Abstract
Global DNA hypomethylation at CpG islands coupled with local hypermethylation is a hallmark for breast cancer, yet the mechanism underlying this change remains elusive. In this study, we showed that DNMT1, which encodes a methylation maintenance enzyme, is a transcriptional target of BRCA1. BRCA1 binds to the promoter of the DNMT1 gene through a potential OCT1 site and the binding is required for maintaining a transcriptional active configuration of the promoter in both mouse and human cells. We further demonstrated that impaired function of BRCA1 leads to global DNA hypomethylation, loss of genomic imprinting, and an open chromatin configuration in several types of tissues examined in a BRCA1 mutant mouse model at premaligant stages. BRCA1 deficiency is also associated with significantly increased expression levels of several protooncogenes, including c-Fos, Ha-Ras, and c-Myc, with a higher expression in tumors, while premalignant mammary epithelial cells displayed an intermediate state between tumors and controls. In human clinical samples, reduced expression of BRCA1 correlates with decreased levels of DNMT1, and reduced methylation of CpG islands. Thus, BRCA1 prevents global DNA hypomethylation through positively regulating DNMT1 expression, and this provides one of mechanisms for BRCA1-associated breast cancer formation.
Keywords: BRCA1, histone modification, DNA methylation, DNMT1, genomic imprinting, tumor formation
Introduction
DNA methylation is an epigenetic change that occurs in cells by adding a methyl group (CH3) to the 5′ position of cytidyl residues in the dinucleotide sequence CpG. Hypomethylation is usually associated with increased gene expression while hypermethylation is linked to gene silencing [1–4]. It has been shown that global DNA hypomethylation is increased in breast cancer, playing an important role in late stages of cancer development, while local promoter hypermethylation of tumor suppressor genes may be responsible for early stages of breast cancer [2, 4–6]. It was also reported that hypomethylation of juxtacentromeric satellite DNA and global DNA hypomethylation were common even in an early stage (i.e. stage-1) of breast cancer, arguing that hypomethylation can be an early event in breast carcinogenesis [7]. DNA is methylated by a group of DNA methyltransferases (DNMTs), including DNMT1, DNMT3a, DNMT3b, and DNMT3L, while DNMT2 is an inactivated isoform [3, 4, 8]. DNMT1 is also called methylation maintenance enzyme. At each cell division, DNMT1 completes the methylation pattern of each nascent replicating DNA strand, allowing maintenance of complete DNA methylation originally hemi-methylated on the template strand. DNMT3a and DNTM3b act as de novo methyltransferases and methylate unmethylated DNA. They are associated with the replication fork in late S phase during the replication of constitutive heterochromatin. DNMT3L is expressed during gametogenesis and establishes maternal genomic imprinting [3, 8]. Despite the important roles of DNMTs in breast cancer formation, it is poorly understood how expression of these factors is regulated.
Functions of DNMTs have been extensively studied in mice by using gene targeting. DNMT1-deficient mice lose about 90% of their DNA methylation and die early in embryogenesis [9]. Mouse embryonic stem (ES) cells deficient for DNMT1 or for both DNMT3a and DNMT3b have dramatically elongated telomeres compared with wild-type controls, highlighting a role for DNA methylation in maintaining telomere integrity [10]. Hypomorphic DNMT1 activity has been related to aggressive T-cell lymphomas in a mouse model showing a potential relationship between expression levels of this enzyme and cancer incidence, probably through regulation of genomic stability [11]. Interestingly, when a DNMT1 hypomorphic mutation was introduced into Apc (Min/+) mice, it suppressed later stages of intestinal tumorigenesis, but promoted early lesions in the colon and liver through a loss of heterozygosity (LOH) mechanism [12]. This observation suggests that DNA hypomethylation may play a dual role in tumorigenesis depending on different interacting environments.
Breast cancer-associated gene 1 (BRCA1) is the first identified breast tumor suppressor gene whose functions have been the subject of extensive research since its cloning [13]. Mounting evidence reveals that BRCA1 plays essential roles in many biological processes, including transcription regulation, cell cycle regulation, DNA damage repair, and centrosome duplication (reviewed in [14–17]). Animal models have confirmed its implication as a tumor suppressor gene [18, 19]. However, mouse embryos carrying BRCA1-null mutations die early in gestation, displaying proliferation defects and massive apoptosis [18, 20–22], and embryos carrying a homozygous deletion of Brca1 exon 11 (Brca1Δ11/Δ11), which encodes 60% of the amino acids of the protein, die at later stages of gestation because of widespread apoptosis [23]. Of note, haploinsufficiency of p53 (+/−) can suppress apoptosis in Brca1Δ11/Δ11 embryos and allow them to develop to adulthood. However, the survivors (Brca1Δ11/Δ11;p53+/−) exhibit abnormalities in multiple organs/tissues, and eventually die of premature aging and tumorigenesis [16, 17, 23–27]. The pleiotropic effect of BRCA1 deficiency correlates well with the fact that BRCA1 interacts with many proteins with important functions, and that BRCA1 acts as a transcription factor, which regulates expression of many genes involved in many biological processes [28, 29]. It was recently shown that methylation is less abundant in BRCA1-associated breast cancers compared with sporadic cases [30, 31]. However, it is unclear if BRCA1 deficiency is directly related to this DNA hypomethylation, and whether BRCA1 affects expression of genes involved in DNA methylation, considering that global hypomethylation is a general hallmark of most cancer cells.
Here, we investigate a role of BRCA1 in DNA methylation. We show both in vitro and in vivo that BRCA1 deficiency leads to global DNA hypomethylation and chromatin abnormalities related to cancer hallmarks. We also identify the mechanism related to this potential tumor predisposing phenotype: DNMT1, the methylation maintenance enzyme, is a transcriptional target of BRCA1.
Results
BRCA1 deficiency is associated with global DNA hypomethylation
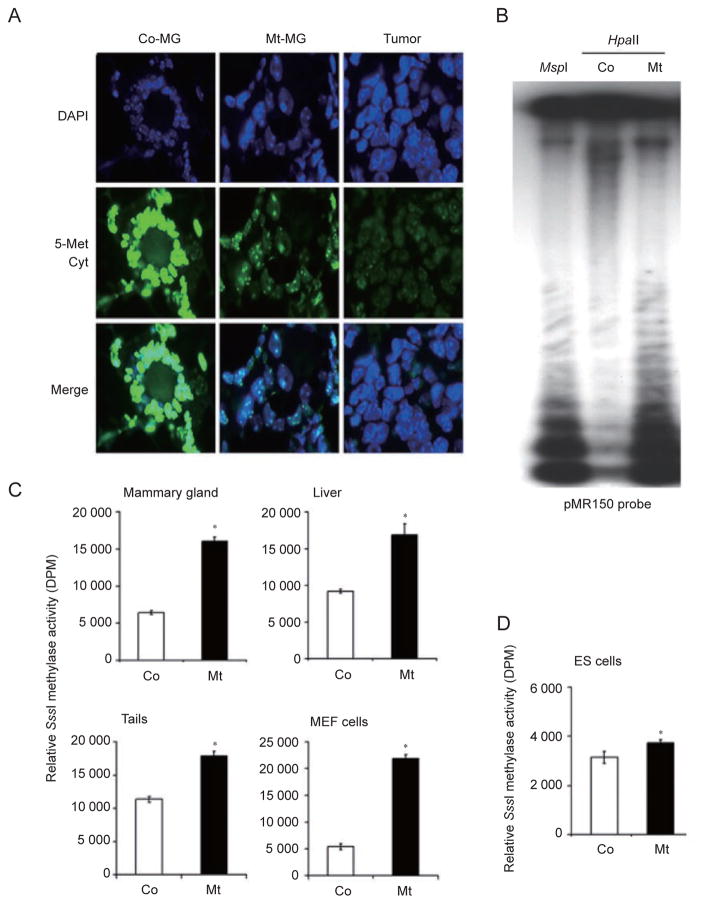
To investigate whether BRCA1 deficiency can affect DNA methylation, we stained mammary tissues isolated from Brca1Δ11/Δ11;p53+/− and Brca1+/+;p53+/− (referred as p53+/−) control mice, and mammary tumors developed in the Brca1Δ11/Δ11;p53+/− mice using an antibody against 5-methylcytosine, a marker of methylated DNA. We showed that Brca1Δ11/Δ11;p53+/− mammary tissues displayed a significant reduction in methylation compared to control tissues, while a further reduction was observed in mammary tumors (Figure 1A and Supplementary information, Figure S1). To provide an independent verification whether the staining of 5-methylcytosine indeed reflected the changes of DNA methylation, we isolated genomic DNA from tails of Brca1Δ11/Δ11;p53+/− and p53+/− mice, and digested it with the methylation-sensitive restriction enzyme, HpaII, followed by Southern blot analysis using a probe for minor satellite repeat (pMR150), which is highly methylated in normal cells [32]. We found that control genomic DNA was resistant to HpaII digestion, while Brca1Δ11/Δ11;p53+/− DNA displayed a similar digestion pattern comparable to MspI, a nonsensitive enzyme that recognizes the same site as HpaII. This data indicates that DNA isolated from Brca1Δ11/Δ11;p53+/− mice is significantly hypomethylated on minor satellite repeats (Figure 1B).
Figure 1.
Absence of BRCA1 leads to global DNA hypomethylation. (A) Methylation status of mammary gland tissues (MG) and mammary tumors isolated from Brca1+/+; p53+/− control (Co), and Brca1Δ11/Δ11;p53+/−-mutant (Mt) mice using an antibody against 5-methylcytosine. (B) Methylation analysis with a pMR150 probe using DNA digested with the methylation-sensitive restriction enzyme, HpaII, and methylation-insensitive restriction enzyme, MspI. (C) Methylation status of multiple tissues revealed by relative SssI methylase activity reflected by DPM. (D) SssI methylase activity in ES cells that carry an acute deletion of Brca1 exon 11 (Brca1Co/D;Cre-ERT2) using a 4-hydroxytamoxifen (4-HT)-inducible Cre-LoxP system. Those cells contain one mutant copy (D) and one conditional copy (Co) of Brca1. The Brca1Co allele can be deleted through 4-HT treatment, which activates an inducible recombinase, Cre-ERT2. Analysis was performed 4 days after 4-HT treatment, when about 90% of BRCA1 was deleted. * Represents P < 0.05 of Student’s t-test.
Next, we measured and quantified genomic DNA methylation using incorporation of a radioactive methyl group in naked genomic DNA using SssI methylase and radioactive substrate SAM, which catalyzes incorporation of a methyl group only into unmethylated DNA at the CpG repeats [33]. In the presence of SssI methylase, hypomethylated DNA can incorporate more radioactive methyl groups than hypermethylated DNA. We detected significantly higher methyl group incorporation in BRCA1 mutant mammary gland, liver, tail, and mouse embryonic fibroblasts (MEFs) compared with that of p53+/− controls (Figure 1C). A smaller, yet statistically significant, decrease of global DNA methylation was also observed in ES cells carrying a tamoxifen-inducible Cre-loxP-mediated acute deletion of BRCA1 (Brca1Co/D) (Figure 1D). The different level of DNA methylation between ES cells and tissues might reflect a difference between naive and differentiated cells or between chronic BRCA1 mutation and acute deletion, or both. Consistent with this view, a previous study indicated that DNA methylation level was much lower in DNMT1−/− ES cells after they were differentiated [32]. A similar gradually decreased methylation was also observed in ES cells that were defective for both DNMT3A and DNMT3B at different passages [34], that is, the double mutant cells (DNMT3aa/bb) exhibited a comparable degree of methylation at passage five with wild-type cells, and the difference became significant at P15 and more profound at P40. Thus, our work uncovers a novel finding that BRCA1 deficiency causes a progressive trend of DNA hypomethylation compared with wild-type cells in the following order: control < mutant ES cells < mutant differentiated somatic cells (tissues) < tumors.
BRCA1 deficiency affects posttranscriptional patterns of histone H3 and H4
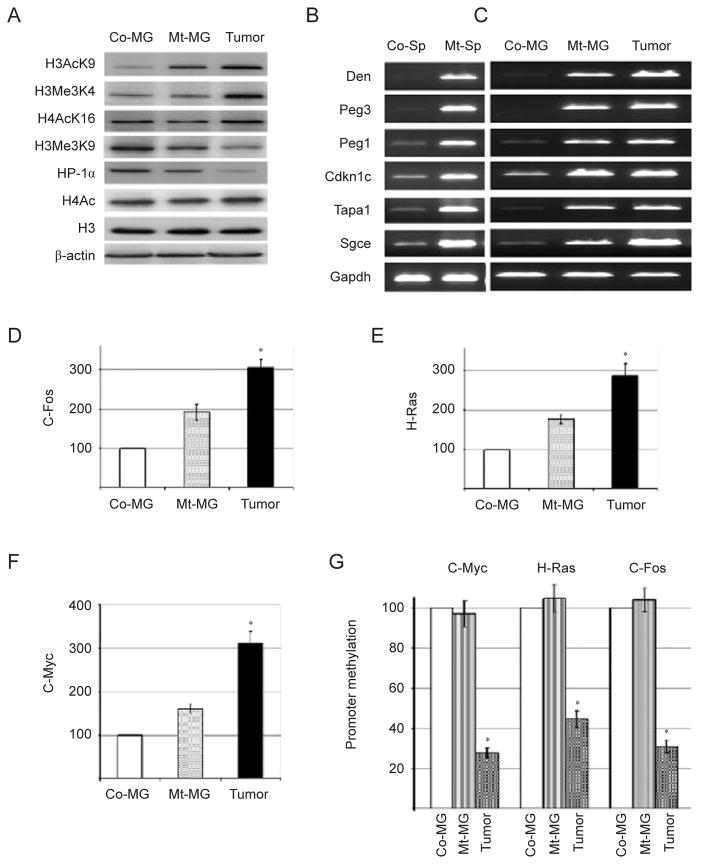
DNA methylation is often related to posttranslational modification of histones [35]. Next, we checked whether BRCA1 deficiency triggered the posttranscriptional modifications that occur on histones H3 and H4, in mammary gland extracts from control and BRCA1-mutant mice. Acetylation of H3-lysine9 (H3AcK9), H4AcK16, and trimethylation of H3-K4 (H3Me3K4) are hallmarks of transcriptionally active chromatin [36]. We showed that all these modifications of transcriptionally active chromatin were significantly upregulated in Brca1Δ11/Δ11;p53+/− mammary tumors, and levels of H3AcK9 and H3Me3K4 were slightly increased in the premalignant Brca1Δ11/Δ11;p53+/− mammary tissues compared with control mammary tissues (Figure 2A and Supplementary information, Figure S2). On the other hand, trimethylation of H3-K9 (H3Me3K9), a modification related to transcriptional repression, was significantly decreased in BRCA1 mutant mammary tissues and tumors. HP-1α (a heterochromatin marker) expression also significantly decreased in Brca1Δ11/Δ11;p53+/− samples (Figure 2A and Supplementary information, Figure S2). These results indicate that lack of BRCA1 affects several DNA epigenetic modifications and results in an open chromatin configuration, which is more prone to genomic instability.
Figure 2.
Absence of BRCA1 favors open chromatin structure and increases expression of protooncogenes. (A) Altered epigenetic modifications in BRCA1-mutant mammary gland and mammary tumors revealed by western blots. (B, C) BRCA1 deficiency causes abnormal expression of genes involved in genomic imprinting in spermatocytes (Sp) (B) and mammary tissue and tumor (C) revealed by RT-PCR. (D–F) Relative expression of protooncogenes C-Fos (D), Ha-Ras (E), and C-Myc (F) in BRCA1-mutant mammary gland (MG) and mammary tumors revealed by real-time PCR. (G) Semiquantitative analysis of promoter methylation of protooncogenes by methylation-specific PCR. * Represents P < 0.05 of Student’s t-test.
BRCA1 deficiency reduces genomic imprinting
In mammals, genomic imprinting is a physiological phenomenon responsible for functional inequality between two parental alleles of a gene [37]. Two major mechanisms are involved in establishing the imprint, that is, DNA methylation and histone modifications [38]. Because loss of BRCA1 caused abnormalities in both processes, we checked the expression of several imprinting genes including maternally (Peg3, Sgce, and Peg1) and paternally (Cdkn1c, Dcn, and Tapa1) regulated genes. Analyzing RNA extracted from spleen of Brca1Δ11/Δ11;p53+/− mice by RT-PCR, we detected a gradual upregulation of these genes during the first wave of spermatogenesis in Brca1Δ11/Δ11;p53+/− mice (Figure 2B). We also found that all the tested imprinting genes were overexpressed in mammary tissues of mutant mice compared with controls, and the expression levels were even higher in mammary tumors (Figure 2C). These data indicate that absence of BRCA1 reduces genomic imprinting.
BRCA1 deficiency leads to increased expression of several protooncogenes
Considering the influence of epigenetic changes on gene regulation and the consequence of BRCA1 deficiency in triggering tumorigenesis, we determined expression levels of several protooncogenes in the mammary gland of Brca1Δ11/Δ11;p53+/− mice. We found that BRCA1 deficiency was associated with significantly increased expression levels of c-Fos, Ha-Ras, and c-Myc in Brca1Δ11/Δ11;p53+/− samples, with a higher expression in tumors, while premalignant mutant tissue displayed an intermediate state between tumors and controls (Figure 2D–2F). We next examined the promoter of c-Fos, Ha-Ras, and c-Myc using methylation-sensitive semiquantitative PCR. We detected a marked reduction of promoter methylation of c-Myc, Ha-Ras, and c-Fos in BRCA1-mutant tumors compared with control mammary tissues (Figure 2G), which is consistent with the high level of expression of these genes. Of note, our semiquantitative PCR detected no change of promoter methylation between control and mutant mammary tissues, although increased gene expression was observed (Figure 2D–2F). This observation suggests that in addition to methylation, some other factors may also contribute to altered gene expression associated with BRCA1 deficiency. In this case, we have demonstrated that loss of BRCA1 also changes histone code, which could be one of the other factors involved in gene regulation. Altogether, these results suggest that chromatin methylation and several posttranslational histone modifications are affected in BRCA1-deficient cells or organs, resulting in deregulation of gene expression including protooncogenes.
BRCA1 regulates expression of the DNMT1 gene
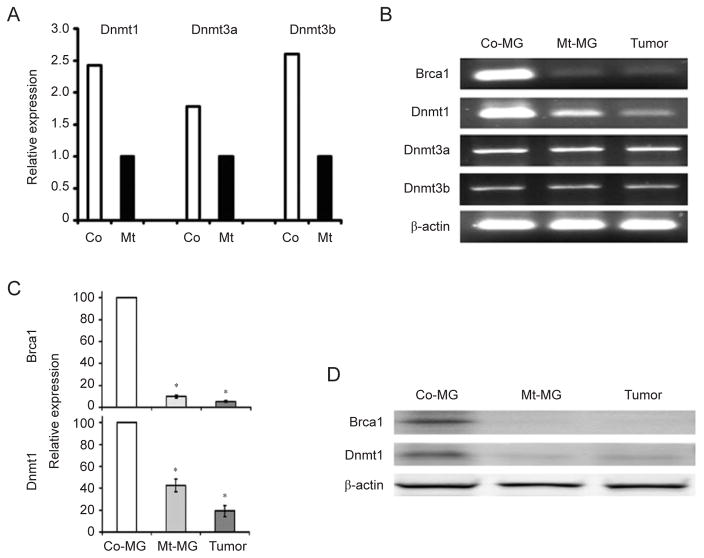
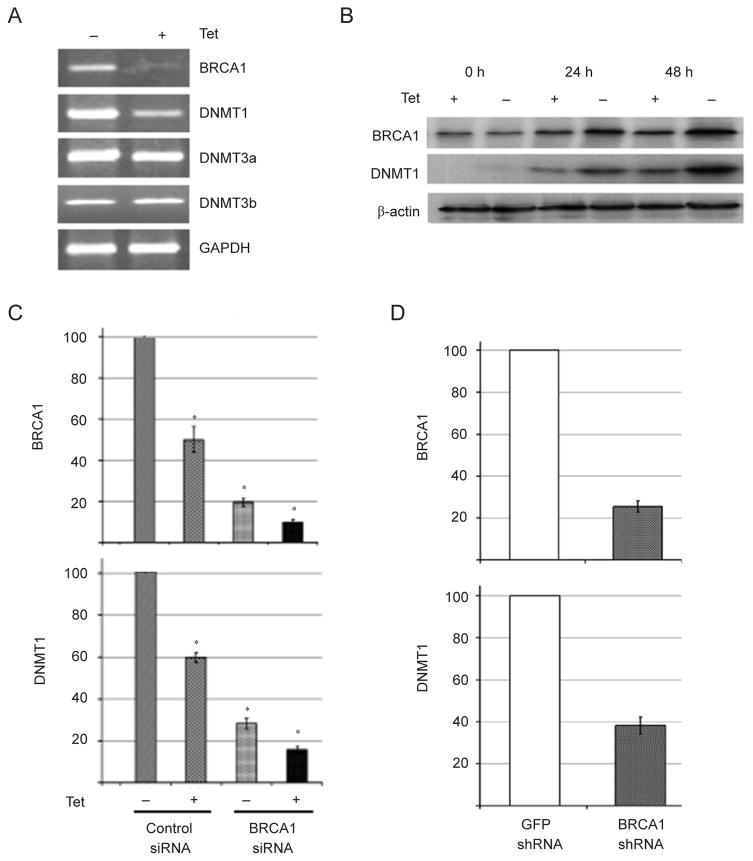
The global hypomethylation observed in BRCA1 mutant tissues/cells prompted us to hypothesize that BRCA1 might regulate expression of DNMT genes. Analyzing our previously published microarray data obtained from Brca1Δ11/Δ11;p53+/− liver samples [39], we found that Dn-mt1, Dnmt3a, and Dnmt3b mRNAs were downregulated in Brca1Δ11/Δ11;p53+/− samples compared with p53+/− controls, while no significant changes were detected for Dn-mt3L (Figure 3A). This expression pattern was confirmed by our RT-PCR analysis (Supplementary information, Figure S3). We then performed an analysis of Dnmt expression in mammary tissues through RT-PCR and quantitative real-time PCR experiments, but we only detected reduced Dnmt1 mRNA expression in Brca1Δ11/Δ11;p53+/− mammary tissue, but not Dnmt3a and Dnmt3b expression (Figure 3B and 3C). This might result from differential tissue context between the liver and the mammary gland. Considering that the DNMT1 enzymatic activity relies on protein expression, a critical feature related to DNA methylation maintenance, we performed a western blot analysis and confirmed that DNMT1 protein level dramatically decreased in Brca1Δ11/Δ11;p53+/− mammary gland samples (Figure 3D). Next, we investigated the link between BRCA1 and DNMT1 expression in human samples. We chose human UBR60 cells where BRCA1 levels could be increased using a tetracycline (Tet)-off system [40]. UBR60 cells cultured with Tet express basal endogenous levels of BRCA1 while Tet withdrawal induces expression of a transgenic BRCA1. We showed that overexpression of BRCA1 was correlated with increased DNMT1 mRNA expression while DNMT3a and DNMT3b levels remained unchanged (Figure 4A). The induction of DNMT1 by BRCA1, but not DNMT3a and DNMT3b (data not shown), was also confirmed at the protein level by western blot analysis (Figure 4B).
Figure 3.
Expression of DNMT1 in liver and mammary glands. (A) Gene expression revealed by microarray analysis of liver tissue samples. (B–D) Gene expression revealed by RT-PCR (B), real-time PCR (C), and western blot (D) of samples isolated from mammary glands and mammary tumors. * Represents P < 0.05 of Student’s t-test.
Figure 4.
Analysis of DNMT1 expression in vitro. (A) RT-PCR analysis of Tet regulated expression of BRCA1 and DNMTs in UBR-60 cells. (B) Western blot analysis of Tet-regulated BRCA1 and DNMT1 expression in UBR-60 cells. (C) Real-time PCR analysis of BRCA1 and DNMT1 expression in UBR-60 cells after downregulation of BRCA1 by RNAi compared with scrambled RNAi. (D) Real-time PCR analysis of BRCA1 and DNMT1 expression from a murine mammary tumor cell line derived from MMTV-cNeu transgenic mice after downregulation of BRCA1 by shRNA compared with shRNA against GFP. * Represents P < 0.05 of Student’s t-test.
In addition, instead of increasing BRCA1 level, we acutely depleted it in UBR60 cells using siRNA. We showed that siRNA specific to BRCA1 decreased both BRCA1 and DNMT1 mRNA levels by about 4- to 5-fold in both conditions with or without Tet (Figure 4C). Moreover, we were able to reproduce this effect on both endogenous BRCA1 and DNMT1 in other cell lines, including murine mammary tumors cells derived from MMTV-cNeu mice (Figure 4D), estrogen receptor-positive human breast cancer MCF-7 cells and estrogen receptor-negative BT-20 human breast cancer cells using control- and BRCA1-siRNA (Supplementary information, Figure S4).
BRCA1 regulates DNMT1 promoter activity
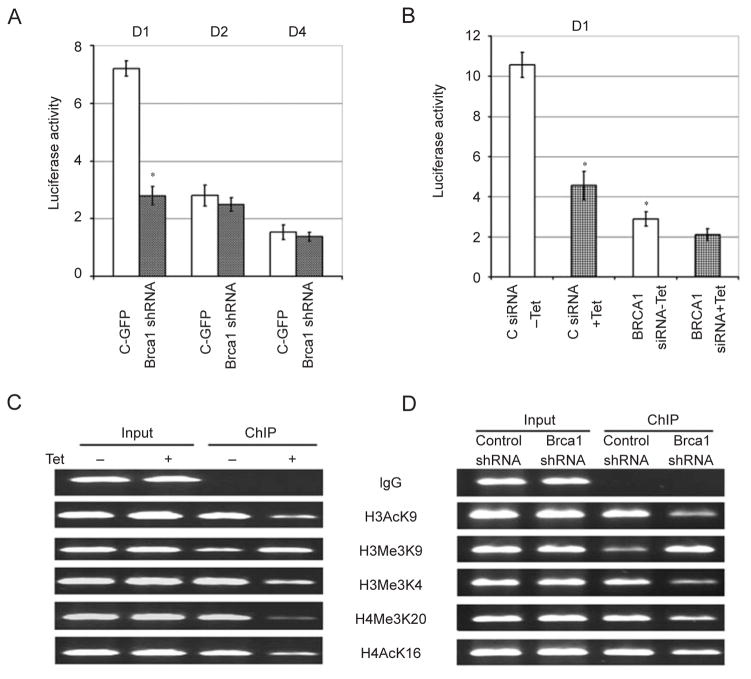
Next, we were interested in understanding how BR-CA1 regulates DNMT1 mRNA and protein levels. We first transfected three Dnmt1 promoter luciferase reporters of varying lengths ranging between 2 079 bp (D1), 299 bp (D2), and 199 bp (D4) (Supplementary information, Figure S5A) [41] into cultured murine c ells to check whether they have reporter activity. We found that all three constructs displayed higher luciferase activity compared with the pGL3 vector, and that the activity of the longest construct, D1, is the highest (Figure 5A). We also found that transfection of a Brca1-specific shRNA, but not a control shRNA (against GFP), significantly reduced the D1 reporter activity while it had very little effect on the D2 and D4 reporters (Figure 5A), suggesting that the D1 fragment bears a BRCA1-dependent regulatory region.
Figure 5.
BRCA1 regulates DNMT1 expression on its promoter. (A) Activities of three Dnmt1 luciferase reporters (D1, D2, and D4) in relation to pGL3 vector, which were set at 1, in neu cells upon shRNA-mediated downregulation of BRCA1. (B) Luciferase activity of D1 reporter in Tet-regulated UBR-60 cells. (C, D) ChIP analysis of histone modifications of the human (C) and mouse (D) DNMT1 promoter. * Represents P < 0.05 of Student’s t-test.
Next, we performed the promoter reporter assay in human UBR60 cells. Our data indicates that induction of BRCA1 by tetracycline withdrawal increased D1 activity in control siRNA-treated cells while BRCA1 siRNA reduced D1 basal activity and tetracycline-dependent induction (Figure 5B). Induction of BRCA1 upon tetracycline withdrawal did not have obvious effects on the luciferase activities of D2 and D4 constructs (Supplementary information, Figure S5B and S5C). This observation that the human BRCA1 can regulate the mouse Dnmt1 promoter suggests that regulation of the DNMT1 promoter by BRCA1 is evolutionarily conserved.
BRCA1 binds to the DNMT1 promoter and converts it to a transcriptionally active configuration
We then performed chromatin immunoprecipitation (ChIP) experiments to investigate whether BRCA1 binds to the human and murine DNMT1 promoters. PCR assays using primers that cover five fragments in the 5′ regulatory region of the endogenous DNMT1 promoter in UBR60 cells showed that BRCA1 bound to a fragment from −2 440 to −2 843 (403 bp) upstream of the translation start site (ATG), but not to the other regions in the human DNMT1 promoter in both the presence of Tet (expression of endogenous BRCA1) and the absence of Tet (expression of both endogenous and transgenic BRCA1), with a higher intensity in the latter condition (Supplementary information, Figure S6A). We have also performed a similar experiment with the mouse Dnmt1 promoter and found BRCA1 bound to a fragment from −7 to −506 (499 bp) upstream of the start codon (Supplementary information, Figure S6B).
Next, we performed ChIP analysis to examine histone modifications in the DNMT1 promoter (−2 440 to −2 843) using antibodies specific to H3AcK9, H3Me3K9, H3Me3K4, H4Me3K20, and H4AcK16. We observed a marked increase in human DNMT1 promoter occupancy, as reflected by increased levels of transcriptionally active histone marks (H3AcK9, H3Me3K4, H4Me3K20, H4AcK16) and decreased levels of transcriptionally inactive histone marks (H3Me3K9) when BRCA1 was induced in UBR60 cells (Figure 5C and Supplementary information, Figure S7A). Similar results were observed with the mouse Dnmt1 promoter (−7 to −506) when BRCA1 was knocked down in neu cells by shBrca1 compared with control shRNA (Figure 5D and Supplementary information, Figure S7B). These data suggest that BRCA1 binds to and causes transcriptionally active configuration of the DNMT1 promoter, both in humans and mice.
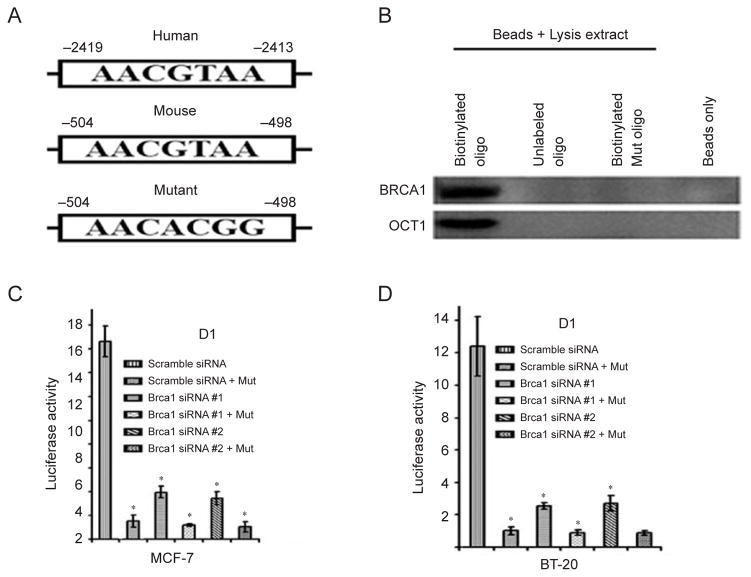
BRCA1 binds to the DNMT1 promoter through a potential OCT1 site in both human and mouse cells
We then compared the corresponding regions of both the mouse and human DNMT1 promoter using Genomatix software, which allows identification of putative transcription factor-binding sites. We identified a common motif (AACGTAA) in both mouse and human fragments that are potentially responsive to OCT1, a well-studied transcription factor (Figure 6A). Previous investigations indicated that BRCA1 and OCT1 interact with each other and regulate downstream gene expression through binding to the OCT1 consensus sequence [42], and Oct1 deficiency impairs transcriptional activity of BRCA1 [43]. Next, we performed following experiments to investigate whether the regulation of DNMT1 by BR-CA1 is indeed mediated by the OCT1 consensus site. We first performed a biotinylated oligonucleotide pull-down experiment and found that both BRCA1 and OCT1 interacted with the OCT1-responsive element, as point mutation in the consensus site abolished the binding of both BRCA1 and OCT1 (Figure 6B). Next, we performed human DNMT1 promoter reporter assays comparing mutated D1 and wild-type D1 fragments upon siRNA-mediated knockdown of BRCA1 in human MCF-7 (Figure 6C) and BT-20 cells (Figure 6D). Brca1 knockdown (using two Brca1 siRNA targeting two different sequences on Brca1 mRNA) significantly reduced luciferase activity in both cell lines. Moreover, mutation of the OCT1-binding site significantly reduced basal luciferase activity, while the promoter becomes insensible to BRCA1 knockdown. These results clearly show that the OCT1 binding site is critical to BRCA1 transcriptional regulation of DNMT1.
Figure 6.
BRCA1 regulates DNMT1 through the OCT1-binding site on the promoter of DNMT1. (A) Both the human and mouse DNMT1 promoter contain a common motif (AACGTAA) for OCT1. (B) Biotinylated oligo pull-down assay showing that BRCA1 and OCT1 bind to wild-type oligo but not to mutant oligo. (C, D) Reduced DNMT1 luciferase activity in MCF-7 (C) and BT-20 (D) cells after mutation of the OCT1-binding site in the D1 reporter. * Represents P < 0.05 of Student’s t-test.
Levels of global DNA methylation positively correlate with expression levels of BRCA1 and DNMT1 in human clinical samples
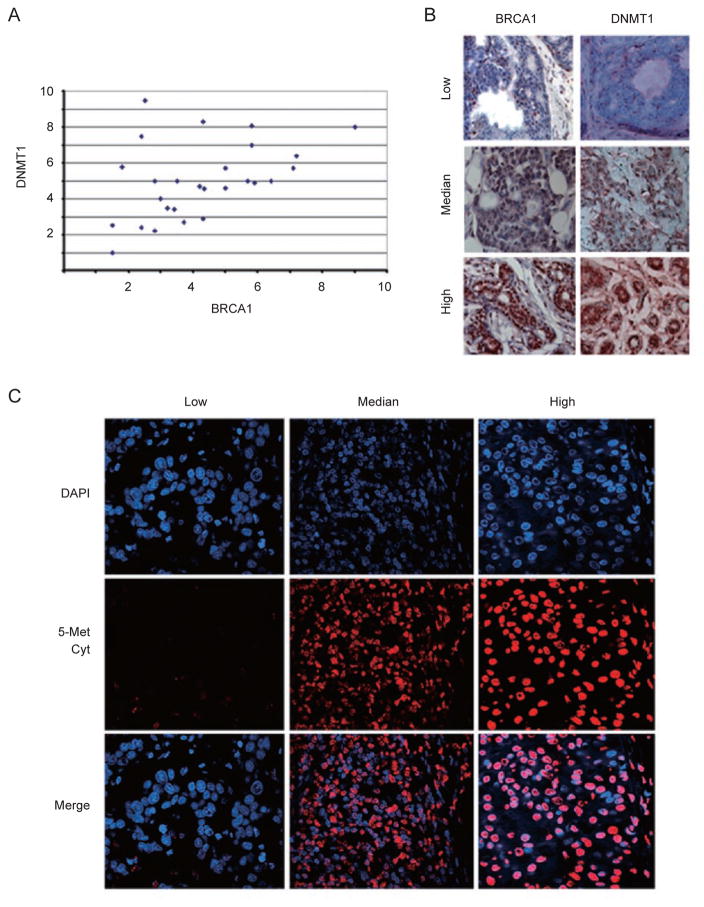
To provide clinical relevance regarding relationship between BRCA1 and DNMT1, we blindly studied levels of these proteins in 31 human breast cancers by immunohistochemical staining using antibodies against BRCA1 and DNMT1. Our data revealed a positive correlation between BRCA1 and DNMT1 expression in these cancer samples (Figure 7A). Examples of BRCA1 and DNMT1 expression levels from negative to high are shown in Figure 7B. This data is consistent with our finding that BR-CA1 positively regulates DNMT1 expression. Reduced level of DNMT1 is predicted to impair DNA methylation. To investigate whether this is the case in this set of samples, we studied their global DNA methylation using an antibody against 5-methylcytosine. Our data revealed that the intensity of methylation in these samples (Figure 7C and Supplementary information, Figure S8) correlated well with expression levels of BRCA1 and DNMT1 shown in Figure 7A, that is, when levels of BRCA1 and DNMT1 were low, the antibody staining was low, while when BRCA1 and DNMT1 were high, the staining intensity was high. Together with the data shown earlier, our study demonstrates that BRCA1 affects global DNA methylation through its positive regulation of DNMT1 in breast tissue and cancer.
Figure 7.
Expression of DNMT1 and BRCA1 in human primary breast cancers. (A) BRCA1 and DNMT1 protein levels in 31 human sporadic breast cancers. Immunohistochemical staining of BRCA1 and DNMT1 protein levels were detected by using antibodies against BRCA1 and DNMT1. Intensity of the staining was scored by division into 10 arbitrary units based on the graded intensities. The x axis and y axis are for DNMT1 and BRCA1 levels, respectively. (B) Examples of immunohistochemical staining showing positive correlation between expression levels of BRCA1 and DNMT1 in human breast cancers. (C) Immunofluorescent images of varying methylation levels after 5-methyl cytosine staining, which was also proportional to levels of BRCA1 staining in all analyzed samples.
Discussion
In this study, we investigated the mechanism underlying how BRCA1 deficiency results in reduced global DNA methylation. Our analyses of cultured cells, animal tissues, and cancers revealed that absence of BRCA1 causes a marked decrease of global CpG methylation, loss of imprinting, alteration of histone modification, and hypomethylation of a number of protooncogene promoters. We further demonstrated that BRCA1 is a positive regulator of DNMT1 transcription. As DNMT1 is a potent methylation maintenance enzyme [3, 4, 8], our study uncovers an important role of BRCA1 in maintaining proper methylation status through regulation of DNMT1 expression.
BRCA1 is a well-established transcription factor because it regulates expression of many genes, some of which include p21 [44], Gadd45 [40, 45], Mad2 [42], Sirt1 [46], and Igf axis members [39]. The C-terminal region of BRCA1 contains two BRCT domains that interact with multiple transcription activators and corepressors. Recently, the BRCA1 RING finger domain has been found to be involved in critical interactions with another RING domain-containing protein called BARD1. The BRCA1-BARD1 complex has an E3-ubiquitin ligase activity that might be critical to ensure BRCA1 protective functions [13, 28, 47]. Here, we show that BRCA1 and OCT1 bind to the promoter of DNMT1 through a potential OCT1-binding site, AACGTAA, which is identical in both the mouse and human DNMT1 promoter.
BRCA1 plays an essential role in maintaining genome integrity, as its absence results in profound chromosome abnormalities [14, 15, 29, 48]. Genomic stability highly depends on DNA structure, which is called chromatin during interphase [49, 50]. Since its discovery, chromatin has been intensively studied and the first microscopic observations identified eu- and heterochromatin [49–51]. While the euchromatin is the predominant site of active transcription, heterochromatin contains “inactive” DNA, which is very compacted with the presence of highly methylated DNA, notably in untranscribed regions [49–51]. Hypomethylation of those regions has been related to genomic instability, aging, and tumorigenesis [11, 52, 53]. Moreover, changes in DNA methylation levels have also been related to transcriptional regulation. The human genome conta ins 29 000 CpG islands and almost half of gene promoters contain those particular CG-rich domains that, in the case of high methylation, leads to transcriptional inactivity of these regions, which when specifically occurring in tumor suppressor genes, drives tumorigenesis [54, 55]. On the other hand, hypomethylation of CpG islands in the promoter of oncogenes, which results in transcriptional activation, could also promote tumorigenesis.
The status of DNA methylation is primarily maintained by DNMT1, although recent results have shown that both DNMT3a and DNMT3b could also play a minor role in this process through completion of the few “holes” missed by DNMT1. It has been shown that DNMT1 deficiency in mice causes profound DNA hypomethylation that compromises embryogenesis [56], highlighting an essential role of DNMT1 in mammalian development. Severely reduced DNMT1 activity (to about 10% of wild-type level) was also reported to cause abnormal telomere recombination, aggressive T-cell lymphomas, and early lesions in the colon and liver, probably through increased genomic instability [11, 12]. Complete knockout of DNMT1 in the human colorectal carcinoma cell line HCT116 resulted in mitotic catastrophe and genetic instability [57]. Our finding that BRCA1 regulates expression of DNMT1 is important in that it links many of BRCA1 functions to DNA methylation. We showed previously that BRCA1 deficiency in mice causes X-chromosome inactivation during spermatogenesis because of an unknown mechanism [58]. We also show here that the absence of BRCA1 causes reduced genomic imprinting. As both events are primarily mediated by hypermethylation of CpG islands, reduced DN-MT1 expression provides a reasonable explanation for these events.
In addition, BRCA1 plays many other important roles, including transcription, DNA damage repair, cell cycle regulation, centrosome duplication, spindle checkpoint control, and tumor suppression [14, 15, 29, 48, 59–61].
Most of those events may have a direct or indirect link to global DNA hypomethylation. It was recently shown that the overall frequency of CpG island promoter methylation is less abundant in BRCA1-associated breast cancers compared with sporadic cases [30, 31]. These studies, which used quantitative multiplex methylation-specific PCR, were restricted to a dozen of genes (10 and 11, respectively) involved in breast carcinogenesis. They notably showed that women with BRCA1/2 mutations had a lower frequency of CpG island promoter methylation. In contrast to those studies focusing mainly on tumor suppressors genes, we show that BRCA1 samples display a higher expression of several protooncogenes associated with a global decrease in their promoter methylation restricted to the tumor samples. In the light of our findings, we believe that this could be attributed, at least in part, to decreased DNMT1 activity. Our data demonstrated that in about 30 sporadic breast cancers examined, BRCA1 expression levels positively correlate with levels of DNMT1 and global methylation status. Decreased CpG methylation of regulatory regions in promoters due to DNMT1 deficiency may cause a catastrophic effect on cell viability and genomic stability, which are similarly observed in BRCA1-deficient cells [29, 56, 57]. On the other hand, the genetic instability coupled with decreased CpG methylation may eventually cause inactivation of tumor suppressor genes or activation of protooncogenes, events that are gradually selected by proliferating cells, which in turn enriches for favorable alterations for tumor maintenance and progression.
Our work raises several important questions on BR-CA1-dependant pathogenesis and potential preventive issues. We have positively linked BRCA1 and DNMT1 expression. Women, who have a BRCA1 mutation in one allele, have probably lower levels of BRCA1 protein in their mammary epithelium, and consequently, might express less DNMT1 enzyme. While DNMT1 complete deficiency is lethal, hypomorphic DNMT1 activity results in substantial genome-wide hypomethylation in all tissues and development of aggressive T-cell lymphomas [11, 56]. Thus, women with only one remaining BRCA1 allele are at risk of reduced DNMT1 levels, and subsequent higher DNA hypomethylation and genomic instability. The genomic instability should, in theory, facilitate further genetic alterations including losing the second functional BRCA1 allele, increasing risk of breast and/or ovarian tumorigenesis (model in Supplementary information, Figure S9). Alternatively, reduced DNMT1 levels could also cause CpG islands hypermethylation of BRCA1 promoter, leading to its inactivation. It would be interesting to evaluate DNMT1 levels, methylation status, and genomic instability in BRCA1 mutant allele carriers at different ages to see whether this is the case. Moreover, this could open a preventive window considering the dependency of DNMT1 on methyl donors to stabilize the DNA. Supplementation with methyl donor substrates might represent a preventive way to increase DNMT1 activity and to maintain the normal methylation levels in BRCA1 carriers.
Materials and methods
Mice, MEFs, and cell lines
Brca1Δ11/Δ11;p53+/− mice were generated as described [23]. MEF cells were derived from E14.5 embryos generated from intercrosses of Brca1+/311 mice. MEFs were cultured in DMEM supplemented with 10% FBS. BRCA1 inducible knockout ES cells contain only one copy of functional BRCA1 that can be deleted through 4-hydroxytamoxifen (4-HT) treatment by activation of an inducible recombinase, Cre-ERT2 [62].
Total RNA preparation and RT-PCR
Total RNA extraction was performed using RNA stat 60 (Tel-Test Inc). Reverse transcription was performed using the Cells-to-cDNA II Kit (Ambion Inc.). PCR reactions were performed using the Taq polymerase from GeneChoice following a regular program. All primers used in this study are listed in the Supplementary information, Table S1.
Western blot analysis
Western blot analysis was performed following a regular ECL procedure kit (Amersham Biosciences). Abcam antibodies were used for di- and tri-methyl histone H3 (Abcam Inc., MA, USA), Upstate antibodies for other proteins (Upstate, Lake Placid, NY, USA). Membranes were stripped and reblotted again for a maximum of three times.
Global methylation experiments
After purification from cells or tissues, 200 ng of DNA is combined with 4U of SssI methylase (New England Biolabs Inc.), 1.5 μM S-adenosyl-L-(methyl-3H)methionine (Amersham Pharmacia) and 1.5 μM nonradioactive S-adenosylmethionine (+ 1× SssI buffer). The reactions were incubated at 37 °C for 4 h, allowing incorporation of radioactive methyl group only in hypomethylated DNA. The reactions were stopped by adding 25 μl of 1 mM nonradioactive SAM and filtered using Millipore Ultrafree-MC 100.000 NMWL filter units according to manufacturer’s protocol. The units were then transferred to a scintillation vial containing 10 ml of scintillation cocktail and a liquid scintillation counter was used to measure radioactivity.
Southern blot analysis
Genomic DNA isolated from various cell lines was digested with methylation-sensitive restriction enzymes (HpaII: sensitive; MspI: nonsensitive) and analyzed by Southern hybridization as previously described [32]. The probe used for methylation analysis is pMR150 for minor satellite repeats (accession no. X14469 and accession no. X07949).
Immunohistochemistry
Normal mammary glands or mammary tumors were harvested and fixed with 4% paraformaldehyde. Slides were stained with a primary antibody against 5′-methylcytosine (a marker of DNA methylation) from Abcam overnight followed by staining with the HistoMouse-SP Broad spectrum kit from Zymed Laboratories Inc. BRCA1 antibody was from Upstate and DNMT1 antibody (ab19905) was from Abcam.
Biotin pull-down assay
Biotin pull-down assay was carried out as described in [42]. Briefly, nuclear extract was made from UBR60 cells. The extracts were incubated with biotin-labeled wild-type or Dnmt1 mutant double-stranded oligos. Streptavidin-sepharose beads (Sigma) were used to pull down biotin-associated complex.
Supplementary Material
Acknowledgments
I thank Dr E Li for providing the probe for Southern blots and members of Deng lab for their critical reading and discussion. This work was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA(C-X D), and CA79892, CA90631, and Susan G Komen Foundation Grant (TO).
Footnotes
Supplementary information is linked to the online version of the paper on the Cell Research website.
References
- 1.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20:711–721. doi: 10.1038/modpathol.3800822. [DOI] [PubMed] [Google Scholar]
- 3.Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006;53:245–256. [PubMed] [Google Scholar]
- 4.Lafon-Hughes L, Di Tomaso MV, Mendez-Acuna L, Martinez-Lopez W. Chromatin-remodelling mechanisms in cancer. Mutat Res. 2008;658:191–214. doi: 10.1016/j.mrrev.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003;532:39–49. doi: 10.1007/978-1-4615-0081-0_5. [DOI] [PubMed] [Google Scholar]
- 6.Soares J, Pinto AE, Cunha CV, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- 7.Jackson K, Yu MC, Arakawa K, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 8.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 9.Bonfini L, Karlovich CA, Dasgupta C, Banerjee U. The son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 11.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 12.Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 14.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 15.Zheng L, Li S, Boyer TG, Lee WH. Lessons learned from BRCA1 and BRCA2. Oncogene. 2000;19:6159–6175. doi: 10.1038/sj.onc.1203968. [DOI] [PubMed] [Google Scholar]
- 16.Cao L, Kim S, Xiao C, et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 2006;25:2167–2177. doi: 10.1038/sj.emboj.7601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachelier R, Xu X, Wang X, et al. Normal lymphocyte development and thymic lymphoma formation in Brca1 exon-11-deficient mice. Oncogene. 2003;22:528–537. doi: 10.1038/sj.onc.1206208. [DOI] [PubMed] [Google Scholar]
- 18.Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci USA. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng CX. Tumor formation in Brca1 conditional mutant mice. Envron Mol Mutagen. 2002;39:171–177. doi: 10.1002/em.10069. [DOI] [PubMed] [Google Scholar]
- 20.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 21.Hakem R, de la Pompa JL, Sirard C, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 22.Shen SX, Weaver Z, Xu X, et al. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, Qiao W, Linke SP, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Aprelikova O, Moens P, Deng CX, Furth PA. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- 25.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Xiao C, Vonderhaar BK, Deng CX. A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene. 2007;26:7204–7212. doi: 10.1038/sj.onc.1210527. [DOI] [PubMed] [Google Scholar]
- 27.Cao L, Xu X, Cao LL, et al. Absence of full-length Brca1 sensitizes mice to oxidative stress and carcinogen-induced tumorigenesis in the esophagus and forestomach. Carcinogenesis. 2007;28:1401–1407. doi: 10.1093/carcin/bgm060. [DOI] [PubMed] [Google Scholar]
- 28.Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suijkerbuijk KP, Fackler MJ, Sukumar S, et al. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann Oncol. 2008;19:1870–1874. doi: 10.1093/annonc/mdn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasilatos SN, Broadwater G, Barry WT, et al. CpG island tumor suppressor promoter methylation in non-BRCA-associated early mammary carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2009;18:901–914. doi: 10.1158/1055-9965.EPI-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei H, Oh SP, Okano M, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 33.Watson RE, Curtin GM, Doolittle DJ, Goodman JI. Progressive alterations in global and GC-rich DNA methylation during tumorigenesis. Toxicol Sci. 2003;75:289–299. doi: 10.1093/toxsci/kfg190. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 36.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Sha K. A mechanistic view of genomic imprinting. Annu Rev Genomics Hum Genet. 2008;9:197–216. doi: 10.1146/annurev.genom.122007.110031. [DOI] [PubMed] [Google Scholar]
- 38.Suetake I, Hayata D, Tajima S. The amino-terminus of mouse DNA methyltransferase 1 forms an independent domain and binds to DNA with the sequence involving PCNA binding motif. J Biochem. 2006;140:763–776. doi: 10.1093/jb/mvj210. [DOI] [PubMed] [Google Scholar]
- 39.Shukla V, Coumoul X, Cao L, et al. Absence of the full-length breast cancer-associated gene-1 leads to increased expression of insulin-like growth factor signaling axis members. Cancer Res. 2006;66:7151–7157. doi: 10.1158/0008-5472.CAN-05-4570. [DOI] [PubMed] [Google Scholar]
- 40.Harkin DP, Bean JM, Miklos D, et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 41.Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 2002;269:2961–2970. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci USA. 2004;101:17108–17113. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha T, Rih JK, Roy R, Ballal R, Rosen EM. Transcriptional regulation of the base excision repair pathway by BRCA1. J Biol Chem. 2010;285:19092–19105. doi: 10.1074/jbc.M110.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somasundaram K, Zhang H, Zeng YX, et al. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Wang RH, Li W, et al. Genetic interactions between Brca1 and Gadd45a in centrosome duplication, genetic stability, and neural tube closure. J Biol Chem. 2004;279:29606–29614. doi: 10.1074/jbc.M312279200. [DOI] [PubMed] [Google Scholar]
- 46.Wang RH, Zheng Y, Kim HS, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brzovic PS, Meza JE, King MC, Klevit RE. BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein-protein interactions. J Biol Chem. 2001;276:41399–41406. doi: 10.1074/jbc.M106551200. [DOI] [PubMed] [Google Scholar]
- 48.Deng CX, Wang RH. Roles of BRCA1 in DNA damage repair: a link between development and cancer. Hum Mol Genet. 2003;12(Spec No 1):R113–123. doi: 10.1093/hmg/ddg082. [DOI] [PubMed] [Google Scholar]
- 49.Cremer T, Kreth G, Koester H, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- 50.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 51.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 52.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Luczak MW, Jagodzinski PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 55.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 56.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 57.Chen T, Hevi S, Gay F, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 58.Turner JM, Aprelikova O, Xu X, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 59.Hu Y. BRCA1, hormone, and tissue-specific tumor suppression. Int J Biol Sci. 2009;5:20–27. doi: 10.7150/ijbs.5.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu YM, Pace SM, Allen SR, Deng CX, Hsu LC. A PP1-binding motif present in BRCA1 plays a role in its DNA repair function. Int J Biol Sci. 2008;4:352–361. doi: 10.7150/ijbs.4.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vassilopoulos A, Wang RH, Petrovas C, et al. Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. Int J Biol Sci. 2008;4:133–142. doi: 10.7150/ijbs.4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.