Abstract
Serotonin receptors play central roles in neuromodulation and are critical drug targets for psychiatric disorders. Optical control of serotonin receptor subtypes has the potential to greatly enhance our understanding of the spatiotemporal dynamics of receptor function. While other neuromodulatory receptors have been successfully rendered photoswitchable, reversible photocontrol of serotonin receptors has not been achieved, representing a major gap in GPCR photopharmacology. Herein, we develop the first tools that allow for such control. Azo5HT-2 shows light-dependent 5-HT2AR agonism, with greater activity in the cis-form. Based on docking and test compound analysis, we also develop photoswitchable orthogonal, remotely-tethered ligands (PORTLs). These BG-Azo5HTs provide rapid, reversible, and repeatable optical control following conjugation to SNAP-tagged 5-HT2AR. Overall, this study provides a foundation for the broad extension of photopharmacology to the serotonin receptor family.
Keywords: Serotonin, Photopharmacology, GPCR, Photoswitch, SNAP Tag
Entry for the Table of Contents
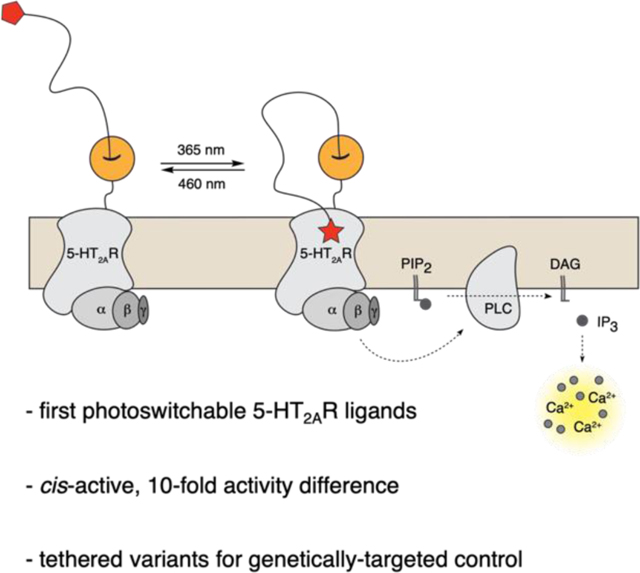
We have developed the first-in-class photoswitchable agonists for the 5HT2A receptor (serotonin family GPCR). Bifunctional, tethered variants (PORTLs) enable genetically targeted optical control.
Introduction
Serotonin (5-hydroxytryptamine; 5-HT) is a neuromodulator that is released in the brain primarily by dorsal raphe nuclei neurons, in the gut by enterochromaffin cells, and in blood platelet cells.[1,2] Serotonin acts through a large family of G protein-coupled receptor (5-HT1Rs, 5-HT2Rs, 5-HT4Rs, 5-HT5Rs, 5-HT6Rs, 5-HT7Rs) and ion channel (5-HT3Rs) subfamilies to regulate a plethora of neuronal and behavioral processes.[1] Given the importance of 5-HT to the regulation of mood, cognition and reward, great effort has been made to harness pharmacology to manipulate 5-HTRs for both basic study and therapeutic applications. Recent developments establishing the potential of 5-HT2AR-targeting psychedelic drugs for the treatment of depression, anxiety, and addiction have further motivated the detailed study of 5-HTR signaling.[3–7] Despite great attention, limitations in the ability of 5-HTR-targeting compounds in terms of subtype-specificity and spatiotemporal precision and their inability to be targeted to genetically defined cell types have hindered progress toward a mechanistic understanding of the physiological and therapeutic effects of 5-HTR signaling. As an alternative to classical pharmacology, photopharmacology has emerged as a means of gaining further precision through the development of photosensitive compounds whose activity can be modified depending on the wavelength of illumination[8–10]. Photopharmacological compounds have enabled the optical control of a variety of GPCRs, including family A (μ-opioid receptor,[11] dopamine receptors,[12,13] histamine receptors,[14] adenosine receptors,[15] muscarinic receptors,[16] adrenergic receptors[17], fatty acid receptors,[18] lysophospholipid receptors,[19,20] and cannabinoid receptors[21,22]), family B (glucagon-like peptide 1 receptor[23,24]), and family C GPCRs (metabotropic glutamate receptors[25–28]). For further precision, including the ability to target the effects of light to genetically-defined cell populations, photoswitches may be covalently tethered to a genetically engineered labeling domain (i.e. SNAP, Halo) as Photoswitchable Orthogonal Remotely Tethered Ligands (PORTLs), as has been demonstrated with metabotropic glutamate receptors (mGluRs).[29,30] mGluR-targeting PORTLs have been applied for both molecular biophysical studies[31] and the in vivo manipulation of mGluR2 in specific cell types[32–34] in mice, providing a template for their development and application in complex systems.
Surprisingly, 5-HTRs have received limited attention in terms of photopharmacology. Photocaged variants of serotonin have enabled light-induced release of 5-HT through removal of photocleavable protecting groups.[38–41] However, these tools do not offer reversible control, lack 5-HTR subtype targeting, and have not been paired with genetic targeting as can be done with PORTLs.[29,30,42] Thus, the development of a photoswitchable ligand platform for the 5-HTR family would enable the study of these receptors with unprecedented spatiotemporal control, which could facilitate new insight into the dynamics of neural signaling. Recently, König and co-workers reported an attempt to develop a photoswitchable ligand for the 5-HT3R, a pentameric ligand-gated ion channel.[43] Herein, we describe the first series of photoswitchable ligands for the 5-HT2 receptors, which belong to family A GPCRs, representing the majority of serotonin receptors. We identify an azobenzene-conjugated 5-HT lead compound, Azo5HT-2, that enables optical control of 5-HT2ARs with activity which is increased in the cis form upon irradiation. Computational structural analysis suggests that the 5-HT moiety of cis-Azo5HT-2 binds with a canonical pose and enables access to the azobenzene ring from the extracellular face of the receptor, motivating the design and synthesis of a first generation of 5-HT PORTLs. Finally, BG-Azo5HTn PORTLs of variable linker length enable repeatable optical control of SNAP-tagged 5-HT2AR, opening the door to genetically targeted, receptor-specific optical control of serotonergic signaling.
Results and Discussion
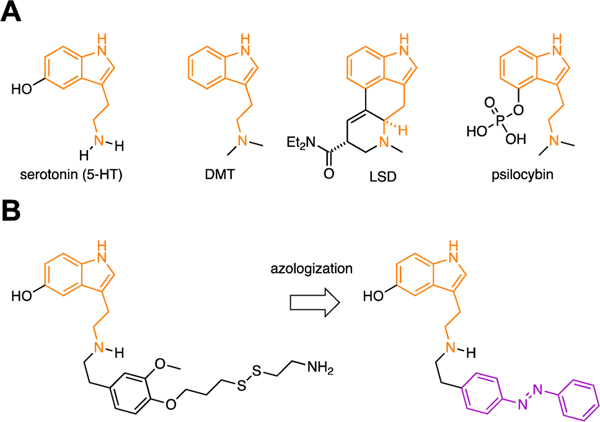
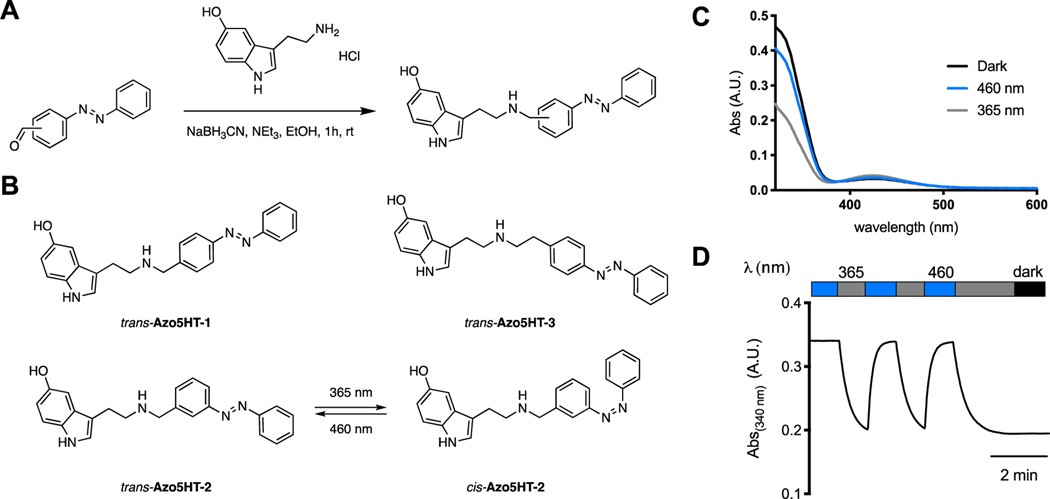
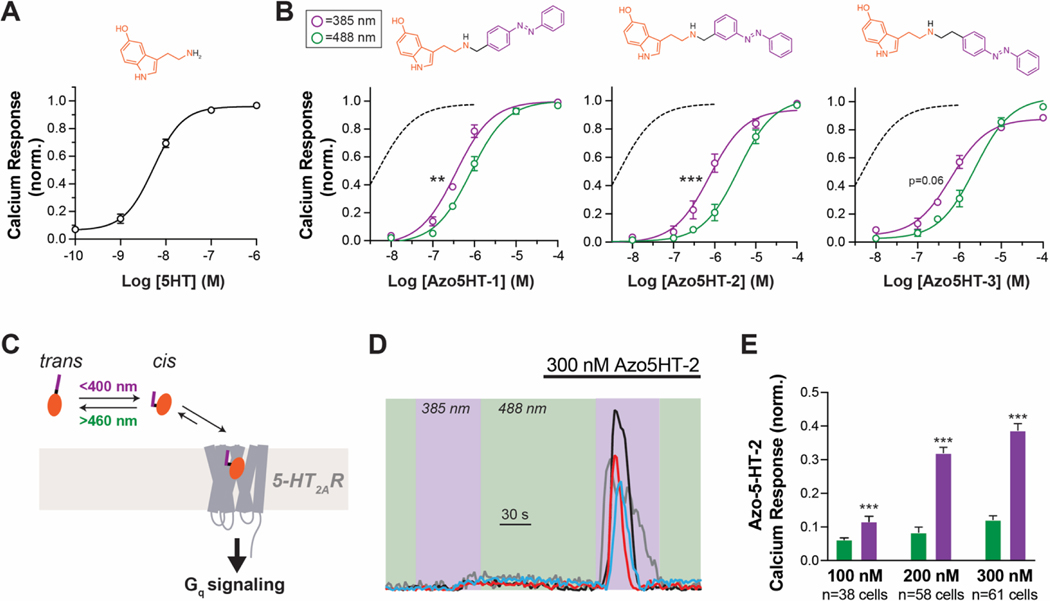
The 5-HT receptor family is targeted by a variety of natural and synthetic agonists, including many with a tryptamine (indolamine) moiety (Fig. 1A). We considered several of these ligands for the design of photochromic agonists but reasoned that serotonin would be best suited because analogs would likely mimic endogenous signaling and derivatives which could be suited for incorporation of azobenzene motifs have been reported previously. These derivatives include covalent 5-HT analogs with an appended benzene ring (Fig. 1B).[37] We considered an ‘azologization’[35,36] approach to install the azobenzene at the matching position (Azo5HT-3) and designed additional derivatives with the azobenzene moved one carbon atom closer to the pharmacophore (Azo5HT-1 and Azo5HT-2). The derivatives were synthesized through reductive amination of 5-HT with the corresponding azobenzene-aldehydes (Fig. 2A,B). Photophysical characterization of Azo5HT-1 to Azo5HT-3 (Fig. 2C,D and Fig. S1) revealed similar properties to classical azobenzenes. All derivatives could be reversibly switched to their respective cis and trans forms with UV-A (365 nm) and blue light (460 nm), respectively, and underwent slow thermal relaxation (t1/2 > 1h). To assess the ability of Azo5HT molecules to serve as 5-HTR agonists, we tested each compound across the 5-HT2R family (5-HT2AR, 5-HT2BR, 5-HT2CR). As these receptors are all Gq-coupled and produce intracellular Ca2+ release via phospholipase C-β activation, we performed live cell Ca2+ imaging with the fluorescent sensor, GCaMP6f. Using this assay, all three receptors showed the expected 5-HT responses with nM EC50 values (Fig. 3A; Table 1; Fig. S2). Compounds Azo5HT-1–3 were tested independently either under standard conditions with 488 nm illumination for GCaMP6f excitation, which maintain them in the trans state, or with interweaved 385 nm illumination to convert them to the cis state. All three compounds showed dose-dependent activation of 5-HT2AR in the trans and cis states with a leftward shift in the cis state (Fig. 3B; Fig. S2). For Azo5HT-1 there was a ~2-fold shift, while a larger 5–10-fold shift was seen for Azo5HT-2 and Azo5HT-3 (Table 1). It is worth noting that 385 nm likely does not maximally occupy the cis state, so the relative difference between cis and trans may be underestimated using this approach. In contrast to the 5-HT2AR, no or very modest differences were observed between cis and trans for each molecule on 5-HT2BR and 5-HT2CR (Table 1; Fig. S3). Next, we asked if Azo5HT-2 photoconversion could be harnessed for optical activation of 5-HT2AR (Fig. 3C). Application of 100–300 nM Azo5HT-2 produced minimal responses under 488 nm illumination but following application of 385 nm light, clear responses were observed that were up to 50% in amplitude relative to saturating 5-HT (Fig. 3D,E; Fig. S4) and blocked by the 5-HT2R antagonist ketanserin (Fig. S4). Together, these data indicate that Azo5HT-2 enables reversible photoagonism of 5-HT2AR with similar signaling properties to the endogenous agonist 5-HT.
Figure 1.
(A) Representative tryptamine-derived agonists of 5-HT receptors: serotonin, PNU 22394, LSD, and psilocybin. Shared tryptamine moiety highlighted in orange. (B) Azologization strategy[35,36] for the design of photoswitchable agonists based on a previously-reported covalent agonist of 5-HT2AR (left).[37]
Figure 2.
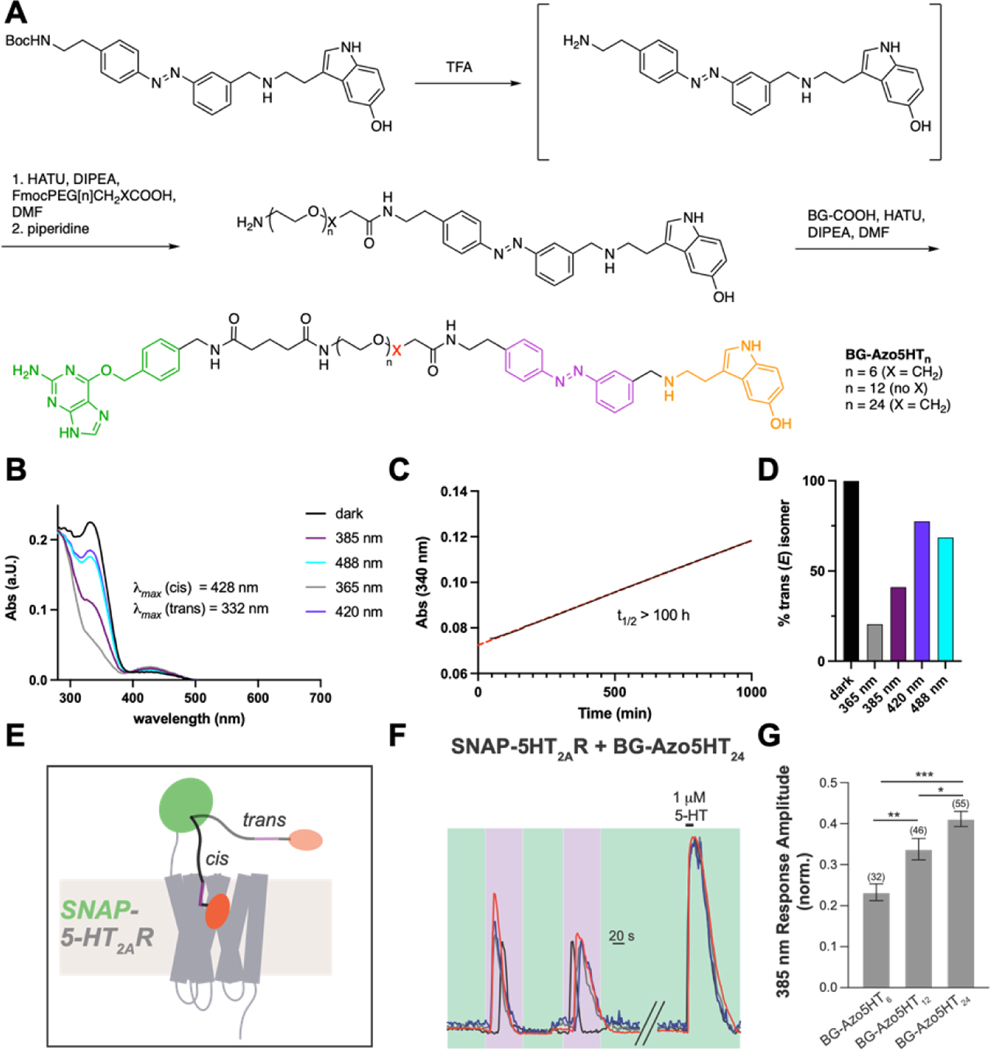
Synthesis and photophysical characterization of photoswitchable 5-HT derivatives Azo5HT-1–3. (A) Synthesis of Azo5HT series. (B) Chemical structures of Azo5HTs. (C) The UV-Vis spectra of Azo5HT-2 in the dark-adapted (black, trans), 365 nm adapted (grey, cis) and 460 nm adapted (blue, trans) photostationary states (50 μM, DMSO, rt). (D) Reversible cycling between Azo5HT-2 photoisomers with alternating illumination at 365/460 nm (50 μM, DMSO, rt)
Figure 3.
Photoactivation of 5-HT2AR by Azo5HT-2. (A) Dose-response curve of 5HT on 5-HT2AR using a Ca2+ imaging assay. (B) Dose-response curves for Azo5HT compounds showing enhanced agonism for cis versus trans for all compounds. ** p<0.01, *** p < 0.001, 2-way ANOVA test (C) Schematic of Azo5HT mediated optical control. (D) Representative Ca2+ imaging traces showing photoactivation of 5-HT2AR by Azo5HT-2. In the absence of Azo5HT-2, no 385 nm light response is seen but a clear response is seen upon irradiation in the presence of 300 nM Azo5HT-2 with similar on and desensitization kinetics compared to 5-HT application (see Fig. S4). (E) Statistical analysis of Azo5HT-2 response at different photoswitch concentrations. *** p<0.001, paired t-test. Error bars represent mean ± SEM.
Table 1.
Summary of Ca2+ imaging dose response data for all compounds tested in this study. All values are normalized to the response to 1 mM 5-HT and the Max is determined by dose-response curve fit. P values are reported for 2-way ANOVA tests between cis and trans dose response curves.
| Receptor | Ligand | N (expts) | n (cells) | EC50 (μM) | 95% CI (μM) | Max ± (μM) | P value (cis vs. trans) | |
|---|---|---|---|---|---|---|---|---|
| 5-HT2AR | 5-HT | 4 | 97 | 0.0053 | 0.0007 to 0.0004 | 0.96 ± 0.02 | ||
| AZo5HT-1 | cis (385 nm) | 5 | 125 | 0.36 | 0.53 to 0.24 | 1.00 ±0.04 | 0.005** | |
| trans (488 nm) | 5 | 157 | 0.82 | 1.15 to 0.59 | 1.00 ±0.04 | |||
| AZo5HT-2 | cis (385 nm) | 7 | 163 | 0.75 | 1.14 to 0.50 | 0.94 ± 0.04 | 0.0008*** | |
| trans (488 nm) | 8 | 265 | 3.75 | 5.78 to 2.43 | 1.02 ±0.06 | |||
| Azo5HT-3 | cis (385 nm) | 4 | 42 | 0.64 | 1.02 to 0.40 | 0.88 ± 0.04 | 0.06 | |
| trans (488 nm) | 5 | 83 | 2.36 | 3.90 to 1.48 | 1.03 ±0.06 | |||
| Boc-Azo5HT-2 | cis (385 nm) | 4 | 103 | 0.84 | 1.12 to 0.63 | 0.80 ± 0.02 | <0.0001*** | |
| trans (488 nm) | 4 | 159 | 3.21 | 4.95 to 2.08 | 0.80 ± 0.04 | |||
| 5-HT2BR | 5-HT | 1 | 15 | 0.0042 | 0.96 ± 0.03 | |||
| Azo5HT-1 | cis (385 nm) | 3 | 45 | 1.99 | 4.01 to 1.13 | 0.69 ± 0.04 | 0.89 | |
| trans (488 nm) | 3 | 28 | 2.28 | 137.4 to 0.68 | 0.70 ±0.10 | |||
| AZ05HT-2 | cis (385 nm) | 3 | 45 | 1.57 | 4.17 to 0.72 | 0.93 ± 0.08 | 0.65 | |
| trans (488 nm) | 4 | 29 | 1.62 | 4.06 to 0.77 | 1.00 ±0.09 | |||
| AZ05HT-3 | cis (385 nm) | 3 | 32 | 1.09 | 1.86 to 0.65 | 0.70 ± 0.04 | 0.04* | |
| trans (488 nm) | 3 | 75 | 0.88 | 2.35 to 0.33 | 0.88 ± 0.08 | |||
| 5-HT2CR | 5-HT | 1 | 45 | 0.0063 | 0.95 ± 0.05 | |||
| Azo5HT-1 | cis (385 nm) | 4 | 52 | 0.53 | 0.91 to 0.30 | 1.16 ±0.06 | 0.13 | |
| trans (488 nm) | 4 | 77 | 0.48 | 0.73 to 0.30 | 1.00 ±0.04 | |||
| Azo5HT-2 | cis (385 nm) | 4 | 124 | 1.04 | 2.51 to 0.44 | 1.02 ±0.10 | 0.57 | |
| trans (488 nm) | 5 | 144 | 1.20 | 2.15 to 0.69 | 1.00 ±0.06 | |||
| Azo5HT-3 | cis (385 nm) | 3 | 46 | 1.56 | 2.65 to 0.97 | 0.89 ± 0.05 | 0.13 | |
| trans (488 nm) | 4 | 100 | 1.24 | 2.18 to 0.73 | 0.87 ±0.05 |
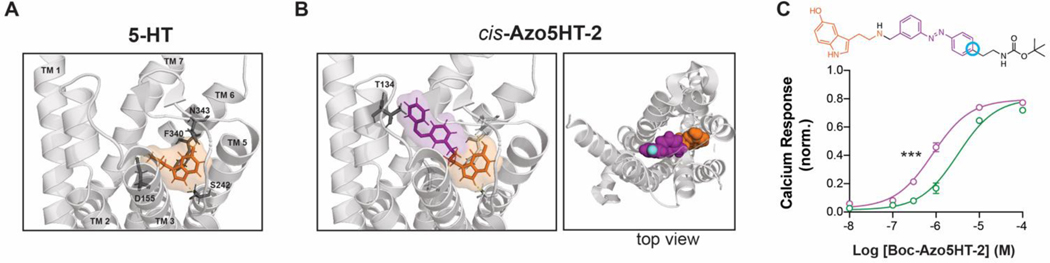
We next used computational ligand docking to gain insight into the binding mode of Azo5HT-2 using the recently reported LSD-bound crystal structure of the 5-HT2AR.[44] Cis-Azo5HT-2 showed similar binding of the 5-HT moiety compared to 5-HT alone with a mix of canonical and non-canonical poses (Fig. 4A, B; Fig. S5A–C; see SI for details). The azobenzene moiety showed occupancy of a pocket toward the extracellular face of the receptor with likely solvent accessibility from the cell surface (Fig. 4B; Fig. S5E, F). In contrast, trans-Azo5HT-2 showed variable docking results with a lower proportion of docks containing a canonical pose (see SI for details) for the 5-HT moiety (Fig. S5C, D). Based on the potential binding pose of cis-Azo5HT-2, we reasoned that extension of this molecule would be tolerated and, ultimately, enable tethering to a labeling site (e.g. SNAP-tag) outside the core of the transmembrane helix bundle and extracellular loops of the receptor. To test this, we synthesized the extended photoswitch Boc-Azo5HT-2 (Fig. 4C) using our established reductive amination conditions. Boc-Azo5HT-2 showed clear agonism of 5-HT2AR and maintained enhanced apparent affinity in the cis state (Fig. 4C; Table 1), enabling photo-activation of Ca2+ responses (Fig. S6). Motivated by our docking and Boc-Azo5HT-2 test compound analysis, we designed PORTLs with the goal of enabling tethered optical control of SNAP-5HT2AR (Fig. 5). To this end, BOC-Azo5HT-2 was deprotected and various PEG-linkers and benzyl guanine (BG) were attached through successive amide couplings, giving rise to a series of PORTLs with varying linker length, termed BG-Azo5HT6,12,24 (Fig. 5A). Photophysical characterization of BG-Azo5HT24 revealed effective switching with UV-A and blue light (Fig. 5B), high thermal stability (Fig. 5C), and typical photostationary states for classic azobenzenes (Fig. 5D). To test the capacity of these probes for genetically targeted control, we first used a previously established fluorophore competition labeling assay[30] to confirm that all PORTLs efficiently label N-terminally SNAP-tagged 5-HT2AR (“SNAP-5HT2AR”) (Fig. S7A, B). We then tested the ability of each PORTL to produce optically-evoked Ca2+ responses following conjugation to SNAP-5HT2AR and 385 nm illumination (Fig. 5E). Reversible and repeatable 385 nm light-evoked Ca2+ transients were seen with all 3 PORTL variants in 10–40% of cells with a higher proportion of cells showing photoactivation with BG-Azo5HT24 and BG-Azo5HT12 compared to BG-Azo5HT6 (Fig. S7C, D). Light responses were not seen in the absence of PORTL labeling (Fig. S7E) labeling and were as large as 60% in amplitude relative to saturating 5-HT for BG-Azo5HT24 and smaller for shorter variants (Fig 5F, G). Importantly, 385 nm light responses were blocked by ketanserin (Fig. S7F, G). A subset (<10%) of cells showed Ca2+ transients in the absence of 385 nm illumination (Fig. S7H), likely indicative of some receptor activation by the PORTL in trans. This potential trans activation was more pronounced in shorter variants, suggesting that the decreased local concentration associated with longer PORTLs enhances the relative cis versus trans agonism via the Azo5HT moiety. Together, these data establish genetically targetable, PORTL-mediated optical control of 5-HT2AR and provide a strong foundation for both further engineering and application.
Figure 4.
Docking and test compound analysis enable linker attachment. (A-B) Docking analysis showing that both 5-HT (A) and the 5-HT moiety of Azo5HT-2 (B) show identical poses, with the azobenzene moiety occupying a water-filled cavity at the extracellular face of 5-HT2AR. Residues associated with canonical 5-HT binding are highlighted in (A) and position T134, which was previously substituted for conjugation of a covalent 5-HT2AR agonist[37], is highlighted in (B). Top view (B, right) shows that the para position (yellow) is positioned facing toward the extracellular solution. C) Chemical structure (top; para position circled in cyan and dose response curve (bottom) showing light-dependent (purple=385 nm illumination; green=488 nm illumination) activation of 5-HT2AR by Boc-Azo5HT-2. *** p < 0.001, 2-way ANOVA test.
Figure 5.
Synthesis and photophysical characterization of Azo5HT-2-based PORTLs BG-Azo5HTn. (A) Synthesis and chemical structures of BG-Azo5HTn PORTLs. (B) The UV-Vis spectra of BG-Azo5HT24 in the dark-adapted (black), 365 nm adapted (grey), 385 nm adapted (purple), 420 nm adapted (turquoise), and 488 nm adapted (turquoise) photostationary states (20 mM, 10% DMSO in PBS, rt). (C) Thermal relaxation of BG-Azo5HT24 after pre-irradiation at 365 nm for 10 min (20 mM, 10% DMSO in PBS, 37 °C). (D) Quantification of trans-isomer of BG-Azo5HT24 in the dark- (black), 365 nm- (grey), 385 nm- (mauve), 420 nm- (violet), and 488 nm-adapted (cyan) photostationary states through LCMS separation and detection at isosbestic point. (E) Schematic of BG-Azo5HTn PORTL-mediated optical control of SNAP-tagged 5-HT2AR. (F-G) Representative traces (F) and summary bar graph (G) showing photoactivation of SNAP-5-HT2AR by BG-Azo5HTn PORTLs. The numbers of cells analyzed are shown in parentheses. *indicates statistical significance (1-way ANOVA with Tukey-Kramer Multiple Comparisons; p=0.0068 for BG-Azo5HT6 vs. BG-Azo5HT12, p<0.0001 for BG-Azo5HT6 vs. BG-Azo5HT24, p=0.036 for BG-Azo5HT12 vs. BG-Azo5HT24).
Conclusion
In summary, we have developed first-in-class photoswitchable analogs of serotonin that allow for the optical control of 5-HT2AR. Interestingly, all three test compounds, Azo5HT1–3 showed preferential agonism in cis over trans on the 5-HT2AR, but no clear difference between states on the 5-HT2BR or 5-HT2CR, providing a powerful chemical lead for further molecular pharmacological analysis. At the appropriate concentrations (100–300 nM), our lead compound Azo5HT-2 is inactive in the dark and becomes an effective agonist for 5-HT2AR following illumination. While Azo5HT-2 offers the advantage of being based on the endogenous 5-HT ligand, the design employed likely also provides a template for azologization of other 5-HTR agonists, including psilocin and LSD, which all contain a shared tryptamine motif. Furthermore, this study establishes the proof-of-principle of photopharmacology for 5-HTRs and should provide a basis for extension of this approach to other 5-HTR subfamilies, including those that are Gi/o (5-HT1Rs, 5-HT5Rs) or Gs (5-HT4R, 5-HT6R, 5-HT7R)-coupled. Most importantly, our screen of azobenzene-conjugated 5-HT analogs lays the groundwork for their proximity photopharmacology. BG-Azo5HT PORTLs enable reversible, repeatable optical control of SNAP-5HT2AR, opening the door to spatiotemporally precise and genetically-targeted control of this biologically important receptor. While PORTLs have recently been extended to the D1 dopamine receptors through a membrane anchored SNAP-tag,[45] the present study represents another key extension of the PORTL approach to a family A GPCR. As an intriguing possibility, the PORTL technique enables incorporation of mutations to the SNAP-tagged receptor that alter transducer coupling (e.g. G protein versus arrestin) or regulation (e.g. phosphorylation or scaffold sites) to test their roles in a biological context. This approach has long-term potential for untangling the pleiotropic antidepressant, anxiolytic, anti-addictive and hallucinogenic effects of 5-HT2AR agonism. Finally, the establishment of a core PORTL for the 5-HT2AR may enable the application of next-generation PORTL approaches including branched PORTLs for dual imaging and manipulation,[33] spectrally fine-tuned PORTLs[34] or PORTL-based strategies for targeting native receptors.[42,46]
Supplementary Material
Acknowledgements
We thank New York University for financial support. We thank Jordana Thibado for preliminary functional studies and SNAP-5HT2AR cloning. NMR spectra were acquired using the TCI cryoprobe supported by the NIH (OD016343). J.M. thanks the New York University for a Margaret and Herman Sokol fellowship, and the NCI for a K00 award (4K00CA253758). G.R. is supported by the Weill Cornell Molecular Biophysics Training Grant (T32GM132081). A.P. gratefully acknowledges support from NSF grant BIGDATA: IA: Collaborative Research: In Situ Data Analytics for Next Generation Molecular Dynamics Workflows (NSF #1740990) and computational resources from (project BIP109) of the Oak Ridge Leadership Computing Facility under Contract DE-AC05-00OR22725. J.L. and D.T. are supported by an R61 (R61 DA051529) grant from NIDA. J.L. is supported by an R35 grant (R35 GM124731) from NIGMS, the Rohr Family Research Scholar Award and the Irma T. Hirschl/Monique Weill-Caulier Research Award. DT thanks the McKnight Foundation for a McKnight Memory and Cognitive Disorders Award.
References
- [1].Nichols DE, Nichols CD, Chem. Rev. 2008, 108, 1614–1641. [DOI] [PubMed] [Google Scholar]
- [2].Berger M, Gray JA, Roth BL, Annu. Rev. Med. 2009, 60, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Halberstadt AL, Geyer MA, Neuropharmacology 2011, 61, 364–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cameron LP, Tombari RJ, Lu J, Pell AJ, Hurley ZQ, Ehinger Y, Vargas MV, McCarroll MN, Taylor JC, Myers-Turnbull D, Liu T, Yaghoobi B, Laskowski LJ, Anderson EI, Zhang G, Viswanathan J, Brown BM, Tjia M, Dunlap LE, Rabow ZT, Fiehn O, Wulff H, McCorvy JD, Lein PJ, Kokel D, Ron D, Peters J, Zuo Y, Olson DE, Nature 2021, 589, 474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nutt D, Erritzoe D, Carhart-Harris R, Cell 2020, 181, 24–28. [DOI] [PubMed] [Google Scholar]
- [6].Dong C, Ly C, Dunlap LE, Vargas MV, Sun J, Hwang I-W, Azinfar A, Oh WC, Wetsel WC, Olson DE, Tian L, Cell 2021, 184, 2779–2792.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shao L-X, Liao C, Gregg I, Davoudian PA, Savalia NK, Delagarza K, Kwan AC, Neuron 2021, S0896–6273(21)00423–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Beharry AA, Woolley GA, Chem. Soc. Rev. 2011, 40, 4422–4437. [DOI] [PubMed] [Google Scholar]
- [9].Szymański W, Beierle JM, Kistemaker HAV, Velema WA, Feringa BL, Chemical Reviews 2013, 113, 6114–6178. [DOI] [PubMed] [Google Scholar]
- [10].Hüll K, Morstein J, Trauner D, Chem. Rev. 2018, 118, 10710–10747. [DOI] [PubMed] [Google Scholar]
- [11].Schönberger M, Trauner D, Angew. Chem. Int. Ed. 2014, 53, 3264–3267. [DOI] [PubMed] [Google Scholar]
- [12].Lachmann D, Studte C, Männel B, Hübner H, Gmeiner P, König B, Chemistry – A European Journal 2017, 23, 13423–13434. [DOI] [PubMed] [Google Scholar]
- [13].Donthamsetti PC, Winter N, Schönberger M, Levitz J, Stanley C, Javitch JA, Isacoff EY, Trauner D, J. Am. Chem. Soc. 2017, 139, 18522–18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hauwert NJ, Mocking TAM, Da D Costa Pereira, Lion K, Huppelschoten Y, Vischer HF, De Esch IJP, Wijtmans M, Leurs R, Angewandte Chemie International Edition 2019, 58, 4531–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bahamonde MI, Taura J, Paoletta S, Gakh AA, Chakraborty S, Hernando J, Fernández-Dueñas V, Jacobson KA, Gorostiza P, Ciruela F, Bioconjugate Chem. 2014, 25, 1847–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Agnetta L, Kauk M, Canizal MCA, Messerer R, Holzgrabe U, Hoffmann C, Decker M, Angewandte Chemie International Edition 2017, 56, 7282–7287. [DOI] [PubMed] [Google Scholar]
- [17].Prischich D, Gomila AMJ, Milla-Navarro S, Sangüesa G, Diez-Alarcia R, Preda B, Matera C, Batlle M, Ramírez L, Giralt E, Hernando J, Guasch E, Meana JJ, de la Villa P, Gorostiza P, Angewandte Chemie International Edition 2021, 60, 3625–3631. [DOI] [PubMed] [Google Scholar]
- [18].Frank JA, Yushchenko DA, Fine NHF, Duca M, Citir M, Broichhagen J, Hodson DJ, Schultz C, Trauner D, Chem. Sci. 2017, 8, 7604–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Morstein J, Hill RZ, Novak AJE, Feng S, Norman DD, Donthamsetti PC, Frank JA, Harayama T, Williams BM, Parrill AL, Tigyi GJ, Riezman H, Isacoff EY, Bautista DM, Trauner D, Nature Chemical Biology 2019, 15, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morstein J, Dacheux MA, Norman DD, Shemet A, Donthamsetti PC, Citir M, Frank JA, Schultz C, Isacoff EY, Parrill AL, Tigyi GJ, Trauner D, J. Am. Chem. Soc. 2020, 142, 10612–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Westphal MV, Schafroth MA, Sarott RC, Imhof MA, Bold CP, Leippe P, Dhopeshwarkar A, Grandner JM, Katritch V, Mackie K, Trauner D, Carreira EM, Frank JA, J. Am. Chem. Soc. 2017, 139, 18206–18212. [DOI] [PubMed] [Google Scholar]
- [22].Sarott RC, Viray AEG, Pfaff P, Sadybekov A, Rajic G, Katritch V, Carreira EM, Frank JA, J. Am. Chem. Soc. 2021, 143, 736–743. [DOI] [PubMed] [Google Scholar]
- [23].Broichhagen J, Johnston NR, von Ohlen Y, Meyer-Berg H, Jones BJ, Bloom SR, Rutter GA, Trauner D, Hodson DJ, Angew Chem Int Ed Engl 2016, 55, 5865–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Broichhagen J, Podewin T, Meyer-Berg H, von Ohlen Y, Johnston NR, Jones BJ, Bloom SR, Rutter GA, Hoffmann-Röder A, Hodson DJ, Trauner D, Angew. Chem. Int. Ed. 2015, 54, 15565–15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pittolo S, Gómez-Santacana X, Eckelt K, Rovira X, Dalton J, Goudet C, Pin J-P, Llobet A, Giraldo J, Llebaria A, Gorostiza P, Nat. Chem. Biol. 2014, 10, 813–815. [DOI] [PubMed] [Google Scholar]
- [26].Rovira X, Trapero A, Pittolo S, Zussy C, Faucherre A, Jopling C, Giraldo J, Pin J-P, Gorostiza P, Goudet C, Llebaria A, Cell Chemical Biology 2016, 23, 929–934. [DOI] [PubMed] [Google Scholar]
- [27].Font J, López-Cano M, Notartomaso S, Scarselli P, Di Pietro P, Bresolí-Obach R, Battaglia G, Malhaire F, Rovira X, Catena J, Giraldo J, Pin J-P, Fernández-Dueñas V, Goudet C, Nonell S, Nicoletti F, Llebaria A, Ciruela F, eLife 2017, 6, e23545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Donthamsetti P, Konrad DB, Hetzler B, Fu Z, Trauner D, Isacoff EY, J. Am. Chem. Soc. 2021, 143, 8951–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, Trauner D, ACS Cent. Sci. 2015, 1, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Levitz J, Broichhagen J, Leippe P, Konrad D, Trauner D, Isacoff EY, PNAS 2017, 114, E3546–E3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, Isacoff EY, Neuron 2016, 92, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Berry MH, Holt A, Salari A, Veit J, Visel M, Levitz J, Aghi K, Gaub BM, Sivyer B, Flannery JG, Isacoff EY, Nat Commun 2019, 10, 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Acosta-Ruiz A, Gutzeit VA, Skelly MJ, Meadows S, Lee J, Parekh P, Orr AG, Liston C, Pleil KE, Broichhagen J, Levitz J, Neuron 2020, 105, 446–463.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gutzeit VA, Acosta-Ruiz A, Munguba H, Häfner S, Landra-Willm A, Mathes B, Mony J, Yarotski D, Börjesson K, Liston C, Sandoz G, Levitz J, Broichhagen J, Cell Chemical Biology 2021, DOI 10.1016/j.chembiol.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Broichhagen J, Frank JA, Trauner D, Accounts of Chemical Research 2015, 48, 1947–1960. [DOI] [PubMed] [Google Scholar]
- [36].Morstein J, Awale M, Reymond J-L, Trauner D, ACS Cent. Sci. 2019, 5, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weichert D, Kruse AC, Manglik A, Hiller C, Zhang C, Hübner H, Kobilka BK, Gmeiner P, PNAS 2014, 111, 10744–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cabrera R, Filevich O, García-Acosta B, Athilingam J, Bender KJ, Poskanzer KE, Etchenique R, ACS Chem. Neurosci. 2017, 8, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rea AC, Vandenberg LN, Ball RE, Snouffer AA, Hudson AG, Zhu Y, McLain DE, Johnston LL, Lauderdale JD, Levin M, Dore TM, Chemistry & Biology 2013, 20, 1536–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zayat L, Salierno M, Etchenique R, Inorg. Chem. 2006, 45, 1728–1731. [DOI] [PubMed] [Google Scholar]
- [41].Breitinger H-GA, Wieboldt R, Ramesh D, Carpenter BK, Hess GP, Biochemistry 2000, 39, 5500–5508. [DOI] [PubMed] [Google Scholar]
- [42].Donthamsetti PC, Broichhagen J, Vyklicky V, Stanley C, Fu Z, Visel M, Levitz JL, Javitch JA, Trauner D, Isacoff EY, J. Am. Chem. Soc. 2019, 141, 11522–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rustler K, Maleeva G, Bregestovski P, König B, Beilstein J. Org. Chem. 2019, 15, 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kim K, Che T, Panova O, DiBerto JF, Lyu J, Krumm BE, Wacker D, Robertson MJ, Seven AB, Nichols DE, Shoichet BK, Skiniotis G, Roth BL, Cell 2020, 182, 1574–1588.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Donthamsetti P, Winter N, Hoagland A, Stanley C, Visel M, Lammel S, Trauner D, Isacoff E, Nat Commun 2021, 12, 4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Farrants H, Gutzeit VA, Acosta-Ruiz A, Trauner D, Johnsson K, Levitz J, Broichhagen J, ACS Chem Biol 2018, 13, 2682–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.