke M, Lyn S, Tan KB, Silverman C, Eichman C, DiPrinzio R, Spampanato J, Porter T, Holmes S, Young PR, Truneh A, Antibodies to TR2 (herpesvirus entry mediator), a new member of the TNF receptor superfamily, block T cell proliferation, expression of activation markers, and production of cytokines. Journal of immunology (Baltimore, Md. : 1950) 161, 1786–1794 (1998). [PubMed] [Google Scholar]
- 44.Liu W, Chou TF, Garrett-Thomson SC, Seo GY, Fedorov E, Ramagopal UA, Bonanno JB, Wang Q, Kim K, Garforth SJ, Kakugawa K, Cheroutre H, Kronenberg M, Almo SC, HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160. The Journal of experimental medicine 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinberg MW, Cheung TC, Ware CF, The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev 244, 169–187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D, Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nature immunology 14, 271–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, Attinger A, Shui JW, Kim G, Lena CJ, Sakaguchi S, Miyamoto C, Wang P, Atarashi K, Park Y, Nakayama T, Honda K, Ellmeier W, Kronenberg M, Taniuchi I, Cheroutre H, Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nature immunology 14, 281–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H, Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity 25, 631–641 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Fiala GJ, Schaffer AM, Merches K, Morath A, Swann J, Herr LA, Hils M, Esser C, Minguet S, Schamel WWA, Proximal Lck Promoter-Driven Cre Function Is Limited in Neonatal and Ineffective in Adult gammadelta T Cell Development. Journal of immunology 203, 569–579 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Fimia GM, Corazzari M, Antonioli M, Piacentini M, Ambra1 at the crossroad between autophagy and cell death. Oncogene 32, 3311–3318 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Che Z, Liu F, Zhang W, McGrath M, Hou D, Chen P, Song C, Yang D, Targeting CAND1 promotes caspase-8/RIP1-dependent apoptosis in liver cancer cells. Am J Transl Res 10, 1357–1372 (2018). [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts AI, Brolin RE, Ebert EC, Integrin alpha1beta1 (VLA-1) mediates adhesion of activated intraepithelial lymphocytes to collagen. Immunology 97, 679–685 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meharra EJ, Schon M, Hassett D, Parker C, Havran W, Gardner H, Reduced gut intraepithelial lymphocytes in VLA1 null mice. Cell Immunol 201, 1–5 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Mao M, Alavi MV, Labelle-Dumais C, Gould DB, Type IV Collagens and Basement Membrane Diseases: Cell Biology and Pathogenic Mechanisms. Curr Top Membr 76, 61–116 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Timpl R, Structure and biological activity of basement membrane proteins. Eur J Biochem 180, 487–502 (1989). [DOI] [PubMed] [Google Scholar]
- 56.Marsal J, Brakebusch C, Bungartz G, Fassler R, Agace WW, beta1 integrins are not required for the maintenance of lymphocytes within intestinal epithelia. European journal of immunology 35, 1805–1811 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Tan CL, Peluso MJ, Drijvers JM, Mera CM, Grande SM, Brown KE, Godec J, Freeman GJ, Sharpe AH, CD160 Stimulates CD8(+) T Cell Responses and Is Required for Optimal Protective Immunity to Listeria monocytogenes. Immunohorizons 2, 238–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadighi Akha AA, McDermott AJ, Theriot CM, Carlson PE Jr., Frank CR, McDonald RA, Falkowski NR, Bergin IL, Young VB, Huffnagle GB, Interleukin-22 and CD160 play additive roles in the host mucosal response to Clostridium difficile infection in mice. Immunology 144, 587–597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez-Santiago G, Robles-Valero J, Morlino G, Cruz-Adalia A, Perez-Martinez M, Zaldivar A, Torres-Torresano M, Chichon FJ, Sorrentino A, Pereiro E, Carrascosa JL, Megias D, Sorzano CO, Sanchez-Madrid F, Veiga E, Clathrin regulates lymphocyte migration by driving actin accumulation at the cellular leading edge. European journal of immunology 46, 2376–2387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, De Juan-Pardo E, Demeyer K, Hole K, Larrea E, Timmerman E, Prieto J, Arnesen T, Sherman F, Gevaert K, Aldabe R, N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proceedings of the National Academy of Sciences of the United States of America 109, 12449–12454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santiago MF, Costa MR, Mendez-Otero R, Immunoblockage of 9-O-acetyl GD3 ganglioside arrests the in vivo migration of cerebellar granule neurons. J Neurosci 24, 474–478 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, Zhang C, Zhou Z, Zhang J, Zhang J, Tian Z, Small intestinal intraepithelial lymphocytes expressing CD8 and T cell receptor gammadelta are involved in bacterial clearance during Salmonella enterica serovar Typhimurium infection. Infection and immunity 80, 565–574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edelblum KL, Singh G, Odenwald MA, Lingaraju A, El Bissati K, McLeod R, Sperling AI, Turner JR, gammadelta Intraepithelial Lymphocyte Migration Limits Transepithelial Pathogen Invasion and Systemic Disease in Mice. Gastroenterology 148, 1417–1426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umesaki Y, Setoyama H, Matsumoto S, Okada Y, Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology 79, 32–37 (1993). [PMC free article] [PubMed] [Google Scholar]
- 65.Bandeira A, Mota-Santos T, Itohara S, Degermann S, Heusser C, Tonegawa S, Coutinho A, Localization of gamma/delta T cells to the intestinal epithelium is independent of normal microbial colonization. The Journal of experimental medicine 172, 239–244 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohlund D, Franklin O, Lundberg E, Lundin C, Sund M, Type IV collagen stimulates pancreatic cancer cell proliferation, migration, and inhibits apoptosis through an autocrine loop. BMC Cancer 13, 154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burnier JV, Wang N, Michel RP, Hassanain M, Li S, Lu Y, Metrakos P, Antecka E, Burnier MN, Ponton A, Gallinger S, Brodt P, Type IV collagen-initiated signals provide survival and growth cues required for liver metastasis. Oncogene 30, 3766–3783 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Barczyk M, Carracedo S, Gullberg D, Integrins. Cell Tissue Res 339, 269–280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eble JA, Golbik R, Mann K, Kuhn K, The alpha 1 beta 1 integrin recognition site of the basement membrane collagen molecule [alpha 1(IV)]2 alpha 2(IV). EMBO J 12, 4795–4802 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray SJ, Franki SN, Pierce RH, Dimitrova S, Koteliansky V, Sprague AG, Doherty PC, de Fougerolles AR, Topham DJ, The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity 20, 167–179 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Gendron S, Couture J, Aoudjit F, Integrin alpha2beta1 inhibits Fas-mediated apoptosis in T lymphocytes by protein phosphatase 2A-dependent activation of the MAPK/ERK pathway. The Journal of biological chemistry 278, 48633–48643 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Chelberg MK, Tsilibary EC, Hauser AR, McCarthy JB, Type IV collagen-mediated melanoma cell adhesion and migration: involvement of multiple, distinct domains of the collagen molecule. Cancer research 49, 4796–4802 (1989). [PubMed] [Google Scholar]
- 73.Herbst TJ, McCarthy JB, Tsilibary EC, Furcht LT, Differential effects of laminin, intact type IV collagen, and specific domains of type IV collagen on endothelial cell adhesion and migration. J Cell Biol 106, 1365–1373 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tu TC, Brown NK, Kim TJ, Wroblewska J, Yang X, Guo X, Lee SH, Kumar V, Lee KM, Fu YX, CD160 is essential for NK-mediated IFN-gamma production. The Journal of experimental medicine 212, 415–429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK, Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity 16, 365–377 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF, Stabilized imaging of immune surveillance in the mouse lung. Nat Methods 8, 91–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R, Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmieder R, Edwards R, Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome S Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 85.Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, Chim N, Nuccio SP, Rathi SG, Mastroianni JR, Edwards RA, Jacobo CM, Cerasi M, Battistoni A, Ouellette AJ, Goulding CW, Chazin WJ, Skaar EP, Raffatellu M, Salmonella Mitigates Oxidative Stress and Thrives in the Inflamed Gut by Evading Calprotectin-Mediated Manganese Sequestration. Cell Host Microbe 19, 814–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD, Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity 71, 2839–2858 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the main text or the Supplementary Materials. All reasonable requests for materials including, mouse strains, will be available upon request to Mitchell Kronenberg (mitch@lji.org). Mouse strain # 030862 with a floxed allele of the gene encoding HVEM (B6;SJL-Tnfrsf14tm1.1Kro/J) has been deposited at the Jackson lab. RNA-seq data are available from GEO under accession code GSE206183.


