Regulation by transcription termination/antitermination, transcription attenuation, is a commonly used strategy. It is most often based on selective formation of either of two alternative base-paired structures in a nascent transcript, one of which causes transcription termination. In this commentary I describe our discovery of transcription attenuation in the trp operon of Escherichia coli. I also relate our excitement as we unravelled the independent sequential events that contribute to this regulatory process. Most importantly, I cite the names and contributions of my many coinvestigators who were responsible for this advance in our knowledge. To illustrate the versatility of this regulatory strategy, I briefly describe several different mechanisms of transcription attenuation. Finally, I comment on its possible evolutionary origins. (The terms terminator, antiterminator, and attenuator are used to describe, respectively, the RNA terminator structure, the RNA antiterminator structure, and the region of an operon that is specifically responsible for termination/antitermination.)
Scientists working on any research project will occasionally obtain results that appear to be inconsistent with their current working hypothesis. What to do? Should you consider these unanticipated findings seriously and perform experiments to examine their significance? Or, should you put them aside until you have additional data that bear on their validity? This is the position I was in, in the early 1970s, when members of my research group were in the midst of our initial regulatory studies with the trp operon of E. coli. Although we were aware of several experimental observations (described below) inconsistent with repression serving as the sole tryptophan-responsive regulatory mechanism controlling transcription of the trp operon, we were principally focused on the repression process. About 10 years earlier Georges Cohen and Francois Jacob had made the groundbreaking observation that E. coli contained a locus, trpR, which, when mutated, increased trp operon expression (12). They proposed that the product of this gene must act much like the lac repressor, and regulate trp operon transcription initiation. Dan Morse, the member of my group who initiated our regulatory studies, carried the Cohen-Jacob observation one step further and isolated nonsense mutants altered in trpR to prove that this gene did encode a protein (44). Subsequent in vivo and in vitro studies on the trp repressor and how it acts by Cathy Squires and Jack Rose of my group, in collaboration with H.-L. Yang and Geoffrey Zubay of Columbia University, yielded the initial results establishing that the trp repressor, like the lac repressor, negatively regulates transcription initiation (54). They also demonstrated in vitro that tryptophan, not charged tRNATrp, was the activator of this repressor (57). This issue was particularly relevant at the time because parallel regulatory studies being conducted by Bruce Ames and his coworkers on the his operon of Salmonella had suggested that tRNAHis, not histidine, serves as the regulatory signal for this biosynthetic operon (38). This point was sufficiently important to us that both Ford Doolittle and Ray Mosteller of my group performed in vivo experiments to rule out tryptophanyl-tRNA serving as the trp corepressor (16, 46). Ironically, we had not considered the possibility that there was a second regulatory mechanism controlling transcription of the trp operon that was responsive to uncharged tRNATrp.
In the early 1970s one of my graduate students, Ethel Jackson, made the crucial observation that alerted us to the probable existence of a second mechanism of transcription regulation (28). She was isolating and examining trp operon deletion mutants with both deletion endpoints within the operon, searching for the location of the operon's internal promoter. In the course of these studies she observed that several deletions with one endpoint just beyond the promoter-operator and the second in one of the first three structural genes increased transcription of the operon four- to eightfold. This increase could not have been due to removal of a second operator site regulated by the trp repressor since mutant strains that lacked the repressor and carried these deletions also exhibited elevated trp operon expression. The most likely explanation was the existence of a previously unrecognized site within the initially transcribed region of the operon which, when deleted, increased operon expression. What type of site could this be? The most reasonable possibility was a site of regulated transcription termination. Studies performed at about the same time with the his operon of Salmonella by T. Kasai led to the same conclusion for this operon; namely, there must be a regulated site of transcription termination in the his operon's leader region (29). Kasai introduced the term “transcription attenuation” to describe this “new” mechanism of regulation.
Looking back, it is clear that our knowledge of the existence of the trp repressor and operator, and our belief that repression would be sufficient to regulate transcription of the operon, inhibited our willingness to pay more attention to the regulatory findings that were inconsistent with this hypothesis. I believe we were also influenced by the prevailing view at the time, that gene regulation was a costly process, and therefore a single regulatory strategy would suffice for each operon. Although we were surprised in the early 1970s to discover that E. coli uses two distinct mechanisms to regulate trp operon transcription, today we appreciate the importance of gene regulation to each organism and readily accept dedication of much genetic information to this purpose.
DISCOVERY OF TRANSCRIPTION ATTENUATION IN THE TRP OPERON OF E. COLI
Prior to Ethel Jackson's observations there were several reported findings that were inconsistent with our presumption that trp repressor-operator interactions were solely responsible for trp operon transcription regulation. For example, studies by Ron Baker of my group, principally designed to determine the polypeptide yield per trp operon transcript under different growth conditions, revealed that strains lacking a functional repressor still responded to a tryptophan deficiency by increasing their rate of trp mRNA synthesis (6). Similarly, while performing analyses of transcription of different regions of the trp operon, Fumio Imamoto of Osaka University observed that upon addition of tryptophan to a tryptophan-starved culture, transcription of the trp operon was prematurely terminated within the operon's leader region (27). This observation was confirmed, and expanded, by Sota Hiraga of my group (25). Retrospectively, these findings established that there must be a second tryptophan-responsive regulatory mechanism that influences the rate of trp mRNA synthesis. However, it was Ethel Jackson's definitive finding that trp mRNA and enzyme levels were increased in strains with leader deletions that left the promoter/operator intact that demanded that we search for a site of tryptophan-regulated transcription termination.
Ethel Jackson's immediate successor on this project, Kevin Bertrand, mapped the presumed regulatory site precisely to the distal end of the 260-bp transcribed leader region that precedes trpE, the first major structural gene in the operon (7, 9). Studies by Bertrand, Frank Lee, Laurence Korn, and Craig and Cathy Squires, showed that transcription was terminated at this site, both in vivo and in vitro (7–9, 36). At the same time, Craig and Cathy Squires, Frank Lee, Kevin Bertrand, and Morley Bronson determined the complete nucleotide sequence of the 5′ end of the trp readthrough transcript and the terminated transcript. Impressively, this was done not by extrapolation from the DNA sequence but by RNA sequencing (56)! Other studies, initially thought to be unrelated, by Terry Platt and Craig Squires revealed that the trp leader transcript contained an unsuspected ribosome binding site and a coding region for a 14-residue leader peptide (49). Most interestingly, the predicted leader peptide would contain tandem tryptophan residues. Frank Lee analyzed the structure of the leader transcript using nuclease digestion and electrophoresis and showed that it could fold to form alternative hairpin structures, one of which was a terminator—of the type we now call an intrinsic terminator (37). He postulated that the alternative RNA structure that preceded and partially overlapped the terminator could theoretically act as an antiterminator and prevent formation of the terminator.
Dan Morse, after he left my group, continued his regulatory studies with the trp operon and obtained the first evidence suggesting that charging of tRNATrp regulated trp operon expression (45). Gerard Zurawski, in studies with Dirk Elseviers and George Stauffer, then established the relationship between tRNATrp charging, translation, and ribosome stalling at the leader peptide Trp codons during attempted synthesis of the tryptophan-containing leader peptide (75, 76). They also demonstrated the importance of the alternative RNA hairpin structures, antiterminator and terminator, that Frank Lee had predicted and detected (75, 76). Collaborative studies with Larry Soll of the University of Colorado confirmed the role of tRNATrp charging on termination regulation (72). Dale Oxender, while on sabbatical leave in my lab, with Zurawski, examined leader RNA secondary structures and explained how ribosome stalling at the Trp codons in the leader peptide coding region would favor formation of the antiterminator structure and thus allow transcription to continue past the regulated site of transcription termination (47). It was also found that leader RNA self-pairing could block the leader ribosome binding site and inhibit continued leader peptide synthesis. This presumably occurs following the cell's decision whether or not to terminate transcription. Iwona Stroynowski, with Mitzi Kuroda, performed extensive deletion analyses with the trp leader region and examined their regulatory effects, both in vivo and in vitro (60, 62). Roberto Kolter focused on generating point mutations to demonstrate the functions of the different leader RNA segments (31). Their combined results showed that leader transcript sequences and structures acted as predicted in regulating transcription termination. Overall, this 10-year period beginning in 1973 was extraordinarily exciting. Findings by many coinvestigators, working on seemingly unrelated projects, combined to reveal many of the features of what turned out to be a fascinating regulatory process.
Transcription pausing couples transcription with translation.
Our observations described above did not explain all the events that must occur during transcription attenuation. Most importantly, they did not reveal how translation of the leader peptide coding region was synchronized with transcription of the leader region. We knew that tryptophan-starved cultures did not terminate transcription at the attenuator. Therefore, some means must exist of ensuring that translation is proceeding on every transcript. This was necessary to facilitate stalling of the translating ribosome at the tandem Trp codons. Stalling would of course be caused by a deficiency of charged tRNATrp. We knew that a third alternative hairpin structure could form in the initial segment of the leader transcript, but we had no reason to suspect that it played any role in attenuation. Studies by Malcolm Winkler and by Bob Fischer, with the help of Anath Das and Roberto Kolter, established that this third, alternative structure, did form and that it forced the transcribing RNA polymerase to pause during transcription (17, 67). Bob Landick and Jannette Carey then demonstrated that the ribosome translating the leader peptide-coding region was responsible for releasing the paused polymerase (33). Thus, two events, transcription pausing and ribosome release of the paused complex, ensure that ribosome movement on the transcript and polymerase movement on the template proceed in unison. The process of transcription pausing and how polymerase recognizes a pause structure are extremely complex and important issues. They have been beautifully addressed in ongoing studies by Landick and his collaborators (43).
Determination of basal level expression.
An important feature of all transcription attenuation mechanisms is how they permit cells to maintain a desired basal level of expression. This is particularly crucial for operons like the trp operon of E. coli, which is subject to a second, more effective transcription regulatory mechanism, in this case, repression. Thus, if transcription termination in the leader region of the trp operon were ever 100% efficient, repressor regulation would be irrelevant. However, termination is only about 90 to 95% complete, even when cells have fully charged tRNATrp. Therefore, despite the presence of adequate levels of tryptophan and charged tRNATrp to sustain rapid protein synthesis, some polymerase molecules must escape repression and attenuation and continue transcription into the structural genes of the operon. These readthrough transcripts would provide sufficient levels of the polypeptide products of the operon to allow recovery following a shift from growth with excess tryptophan to culture conditions where extracellular tryptophan was lacking.
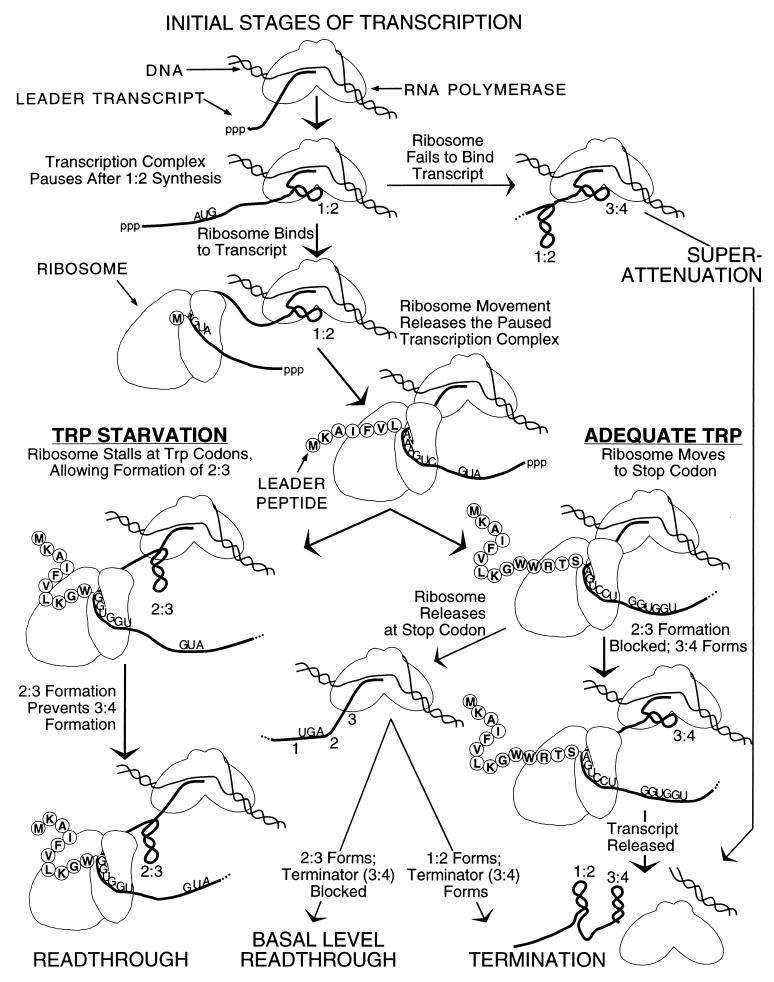
The presence of a low-efficiency constitutive promoter preceding the trpC-trpB-trpA region of the trp operon provides modest levels of the corresponding polypeptides, under all growth conditions. Basal level expression from the principal promoter contributes TrpE and TrpD polypeptides as well. Jim Roesser and Iwona Stroynowski analyzed the features of transcription attenuation that contribute to basal level expression, using the trp operon of E. coli and Serratia marcescens, respectively (52, 53, 61). Roesser identified the segments of the leader region and the events that were responsible for basal level expression when the repressor was fully active and termination at the attenuator was maximal. These features were ribosome release at the leader peptide stop codon, the relative stabilities of the antiterminator and terminator structures, and the position of the transcribing polymerase. Stroynowski showed that synthesis of the leader peptide also affected basal level expression; when translation was prevented, termination at the attenuator increased appreciably. She called this state “super-attenuation” (61). A diagram summarizing all the events in transcription attenuation in the trp operon of E. coli is reproduced in Fig. 1.
FIG. 1.
The stages and events that can occur during transcription of the leader region of the trp operon of E. coli. The paired numbers 1:2, 2:3, and 3:4 refer to the RNA strand pairs that form the three leader RNA hairpin structures: the initial transcription pause structure (1:2), the antiterminator (2:3), and the terminator (3:4). 1, 2, 3, and 4 refer to sequential linear segments of the trp leader transcript. This figure is slightly modified from Landick et al. (34).
All of the efforts of my group benefited from the findings of Bruce Ames and his coworkers in their studies on the mechanism of regulation of the his operon of Salmonella. The Ames group was first to implicate tRNA charging in amino acid biosynthetic operon regulation (38); they also proposed that there was a short coding region in the leader segment of the his operon transcript (R. G. Martin, B. N. Ames, and P. E. Hartman, Abstr. 7th Int. Congress Biochem., p. 261–262, 1967), and they demonstrated that his operon regulation did not involve a repressor.
EXAMPLES OF REGULATION BY TRANSCRIPTION ATTENUATION
In prokaryotes there are two mechanisms of transcription termination, intrinsic termination and factor-dependent termination (see references 22, 34, 48, 51, and 73). In the former, the major event is formation of a stable transcript hairpin structure followed by a series of U's. This hairpin causes the transcribing polymerase to pause on the template; the polymerase subsequently releases both transcript and template. In factor-dependent termination, a protein complex containing Rho factor binds to an unstructured segment of a transcript and surveys that transcript in the 3′ direction, searching for a paused RNA polymerase. If the Rho complex contacts a polymerase, it directs it to terminate transcription. Both mechanisms are used to regulate operon expression, and either or both are used to terminate transcription at the end of an operon.
Ribosome-mediated transcription attenuation.
The trp operon attenuation mechanism described above is typical of the ribosome-mediated mechanisms that regulate expression of many biosynthetic operons of enteric and other bacteria. The regulatory region of each operon is unique, allowing the bacterium to recognize a specific metabolic signal and to respond to it by regulating transcription termination. Most of these operons have been studied thoroughly, by J. Calvo, G. W. Hatfield, J. Gardner, J. R. Roth, H. Umbarger, H. Schachman, and their coworkers and others (see references 34, 68, and 70 for reviews describing these systems).
Ribosome-mediated regulation of pyrimidine biosynthesis in E. coli justifies additional comment. Chuck Turnbough and his coworkers have shown that during transcription of the pyrBI operon of E. coli, availability of pyrimidine nucleoside triphosphates determines the efficiency of polymerase readthrough at a transcription termination site in the operon's leader region (15). This leader region has several important features: AT-rich transcription pause sites, a site of regulated transcription termination, and a leader peptide coding region. The peptide coding region overlaps the sequence of the terminator. Whenever the UTP concentration is insufficient for rapid RNA synthesis the transcribing polymerase pauses at these AT-rich leader transcription pause sites. Pausing allows the translating ribosome to catch up with the transcribing polymerase, permitting this ribosome to prevent formation of the terminator. This promotes readthrough. When the UTP level is sufficient for rapid RNA synthesis the transcribing polymerase pauses only briefly in the AT-rich region and moves well ahead of the ribosome translating the leader peptide coding region. Under these conditions the terminator forms in the transcript, and transcription is terminated. This regulatory mechanism therefore exploits UTP deficiency-dependent transcription pausing and ribosome movement and location in regulating transcription termination (15).
Direct tRNA interaction with a leader transcript.
A very different mechanism of transcription attenuation, in which uncharged tRNA appears to pair directly with a leader transcript, was discovered by Tina Henkin and Frank Grundy (21, 23). This mechanism is widely used in gram-positive bacteria to regulate tRNA synthetase operons and operons concerned with amino acid biosynthesis. As in other examples of transcription attenuation, both antiterminator and terminator structures are essential participants. Regulatory discrimination appears to be achieved by direct interaction of the appropriate uncharged tRNA with specific leader transcript sequences. The uncharged tRNA is believed to pair with a leader transcript sequence, called the specifier, in a codon-anticodon-like manner. This leader RNA codon is located in an unpaired bulge in an RNA hairpin structure. In addition, the 3′ end of the uncharged tRNA is thought to pair with complementary bases in a second unpaired segment of the transcript, called a T box, located in a side bulge in the antiterminator structure. Uncharged tRNA pairing promotes formation of a stable antiterminator, which prevents formation of the terminator. Charged tRNA competes with uncharged tRNA, probably in codon-anticodon pairing, but its attached amino acid probably prevents it from binding to the T box sequence; therefore, it cannot promote antiterminator formation. This design allows the relative concentrations of charged and uncharged forms of a tRNA to provide the basis for the decision whether or not to allow transcription to continue. The trp operon of the gram-positive species Lactococcus lactis is believed to be regulated by this mechanism of transcription attenuation (66).
A second mechanism of transcription attenuation discovered by Grundy and Henkin appears to regulate transcription termination in a group of operons concerned with methionine and cysteine biosynthesis (20). These operons, from gram-positive species, are called members of the S box regulon. Their leader transcripts can fold to form a similar set of secondary structures, two of which are alternative antiterminator and terminator structures. It is not yet known how a defect in sulfur amino acid metabolism leads to termination relief in these operons.
RNA binding protein-mediated transcription attenuation.
RNA-binding proteins also regulate transcription termination (for reviews, see references 22, 34, and 51). These proteins are activated by appropriate signals or events, following which they bind to a specific segment of a transcript, which determines whether or not a terminator structure will form. Examples include the bgl operon of E. coli and the sac, trp, and pyr operons of Bacillus subtilis.
The bgl operon of E. coli.
The features of attenuation regulation of the bgl operon of E. coli have mostly been worked out by Andrew Wright, Orna Amster-Choder, and their coworkers (2, 11). The bgl operon encodes two proteins, BglF and BglG, that are required for utilization of β-glucosides as carbon sources. These proteins function as a membrane-bound sensor (BglF) and a cytoplasmic response regulator (BglG). BglF-mediated phosphorylation/dephosphorylation at His 208 of BglG determines the monomeric versus dimeric state of BglG. The unphosphorylated, dimeric form of BglG has the ability to bind to specific RNA targets and, by so doing, regulate transcription termination. When a β-glucoside is in the environment of the bacterium, the BglF protein binds the sugar, it is phosphorylated, and it is then transported into the cell. The β-glucoside-activated BglF also dephosphorylates BglG, promoting its dimerization. The dimeric BglG then binds to and stabilizes a bgl transcript antiterminator structure, preventing formation of an overlapping terminator. When there is no β-glucoside in the environment of the bacterium, the BglG protein is phosphorylated by BglF; BglG then remains monomeric and inactive. There are two independent sites of regulated transcription termination in the bgl operon, one before bglG, the first gene of the operon, and the second between bglG and bglF. BglG appears to act similarly at the two antiterminators. SacY and SacT of B. subtilis are homologs of BglG; they behave similarly, and when appropriately activated, they relieve transcription termination in the sacB and sacPA operons, respectively (26). The products of these operons are responsible for sucrose utilization.
The trp operon of B. subtilis.
In B. subtilis, seven genes encode the polypeptides required for tryptophan synthesis from chorismate (24). Six of these trp genes are organized as a trp operon, while the seventh, trpG, is in a folate operon. trpG is separate because its polypeptide product forms two enzyme complexes, one that participates in tryptophan synthesis and the other in folate formation. Studies in my lab by Mitzi Kuroda, Paul Gollnick, and Paul Babitzke, in collaboration with Dennis Henner and his coworkers at Genentech, established that expression of the trp operon and trpG is coregulated by a tryptophan-activated RNA-binding protein, named TRAP (for trp RNA-binding attenuation protein) (3, 5, 19, 71). TRAP down-regulates trp operon expression by binding to the trp leader transcript and promoting formation of a terminator. It therefore functions much like the stalled ribosome does in trp operon attenuation in E. coli; however, TRAP binding promotes termination, not antitermination. Elegant structural studies by A. Antson and P. Gollnick and their coworkers on TRAP and a TRAP-RNA complex have shown that TRAP consists of 11 identical polypeptide subunits organized in a doughnut-shaped molecule (3, 4). Each subunit contributes an RNA binding site; these are located at the protein's outer surface (4). A tryptophan binding site is formed by each pair of adjacent subunits (3). Activation of TRAP by tryptophan allows it to bind to an extended segment of the leader transcript that contains 11 (U/G)AG repeats. Most of these (U/G)AG's are located in the 5′ strand of the antiterminator; thus, TRAP binding prevents antiterminator formation. Bound TRAP frees the antiterminator's 3′ basal nucleotides and allows them to pair with a distal complementary set of nucleotides to form a functional terminator. TRAP disrupts the antiterminator by wrapping it around its surface (4, 5, 19, 71). Enrique Merino and Paul Babitzke of my lab also showed that TRAP binding to the trp operon transcript regulates translation of the first gene of the operon, trpE (41). Tryptophan-activated TRAP also regulates translation of trpG (5, 19, 71) by binding to a (U/G)AG-rich sequence that overlaps the trpG ribosome binding site.
The trp operon of B. subtilis is located within a 13-gene supraoperon that contains other genes concerned with aromatic amino acid biosynthesis (24). Accordingly, the upstream aroF promoter of the supraoperon can also be used to produce transcripts encoding the six trp polypeptides. In addition, like E. coli, B. subtilis has the ability to increase trp operon expression in response to a deficiency of tryptophan-charged tRNATrp (59). Joe Sarsero and Alfred Lee of my group have shown that when cells lack this charged tRNA, the TRAP protein appears to become inactivated and unavailable for down-regulating trp operon transcription and trpG translation (35). The rate of tryptophan biosynthesis is then increased, as the organism attempts to overcome the charged tRNATrp deficiency.
The pyr operon of B. subtilis.
Studies by Bob Switzer and his coworkers have shown that transcription termination in the pyr operon of B. subtilis is regulated by an RNA-binding protein, PyrR (63). PyrR also functions enzymatically, as a uracil phosphoribosyltransferase (65). UMP (product) and 5-phosphoribosyl-1-pyrophosphate (substrate) are coregulators of PyrR function (39). PyrR can promote transcription termination at sites located in three ca. 150-nucleotide untranslated regions, each preceding a different gene of the pyr operon (39). Each untranslated transcript segment can fold to form three alternative structures, an antiterminator, a terminator, and an anti-antiterminator that precedes the antiterminator (40). The PyrR protein, when activated by UMP, can bind to, and stabilize, the anti-antiterminator (40). This stabilization prevents formation of the antiterminator, resulting in terminator formation and termination. When cells are pyrimidine deficient, PyrR is inactive, the antiterminator forms, and termination is prevented. UMP activation of PyrR is antagonized by 5-phosphoribosyl-1-pyrophosphate, a precursor of UMP. The structure of PyrR of B. subtilis has been determined in studies with Janet Smith and her coworkers; the transcript sites at which Pyr binds have also been identified (40, 64). Other bacterial species have homologs of PyrR that may regulate their pyr operons similarly (65).
The S10 operon of E. coli.
The S10 ribosomal protein operon of E. coli is regulated transcriptionally and translationally by protein L4, the product of one of its structural genes (55, 74). L. Lindahl and J. Zengel have shown that whenever the L4 concentration exceeds the level necessary for ribosome synthesis, this protein binds to a leader hairpin structure in the S10 transcript and promotes transcription termination (74). M. Nomura's group had shown that L4 binding to S10 leader RNA also reduced translation of the coding regions of the operon by inhibiting translation initiation and by translational coupling (30). The transcript of the leader region of the S10 operon forms multiple hairpin structures, one of which is an intrinsic terminator. Zengel and Lindahl have shown that binding of the L4 and NusA proteins during transcription of the leader region stabilizes a transcription pause structure that precedes the terminator (55). This stabilization leads to efficient transcription termination. The roles of the various RNA structures, and the possibility that other factors participate in the termination process, remain to be addressed.
N protein-mediated antitermination in bacteriophage lambda.
The initial discovery of regulation by transcription termination/antitermination was made by Jeffrey Roberts while studying transcription of the so-called “early region” of bacteriophage lambda (50). Rho factor was identified as a bacterial protein that prevents transcription from proceeding beyond the phage early region. The phage genome specifies a protein, N, that can interact with RNA polymerase as it transcribes the early region of the phage genome and form an antitermination complex that ignores sites of Rho-dependent termination (51). To act, N protein requires cis-acting transcript sites, called nut sites, and trans-acting protein factors, called Nus proteins (51). N protein associates with the nut site, it complexes with Nus proteins, and it renders the transcribing RNA polymerase termination resistant (10, 42). Phage lambda encodes a second antitermination factor, the Q protein. Q acts differently than N; it binds to a promoter region and alters the initiating RNA polymerase, rendering it immune to Rho-dependent terminators as well as intrinsic terminators (51).
Antitermination in rRNA operons.
It is essential to E. coli to carefully regulate the synthesis of rRNA. Cathy Squires and her coworkers have discovered an antitermination mechanism employed by E. coli that prevents premature termination of transcription in rrn operons (13). The sensitive rrn leader regulatory region has features resembling the early region of phage lambda. A nut site serves as the assembly point for a polymerase antitermination complex that is resistant to Rho action (13), and NusA and NusB proteins have been shown to be components of the rrn antitermination complex. NusE and NusG proteins may also participate (13, 58). The identity of the presumed additional protein or proteins that confer antitermination behavior is under investigation (13).
Translation-mediated inhibition of Rho-dependent termination in the tna operon.
E. coli and some other bacterial species use transcription attenuation to regulate transcription of the tryptophanase (tna) operon, a catabolic operon that encodes proteins that can degrade tryptophan to utilizable carbon and nitrogen sources (71). The novel feature of this mechanism of transcription attenuation is that a tryptophan-containing leader peptide, in the presence of excess tryptophan, can prevent Rho factor-dependent termination at termination sites located in the leader region of the operon (71). It is thought that the leader peptide acts in cis on the translating ribosome to block sites in the transcript needed for Rho binding or action (32). Most of the known features of tna operon regulation were determined by M. Deeley, V. Stewart, R. Landick, P. Gollnick, A. Kamath, K. Gish, and V. Konan, while members of my group (see reference 71).
ADVANTAGES AND POSSIBLE ORIGINS OF REGULATION BY TRANSCRIPTION ATTENUATION
It is evident from the examples described above that transcription attenuation is an often-used, effective, regulatory strategy. It can be an economical process as well, for relatively little unique DNA/RNA sequence information is needed to form an intrinsic terminator, antiterminator, or anti-antiterminator or when termination is mediated by a translating ribosome or tRNA. However, if a specific protein participates in the termination decision, as it often does, attenuation is clearly more costly than repression. Nonetheless, by using RNA as the principal element in a regulatory decision, strategies become available that are not possible with DNA as the target. The overriding conclusion that can be reached from the available data is that transcription attenuation can be an economical process and that it can be adapted to almost any regulatory need. There are several examples in eukaryotes with features resembling those of prokaryote transcription attenuation mechanisms; however this form of regulation has not been extensively studied in higher organisms (see references 34, 48, and 70).
When investigating any biological process one would ultimately like to know its evolutionary origins. The genetic makeup and capabilities of each present-day organism reflect each species' ancestry and experiences. Transcription attenuation mechanisms rely on RNA. With the discovery of self-splicing RNA, it now seems likely that the DNA world was preceded by a period during which RNA served both as catalyst and as genetic material (18). If mechanisms of protein synthesis appeared first in this RNA world, it would seem logical that mechanisms of transcription attenuation somewhat analogous to those in use today would have been developed to regulate the events in protein synthesis. Obviously DNA-dependent transcription termination would not have been involved, but regulation of complementary RNA synthesis, or RNA translation, could have been the objective. Translational regulation is particularly attractive as the potential ancestral process since there are many examples of translational attenuation (a description of these is not included in this article). During translational attenuation RNA sequences and structures resembling those used in transcription attenuation participate in translational regulatory decisions. As organisms became more complex, and DNA assumed its role as the primary genetic material, some of these RNA-based regulatory mechanisms could have been slightly modified and adopted. All that would be needed to facilitate regulation by transcription termination would be the addition of a sequence specifying a transcription terminator.
What might have been the origin of the RNA structures used to regulate translation, and subsequently transcription? There are striking similarities between the structures formed by leader transcripts involved in attenuation and the structures of some tRNAs. These similarities were pointed out many years ago by Ames et al. (1) in a comparison of the sequences and predicted structures of the leader transcript of the his operon of Salmonella and tRNAHis. It is conceivable that the tRNAs themselves, or DNA copies of tRNAs, served as sources of leader regulatory material. With regard to the origin of terminators, they could have been derived from the terminators that function at the end of operons. As additional sequence information becomes available for leader RNAs from different species, it will be interesting to explore the relatedness of the sequences and structures participating in different forms of attenuation.
Considerable comparative information already exists that bears on the evolutionary origins of the genes, proteins, and operons of tryptophan biosynthesis and the regulatory processes that control their expression (14, 69). These compilations suggest that the genes specifying the seven polypeptide domains required for tryptophan synthesis evolved from a common set of ancestral genes encoding these domains. However, the organization of these genes within transcriptional units differs among the species. In some organisms different trp genes are present in the same operon, and in many organisms the order of trp genes within an operon varies. In addition, trp genes are sometimes fused and encode bi- or multifunctional polypeptides. In general, the findings obtained in evolutionary comparisons suggest that gene content and organization between and within operons reflect metabolic differences between species and the need to develop independently regulated transcriptional units and appropriate regulatory strategies. For example, as described in this Commentary, three very different mechanisms of transcription attenuation are used by different organisms to regulate expression of the genes of tryptophan biosynthesis. A fourth attenuation mechanism regulates expression of the tryptophan degradative operon.
An oft-told tale in science is that newly discovered processes that first seem novel turn out to be common, and subsequently expected. Transcription attenuation is one such example.
ACKNOWLEDGMENTS
I would like to express my sincere appreciation to all the dedicated individuals who have contributed to the output of my laboratory. I offer my special thanks to Cathy Squires and Bob Landick, former coworkers and good friends, who made many helpful suggestions regarding this article. Ginny Horn deserves special recognition: she participated in all the studies performed in my laboratory.
I also wish to acknowledge that the research performed in my laboratory was supported by funds provided by the NIH, NSF, American Heart Association, and American Cancer Society.
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Ames B N, Tsang T H, Buck M, Christman M F. The leader mRNA of the histidine attenuator region resembles tRNAHis: possible general regulatory implications. Proc Natl Acad Sci USA. 1983;80:5240–5242. doi: 10.1073/pnas.80.17.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amster-Choder O, Wright A. BglG, the response regulator of the Escherichia coli bgl operon, is phosphorylated on a histidine residue. J Bacteriol. 1997;179:5621–5624. doi: 10.1128/jb.179.17.5621-5624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 4.Antson A A, Dodson E J, Dodson G, Greaves R B, Chen X-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 6.Baker R, Yanofsky C. Transcription initiation frequency and translational yield for the tryptophan operon of Escherichia coli. J Mol Biol. 1972;69:89–102. doi: 10.1016/0022-2836(72)90025-3. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand K, Korn L J, Lee F, Yanofsky C. The attenuator of the tryptophan operon on Escherichia coli: heterogenous 3′-OH termini in vivo and deletion mapping of functions. J Mol Biol. 1977;117:227–247. doi: 10.1016/0022-2836(77)90032-8. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand K, Squires C, Yanofsky C. Transcription termination in vivo in the leader region of the tryptophan operon of Escherichia coli. J Mol Biol. 1976;103:319–337. doi: 10.1016/0022-2836(76)90315-6. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand K, Yanofsky C. Regulation of transcription termination in the leader region of the tryptophan operon of Escherichia coli involves tryptophan or its metabolic product. J Mol Biol. 1976;103:339–349. doi: 10.1016/0022-2836(76)90316-8. [DOI] [PubMed] [Google Scholar]
- 10.Chattopadhyay S, Garcia-Mena J, DeVito J, Wolska K, Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci USA. 1995;92:4061–4065. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Engelberg-Kulka H, Amster-Choder O. The phosphorylation site of BglG, the response regulator of the Escherichia coli bgl sensory system. J Biol Chem. 1997;272:17263–17268. doi: 10.1074/jbc.272.28.17263. [DOI] [PubMed] [Google Scholar]
- 12.Cohen G N, Jacob F. Sur la repression de la synthese des enzymes intervenant dans la formation du tryptophane chez Escherichia coli. C R Acad Sci. 1959;248:3490–3492. [PubMed] [Google Scholar]
- 13.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford I P. Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu Rev Microbiol. 1989;43:567–600. doi: 10.1146/annurev.mi.43.100189.003031. [DOI] [PubMed] [Google Scholar]
- 15.Donahue J P, Turnbough C L., Jr Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12. J Biol Chem. 1994;269:18185–18191. [PubMed] [Google Scholar]
- 16.Doolittle W F, Yanofsky C. Mutants of Escherichia coli with an altered tryptophanyl-transfer ribonucleic acid synthetase. J Bacteriol. 1968;95:1283–1294. doi: 10.1128/jb.95.4.1283-1294.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher R F, Das A, Kolter R, Winkler M E, Yanofsky C. Analysis of the requirements for transcription pausing in the tryptophan operon. J Mol Biol. 1985;182:397–409. doi: 10.1016/0022-2836(85)90199-8. [DOI] [PubMed] [Google Scholar]
- 18.Gesteland R F, Cech R F, Atkins T T, Atkins J F. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 19.Gollnick P. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol. 1994;11:991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 20.Grundy F J, Henkin T M. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 21.Grundy F J, Hodil S E, Rollins S M, Henkin T M. Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J Bacteriol. 1997;179:2587–2594. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henkin T M. Control of transcription termination in prokaryotes. Annu Rev Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 23.Henkin T M. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 24.Henner D, Yanofsky C. Biosynthesis of aromatic amino acids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 269–280. [Google Scholar]
- 25.Hiraga S, Yanofsky C. Inhibition of the progress of transcription on the tryptophan operon of Escherichia coli. J Mol Biol. 1973;79:339–349. doi: 10.1016/0022-2836(73)90010-7. [DOI] [PubMed] [Google Scholar]
- 26.Idelson M, Amster-Choder O. SacY, a transcriptional antiterminator from Bacillus subtilis, is regulated by phosphorylation in vivo. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamoto F. Immediate cessation of transcription of the operator-proximal region of the tryptophan operon in E. coli after repression of the operon. Nature. 1968;220:31–34. doi: 10.1038/220031a0. [DOI] [PubMed] [Google Scholar]
- 28.Jackson E N, Yanofsky C. The region between the operator and first structural gene of the tryptophan operon of Escherichia coli may have a regulatory function. J Mol Biol. 1973;76:89–101. doi: 10.1016/0022-2836(73)90082-x. [DOI] [PubMed] [Google Scholar]
- 29.Kasai T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature. 1974;249:523–527. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- 30.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- 31.Kolter R L, Yanofsky C. Genetic analysis of the tryptophan operon regulatory region using site-directed mutagenesis. J Mol Biol. 1984;175:299–312. doi: 10.1016/0022-2836(84)90350-4. [DOI] [PubMed] [Google Scholar]
- 32.Konan K V, Yanofsky C. Role of ribosome release in regulation of tna operon expression in Escherichia coli. J Bacteriol. 1999;181:1530–1536. doi: 10.1128/jb.181.5.1530-1536.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci USA. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landick R, Turnbough C L, Jr, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 35.Lee A I, Sarsero J P, Yanofsky C. A temperature-sensitive trpS mutation interferes with TRAP regulation of trp gene expression in Bacillus subtilis. J Bacteriol. 1996;178:6518–6524. doi: 10.1128/jb.178.22.6518-6524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee F, Squires C L, Squires C, Yanofsky C. Termination of transcription in vitro in the Escherichia coli tryptophan operon leader region. J Mol Biol. 1976;103:383–393. doi: 10.1016/0022-2836(76)90318-1. [DOI] [PubMed] [Google Scholar]
- 37.Lee F, Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci USA. 1977;74:4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis J A, Ames B N. Histidine regulation in Salmonella typhimurium. J Mol Biol. 1972;66:131–142. doi: 10.1016/s0022-2836(72)80011-1. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Switzer R L. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J Bacteriol. 1996;178:7206–7211. doi: 10.1128/jb.178.24.7206-7211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Turner R J, Switzer R L. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc Natl Acad Sci USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogridge J, Mah T F, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2844. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 43.Mooney R A, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morse D, Yanofsky C. Amber mutants of the trpR regulatory gene. J Mol Biol. 1969;44:185–193. doi: 10.1016/0022-2836(69)90413-6. [DOI] [PubMed] [Google Scholar]
- 45.Morse D E, Morse E. Dual-control of the tryptophan operon is mediated by both tryptophanyl-tRNA synthetase and the repressor. J Mol Biol. 1976;103:209–222. doi: 10.1016/0022-2836(76)90310-7. [DOI] [PubMed] [Google Scholar]
- 46.Mosteller R D, Yanofsky C. Evidence that tryptophanyl transfer ribonucleic acid is not the corepressor of the tryptophan operon of Escherichia coli. J Bacteriol. 1971;105:268–275. doi: 10.1128/jb.105.1.268-275.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oxender D L, Zurawski G, Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci USA. 1979;76:5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Platt T. RNA structure in transcription elongation, termination, and antitermination. In: Simons R W, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1998. pp. 541–574. [Google Scholar]
- 49.Platt T, Squires C, Yanofsky C. Ribosome-protected regions of the leader-trpE sequence of Escherichia coli tryptophan operon messenger RNA. J Mol Biol. 1976;103:411–420. doi: 10.1016/0022-2836(76)90320-x. [DOI] [PubMed] [Google Scholar]
- 50.Roberts J W. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 51.Roberts J W. Transcription termination and its control. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 27–45. [Google Scholar]
- 52.Roesser J R, Yanofsky C. The effects of leader peptide sequence and length on attenuation control of the trp operon of Escherichia coli. Nucleic Acids Res. 1991;19:795–800. doi: 10.1093/nar/19.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roesser J R, Yanofsky C. Ribosome release modulates basal level expression of the trp operon of Escherichia coli. J Biol Chem. 1988;263:14251–14255. [PubMed] [Google Scholar]
- 54.Rose J K, Squires C L, Yanofsky C, Yang H L, Zubay G. Regulation in in vitro transcription of the tryptophan operon by purified RNA polymerase in the presence of partially purified repressor and tryptophan. Nature. 1973;245:133–137. doi: 10.1038/newbio245133a0. [DOI] [PubMed] [Google Scholar]
- 55.Sha Y, Lindahl L, Zengel J R. RNA determinants required for L4-mediated attenuation control of the S10 r-protein operon of Escherichia coli. J Mol Biol. 1995;245:486–498. doi: 10.1006/jmbi.1994.0040. [DOI] [PubMed] [Google Scholar]
- 56.Squires C, Lee F, Bertrand K, Squires C L, Bronson M J, Yanofsky C. Nucleotide sequence of the 5′ end of tryptophan messenger of Escherichia coli. J Mol Biol. 1976;103:351–380. doi: 10.1016/0022-2836(76)90317-x. [DOI] [PubMed] [Google Scholar]
- 57.Squires C L, Rose J K, Yanofsky C. Tryptophanyl-tRNA and tryptophanyl-tRNA synthetase are not required for in vitro repression of the tryptophan operon. Nature. 1973;245:131–133. doi: 10.1038/newbio245131a0. [DOI] [PubMed] [Google Scholar]
- 58.Squires C L, Greenblatt J, Li J, Condon C, Squires C L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinberg W. Temperature-induced derepression of tryptophan biosynthesis in a tryptophanyl-transfer ribonucleic acid synthetase mutant of Bacillus subtilis. J Bacteriol. 1974;117:1023–1034. doi: 10.1128/jb.117.3.1023-1034.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroynowski I, Kuroda M I, Yanofsky C. Transcription termination in vitro at the tryptophan operon attenuator is controlled by secondary structures in the leader transcript. Proc Natl Acad Sci USA. 1982;80:2206–2210. doi: 10.1073/pnas.80.8.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stroynowski I, van Cleemput C M, Yanofsky C. Superattenuation in the tryptophan operon of Serratia marcescens. Nature. 1982;298:38–41. doi: 10.1038/298038a0. [DOI] [PubMed] [Google Scholar]
- 62.Stroynowski I, Yanofsky C. Transcript secondary structures regulate transcription termination at the attenuator in Serratia marcescens. Nature. 1982;298:34–38. doi: 10.1038/298034a0. [DOI] [PubMed] [Google Scholar]
- 63.Switzer R L, Turner R J, Lu Y. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog Nucleic Acid Res Mol Biol. 1996;62:329–367. doi: 10.1016/s0079-6603(08)60512-7. [DOI] [PubMed] [Google Scholar]
- 64.Tomchick D R, Turner R J, Switzer R L, Smith J I. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure. 1998;6:337–350. doi: 10.1016/s0969-2126(98)00036-7. [DOI] [PubMed] [Google Scholar]
- 65.Turner R J, Bonner E R, Grabner G K, Switzer R L. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J Biol Chem. 1998;273:5932–5938. doi: 10.1074/jbc.273.10.5932. [DOI] [PubMed] [Google Scholar]
- 66.Van de Guchte M, Ehrlich S D, Chopin A. tRNATrp as a key element of antitermination in the Lactococcus lactis trp operon. Mol Microbiol. 1998;29:61–74. doi: 10.1046/j.1365-2958.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 67.Winkler M E, Yanofsky C. Pausing the RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry. 1981;20:3738–3744. doi: 10.1021/bi00516a011. [DOI] [PubMed] [Google Scholar]
- 68.Yanofsky C. Attenuation in the control of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 69.Yanofsky C. Comparison of regulatory and structural regions of genes of tryptophan metabolism. Mol Biol Evol. 1984;1:143–161. doi: 10.1093/oxfordjournals.molbev.a040307. [DOI] [PubMed] [Google Scholar]
- 70.Yanofsky C. Transcription attenuation—a minireview. J Biol Chem. 1988;263:609–612. [PubMed] [Google Scholar]
- 71.Yanofsky C, Konan K V, Sarsero J P. Some novel transcription attenuation mechanisms used by bacteria. Biochimie. 1996;78:1017–1024. doi: 10.1016/s0300-9084(97)86725-9. [DOI] [PubMed] [Google Scholar]
- 72.Yanofsky C, Soll L. Mutations affecting tRNATrp and its charging and their effect on regulation of transcription termination at the attenuator of the tryptophan operon. J Mol Biol. 1977;113:663–677. doi: 10.1016/0022-2836(77)90229-7. [DOI] [PubMed] [Google Scholar]
- 73.Yarnell W S, Roberts J W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–616. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 74.Zengel J M, Lindahl L. A hairpin structure upstream of the terminator hairpin required for ribosomal protein L4-mediated attenuation control of the S10 operon of Escherichia coli. J Bacteriol. 1996;178:2383–2387. doi: 10.1128/jb.178.8.2383-2387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zurawski G, Yanofsky C. Escherichia coli tryptophan operon leader mutations which relieve transcription termination are cis-dominant to trp leader mutations which increase transcription termination. J Mol Biol. 1980;142:123–129. doi: 10.1016/0022-2836(80)90210-7. [DOI] [PubMed] [Google Scholar]
- 76.Zurawski G, Elseviers D, Stauffer G V, Yanofsky C. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc Natl Acad Sci USA. 1978;75:5988–5992. doi: 10.1073/pnas.75.12.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]