Abstract
A large number of bacteria regulate chaperone gene expression by the CIRCE-HrcA system in which a DNA element called CIRCE serves as binding site for the repressor protein HrcA under non-heat-shock conditions. We have cloned the two consecutive genes hrcA and grpE of Bradyrhizobium japonicum by using a complementation approach that screened for GrpE function. In vivo and in vitro transcript mapping demonstrated that both genes are transcribed separately from RpoH (ς32)-dependent promoters. To investigate the supposed negative regulatory function of HrcA, we compared the expression of putative target genes in the wild type with that in an hrcA mutant. Transcription of the CIRCE-associated chaperonin operons groESL4 and groESL5, as well as the β-galactosidase activity derived from corresponding groE-lacZ fusions, was strongly elevated in the hrcA mutant even at physiological temperatures. Expression of other heat shock regulons (RpoH or ROSE dependent) was not affected. To study the activity of HrcA in vitro, we purified a histidine-tagged version of the protein under nondenaturing conditions. Specific binding to the CIRCE element was obtained with a soluble fraction of HrcA in gel retardation experiments.
The survival of a cell depends on its ability to adapt to changing environmental conditions. For a sudden temperature upshift, all organisms cope with the increased amount of misfolded proteins by an enhanced synthesis of so-called heat shock proteins (Hsps). This collection of proteins consists mainly of chaperones and proteases, which are involved in protein (re)folding and protein degradation, respectively (for reviews see references 7 and 18).
Both eukaryotic and prokaryotic cells have established a number of complex regulatory strategies to tightly control the transcription of heat shock genes under any given condition (21, 26, 27). Coordinate expression of heat shock genes in Escherichia coli is mediated by alternative sigma factors which direct the RNA polymerase to specific promoter sequences upstream of these genes. An increase in the cellular concentration of ς32 (RpoH) after a sudden temperature upshift results in elevated transcription of more than 30 genes of the ς32 regulon. The activity and stability of ς32 are subject to feedback control by the DnaK machinery, which sequesters the sigma factor under low-temperature conditions. Binding of ς32 to the DnaK system prevents association with the RNA polymerase core enzyme and promotes degradation of ς32 by the FtsH protease. After a heat shock, denatured proteins presumably titrate the DnaK chaperones away from ς32, leaving the latter stable and competent for complex formation with RNA polymerase (for recent reviews see references 14 and 53).
In contrast to the positive regulation by alternative sigma factors, heat shock expression in a majority of bacteria was found to be controlled by negative regulation (summarized in reference 27). Our knowledge of such systems is far less advanced than that of the E. coli-type regulation. The regulatory principle is often based on a specific interaction between a repressor protein and a DNA element that is located in the promoter regions of heat shock genes. Repression is relieved upon a temperature upshift, thereby allowing transcription of the downstream genes. A widespread negative control mechanism consists of the repressor protein HrcA and a DNA element called CIRCE (for controlling inverted repeat of chaperone expression [54]). The first evidence that this highly conserved inverted repeat might act as a negative cis element was obtained by the observation that mutations in one or both arms of the inverted repeat resulted in elevated transcription of the downstream genes even at normal growth temperatures (47, 54). Chromosomal mutations in Bacillus subtilis which affected the expression of CIRCE-dependent genes were localized in orf39, the first gene of the dnaK operon (44, 51). Disruption of orf39 in B. subtilis and of the equivalent gene in Caulobacter crescentus confirmed the function of the Orf39 protein as a negative regulator of CIRCE-dependent genes and led to its designation as HrcA (for heat regulation at CIRCE) (38, 43). By analogy with the feedback control of ς32 activity and stability by the DnaK machinery in E. coli, the activity of B. subtilis HrcA was found to be modulated by the GroE chaperonin system (25). A direct correlation between the cellular GroESL level and HrcA activity suggested that the repressor requires chaperonins for proper function in vivo (3, 25). Accordingly, heat stress would render HrcA inactive because GroES and GroEL become engaged in the refolding of denatured proteins.
Unfortunately, attempts to confirm the proposed HrcA-CIRCE and HrcA-GroEL interactions in vitro were hampered by the insolubility of overproduced HrcA, which tends to form inclusion bodies, requiring purification under denaturing conditions. Despite this shortcoming, it has been possible to visualize the binding of purified HrcA of Staphylococcus aureus to the CIRCE element by atomic force microscopy (35) and to document CIRCE binding of purified Bacillus stearothermophilus HrcA in gel retardation experiments (25).
An unusually complex network of both positive and negative regulatory mechanisms controls transcription of heat shock genes in Bradyrhizobium japonicum, the nitrogen-fixing root nodule symbiont of soybeans (27). Three different regulatory systems have been identified so far. One class of heat shock genes is transcribed from ς32-dependent promoters. The search for the corresponding sigma factor revealed the existence of three disparately regulated rpoH genes whose ς32-like products have different promoter specificities (30, 31). The expression of a second class of heat shock genes is negatively controlled by a highly conserved DNA element called ROSE (for repression of heat shock gene expression) (29). Evidence for a third class of heat shock genes was deduced from the presence of a CIRCE element in the promoter region of the heat shock-inducible chaperonin operons groESL4 and groESL5 (3). Elevated transcription from the groESL4 promoter at normal temperatures, caused either by a deletion of 4 bp within the corresponding CIRCE element or by disruption of groEL4, had clearly demonstrated the regulatory function of CIRCE and, moreover, had indicated the presence of a feedback regulatory mechanism. According to these observations, the existence of a CIRCE-specific repressor protein whose stability or activity or both are modulated by chaperonins was suggested.
Here, we report the cloning of the B. japonicum hrcA gene and the assignment of its position in the complex heat shock network of this organism. The function of the HrcA protein as a CIRCE-specific repressor protein is demonstrated by a complementary set of in vivo and in vitro experiments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli cells were grown in Luria-Bertani medium (22) supplemented with ampicillin (200 μg/ml), chloramphenicol (20 μg/ml), kanamycin (30 μg/ml), or tetracycline (10 μg/ml) if required. The growth temperature for E. coli strains was 37°C except for E. coli DA259 (grpE mutant), which was grown at 30°C. B. japonicum strains were propagated aerobically at 30°C in PSY medium (37) supplemented with 0.1% (wt/vol) arabinose. If appropriate, antibiotics were added at the following concentrations: chloramphenicol, 20 μg/ml (for counterselection against E. coli donor strains); kanamycin, 100 μg/ml; spectinomycin, 100 μg/ml; and tetracycline, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Reference or origin |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | Gibco-BRL, Gaithersburg, Md. |
| S17-1 | Smr SprhdsR RP4-2 kan::Tn7 tet::Mu; integrated in the chromosome | 45 |
| C600 | supE44 hsdR thi-1 leuB6 lacY1 tonA21 | 49 |
| DA258 | Cmr Kmr Tcr (C600) grpE+::Ω-Cm thr::Tn10 | 2 |
| DA259 | Cmr Kmr Tcr (C600) ΔgrpE::Ω-Cm thr::Tn10 | 2 |
| BL21/pLysS | Cmr F−ompT (rB− mB−) T7lys | 46 |
| B. japonicum strainsa | ||
| 110spc4 | Spr (wild type) | 37 |
| 5549 | Spr KmrhrcA::kan; hrcA and kan oriented in opposite directions | This study |
| Plasmids | ||
| pBluescript II KS+ | Apr | Stratagene |
| pBluescript II SK+ | Apr | Stratagene |
| pBSL15 | Apr Kmr | 1 |
| pUC18 | Apr | 34 |
| pSUP202 | Apr Cmr TcroriT from RP4 | 45 |
| pSUP202pol4 | Tcr (pSUP202); part of polylinker from pBluescript II KS+ between EcoRI and PstI | 10 |
| pNM481 | Apr (pUC8) ′lacZ | 24 |
| pNM482 | Apr (pUC8) ′lacZ | 24 |
| pSUP481 | Tcr (pSUP202pol4) ′lacZ (5.1-kb EcoRI-StuI fragment from pNM481) | H. M. Fischer, unpublished data |
| pSUP482 | Tcr (pSUP202pol4) ′lacZ (5.1-kb EcoRI-StuI fragment from pNM482) | H. M. Fischer, unpublished data |
| pET28a(+) | Kmr | Novagene, Madison, Wis. |
| pWSK29 | Apr; low-copy-number vector | 48 |
| pBW401 | Apr (pWSK29); 1.3-kb SalI-EcoRI fragment containing E. coli grpE | 49 |
| pRJ5099 | Apr (pRJ9519); 0.9-kb BglII-EcoRI fragment containing the B. japonicum dnaKJ promoter region | 30 |
| pRJ5400 | Tcr (pSUP202pol4) hspA′-′lacZ | 29 |
| pRJ5523b | Apr (pUC18); 3.9-kb EcoRI fragment containing B. japonicum ′hrcA, grpE, and ′dnaK opposite to αlacZ orientation | This study |
| pRJ5530b | Apr (pBluescript II SK+); 3.0-kb NotI fragment containing B. japonicum hrcA and grpE opposite to αlacZ orientation | This study |
| pRJ5542 | Apr (pRJ9519); 0.5-kb NotI-NruI fragment from pRJ5530 containing the B. japonicum hrcA promoter region | This study |
| pRJ5543 | Apr (pRJ9519); 0.7-kb SacI-FspI fragment from pRJ5530 containing the B. japonicum grpE promoter region | This study |
| pRJ5549 | Tcr Kmr (pSUP202pol4) hrcA::kan; hrcA and kan oriented in opposite directions | This study |
| pRJ5552 | Kmr [pET28a(+)]; 1.1-kb NotI-NdeI amplification product containing B. japonicum hrcA gene and plasmid for overexpression of H6-HrcA | This study |
| pRJ5554 | Tcr (pSUP482) grpE′-′lacZ; 348-bp MscI-FspI grpE′ fragment from pRJ5530 | This study |
| pRJ5556 | Apr (pUC18); 198-bp amplification product containing the promoter region of B. japonicum groESL4 | This study |
| pRJ5558 | Apr (pUC18); 265-bp amplification product containing the promoter region of B. japonicum groESL5 | This study |
| pRJ5559 | Tcr (pSUP481) hrcA′-′lacZ; 937-bp EcoRI hrcA′ fragment from pRJ5530 | This study |
| pRJ7998 | Tcr (pSUP202pol4) groEL2′-′lacZ | 10 |
| pRJ8067 | Tcr (pSUP202pol4) groEL1′-′-lacZ | 3 |
| pRJ8092 | Tcr (pSUP202pol4) groEL5′-′lacZ | 3 |
| pRJ8174 | Tcr (pSUP202pol4) dnaK′-′lacZ | 23 |
| pRJ8548 | Tcr (pSUP202pol4) groEL4′-′lacZ | 3 |
| pRJ9519 | Apr (pBluescript II SK+) containing the B. japonicum rrn terminator | 30 |
DNA manipulations.
Recombinant DNA techniques were performed according to standard protocols (40). Construction of B. japonicum mutants by cointegrate formation or marker exchange mutagenesis and isolation of chromosomal DNA from B. japonicum were performed as described previously (15). Southern blot hybridizations with digoxigenin-11-dUTP-labeled DNA probes were performed according to the manufacturer's instructions (Boehringer Mannheim). DNA was sequenced by the chain termination method (41) with an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, Calif.). The DNA region was sequenced and the deduced proteins were analyzed with the software package of the Genetics Computer Group of the University of Wisconsin, Madison (version 8.0) and the National Center for Biotechnology Information BLAST network server. The CODONPREFERENCE program was applied by using the B. japonicum codon usage table (36).
Construction of strains and plasmids.
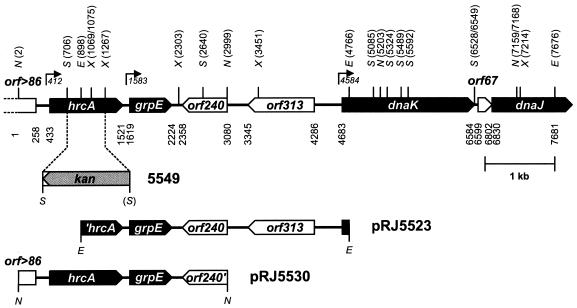
For the construction of hrcA deletion mutants the 2.0-kb NotI (position 2 in Fig. 1)-XmnI (position 2015) fragment of pRJ5530 was ligated into the 6.7-kb NotI-SmaI vector fragment of pSUP202pol4. In the resulting plasmid the 561-bp SalI (position 706)-XhoI (position 1267) internal fragment of hrcA was replaced by the 1.2-kb SalI fragment of pBSL15 containing the neomycin phosphotransferase II cassette (Kmr). Both orientations of the resistance cassette relative to the hrcA gene were obtained, but for the subsequent work we used plasmid pRJ5549, in which the remaining portion of the hrcA gene and the resistance cassette are oriented in opposite directions. Plasmid pRJ5549 was mobilized from E. coli S17-1 into B. japonicum 110spc4 for marker replacement mutagenesis, yielding strain 5549 (Fig. 1).
FIG. 1.
Physical map of the B. japonicum hrcA gene region. Numbers indicate start and stop codon positions of open reading frames, the transcription start sites (horizontal arrows above gene map), the beginning (position 1) and the end (position 7681) of the sequenced gene region, and recognition sites of the following restriction enzymes: E, EcoRI; N, NotI; S, SalI; X, XhoI. The restriction site in parentheses was destroyed during the cloning procedure. The strategy to construct the B. japonicum hrcA deletion resulting in strain 5549 is indicated. The inserts of plasmids pRJ5523 and pRJ5530 are shown below the physical map.
In order to compare the expression of heat shock genes in a wild-type background with that in an hrcA mutant background, suitable translational lacZ fusions present on pSUP202pol4 derivates were cointegrated via homologous recombination into the chromosome of wild-type B. japonicum and 5549 (Table 1). The correct genomic structure of all mutant strains was confirmed by Southern blot hybridization with appropriate digoxigenin-11-dUTP-labeled DNA probes.
Plasmids used as templates for in vitro transcription experiments were based on pRJ9519, which contains the B. japonicum rrn terminator and in which different promoter regions were introduced (30). Plasmid pRJ5542 carries a 537-bp NotI (position 2 in Fig. 1)-NruI (position 538) fragment containing the B. japonicum hrcA promoter region. The promoter region of B. japonicum grpE was introduced as a 691-bp SacI (position 1109)-FspI (position 1796) fragment yielding plasmid pRJ5543.
To construct the overproduction plasmid pRJ5552, the hrcA gene was amplified from plasmid pRJ5530 by PCR with Taq DNA polymerase. The oligonucleotides used as primers were designed such that they introduced an NdeI recognition site overlapping the start codon and a NotI site immediately downstream of the stop codon of hrcA. The PCR-generated fragment was cut with both NdeI and NotI and ligated into pET28a(+) digested with the same enzymes. The encoded recombinant protein H6-HrcA possesses an N-terminal extension of 20 amino acids adding 2.1 kDa to the molecular mass of HrcA (39.2 kDa). The correct nucleotide sequence of the PCR-amplified hrcA fragment was confirmed by sequencing plasmid pRJ5552.
Transcript mapping.
RNA isolation and primer extension analysis were performed as described previously (3). The following oligonucleotides were used to determine the transcription start sites upstream of hrcA and grpE: HrcA10 (hrcA; position 579), 5′-CAGCCGGGAAATATTGCGTGAGCCCACC-3′; HrcA11 (hrcA; position 630), 5′-CAGATCGGCCATGACGTTGCGAACCGAG-3′; GrpE4 (grpE; position 1742), 5′-CTTCCTTCTGCAACAGCTCGACCGAGCC-3′; GrpE5 (grpE; position 1700), 5′-CGGGCATGATGTAGGGCTTCGACACCAC-3′. The numbers in parentheses indicate the positions of the 5′ ends based on the numbering used in the physical map shown in Fig. 1.
In vitro transcription.
Single-round transcription assays with B. japonicum RNA polymerase holoenzyme and core enzyme reconstituted with purified RpoH2 were carried out as described previously (5, 30). Suitable RNA size markers were synthesized in vitro with T7 or T3 RNA polymerase and with linearized pBluescript-based plasmids as templates.
β-Galactosidase assay.
B. japonicum cells were grown aerobically to exponential phase at 30°C in PSY medium with spectinomycin as the only antibiotic. The β-galactosidase assay was performed as described previously (22).
Overproduction and purification of H6-HrcA.
Freshly transformed E. coli BL21/pLysS cells carrying pRJ5552 were used for overproduction of H6-HrcA. The cells were grown at 30°C in 1 liter of Luria-Bertani LB medium containing chloramphenicol and kanamycin. When the cultures had reached an optical density at 600 nm of 0.5, production of the recombinant protein was induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 2 h, the cells were harvested, washed, resuspended in 20 ml of TEPDM buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, 25 mM MgCl2, 100 mM KCl), and disrupted in a French pressure cell. The soluble protein fraction was obtained by removing the cell debris and membranes in two subsequent centrifugation steps at 4°C (1.5 h at 30,000 × g [Sorvall SS-34 rotor] and 1 h at 116,000 × g [Beckman SW55 Ti rotor]).
All of the following purification steps were performed at 4°C. Binding buffer (8×) was added to the supernatant fraction to a final concentration of 500 mM NaCl before loading the fraction onto a 1.5-ml Ni-nitrilotriacetic acid-agarose column (Qiagen, Basel, Switzerland) which had been equilibrated with 1× binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 5 mM imidazole). The column was washed with 15 ml of 1× binding buffer and then with 4.5 ml of 1× binding buffer containing 120 mM imidazole. The H6-HrcA protein was eluted with 4.5 ml of 1× binding buffer containing 300 mM imidazole and collected in fractions of 0.5 ml. The fractions with the highest protein concentrations were pooled, dialyzed for 2 h against storage buffer (20 mM Tris-HCl [pH 8], 200 mM NaCl, 10% glycerol) and centrifuged at 13,000 × g for 1 min. Aliquots of the supernatant were stored at −80°C. The concentration of purified H6-HrcA protein was determined by the Bradford method (6).
Gel retardation assay.
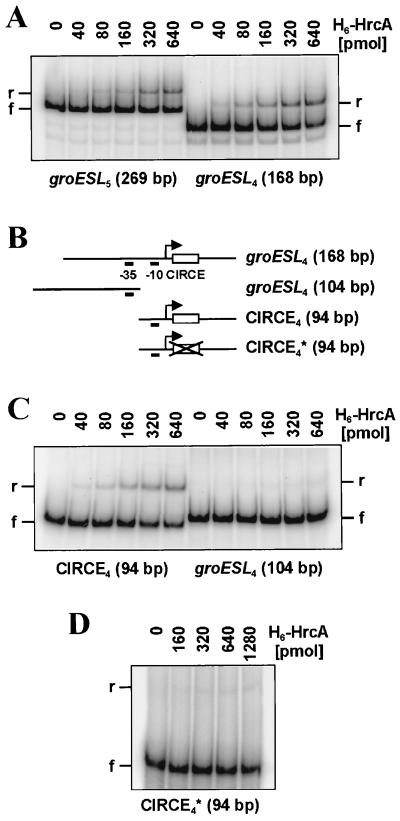
The DNA-binding activity of H6-HrcA was tested in gel shift experiments using a 269-bp PstI-KpnI fragment from pRJ5558 containing the promoter region and the CIRCE element of groESL5, a 168-bp HpaI-KpnI fragment from pRJ5556 containing the promoter region and the CIRCE element of groESL4, a 104-bp PCR amplification product containing the 5′ region of the groESL4 promoter region, or a 94-bp PCR-generated fragment containing either the wild-type CIRCE element of groESL4 (CTAGCACTCgcgggcacaGACTGCTAA [nucleotides in inverted repeat shown in uppercase letters and nucleotides identical to those in consensus sequence are underlined]) or a mutated DNA fragment in which the consensus sequence had been replaced (AGCTACAGAgcgggcacaAGACATCGA) (see Fig. 6B; CIRCE consensus sequence: TTAGCACTC-N9-GAGTGCTAA [17]). These DNA fragments were purified from agarose gels and end labeled with [γ-32P]ATP according to standard protocols. Labeled fragments (25,000 cpm; approximately 10 to 20 fmol of DNA) were mixed with purified H6-HrcA protein in DNA binding buffer (12 mM HEPES [pH 7.9], 4 mM Tris-HCl [pH 8], 6 mM KCl, 3 mM MgCl2, 0.5 mM dithiothreitol, 6 mM EDTA; Stratagene, La Jolla, Calif.) in a final volume of 25 μl. If appropriate, the E. coli chaperones DnaK, DnaJ, and GrpE or GroES and GroEL (Epicentre Technologies, Madison, Wis.) were added to H6-HrcA, and the mixture was preincubated at room temperature for 10 min in DNA binding buffer containing ATP (1.2 mM) and magnesium acetate (23 mM) before the binding reaction was started by the addition of DNA. The DNA-protein mixtures were incubated for 5 or 10 min at room temperature, mixed with 5 μl of loading dye (30% glycerol, 0.02% bromphenol blue in water), and then loaded onto 6% nondenaturing polyacrylamide gels (cross-linker ratio of 29:1 in 1× TBE buffer [pH 8; 89 mM Tris base, 89 mM boric acid, 2.5 mM EDTA]) containing 1 mg of Triton X-100/ml. Gels were run in 1× TBE buffer at 4°C, dried under vacuum, and exposed on a phosphorimager screen. Signal intensities of free DNA and retarded bands were quantified with a phosphorimager and the program ImageQuant (version 3.3; Molecular Dynamics, Sunnyvale, Calif.).
FIG. 6.
Gel retardation experiments with purified B. japonicum HrcA. DNA-binding reactions were performed with DNA fragments derived from the promoter region of groESL5 (A) and groESL4 (A, C, and D). (B) groESL4 fragments represented schematically. CIRCE4* represents a DNA fragment in which the CIRCE element was mutated (see Materials and Methods). The amount of H6-HrcA used is indicated above each lane in all panels. The positions of radiolabeled free DNA (f) and retarded bands (r) are marked.
Plant infection test.
The symbiotic phenotype of the B. japonicum hrcA mutant was determined in a soybean plant infection test as described previously (12, 15).
Nucleotide sequence accession numbers.
The nucleotide sequence of the B. japonicum hrcA and dnaK gene region has been deposited in the GenBank-EMBL database under accession no. Y09633.
RESULTS
Cloning the B. japonicum hrcA gene region.
In contrast to those of several other organisms, the B. japonicum hrcA gene is not part of the dnaK operon (23). Our initial attempts to identify the gene by heterologous hybridization with DNA or oligonucleotide probes derived from hrcA homologs failed. The fact that the C. crescentus hrcA gene is closely linked to grpE (38) prompted us to search first for a grpE homolog of B. japonicum by a complementation approach. Assuming that B. japonicum GrpE would be functional in E. coli, we attempted to rescue the heat-sensitive grpE-deficient E. coli strain DA259 (kindly provided by D. Ang, Geneva, Switzerland) with B. japonicum DNA cloned into pUC18. The inability of this strain to grow at 39 or 42°C could be fully complemented by the presence of plasmid pRJ5523, which contains a 3.9-kb EcoRI fragment of B. japonicum (Table 2; Fig. 1). DNA sequence analysis revealed an open reading frame coding for a protein with a high degree of similarity to all known GrpE proteins. As we had hoped, an open reading frame (3′ end) coding for the C terminus of an HrcA-like protein was present upstream of grpE. Incidentally, the 5′ end of B. japonicum dnaK was found only 2.4 kb downstream of the hrcA-grpE cluster (Fig. 1). The complete B. japonicum hrcA region was subcloned as a 3-kb NotI fragment originating from a suitable cosmid, resulting in plasmid pRJ5530 (Fig. 1).
TABLE 2.
Temperature-dependent growth of E. coli DA259 transformed with different plasmids
| E. coli strain | Plasmid | Growth at:
|
||
|---|---|---|---|---|
| 30°C | 39°C | 42°C | ||
| DA258 (wild type) | + | + | + | |
| DA259 (grpE mutant) | + | − | − | |
| DA259 (grpE mutant) | pUC18 | + | − | − |
| DA259 (grpE mutant) | pBW401 (E. coli grpE+) | + | + | + |
| DA259 (grpE mutant) | pRJ5523 (B. japonicum grpE+) | + | + | + |
To address the question of whether there is more than one copy of the hrcA or grpE gene in the B. japonicum chromosome, as it is the case for groESL (10) and rpoH (32), suitable hrcA and grpE probes were hybridized under low-stringency conditions (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 56°C) with B. japonicum chromosomal DNA that had been digested with different restriction enzymes. No evidence for an hrcA or grpE gene family was obtained.
Nucleotide sequence of the hrcA gene region.
The DNA sequence of the B. japonicum hrcA gene region, extending from the NotI site at position 2 to the XhoI site at position 3451, was established (Fig. 1), expanding the previously described sequence of the dnaKJ gene region (23). Significant similarities of the deduced amino acid sequences to sequences of C. crescentus HrcA (54% identical amino acids) and GrpE (44%) (38) led to the more precise assignment of B. japonicum hrcA (position 433 to 1521) and grpE (position 1619 to 2224). The hrcA gene begins with the alternative start codon GTG and encodes a predicted protein of 362 amino acids (Mr, 39,165). An alignment of the B. japonicum hrcA product with 18 HrcA-like proteins deposited in the publicly available databases displayed only limited sequence similarity (overall sequence identity generally around 30% with the exception of HrcA proteins from related organisms, such as B. japonicum, Agrobacterium tumefaciens, and C. crescentus, that have about 50% identical amino acids). The similarity is restricted mainly to three previously described conserved regions (43).
The deduced gene product of grpE consists of 201 amino acids and has a molecular weight of 21,655. Further investigation of the DNA region upstream of dnaK and hrcA revealed three additional open reading frames, all divergently oriented to hrcA and dnaK. The first open reading frame (orf>86) starts at position 258 and codes for the amino-terminal end of a polypeptide of at least 86 amino acids with a high degree of sequence similarity to the putative tRNA nucleotidyltransferase RnpH of C. crescentus (67% identical amino acids) encoded by the rph gene (38). Moreover, the genetic organization of orf>86, hrcA, and grpE is similar to that of C. crescentus. The gene product of orf240 (Mr, 25,406) shows a high degree of sequence similarity to pyrazin nicotinamidases (e.g., 41% of the amino acids are identical to those of the putative pncA gene product of Aquifex aeolicus [8]). The orf313 product (Mr, 33,241) exhibits no significant similarity to any known protein sequence in the database.
Transcriptional analysis of the hrcA gene region.
The transcription start sites within the B. japonicum hrcA gene region were determined by primer extension analysis with oligonucleotides complementary to the 5′ ends of hrcA and grpE. A single start site was detected for each gene: 21 nucleotides upstream of the proposed translational start site of hrcA (Fig. 2A) and 36 nucleotides upstream of grpE (Fig. 2B). Both transcripts were clearly heat inducible, as indicated by 2.3- and 19-fold increases of the reverse transcription products for hrcA and grpE, respectively, after a 30-min temperature shift of the cells from 30 to 43°C. The deduced promoter regions displayed characteristic sequence motifs of heat-inducible ς32-dependent promoters (Fig. 2C).
FIG. 2.
Determination of the transcription start sites of hrcA (A) and grpE (B) by primer extension mapping. Total RNA was isolated from B. japonicum 110spc4 cells harvested before (0) and 30 min after a heat shock from 30 to 43°C (30). The extension and sequencing reactions (TCGA) were performed with the primers HrcA11 (A) and GrpE4 (B). The transcription start sites are marked with arrows, and the deduced promoter sequences are shown below (C). Nucleotides matching those of the E. coli ς32 consensus promoter (52) and transcriptional start sites are in boldface.
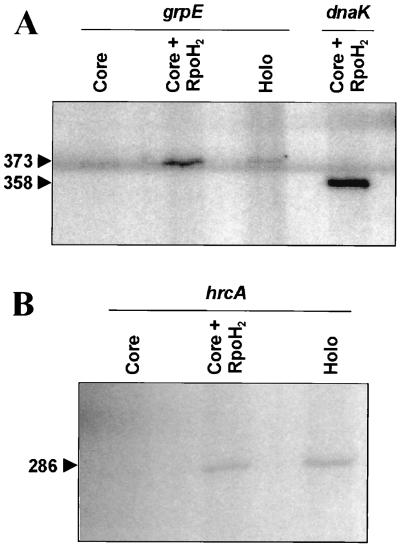
The presumed ς32-dependent transcription of hrcA and grpE was further confirmed by in vitro transcription of both genes with the B. japonicum core RNA polymerase reconstituted with purified RpoH2-H6 protein, a C-terminally histidine-tagged version of this protein. The RpoH2 factor is responsible for expression of ς32-dependent genes under normal growth conditions. The resulting hrcA and grpE transcripts derived from plasmids pRJ5542 and pRJ5543, respectively, are of the expected lengths (286 and 373 nucleotides, respectively; Fig. 3), as judged from a comparison with RNA size markers (not shown) and the dnaKJ transcript (358 nucleotides), which was previously shown to be synthesized efficiently by core RNA polymerase reconstituted with RpoH2-H6 (Fig. 3A) (30). The core enzyme alone produced only small amounts of transcript (Fig. 3A), most probably due to some residual ς32 protein present in the preparation, as described elsewhere (30). Similarly, hrcA and grpE transcripts were obtained with the B. japonicum RNA polymerase holoenzyme, which contains a significant amount of RpoH protein (30). According to these results we conclude that hrcA and grpE belong to the rpoH regulon of B. japonicum together with the genes coding for the DnaK machinery.
FIG. 3.
In vitro transcription from the B. japonicum grpE and dnaK (A) and hrcA (B) promoters. The enzymes used were B. japonicum RNA polymerase core enzyme (Core), purified RpoH2-H6 protein (RpoH2), and B. japonicum RNA polymerase holoenzyme (Holo). Plasmids pRJ5099, pRJ5542, and pRJ5543 were used as templates to transcribe the 5′ ends of dnaK, hrcA, and grpE, respectively. Numbers on the left mark transcript lengths (in nucleotides) of the expected transcripts; lengths were confirmed with suitable in vitro-synthesized RNA size markers (not shown). The signal extending across the entire gel (A) was caused by overloaded RNA size markers on the left.
Heat shock gene expression in an hrcA deletion mutant.
The B. japonicum hrcA mutant 5549 was constructed by replacing an internal hrcA fragment with a kanamycin resistance cassette (Fig. 1; see also Materials and Methods). The effect of the hrcA deletion on the expression of heat shock genes was determined by both primer extension analysis and β-galactosidase measurements of suitable translational lacZ fusions.
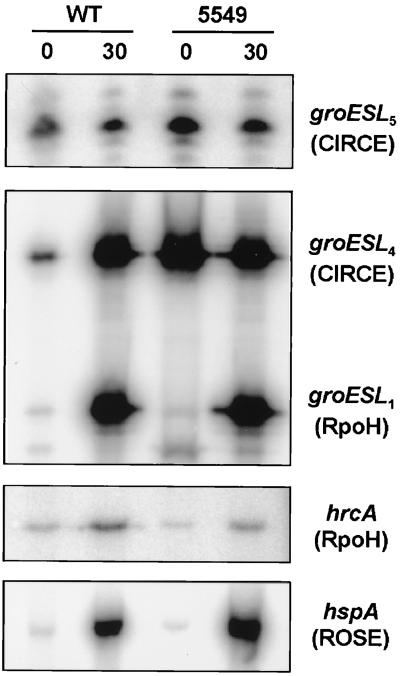
The analysis of RNA isolated from wild-type B. japonicum and strain 5549 revealed strongly derepressed transcription of the CIRCE-dependent genes groESL4 and groESL5 under non-heat-shock conditions (Fig. 4). The transcription intensity slightly surpassed even the intensity of heat-induced transcription after a 30-min heat shock from 30 to 43°C. By contrast, transcription of ς32-dependent genes (e.g., groESL1 and hrcA) and heat shock genes regulated by the ROSE element (e.g., hspA) was not influenced by the lack of HrcA. In line with the absence of a CIRCE element upstream of B. japonicum hrcA, the gene is not subject to autoregulation as it is in B. subtilis (43).
FIG. 4.
Effect of an hrcA mutation on the transcription of different heat shock genes in B. japonicum. Total RNA was isolated from wild-type (WT) B. japonicum and 5549 (hrcA mutant) cells harvested before (0) and 30 min after a heat shock from 30 to 43°C (30) and analyzed by primer extension experiments. The extension reactions were performed with the primers ES5UP2 (groESL5) (3), 702 (groESL4 and groESL1) (3), HrcA11 (hrcA), and Sig107 (hspA) (29). The mode of regulation of each gene or operon is indicated.
Table 3 shows the expression of different lacZ fusions that were integrated into the chromosome of wild-type B. japonicum and strain 5549 (see Materials and Methods for the construction of the strains). The two CIRCE-regulated fusions (groEL4′-′lacZ and groEL5′-′lacZ) exhibited an increased expression in the absence of HrcA, which supports the primer extension results. However, the calculated induction factors (3.2 for groEL4′-′lacZ and 2.3 for groEL5′-′lacZ) appeared to be lower as compared with the strongly increased transcription of the corresponding genes in strain 5549 (Fig. 4). This is probably due to a posttranscriptional effect of CIRCE, which functions as an mRNA destabilizer promoting the rapid turnover of CIRCE-containing transcripts (19, 50).
TABLE 3.
Effect of an hrcA deletion on the expression of chromosomally integrated ′lacZ fusions in B. japonicum
| B. japonicum strainsa | ′lacZ fusion | Induction factorb | Mode of regulation |
|---|---|---|---|
| H8092/8092 | groEL5′-′lacZ | 2.3 ± 0.6 | CIRCE |
| H8548/8548 | groEL4′-′lacZ | 3.2 ± 0.7 | CIRCE |
| H8067/8067 | groEL1′-′lacZ | 1.2 ± 0.3 | RpoH |
| H8174/8174 | dnaK′-′lacZ | 0.9 ± 0.1 | RpoH |
| H5554/5554 | grpE′-′lacZ | 0.9 ± 0.1 | RpoH |
| H5559/5559 | hrcA′-′lacZ | 2.8 ± 0.6 | RpoH |
| H5400/5400 | hspA′-′lacZ | 1.1 ± 0.3 | ROSE |
| H7998/7998 | groEL2′-′lacZ | 1.3 ± 0.7 | Constitutive |
The indicated ′lacZ fusion (see also Table 1) was chromosomally integrated into wild-type B. japonicum (resulting strains without prefix) and into hrcA mutant 5549 (strains with prefix “H”).
The induction factor is the ratio of the measured β-galactosidase activity in the hrcA mutant to that for the wild-type background. Values are means ± standard deviations and are based on at least three independent measurements.
The lack of HrcA did not affect the expression of the lacZ fusions to groEL1, grpE and dnaK (ς32 regulated), hspA (ROSE regulated), and groEL2 (constitutively expressed) (Table 3). The finding for groEL2 supports an earlier notion that the putative element homologous to CIRCE in the promoter region of groESL2 might not be functional (3). Although transcription from the hrcA promoter was not influenced by a deletion of the corresponding gene (Fig. 4), the expression of the hrcA′-′lacZ fusion was increased 2.8-fold in strain H5559 (Table 3). Posttranscriptional events, not further investigated here, might be responsible for this effect.
Complementary evidence that derepression of the groESL4 and groESL5 operons in the hrcA mutant raised the cellular GroEL pool was obtained from immunoblots performed with anti-E. coli GroEL serum and by two-dimensional gel analysis (data not shown).
Phenotypic characterization of an hrcA deletion mutant under symbiotic and heat shock conditions.
As it was known that GroEL is critical for nitrogen fixation in B. japonicum (11), we tested whether the elevated GroESL4 and GroESL5 concentrations in the hrcA mutant would affect the performance of B. japonicum in root nodule symbiosis and under heat stress conditions. Strain 5549 (hrcA mutant) was indistinguishable from the wild-type strain with respect to the ability to nodulate soybean roots and to fix nitrogen. Strain 5549 was further analyzed for its ability to survive a temperature upshift from 30 to 48°C, which is lethal to wild-type B. japonicum (33). The survival rate of strain 5549 after exposure to the nonpermissive temperature showed no significant deviation from that for the wild type. Since the elevated GroEL concentration caused by the hrcA deletion did not improve the heat tolerance of B. japonicum, we conclude that the GroEL pool in wild-type B. japonicum cells is not limiting under the conditions tested.
Purification of soluble H6-HrcA.
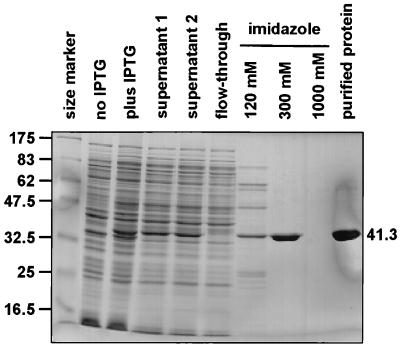
To obtain in vitro evidence for a physical interaction between CIRCE and HrcA we aimed at performing gel retardation assays with purified components. To this end, an amino-terminally histidine-tagged version of B. japonicum HrcA (H6-HrcA) was expressed in E. coli. In contrast to overproduced HrcA from other organisms, a large fraction of B. japonicum H6-HrcA remained in the supernatant even after ultracentrifugation (Fig. 5). This allowed purification under nondenaturing conditions, yielding approximately 4 mg of purified protein per liter of E. coli culture.
FIG. 5.
Overexpression and purification of H6-HrcA. Crude extracts of the H6-HrcA-overproducing E. coli strain harvested before and after induction with IPTG, aliquots from each purification step, and 2 μl of the dialyzed protein fraction, which was subsequently used for in vitro experiments, were separated on an SDS–12% polyacrylamide gel. The proteins were stained with Coomassie blue. Supernatants 1 and 2 mark aliquots of the soluble protein fraction after centrifugation at 30,000 and 116,000 × g, respectively (see Materials and Methods). The apparent molecular masses (in kilodaltons) of H6-HrcA (41.3) and the marker proteins are indicated.
Specific binding of purified HrcA to CIRCE.
Gel retardation experiments with increasing amounts of H6-HrcA and with radioactively labeled DNA fragments containing CIRCE and the promoter region of groESL4 or groESL5 showed a protein-dependent DNA retardation (Fig. 6A). The groESL4 fragment was used to further delineate the DNA region responsible for HrcA binding (Fig. 6B). Clear DNA binding of H6-HrcA was observed only with the 94-bp subfragment that contained the −10 promoter region and CIRCE (Fig. 6C), not with a comparable fragment in which the CIRCE element was replaced by a sequence that had no similarity to CIRCE (Fig. 6D; for details see Materials and Methods). The fact that even very high concentrations of the repressor (approximately a 100,000-fold molar excess of protein over DNA) did not result in an appreciable band shift with CIRCE4* clearly demonstrates the requirement of a CIRCE sequence for HrcA binding.
In an additional set of gel retardation experiments, we attempted to lend support to the concept that the HrcA DNA-binding activity might be chaperonin dependent. H6-HrcA was preincubated in binding buffer containing either E. coli GroEL and GroES (approximate molar ratio of protomers of HrcA, GroEL, and GroES, 3:10:1 or 3:40:4) or E. coli DnaK, DnaJ, and GrpE (approximate molar ratio of protomers of HrcA, DnaK, DnaJ, and GrpE, 10:10:1:1 or 10:40:4:4). Each DNA binding assay was started by the addition of a radiolabeled 168-bp DNA fragment containing the promoter region and the CIRCE element of groESL4. The DNA-binding activity of our soluble HrcA preparation was not improved by the presence of either the DnaK or GroE chaperone machinery. In fact, the addition of GroESL led to a reduction of HrcA-CIRCE complexes (data not shown). In control experiments we confirmed that the GroE preparation had chaperonin activity in the buffer system we used for the gel retardation experiments. This was done by measuring its ability to reactivate heat-denatured malate dehydrogenase (data not shown).
DISCUSSION
This work presents an in-depth in vivo and in vitro investigation of a representative CIRCE-HrcA system, which is responsible for a widespread heat shock control mechanism in bacteria. It has become evident during the last decade that repressor mechanisms are probably much more abundant among prokaryotes than the E. coli-type ς32 control of heat shock genes (27). In B. japonicum, the CIRCE system is embedded in a complex regulatory network. To elucidate the role of CIRCE in this network, it was imperative to clone the structural gene of HrcA, the putative repressor binding to this DNA element. The amino acid sequences of known HrcA proteins deviate greatly despite the high degree of sequence conservation of the CIRCE elements to which they bind (38, 43). This makes the search for an hrcA gene with nucleotide probes rather difficult (13). Here, we made use of the close genetic linkage between hrcA and grpE that is often observed and established a novel approach that facilitated the isolation of the B. japonicum hrcA gene by complementing a temperature-sensitive, grpE-deficient E. coli strain.
Both hrcA and grpE are transcribed separately from heat-inducible ς32-dependent promoters. The control of the HrcA repressor by a heat shock sigma factor demonstrates that different regulatory mechanisms involved in the B. japonicum heat shock response do not act independently but form a complex network. Evidence for a similar network of positive and negative heat shock regulatory mechanisms was also obtained from C. crescentus, in which hrcA transcription depends on a heat-inducible ς32-like promoter (38). Moreover, its groESL operon is subject to dual control by a heat-inducible ς32-dependent promoter and a CIRCE element which is involved in the cell cycle-regulated gene expression (4). Importantly, basal expression of B. japonicum hrcA at normal growth temperatures was observed, thus guaranteeing a sufficient amount of HrcA protein to repress the CIRCE regulon. The sigma factor RpoH2 appears to be responsible for this basal transcription, as it is for a certain threshold level of DnaK (30). Although not proven rigorously, it is reasonable to predict that RpoH1, the heat shock-induced RpoH factor of B. japonicum, is responsible for temperature induction of hrcA expression.
Elevated levels of GroESL in an hrcA mutant had no influence on free-living growth and symbiotic nitrogen fixation of B. japonicum. Similarly, no obvious effect with regard to growth under normal and stress conditions was observed for C. crescentus and Streptomyces albus hrcA mutants (13, 38). However, comparable studies of B. subtilis showed that the hrcA mutant was able to recover from a shift to lethal temperatures whereas the wild type was not (51). A possible reason for this discrepancy between the bacterial species lies in the fact that B. japonicum, C. crescentus, and S. albus hrcA mutants enhance only the concentration of GroE chaperonins whereas the lack of HrcA in B. subtilis leads to an overexpression of both chaperone complexes, DnaKJ-GrpE and GroESL. Probably, only increased levels of the complete set of major chaperones improve the capacity of bacterial cells to survive temperature upshifts. This interpretation is also supported by the model in which DnaK and GroE chaperone complexes interact with each other in order to cope with heat shock situations (16).
Previous attempts to perform in vitro studies with HrcA have been impeded by the insolubility of this protein. Gel retardation experiments could only be performed with cell extracts containing HrcA (9, 20) or with renatured HrcA that had been purified under denaturing conditions (25). By contrast, we found that a large fraction of B. japonicum H6-HrcA remained in a soluble form upon overproduction and during purification. The reason for the comparatively high solubility of B. japonicum HrcA remains unknown at present. Gel mobility experiments performed with this preparation corroborated the in vivo results in showing a sequence-specific binding of HrcA to CIRCE. However, a large excess of HrcA over DNA was necessary to obtain a band shift. This might be due to an intrinsic tendency of the repressor to aggregate or to reach a nonfunctional conformation. It should be noted that our B. japonicum HrcA preparation also tended to precipitate from solution over time. Thus, only a small fraction of the purified protein might really be active. Another, physiologically more relevant reason for the poor retardation activity might be an inherently weak or transient interaction between CIRCE and its repressor. This could explain why both CIRCE-dependent operons in B. japonicum are only partially repressed in vivo at normal growth temperatures, resulting in an appreciable basal transcription of the corresponding genes (Fig. 4) (3). Leaky transcription from CIRCE-regulated promoters was also reported for B. subtilis, Clostridium acetobutylicum, C. crescentus, and other organisms (28, 38, 42), indicating that the CIRCE binding site is not permanently occupied by HrcA. It thus appears that the CIRCE-HrcA system is not designed for complete repression of heat shock genes under physiological temperatures because major chaperones are also required for folding processes during normal growth.
There is compelling in vivo evidence that HrcA activity depends on the presence of GroESL (3, 25). The interpretation of these findings was an appealing titration model in which the availability of GroE chaperones controls HrcA activity. Our failure to simulate this feedback control in vitro appears incompatible with this model. While gel retardation of a CIRCE fragment with B. stearothermophilus HrcA was significantly improved in the presence of GroEL (25), our gel shift experiments showed that the addition of the complete GroE machinery rather impaired the formation of B. japonicum HrcA-CIRCE complexes. One obvious difference between our experiments with B. japonicum HrcA and those with B. stearothermophilus HrcA is that the latter were performed in the presence of 0.5 M urea after it had been completely denatured during purification (25). It is thus conceivable that GroEL conferred a rather general chaperone activity on this partially denatured HrcA preparation, as was indicated by the prevention of HrcA aggregation in the presence of GroEL. Moreover, it remains unclear why stimulation of gel retardation was observed even in the absence of GroES and ATP (25). The prevention of aggregate formation in the presence of urea might enable partially unfolded HrcA protein to reach an active DNA-binding conformation and, hence, a measurable gel retardation activity with a CIRCE fragment. Experiments with C. acetobutylicum HrcA (OrfA), which had been purified under denaturing conditions, revealed that HrcA aggregation was prevented by the addition of DnaK, DnaJ, or the whole DnaK chaperone machinery (39), indicating that urea-denatured HrcA is a substrate not only for the GroE chaperone machinery but also for the DnaK system. While the specific interaction between the CIRCE element and HrcA is well established now, it becomes evident that resolution of the more-debated matters of a physical interaction between HrcA and GroEL and the thermosensing by HrcA must await further studies.
ACKNOWLEDGMENTS
We are indebted to Debbie Ang for providing plasmid pBW401 and the E. coli strains DA258 and DA259, to Christoph Beck for B. japonicum RNA polymerase, and to Michael Kowarik for purified B. japonicum RpoH2-H6 protein. We thank Christopher Kaestner for help in automatic DNA sequencing and Michael Spring, Stephanie Häussler, and Domenic Graf for technical assistance. We thank Martin Münchback and Peter James for performing two-dimensional gel electrophoresis.
This work was supported by grants from the Swiss National Foundation for Scientific Research and the Federal Institute of Technology, Zürich, Switzerland.
REFERENCES
- 1.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–54. [PubMed] [Google Scholar]
- 2.Ang D, Georgopoulos C. The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J Bacteriol. 1989;171:2748–2755. doi: 10.1128/jb.171.5.2748-2755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 4.Baldini R L, Avedissian M, Gomes S L. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J Bacteriol. 1998;180:1632–1641. doi: 10.1128/jb.180.7.1632-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck C, Marty R, Kläusli S, Hennecke H, Göttfert M. Dissection of the transcription machinery for housekeeping genes of Bradyrhizobium japonicum. J Bacteriol. 1997;179:364–369. doi: 10.1128/jb.179.2.364-369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 8.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 9.Duchêne A M, Thompson C J, Mazodier P. Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2) Mol Gen Genet. 1994;245:61–68. doi: 10.1007/BF00279751. [DOI] [PubMed] [Google Scholar]
- 10.Fischer H M, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer H M, Schneider K, Babst M, Hennecke H. GroEL chaperonins are required for the formation of a functional nitrogenase in Bradyrhizobium japonicum. Arch Microbiol. 1999;171:279–289. [Google Scholar]
- 12.Göttfert G, Hitz S, Hennecke H. Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol Plant-Microbe Interact. 1990;3:308–316. doi: 10.1094/mpmi-3-308. [DOI] [PubMed] [Google Scholar]
- 13.Grandvalet C, Rapoport G, Mazodier P. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J Bacteriol. 1998;180:5129–5134. doi: 10.1128/jb.180.19.5129-5134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 15.Hahn M, Hennecke H. Localized mutagenesis in Rhizobium japonicum. Mol Gen Genet. 1984;193:46–52. [Google Scholar]
- 16.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 17.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 18.Herman C, D'Ari R. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr Opin Microbiol. 1998;1:204–209. doi: 10.1016/s1369-5274(98)80012-x. [DOI] [PubMed] [Google Scholar]
- 19.Homuth G, Mogk A, Schumann W. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol Microbiol. 1999;32:1183–1197. doi: 10.1046/j.1365-2958.1999.01428.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda M, Kobayashi D, Honda K, Hayashi H, Ohta T. The hsp operons are repressed by the hrc37 of the hsp70 operon in Staphylococcus aureus. Microbiol Immunol. 1999;43:19–27. doi: 10.1111/j.1348-0421.1999.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 21.Mager W H, De Kruijff A J J. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 23.Minder A C, Narberhaus F, Babst M, Hennecke H, Fischer H M. The dnaKJ operon belongs to the ς32-dependent class of heat shock genes in Bradyrhizobium japonicum. Mol Gen Genet. 1997;254:195–206. doi: 10.1007/s004380050408. [DOI] [PubMed] [Google Scholar]
- 24.Minton N P. Improved plasmid vectors for the isolation of translational lac gene fusion. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 25.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morimoto R I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 27.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 28.Narberhaus F, Bahl H. Cloning, sequencing, and molecular analysis of the groESL operon of Clostridium acetobutylicum. J Bacteriol. 1992;174:3282–3289. doi: 10.1128/jb.174.10.3282-3289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narberhaus F, Käser R, Nocker A, Hennecke H. A novel DNA element that controls bacterial heat shock gene expression. Mol Microbiol. 1998;28:315–323. doi: 10.1046/j.1365-2958.1998.00794.x. [DOI] [PubMed] [Google Scholar]
- 30.Narberhaus F, Kowarik M, Beck C, Hennecke H. Promoter selectivity of the Bradyrhizobium japonicum RpoH transcription factors in vivo and in vitro. J Bacteriol. 1998;180:2395–2401. doi: 10.1128/jb.180.9.2395-2401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narberhaus F, Krummenacher P, Fischer H M, Hennecke H. Three disparately regulated genes for ς32-like transcription factors in Bradyrhizobium japonicum. Mol Microbiol. 1997;24:93–104. doi: 10.1046/j.1365-2958.1997.3141685.x. [DOI] [PubMed] [Google Scholar]
- 32.Narberhaus F, Weiglhofer W, Fischer H M, Hennecke H. The Bradyrhizobium japonicum rpoH1 gene encoding a ς32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J Bacteriol. 1996;178:5337–5346. doi: 10.1128/jb.178.18.5337-5346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narberhaus F, Weiglhofer W, Fischer H M, Hennecke H. Identification of the Bradyrhizobium japonicum degP gene as part of an operon containing small heat-shock protein genes. Arch Microbiol. 1998;169:89–97. doi: 10.1007/s002030050547. [DOI] [PubMed] [Google Scholar]
- 34.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 35.Ohta T, Nettikadan S, Tokumasu F, Ideno H, Abe Y, Kuroda M, Hayashi H, Takeyasu K. Atomic force microscopy proposes a novel model for stem-loop structure that binds a heat shock protein in the Staphylococcus aureus HSP70 operon. Biochem Biophys Res Commun. 1996;226:730–734. doi: 10.1006/bbrc.1996.1421. [DOI] [PubMed] [Google Scholar]
- 36.Ramseier T, Göttfert M. Codon usage and G+C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156:270–276. doi: 10.1007/BF00262997. [DOI] [PubMed] [Google Scholar]
- 37.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 38.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rüngeling E, Laufen T, Bahl H. Functional characterisation of the chaperones DnaK, DnaJ and GrpE from Clostridium acetobutylicum. FEMS Microbiol Lett. 1999;170:119–123. doi: 10.1111/j.1574-6968.1999.tb13363.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schön U, Schumann W. Molecular cloning, sequencing, and transcriptional analysis of the groESL operon from Bacillus stearothermophilus. J Bacteriol. 1993;175:2465–2469. doi: 10.1128/jb.175.8.2465-2469.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulations of Gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria plant interaction. Heidelberg, Germany: Springer Verlag; 1983. pp. 98–106. [Google Scholar]
- 46.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 47.van Asseldonk M, Simons A, Visser H, de Vos W M, Simons G. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J Bacteriol. 1993;175:1637–1644. doi: 10.1128/jb.175.6.1637-1644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R F, Kushner S R. Construction of versatile low-copy number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 49.Wu B, Ang D, Snavely M, Georgopoulos C. Isolation and characterization of point mutations in the Escherichia coli grpE heat shock gene. J Bacteriol. 1994;176:6965–6973. doi: 10.1128/jb.176.22.6965-6973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan G, Wong S-L. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 53.Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
- 54.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]