Abstract
Salmonella enterica serovar Typhimurium responds to superoxide-generating agents through soxR-mediated activation of the soxS gene, whose product, SoxS, is necessary for resistance to oxidative stress. The S. enterica serovar Typhimurium soxRS system also mediates redox-inducible resistance to diverse antibiotics, which may be relevant to clinical infections. In order to identify SoxS-regulated genes in S. enterica serovar Typhimurium, a lacI-regulated expression system for the S. enterica serovar Typhimurium soxS gene was developed. This system was used to demonstrate that soxS expression is sufficient for the induction of resistance to the superoxide-generating drug paraquat and for the transcriptional activation of the sodA and micF genes. In addition, a library of random lacZ insertions was generated and screened for clones displaying differential β-galactosidase activity in the presence or absence of SoxS. This selection yielded six independent chromosomal lacZ transcriptional fusions that were activated by either artificial expression of SoxS or exposure of wild-type cells to micromolar concentrations of paraquat. Moreover, disruption of the inducible genes by the insertions rendered S. enterica serovar Typhimurium hypersensitive to millimolar concentrations of paraquat. Nucleotide sequence determination identified the disrupted genes as sodA (Mn-containing superoxide dismutase), fpr (NADPH:ferredoxin oxidoreductase), and ydbK (a putative Fe-S-containing reductase).
Aerobic organisms obtain energy by the oxidation of organic compounds, with oxygen as the final electron acceptor. As a by-product of this process, reactive oxygen species are generated, with potentially damaging consequences for the cell. Escherichia coli responds to the intracellular increase in reactive oxygen species by inducing sets of genes whose products either avert or repair the eventual oxidative damage (19). The response to increased levels of superoxide is regulated by the soxRS system, a pair of regulatory genes that, together with their downstream-regulated genes, define the soxRS regulon (3, 37).
The SoxR protein is expressed constitutively and is a homodimeric transcriptional regulator that contains redox-active iron-sulfur clusters (18, 38). The oxidation state of the iron-sulfur clusters in SoxR regulates the transcriptional activity of the protein (20): while reduced SoxR does not affect transcription, oxidized SoxR dramatically enhances the transcription rate of soxS, a gene that codes for a second transcriptional activator (22, 23). The SoxS protein is a member of the AraC/XylS family of transcriptional regulators, and enhanced expression of SoxS activates at least 15 genes (20, 24), including sodA (Mn-containing superoxide dismutase), zwf (glucose-6-phosphate dehydrogenase), micF (antisense RNA to the porin OmpF mRNA), nfo (DNA repair endonuclease IV), fpr (NADPH:ferredoxin oxidoreductase), acrAB (efflux pump), acn (aconitase), fumC (heat-resistant fumarase), and nfsA (nitroreductase A). The induction of SoxS also results in enhanced resistance to multiple antibiotics (3, 4), dependent on micF (11) and acrAB (25).
Recently, the soxRS genes from Salmonella enterica serovar Typhimurium were cloned (B. L. Martins, P. J. Pomposiello, Z. Li, and B. Demple, unpublished data). Their nucleotide sequence (GenBank accession no. U61147) showed 97% identity of the predicted polypeptides with SoxR and SoxS of E. coli. Moreover, the regulatory region between the two genes is nearly identical to that of E. coli. The cloned soxRS genes were used to generate soxRS deletion mutants by allelic exchange (Martins et al., unpublished data). These ΔsoxRS strains are hypersensitive to the superoxide-generating agent paraquat (PQ), fail to induce the expression of nfo and sodA, and do not show increased resistance to antibiotics in response to treatment with PQ. Additionally, an independent study (16) showed that the soxS gene from S. enterica serovar Typhimurium is required for activation of sodA expression and resistance to redoxcycling agents.
Despite the similarity of the soxRS genes between S. enterica serovar Typhimurium and E. coli, enzyme activity measurements suggested that the soxRS regulons from E. coli and S. enterica serovar Typhimurium are different: while incubation with PQ induced the SodA protein and endonuclease IV, it failed to induce glucose-6-phosphate dehydrogenase or fumarase C in wild-type S. enterica serovar Typhimurium (Martins et al., unpublished data). The evolution of E. coli and S. enterica serovar Typhimurium under different environmental pressures might have led to differential recruitment of genes into the soxRS regulon. Therefore, a detailed description of the S. enterica serovar Typhimurium soxRS regulon might yield insights into the mechanisms of resistance to antibiotics and antimicrobial compounds generated by the immune system.
In order to characterize the soxRS regulon of S. enterica serovar Typhimurium, we built an inducible soxS expression system that is uncoupled from oxidative stress. We used this system both to measure the effect of the expression of SoxS on genes known to be under soxRS control in E. coli and to identify novel targets of soxS regulation in S. enterica serovar Typhimurium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Table 1 shows the bacterial strains and plasmids used in this work. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) broth (27) with vigorous aeration by shaking at 250 rpm. Ampicillin (100 μg/ml) and kanamycin (50 μg/ml) were added when necessary. The lacZ inducer isopropyl-β-d-thiogalactopyranoside (IPTG) and PQ were added at the final concentrations described in the figure legends. The indicator 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to plates at a final concentration of 25 μg/ml.
TABLE 1.
Strains and plasmids
| Straina or plasmid | Genotype or characteristic | Reference or source |
|---|---|---|
| Strains | ||
| ATCC 14028 | Wild type | American Type Culture Collection |
| SL4213 | Restriction deficient | J. Mekalanos |
| EM1 | ATCC 14028 ΔsoxRS, Tcr | UDb |
| PP120 | ATCC 14028 ΔsoxRS | UD |
| TT10288 | hisD9953::MudJ hisA9944::MudI | 21 |
| PP101 | ATCC 14028, Sin1 | This work |
| PP102 | ATCC 14028, Sin2 | This work |
| PP103 | ATCC 14028, Sin3 | This work |
| PP104 | ATCC 14028, Sin4 | This work |
| PP105 | ATCC 14028, Sin5 | This work |
| PP106 | ATCC 14028, Sin6 | This work |
| PP121 | PP120, Sin1 | This work |
| PP122 | PP120, Sin2 | This work |
| PP123 | PP120, Sin3 | This work |
| PP124 | PP120, Sin4 | This work |
| PP125 | PP120, Sin5 | This work |
| PP126 | PP120, Sin6 | This work |
| Plasmids | ||
| pBR322 | ColE1 Apr Tcr | 9 |
| pSE380 | ColE1 Amp lacI | Novagen |
| pET-21 | ColE1 Amp lacI | Invitrogen |
| pRZ4004 | ColE1 Amp lacZ promoter | W. Reznikoff |
| pJP103 | pBR322 lacI | This work |
| pJP104 | pJP103 soxS | This work |
| pJP105 | pJP104 laZp::soxSSt | This work |
| pDT1-5 | sodA from E. coli Apr | 11 |
| pBCKpn | soxR and soxS genes from S. enterica serovar Typhimurium Cmr | UD |
All strains are from S. enterica serovar Typhimurium.
UD, B. L. Martins, P. J. Pomposiello, Z. Li, and B. Demple, unpublished data.
DNA manipulation and construction of pJP105.
DNA purification, incubation with restriction enzymes, electrophoresis in agarose gels, ligation, and bacterial transformation were performed according to standard protocols (5). Phage P22 transductions were performed by standard methods (26). To construct plasmid pJP105, the S. enterica serovar Typhimurium soxS gene was subcloned from pBCKpn (Martins et al., unpublished data) as an HpaI-SalI fragment into pBR322 (9) digested with EcoRV and SalI, generating plasmid pJP103. The lacI gene from pSE380 (Novagen) was excised as a PshAI-SalI fragment and ligated to pJP103 previously digested with AvaI, followed by treatment with T4 DNA polymerase and SalI, to generate plasmid pJP104. The lacZ promoter from pRZ4004 (32) was liberated by digestion with EcoRI, filled in by Klenow fragment DNA polymerase, and digested with HindIII. Plasmid pJP104 was digested with ClaI, followed by incubation with T4 DNA polymerase and HindIII, and ligated to the lacZ promoter fragment from pRZ4004 to generate plasmid pJP105 (5.4 kbp). This construct was used to transform the intermediary S. enterica serovar Typhimurium strain SL4213 (restriction deficient), and plasmid DNA extracted from this transformant was used to transform strain EM1 (ΔsoxRS) to ampicillin resistance.
Northern blot analysis.
Bacterial cultures were grown overnight in LB broth at 37°C with the appropriate antibiotics, diluted to 1/100 in 2 ml of LB broth, and grown at 250 rpm in 18-mm-diameter tubes to mid-log phase. At an optical density at 600 nm of ∼0.5, growing cultures were exposed to different concentrations of PQ or IPTG for 30 min. Total RNA was extracted with an RNAeasy kit (Qiagen) and resuspended in RNAse-free water. The concentration of the total RNA preparations was determined by measuring the absorbance at 260 nm, and 2 μg of total RNA was run per lane in 1.25% agarose gels containing formaldehyde and transferred to Nytran membranes with a Turboblotter setup (Schleicher & Schuell). The RNA was cross-linked to the membrane by UV irradiation, and the membranes were then hybridized at 68°C with radioactively labeled DNA fragments in cylindrical tubes by using QuickHyb solution (Stratagene). The membranes were washed according to the instructions from the manufacturer. X-ray films were exposed to the membranes at −70°C and developed with a Fuji automatic developer. The radioactive signals were quantitated with an Applied Biosystems phosphorimager.
MudJ mutagenesis.
The MudJ (promoterless lacZ, Kanr) phage was delivered to the ΔsoxRS/pJP105 strain by P22 transduction from strain TT10288 (hisD::MudJ hisA::MudI) as described by Hughes and Roth (21). Kanamycin-resistant transductants were isolated on LB agar plates containing kanamycin and ampicillin. Each selection plate was replica plated onto LB agar plates containing kanamycin, ampicillin, and X-Gal. One of these plates also contained 1 mM IPTG to promote expression of SoxS from pJP105. Colonies displaying differences in blue coloration between the two plates were selected as candidates for harboring lacZ insertions in SoxS-regulated genes.
PQ sensitivity test on gradient plates.
The sensitivities to PQ of different strains were determined by the extent of linear growth on LB agar plates containing a gradient of PQ (13). Briefly, overnight cultures of the different strains were diluted 1/100 and grown in LB broth with aeration for 8 h and stamped on gradient plates containing different maximal concentrations of PQ. The gradient plates were incubated at 37°C for 16 h, and the length of confluent growth along the gradient was measured.
β-Galactosidase assay.
β-Galactosidase-specific activity in chloroform-treated cells was determined as described by Miller (27). Samples were taken from bacterial cultures grown overnight in LB broth, diluted 1/100, and grown with aeration for 2 h to an optical density at 600 nm of ∼0.5. At that point, the cultures were either exposed to PQ or IPTG for 30 min or left untreated.
Nonspecific PCR amplification.
Chromosomal DNA was extracted from each strain carrying a Mu insertion and used as a template in a PCR that included primer MuLEFT (5′-ATAATCCAATGTCCTCCCGG-3′), derived from the sequence of the left end of phage Mu, and a primer of nonspecific sequence (5′-ATTGGAGCGCAGGTTAGTCG-3′). Conditions for these first-round PCRs were as follows: 1.5 mM MgCl2, 0.5 U of Taq DNA polymerase (Sigma), 2 mM deoxynucleoside triphosphates (dNTPs), ∼0.2 μg of chromosomal DNA, and 50 pmol of each primer. Reaction mixtures were incubated for 10 cycles of 1 min at 40°C (annealing), 30 s at 72°C (extension), and 30 s at 95°C (denaturation), followed by 30 cycles of the same steps but with an annealing temperature of 55°C. A second PCR was conducted with 5 μl of the first reaction mixture as the template, the nested primer MuEND (5′-TACTTCAAGTGAATCAATACA-3′), derived from the sequence of the left end of phage Mu, and the same nonspecific primer used in the first reaction. Conditions for these second-round PCRs were as follows: 1.5 mM MgCl2, 0.5 U of Taq DNA polymerase (Sigma), 2 mM dNTPs, ∼0.2 μg of chromosomal DNA, and 50 pmol of each primer. Reaction mixtures were incubated for 30 cycles of 1 min at 55°C (annealing), 30 s at 72°C (extension), and 30 s at 95°C (denaturation). The extension products from these reactions were isolated from agarose gels and sequenced at the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute.
RESULTS
An engineered soxS expression system: SoxS is sufficient for enhanced resistance to PQ.
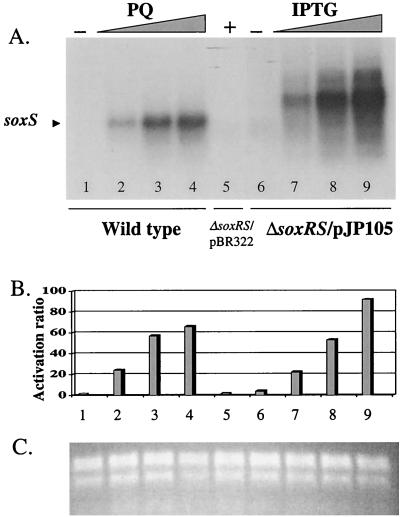
In order to analyze the direct contribution of SoxS to the activation of antioxidant defenses, we built a soxS expression system that uncoupled soxS transcription from oxidative stress. In plasmid pJP105, the lacZ promoter (33) drives the transcription of the soxS cistron. As this plasmid also bears the lacI gene, the lacZ promoter is expected to be repressed in the absence of the inducer IPTG. When pJP105 was used to transform S. enterica serovar Typhimurium EM1 (ΔsoxRS) and the expression of the plasmid-borne soxS gene was monitored by Northern analysis, soxS mRNA showed strong induction (up to 90-fold) by the gratuitous lac inducer IPTG (Fig. 1). As expected, the soxS message from pJP105 is longer than the wild-type message, due to the displaced transcriptional start site provided by the lacZ promoter and intervening cloning fragments, which add a total of 273 nucleotides. The induction level of the IPTG-regulated soxS transcript was comparable to that of the soxS transcript from wild-type strain ATCC 14028 treated with increasing concentrations of PQ (Fig. 1). In control experiments, untransformed strain EM1 showed no soxS message either in the presence or the absence of PQ (data not shown). We conclude from these results that the IPTG-induced expression of the soxS gene from plasmid pJP105 mimics the induction of wild-type soxS by PQ and that this heterologous expression can be regulated by the concentration of inducer in the growth medium.
FIG. 1.
Northern blotting analysis of soxS expression from strains ATCC 14028 and EM1/pJP105. (A) Northern blotting. A radioactively labeled 450-bp EcoRI-HpaI fragment of plasmid pBCKpn (Martins et al., unpublished data) containing the S. enterica serovar Typhimurium soxS coding region was hybridized with total RNA from strain ATCC 14028 (wild type) treated with increasing concentrations of PQ (0, 25, 100, or 250 μM; lanes 1 to 4, respectively) or strain EM1(ΔsoxRS)/pJP105 treated with increasing concentrations of IPTG (0, 0.125, 0.50, or 1.00 mM; lanes 6 to 9, respectively). The control strain, EM1/pBR322, was treated with 1.00 mM IPTG (lane 5). (B) Quantitation of the soxS radioactive signal. The radioactivity for the soxS band in each lane was quantitated by phosphorimaging. The values are the relative activations for the treatments, normalized to the signal in the absence of treatment in the wild-type samples and to the signal from lane 5 in panel A. The lane numbers correspond to those in panel A. (C) Total RNA loading. Shown is an ethidium bromide stain of a duplicate gel to that used for panel A.
In order to test the correlation between the heterologous soxS expression from pJP105 and the cellular resistance to oxidative stress, we assayed the ability of plasmid pJP105 to complement the PQ-sensitive phenotype of a ΔsoxRS strain. Figure 2 shows the growth of strains ATCC 14028 (wild-type), EM1 (ΔsoxRS), and EM1/pJP105 on PQ gradient plates (maximum concentration, 2 mM PQ) at increasing concentrations of IPTG. While the growth of strains ATCC 14028 and EM1 was unaffected by the IPTG concentrations, the resistance to PQ of strain EM1/pJP105 was increased by IPTG. In the absence of IPTG, the resistance of EM1/pJP105 was similar to that of EM1, consistent with the low levels of soxS message (Fig. 1A). As the IPTG concentration in the plates was increased, the resistance to PQ of strain EM1/pJP105 increased in parallel, reaching the wild-type level at 0.2 mM IPTG (Fig. 2). These results demonstrate that the regulated expression of soxS from pJP105 directly controls the phenotypic resistance to PQ in S. enterica serovar Typhimurium; thus soxS expression is not only necessary, as shown previously (16; Martins et al., unpublished data), but also sufficient for full-scale resistance to PQ in this organism, as it is in E. coli (3).
FIG. 2.
Complementation of the sensitivity to PQ by pJP105. Cultures of strains ATCC 14028, EM1, and EM1/pJP105 were grown overnight in LB broth and stamped onto PQ gradient plates with increasing concentrations of IPTG (0 to 2 mM) as indicated. The plates were incubated at 37°C for 14 h, and the extent of growth of each strain was measured. The values are the averages of one representative experiment done in duplicate, and the error bars are the standard deviations.
Induction of soxS and transcriptional activation of sodA and micF.
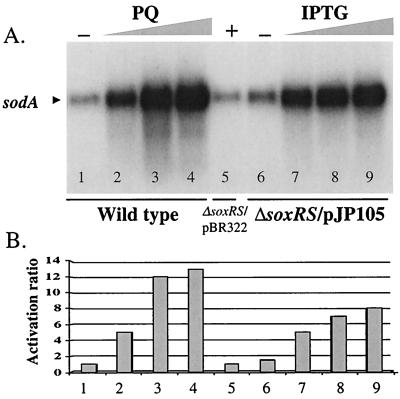
Enzyme assays have shown that SodA activity is induced in S. enterica serovar Typhimurium upon exposure to PQ in a soxRS-dependent manner (16; Martins et al., unpublished data). We probed the activation of the sodA gene in S. enterica serovar Typhimurium either by treatment of cells with PQ or by the regulated expression of SoxS in the absence of oxidative stress. Northern blotting shows a direct correlation between sodA induction (Fig. 3) and soxS expression (Fig. 1), either in response to PQ in the wild-type strain or by IPTG induction in EM1/pJP105. The maximal induction ratio for sodA expression in the wild-type strain after exposure to PQ was 13-fold, while the maximum in the ΔsoxRS/pJP105 strain treated with IPTG was 8-fold (Fig. 3). Thus, other pathways such as the marAB system (1) may contribute to the PQ-induced expression of sodA in S. enterica serovar Typhimurium. In fact, the induction ratio for sodA in a ΔsoxRS strain exposed to 0.25 mM PQ was threefold (data not shown). Nevertheless, transcriptional induction of sodA expression by SoxS is sufficient to account for most of the PQ-inducible SodA activity.
FIG. 3.
Northern blotting analysis of sodA transcription. (A) Northern blotting. The membrane used for Fig. 1 was stripped and rehybridized with a ∼1-kb AvaI fragment from plasmid pDT1.5 (12) containing the coding region of the sodA gene from E. coli. Strain ATCC 14028 (wild type) was treated with increasing concentrations of PQ (0, 25, 100, or 250 μM; lanes 1 to 4, respectively), and strain EM1(ΔsoxRS)/pJP105 was treated with increasing concentrations of IPTG (0, 0.125, 0.50, or 1.00 mM; lanes 6 to 9, respectively). The control strain, EM1/pBR322, was treated with 1.00 mM IPTG (lane 5). (B) Quantitation of the radioactive signals. The radioactivity for the soxS band in each lane was quantitated by phosphorimaging. The values are the relative activations for the treatments as described in the legend for Fig. 1. The lane numbers correspond to those used for panel A.
The periplasmic, Cu-Zn-containing superoxide dismutase encoded by the sodC gene protects S. enterica serovar Typhimurium from extracellular superoxide, and sodC mutants are less virulent than isogenic wild-type strains (14, 17). We tested whether sodC transcription is induced by oxidative stress or expression of soxS under normal growth. We performed Northern blotting of total RNA with a labeled fragment of the S. enterica serovar Typhimurium sodC gene (17). No consistent activation of sodC expression was observed, either in the wild-type strain treated with up to 0.25 mM PQ or in strain EM1/pJP105 treated with up to 1 mM IPTG (data not shown).
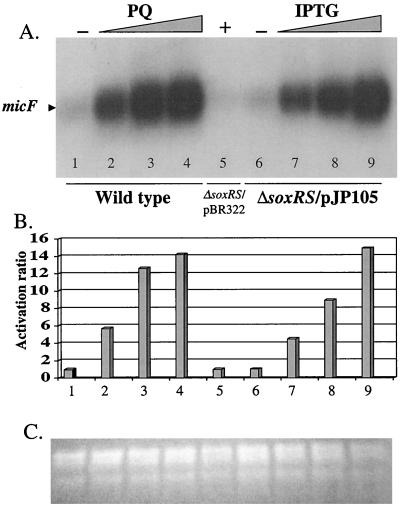
The micF gene is transcriptionally regulated by SoxS in E. coli. We tested for SoxS regulation of this gene in S. enterica serovar Typhimurium by analyzing the transcriptional activity of micF by Northern blotting. Preliminary studies showed that strain EM1, which we used in previous experiments, had an elevated endogenous level of micF RNA (data not shown). We hypothesized that this increase might be related to the tet cassette used to replace the soxRS region in EM1. We therefore turned to PP120, a different ΔsoxRS derivative of strain ATCC 14028 without a drug resistance cassette (Martins et al., unpublished data). This strain had a micF RNA level similar to that of the isogenic wild-type strain (Fig. 4, lane 1 versus lane 6). The transcription of micF increased dramatically with increasing concentration of PQ in a wild-type strain and with increasing IPTG concentration in strain PP120/pJP105. The maximal induction ratio for micF expression in the wild-type strain after exposure to PQ was 15-fold, and after treatment of PP120/pJP105 with IPTG it was also 15-fold (Fig. 4B). In control experiments, the activation ratio of micF expression in a ΔsoxRS strain exposed to PQ was only twofold (data not shown), again consistent with a small contribution by some other regulatory system. This result shows that SoxS is sufficient to account for nearly all of the PQ-inducible micF expression.
FIG. 4.
Northern blotting analysis of micF transcription. (A) Northern blotting. A 51-mer synthetic oligonucleotide derived from the sequence of the micF gene from S. enterica serovar Typhimurium (15) was labeled with T4 polynucleotide kinase and [32P]dATP. The probe was hybridized with total RNA from strain ATCC 14028 (wild type) treated with increasing concentrations of PQ (0, 25, 100, or 250 μM; lanes 1 to 4, respectively) or strain PP120(ΔsoxRS)/pJP105 treated with increasing concentrations of IPTG (0, 0.125, 0.50, or 1.00 mM; lanes 6 to 9, respectively). The control strain, PP120/pBR322, was treated with 1.00 mM IPTG (lane 5). (B) Quantitation of the radioactive signals. The radioactivity for the micF band in each lane was quantitated by phosphorimaging. The values are the relative activations for the treatments as described in the legend for Fig. 1. The lane numbers correspond to those used for panel A. (C) Total RNA loading. Shown is an ethidium bromide stain of a duplicate gel of total RNA.
MudJ mutagenesis and screening for SoxS-regulated genes.
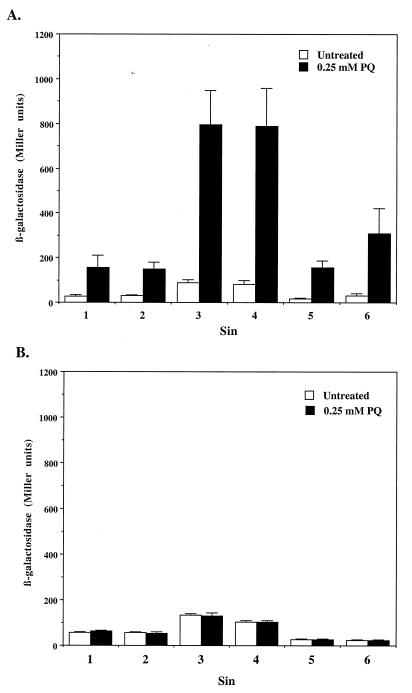
In order to identify novel SoxS-regulated genes, we screened a library of random lacZ insertions for differential expression in the presence or absence of SoxS. A ΔsoxRS strain was transformed with pJP105 and then mutagenized with MudJ, a bacteriophage Mu derivative with a promoterless lacZ operon for the generation of transcriptional fusions (10). Primary candidates for SoxS-regulated insertions were selected as described in Materials and Methods. From approximately 56,000 colonies, 114 candidates were picked, purified by single-colony isolation, and tested for β-galactosidase activity in liquid culture in either the absence or the presence of IPTG (1 mM). We disregarded any transductant showing less than twofold activation by IPTG. From the group of 114 candidates, 32 Kanr transductants showed increased β-galactosidase activity after treatment with IPTG. In order to confirm regulation by SoxS in a physiologically relevant setting, the MudJ (Kanr) insertion from each of these 32 mutants was transduced into a wild-type S. enterica serovar Typhimurium strain and the β-galactosidase activity of each transductant was measured from LB cultures in the presence or absence of 0.25 mM PQ. Only six transductants showed increased β-galactosidase activity after treatment with PQ (Fig. 5A). The rest of the transduced insertions were confirmed as Mu transpositions onto plasmid pJP105, as evidenced by three observations: first, the cotransduction of the plasmid-encoded resistance to ampicillin together with the Mu-encoded resistance to kanamycin; second, the presence of plasmid DNA in the Kanr transductants; third, the increased sizes of these plasmids with respect to pJP105, as revealed by restriction analysis, consistent with a MudJ insertion (data not shown).
FIG. 5.
Induction of β-galactosidase activity by PQ in six independent lacZ insertions. Liquid cultures of the mutant strains were assayed for β-galactosidase activity as explained in Materials and Methods. (A) Sin fusions in a soxRS+ background. (B) Sin fusions in a ΔsoxRS background. The results are the averages of three independent experiments, and the error bars correspond to the standard deviations.
The insertional fusions were named Sins, for SoxS induced. The transduction of these Sin fusions into a ΔsoxRS strain abolished the PQ-induced activation of β-galactosidase activity (Fig. 5B). These results demonstrate the dependence on a functional soxRS system for the inducible activation of lacZ expression from all six Sin fusions.
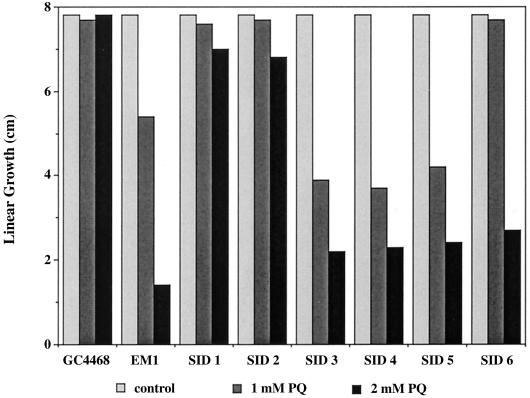
To test the possibility that the MudJ insertions might have disrupted genes involved in resistance to oxidative stress, we measured the relative sensitivities of wild-type, ΔsoxRS, and the six Sin strains with PQ gradient plates. The result of a representative experiment (Fig. 6) showed that all six Sin strains were more sensitive to PQ than the wild-type strain. Thus, all the genes disrupted by the Sin insertions have roles in resistance to oxidative stress.
FIG. 6.
Sensitivity to PQ of six independent lacZ insertions. Linear growth of different strains of S. enterica serovar Typhimurium on PQ gradient plates is shown. Maximum concentrations are shown below the graph. The maximum possible growth was 8 cm.
Molecular characterization of SoxS-regulated genes.
To establish the identity of the soxS-regulated loci, we obtained the nucleotide sequence of the regions adjacent to the left end of the MudJ insertions by a nonspecific PCR method (see Materials and Methods). We then compared these nucleotide sequences with DNA databases by using the BLAST program (2). The Sin1 and Sin2 insertions were located in the sodA gene, which encodes a polypeptide with 97% identity to E. coli SodA (36). Both insertions were located at the 3′-untranslated end of the reported sodA sequence.
The Sin3, Sin4, and Sin5 insertions were located inside an open reading frame (ORF) homologous to that of the fpr gene of E. coli, which encodes the NADPH:ferredoxin oxidoreductase (7). The partial nucleotide sequence obtained from these insertions predicts a polypeptide 80% identical to the one of E. coli (data not shown).
The Sin6 insertion was located within an ORF homologous to that of E. coli ydkB. The E. coli ORF codes for a predicted polypeptide product that is 1,174 residues long; the product has no known function but has been annotated as a putative Fe-S-containing oxidoreductase (8). This predicted E. coli protein is homologous to the NifJ oxidoreductase from Anabaena spp., which shuttles electrons from pyruvate to reduce nitrogenase (6).
These three SoxS-regulated genes identified by the random-insertion approach have either proven or arguable roles in cellular defense against oxidative stress, confirming the pivotal role of SoxS as a transcriptional regulator of antioxidant defenses in S. enterica serovar Typhimurium.
DISCUSSION
The structural similarity between the soxRS genes from E. coli and those from S. enterica serovar Typhimurium (GenBank accession no., U61147) predicted that the soxRS regulon serves similar functions in both species. We constructed an IPTG-regulated expression system that uncouples soxS expression from oxidative stress in order to study the direct contribution of SoxS protein to the activation of antioxidant defenses. By using this expression system, the steady-state levels of soxS message could be titrated by increasing the concentration of IPTG, which mimicked the physiological range of soxS expression in response to oxidative stress. The PQ-induced expression of soxS from wild-type strain ATCC 14028 seemed to saturate at an induction ratio of ∼70-fold, while IPTG-induced expression of soxS from pJP105 was linear up to ∼100-fold. The somewhat higher level attained for the artificial construct could be due to the absence of soxS autorepression (30) in the IPTG-driven lacZ promoter.
The heterologous expression of SoxS in S. enterica serovar Typhimurium resulted in enhanced resistance to PQ (Fig. 2). This result demonstrates that, as in E. coli, an elevated level of SoxS protein is sufficient for activation of important antioxidant genes in S. enterica serovar Typhimurium. Expression of SoxS in the absence of oxidative stress was sufficient for transcriptional activation of sodA to a high level. Interestingly, exposure of a ΔsoxRS strain to PQ elicited a small but reproducible increase in sodA transcription (data not shown). A modest, PQ-dependent, soxRS-independent induction of sodA was also observed by Martins et al. (unpublished data) and might be due to MarA, a transcriptional activator closely related to SoxS (1). The MarA protein activates sodA expression in E. coli (1) and may also do so in S. enterica serovar Typhimurium. The synthesis of MarA in E. coli was reported to be induced slightly by PQ (34), an effect that could underlie SoxS-independent induction of sodA in both organisms. The S. enterica serovar Typhimurium marA gene (35) codes for a predicted polypeptide 86% identical to E. coli MarA. Additionally, sodA in E. coli is regulated by ArcA, Fnr, and Fur (12). Of these, Fur is a reasonable candidate for contributing to PQ-induced sodA expression under aerobic growth. Expression of SoxS in ΔsoxRS strains of S. enterica serovar Typhimurium in the absence of oxidative stress was sufficient for transcriptional activation of micF, which reached the same maximum level as that observed in wild-type cells exposed to PQ. This result demonstrates that SoxS is sufficient for full, PQ-induced expression of micF. The induction of micF has not been directly connected to oxidative stress resistance. However, induction of micF in E. coli is necessary for enhanced resistance to multiple antibiotics. This phenotype depends on repression of the synthesis of porin OmpF, with a consequent reduction in permeability to small hydrophilic molecules (29). The apparent existence of the same mechanism in S. enterica serovar Typhimurium could be relevant in the development of antibiotic resistance during bacterial infection (see below).
SoxS also activates antibiotic resistance mechanisms in other species. In preliminary experiments, Klebsiella aerogenes transformed with pJP105 displayed increased resistance to chloramphenicol, nalidixic acid, tetracycline, and kanamycin in the presence of IPTG (P. J. Pomposiello and B. Demple, unpublished data). This result also suggests the existence of a soxRS regulon in Klebsiella. There is also evidence for a soxRS regulon in clinical strains of Enterobacter agglomerans, Shigella flexneri, and Shigella dysenteriae (B. L. Martins, A. Koutsolioutsou, and B. Demple, unpublished data).
Our screening for Sox-regulated genes with MudJ yielded lacZ insertions in three different genes. Of these, sodA was the only gene already characterized in S. enterica serovar Typhimurium (36), but its role in antioxidant defense is not completely understood. However, we have shown that the insertion of MudJ in sodA and the other two genes rendered the corresponding strains hypersensitive to oxidative stress in comparison to the isogenic wild-type strain. These results suggest strongly that all three genes have roles in protecting the cell against PQ-elicited oxidative stress. The relative PQ resistances of the Sin1 and Sin2 insertions (Fig. 6) are likely due to their locations in the 3′-untranslated region of sodA, which are expected to preserve substantial sodA function.
The remaining Mu insertions map to two uncharacterized S. enterica serovar Typhimurium ORFs. For Sin3, Sin4, and Sin5, the strong homology of the disrupted gene with the E. coli fpr gene and the high sensitivity of the Sin3, Sin4, and Sin5 strains to PQ are consistent with the identification of this locus as the S. enterica serovar Typhimurium fpr gene. The E. coli fpr gene was originally isolated in a screening for PQ-sensitive mutants (28). The E. coli fpr is a NADPH:ferredoxin oxidoreductase required for anaerobic ribonucleotide reduction (7). Finally, Sin6 maps to a gene with a sequence similar to that of the E. coli ydbK ORF. The annotation of this ORF as a putative iron-sulfur-containing oxidoreductase, together with the hypersensitivity to PQ of the corresponding mutant, argues in favor of a role for this S. enterica serovar Typhimurium gene in protection against superoxide stress. Oxidoreductases contribute to resistance to oxidative damage by redirecting electron transfer, rereducing oxidized iron-sulfur centers, and replacing damaged components (20, 24).
There are 15 known genes in E. coli under soxS regulation (20, 24). This figure led us to expect a higher yield of SoxS-regulated genes for our mutagenesis and screening in S. enterica serovar Typhimurium. There are at least three possible reasons for this discrepancy. First, ∼80% of the insertions identified as IPTG responsive were in plasmid pJP105 rather than in chromosomal sites. Thus, the MudJ mutagenesis was not saturating. Second, the strain used in the screening, EM1, has increased basal expression of micF for unknown reasons (unpublished data). If this effect extends to other genes in EM1, the difference between the basal and SoxS-activated levels of expression for various genes might be obscured. Third, it is possible that S. enterica serovar Typhimurium has fewer genes under soxRS control than does E. coli. As noted earlier, glucose-6-phosphate dehydrogenase and fumarase C are regulated by SoxS in E. coli but do not seem to be members of the soxRS regulon in S. enterica serovar Typhimurium (Martins et al., unpublished data).
SoxS does not seem to be a virulence factor in S. enterica serovar Typhimurium. Both ΔsoxRS (Martins et al., unpublished data) and ΔsoxS (16) strains were not attenuated in mouse infection experiments and had no disadvantage in survival inside activated macrophages. Of the S. enterica serovar Typhimurium genes under soxRS control, sodA confers some protection against early killing by macrophages, but a sodA strain of S. enterica serovar Typhimurium was not found to be attenuated in virulence assays (36). However, there is evidence that the soxRS regulon could be contributing to the development of infection in humans through conferring elevated levels of multiple antibiotic resistance. Preliminary results show that expression of SoxS in S. enterica serovar Typhimurium is sufficient to enhance resistance to nalidixic acid and chloramphenicol (Pomposiello and Demple, unpublished data). The enhancement of antibiotic resistance via activation of the soxRS regulon could be a stepping-stone in the acquisition of clinically relevant antibiotic resistance, as has been proposed for the marA regulon (1). This hypothesis is supported by the isolation of soxS-constitutive S. enterica serovar Typhimurium (Martins et al., unpublished data) and pathogenic E. coli strains (31) from human patients with systemic infections. In one case of S. enterica serovar Typhimurium that developed increased antibiotic resistance during an infection, a mutation that contributes to the phenotype and that was mapped to a single-nucleotide change in the soxR gene arose (Martins et al., unpublished data). Thus, activation of the soxRS regulon by redox-active compounds generated by the immune system (19) could paradoxically enhance the antibiotic resistance of enteropathogenic bacteria and complicate the treatment of bacterial infections.
ACKNOWLEDGMENTS
This work was supported by NIH grant CA37831 to Bruce Demple.
We thank Danielle Touati for plasmid pDT1.5, John Mekalanos for phage P22, John Roth for the his::MudJ strain TT12028, W. Reznikoff for plasmid RZ4004, and Bob Bender and Brian Janes for K. aerogenes strains.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amábile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariza R R, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 6.Bauer C C, Scappino L, Haselkorn R. Growth of the cyanobacterium Anabaena on molecular nitrogen: NifJ is required when iron is limited. Proc Natl Acad Sci USA. 1993;90:8812–8816. doi: 10.1073/pnas.90.19.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, Jörnvall H, Haggård-Ljungquist E. Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J Bacteriol. 1993;175:1590–1595. doi: 10.1128/jb.175.6.1590-1595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Bolivar F, Rodriguez R, Greene P, Betlach M, Heynecker H, Boyer H. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 10.Castilho B A, Olfson P, Casabadan M J. Plasmid insertion mutagenesis and lac gene fusions with Mini-Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou J H, Greenberg J T, Demple B. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol. 1993;175:1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella typhimurium from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esterling L, Delihas N. The regulatory RNA gene micF is present in several species of Gram-negative bacteria and is phylogenetically conserved. Mol Microbiol. 1994;12:639–646. doi: 10.1111/j.1365-2958.1994.tb01051.x. [DOI] [PubMed] [Google Scholar]
- 16.Fang F, Vazquez-Torres A, Xu Y. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect Immun. 1997;65:5371–5375. doi: 10.1128/iai.65.12.5371-5375.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 18.Hidalgo E, Bollinger J M, Bradley T M, Walsh C T, Demple B. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J Biol Chem. 1995;270:20908–20914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- 19.Hidalgo E, Demple B. Adaptive responses to oxidative stress: the soxRS and oxyR regulons. In: Lin E C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1997. pp. 433–450. [Google Scholar]
- 20.Hidalgo E, Ding H, Demple B. Redox signal transduction via iron-sulfur clusters in the SoxR transcription factor. Trends Biochem Sci. 1997;22:207–210. doi: 10.1016/s0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- 21.Hughes K T, Roth J R. Transitory cis complementarity: a method for providing transposition functions to defective tranposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jair K W, Fawcett W P, Fujita N, Ishihama A, Wolf R E., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of the Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:306–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 24.Liochev S I, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 26.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 28.Morimyo M. Isolation and characterization of methyl viologen-sensitive mutants of Escherichia coli K-12. J Bacteriol. 1988;170:2136–2142. doi: 10.1128/jb.170.5.2136-2142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido H. Microdermatology: cell surface in the interaction of microbes with the external world. J Bacteriol. 1999;181:4–8. doi: 10.1128/jb.181.1.4-8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunoshiba T, Hidalgo E, Li Z, Demple B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol. 1993;175:7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oethinger M, Podglajen I W, Kern W V, Levy S B. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reznikoff W, McClure W R. E. coli promoters. In: Reznikoff W S, Gold I, editors. Maximizing gene expression. London, United Kingdom: Butterworths Publishers; 1986. pp. 1–33. [Google Scholar]
- 33.Reznikoff W S. Catabolite gene activator protein activation of lac transcription. J Bacteriol. 1992;174:655–658. doi: 10.1128/jb.174.3.655-658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seoane A S, Levy S B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulavik M C, Dazer M, Miller P F. The Salmonella typhimurium mar Locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsolis R M, Baumler A J, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Dunham W R, Weiss B. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative response regulon in Escherichia coli. J Biol Chem. 1995;270:10323–10327. doi: 10.1074/jbc.270.17.10323. [DOI] [PubMed] [Google Scholar]