Abstract
A histidine kinase protein (Cph1) with sequence homology and spectral characteristics very similar to those of the plant phytochrome has been recently identified in the cyanobacterium Synechocystis sp. strain PCC 6803. Cph1 together with Rcp1 (a protein homologue to the response regulator CheY) forms a light-regulated two-component system whose function is presently unknown. Levels of cph1 rcp1 mRNA increase in the dark and decrease upon reillumination. A dark-mediated increase in cph1 rcp1 mRNA levels was inhibited by the presence of glucose, but not by inhibition of the photosynthetic electron flow. The half-life of cph1 rcp1 transcript in the light was about fourfold shorter than in the dark, indicating that control of cph1 rcp1 transcript stability is one of the mechanisms by which light regulates expression of the cyanobacterial phytochrome. After 15 min of darkness, 3-min pulses of red, blue, green, and far-red light were equally efficient in decreasing the cph1 rcp1 mRNA levels. Red light downregulation was not reversed by far-red light, suggesting that cph1 rcp1 mRNA levels are not controlled by a phytochrome-like photoreceptor. Furthermore, a Synechocystis strain containing an H538R Cph1 point mutation, unable to phosphorylate Rcp1, shows normal light-dark regulation of the cph1 rcp1 transcript levels. Our data suggest a role of cyanobacterial phytochrome in the control of processes required for adaptation in light-dark and dark-light transitions.
Photosynthetic organisms must maintain a metabolic homeostasis despite daily variations in incident light. Not only does light provide energy for photosynthesis, but a large number of plant developmental events are also responsive to light cues. Accordingly, photosynthetic organisms have evolved light detection systems (photoreceptors) that control gene expression through signal transduction pathways (25).
Phytochromes are the best characterized of those photoreceptors. Phytochromes exist in two different photoconvertible forms, the red-light-absorbing form (Pr) and the far-red-light-absorbing form (Pfr) (for reviews, see references 27 and 34). In plants, phytochromes are soluble homodimers constituted by two subunits of about 125 kDa, each of which folds into two major structural domains: an amino-terminal domain that binds the chromophore and a carboxy-terminal domain that contains regions necessary for dimerization and biological activity. How the plant phytochrome transduces perceived photosensory information to downstream signaling components remains unclear, although some progress has been made toward determining it (for a review, see reference 10).
The field of phytochrome research has recently been revolutionized by the finding of a phytochrome in the cyanobacterium Synechocystis sp. strain PCC 6803 (16, 18, 20, 37) (for reviews, see references 9, 24, and 26). Cyanobacteria are photosynthetic prokaryotes that carry out oxygenic photosynthesis similar to eukaryotic algae and higher plants. The most exciting aspect of this discovery is that cyanobacterial phytochrome, Cph1, is the sensor component of a typical bacterial two-component signal transduction system (for reviews, see references 15 and 23). The amino-terminal domain of Cph1 shows 30 to 35% amino acid identity to the chromophore-bearing domain of higher plant phytochromes, and it is able to catalyze its own chromophore attachment in vitro, whereas the carboxy terminus contains the consensus sequences of histidine kinases. Immediately downstream of cph1 is found an open reading frame (called rcp1) encoding a protein with striking sequence similarity to the CheY family of response regulators. In fact, Yeh et al. have shown that, in vitro, Cph1 is a light-regulated histidine kinase that mediates red/far-red reversible phosphorylation of the response regulator Rcp1 (37). These findings shed light on the initial step of light signal transduction by phytochrome. However, the function of Cph1 as a phytochrome in vivo has not yet been demonstrated.
Here we characterize the pattern of expression of the cph1 rcp1 operon under different conditions. We demonstrate that the amount of cph1 rcp1 transcript is repressed by light, probably through the concourse of different photoreceptors. Dark-dependent upregulation of cph1 rcp1 transcript levels is abolished by glucose. This pattern of expression suggests a role of cyanobacterial phytochrome in light-dark transitions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Synechocystis sp. strain PCC 6803 was grown photoautotrophically at 30°C in BG11c medium (30) and bubbled with a continuous stream of 1% (vol/vol) CO2 in air under continuous fluorescent illumination (50 μE of white light m−2 s−1) (referred to in the text as “normal illumination conditions”). For mixotrophic growth, glucose was added to a final concentration of 10 mM. Dark conditions were obtained by wrapping culture flasks with aluminum foil. Light intensity was measured with an LI-188B Integrating Quantum/Radiometer/Photometer (LI-COR, Inc). For light quality experiments, Synechocystis cultures were irradiated with 20 μE of light of a specific wavelength m−2 s−1. Selective irradiation was generated with the following narrow-band filters: blue, λmax = 455 nm; green, λmax = 500 nm; red, λmax = 650 nm; far-red, λmax = 725 nm. 3-(3,4-Dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropylbenzoquinone (DBMIB) were used at a final concentration of 5 μM when indicated.
Escherichia coli DH5α (Bethesda Research Laboratories) grown in Luria broth medium was used for plasmid construction and replication. E. coli was supplemented with 100 μg of ampicillin per ml or 50 μg of kanamycin per ml when required.
RNA isolation and Northern blot hybridization.
Total RNA was isolated from 25-ml samples of Synechocystis sp. strain PCC 6803 cultures at the mid-exponential phase (3 to 5 μg of chlorophyll/ml). Extractions were performed by vortexing cells in the presence of phenol-chloroform and acid-washed baked glass beads (0.25 to 0.3 mm in diameter; Braun, Melsungen, Germany) as previously described (12).
For Northern blotting, 15 μg of total RNA was loaded per lane and electrophoresed in 1.2% agarose denaturing formaldehyde gels. Transfer to nylon membranes (Hybond N-plus; Amersham), prehybridization, hybridization, and washes were performed in accordance with Amersham instruction manuals, with hybridization taking place at 42°C in the presence of 50% formamide. The 1,150-bp DNA fragment obtained by PCR amplification with oligonucleotides pht1 (5′ GATCCCATCCAGAGTCGCCTAACG 3′) (from nucleotide +223 to nucleotide +246, considering the first nucleotide of the cph1 gene translation start codon as +1) and pht2 (5′ AAGCATGATTTGGGTCACCGCCCC 3′) (from nucleotide +1372 to nucleotide +1349) and the 467-bp DNA fragment obtained with oligonucleotides pht5 (5′ GGTATTGAACCATGTCCGACG 3′) (from nucleotide −11 to nucleotide +10, considering the first nucleotide of the rcp1 gene translation start codon as +1) and pht6 (5′ GGAGGATGCCAATTAAGCTGC 3′) (from nucleotide +456 to nucleotide +436) were used as the cph1 and rcp1 probes, respectively. As a control, in all cases, the filters were reprobed with a HindIII-BamHI 580-bp probe from plasmid pAV1100 that contains the constitutively expressed RNase P RNA gene from Synechocystis sp. strain PCC 6803 (36). To determine the cpm of radioactive areas in Northern blot hybridizations, an InstantImager Electronic Autoradiography apparatus (Packard Instrument Company, Meriden, Conn.) was used.
Primer extension analysis.
Oligonucleotide pht3 (5′ GGTCGCTGAGTTGTACGG 3′) (from nucleotide +28 to nucleotide +11) end labeled with T4 polynucleotide kinase and [γ-32P]dATP (3,000 Ci/mmol) following standard procedures (31) was used for primer extension analysis. For annealing, a 10-μl mixture containing 0.15 M NaCl, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 20 μg of total RNA, and about 2 pmol of oligonucleotide (106 cpm) was prepared. The annealing mixture was heated for 2 min at 90°C in a water bath that was subsequently kept at room temperature to reach 50°C. For extension, a 10-μl mixture was prepared with one-half of the annealing mixture, 10 mM dithiothreitol, 0.5 mM each deoxynucleoside triphosphate (dNTP), 2 μg of actinomycin D, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 100 U of Superscript II RNase H-reverse transcriptase (Gibco BRL, Gaithersburg, Md.). The mixture was incubated for 45 min at 45°C, and the reaction was stopped by adding 4 μl of formamide-loading buffer. One-half of the reaction mixture was electrophoresed on a 6% polyacrylamide sequencing gel together with a sequencing reaction mixture of the cph1 gene 5′ region by using the pht3 oligonucleotide.
Transcriptional gene fusion.
A transcriptional gene fusion was constructed in the plasmid pFF11, a promoter-probe vector based on the chloramphenicol acetyltransferase (CAT) reporter gene (cat) (11). A 413-bp DNA fragment from nucleotide −245 to +168 bp with respect to the cph1 transcription start point was subcloned into pFF11, yielding the plasmid pFF11-cph. This reporter plasmid was used to transform the SFCΩ5 variant of Synechocystis sp. strain PCC 6803 (4).
CAT activity was assayed in vitro at 37°C by the colorimetric procedure (33). One unit of CAT represents 1 μmol of chloramphenicol acetylated per min per mg of protein. Crude extracts from Synechocystis strains were prepared with glass beads as described in reference 28, except that the buffer was substituted for by 50 mM Tris-HCl (pH 8.0).
The strains SFF16 (11), which contains a promoterless cat gene, and SFC57 (5), which contains the cat gene under the control of its own promoter, were used as controls.
The amount of protein in cell extracts was determined by the method of Bradford (2) with ovalbumin as a standard.
Construction of Synechocystis sp. strain PCC 6803 cph1 mutants.
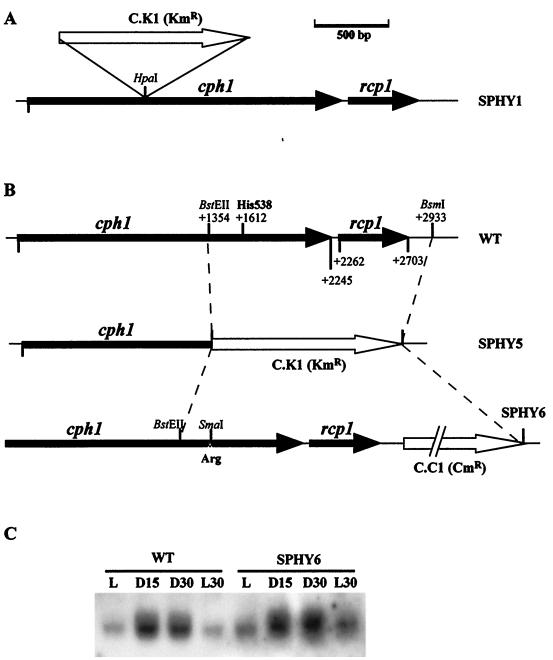
The Synechocystis sp. mutant strain SPHY1 was created by interrupting the cph1 gene with a neomycin phosphotransferase (npt)-containing cassette (C.K1) (8), which confers kanamycin resistance (Kmr). The C.K1 cassette was isolated as a 1.3-kb HincII DNA fragment and inserted into the unique HpaI site of cph1 in both orientations. Transformation of Synechocystis sp. strain PCC 6803 cells was carried out as previously described (4).
A His538-to-Arg site-directed mutant of Cph1 was created by a two-step strategy. First, a region of the Synechocystis sp. strain PCC 6803 cph1 rcp1 operon was deleted and replaced by a kanamycin resistance cassette, to generate strain SPHY5 (see Fig. 6B). The site-directed mutant strain (SPHY6) was then generated by replacing the kanamycin resistance cassette with a construct containing the previously deleted region with the site-directed mutation. A chloramphenicol resistance (Cmr) gene was also introduced downstream of the rcp1 coding region in order to be used as selectable marker (see Fig. 6B).
FIG. 6.
Inactivation of cph1 in Synechocystis sp. strain 6803. (A) Structure of the cph1 rcp1 operon genomic region in the SPHY1 mutant strain. (B) Structure of the cph1 rcp1 operon genomic region in the wild-type (WT), SPHY5, and SPHY6 mutant strains. Restriction sites used for the construction of the mutants are marked. The position of the CAT codon corresponding to the His538 residue is also marked. Nucleotides are numbered with respect to the translation start site. (C) Total RNA was isolated from mid-log-phase Synechocystis wild-type and SPHY6 mutant cells growing under normal illumination conditions (L) or subjected to 15 min (L15) or 30 min (L30) in the dark or 30 min after reillumination (L30). RNA was subjected to Northern blot analysis by using an internal cph1 probe (see Materials and Methods).
The SPHY5 strain was created by replacing a 1,580-bp BstEII-BsmI DNA fragment with a C.K1 cassette (Kmr). The site-directed mutation plasmid pHR3 was generated by changing the His538 CAT codon to an Arg CGG codon by standard PCR techniques. The point mutation created a SmaI site, which was used to test the mutants. The C.C1 cassette (Cmr) (8) was then inserted at the BsmI site located 228 bp downstream of the rcp1 STOP codon, in the same orientation as the operon. Plasmid pHR3 was used to transform Synechocystis sp. strain PCC 6803 SPHY5 cells. Cmr Kms cells were selected. Recombinants that have incorporated the C.C1 cassette in strain SPHY5 also introduce the His538Arg mutation, involving the loss of the C.K1 cassette. Whole segregation of the mutants was checked by Southern blotting.
RESULTS
Expression of cph1 rcp1 mRNA is upregulated by darkness.
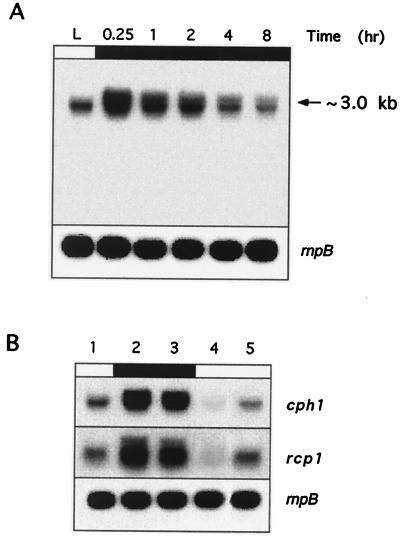
In higher plants, the phytochromes are encoded by a gene family of up to five members (named PHYA–E) (6, 32). In most plants, PHYA mRNA is synthesized in etiolated tissue and down-regulated rapidly in the light, whereas PHYB–E mRNAs are not affected by light (for example, see reference 6). To determine if light-dark transitions also affect the cyanobacterial phytochrome transcript levels, Northern blot hybridizations of total RNA from exponentially growing Synechocystis sp. strain PCC 6803 cultures under illumination conditions or after 0.25, 1, 2, 4, or 8 h of darkness were carried out. Only one band of about 3 kb was observed when filters were hybridized with probes of cph1 or rcp1 genes, demonstrating that both genes are cotranscribed, forming an operon. As shown in Fig. 1A, cph1 rcp1 mRNA levels were upregulated (about a fivefold increase) after transfer of the cultures to the dark. Maximal levels of expression were obtained after 15 min of darkness. Thereafter, the quantity of the transcript decreased slowly, reaching, after 8 h, levels similar to those present under continuous illumination. When cultures that have been maintained in the dark for 1 h were reilluminated, cph1 rcp1 mRNA levels decreased dramatically, becoming almost undetectable (Fig. 1B, lane 4). However, 30 min after reillumination, cph1 rcp1 mRNA steady-state light levels were restored (Fig. 1B, lanes 1 and 5). The same pattern was observed when an rcp1 gene probe was used (Fig. 1B).
FIG. 1.
Dark-dependent upregulation of cph1 rcp1 mRNA levels. (A) Total RNA was isolated from mid-log-phase Synechocystis sp. strain 6803 cells growing under normal illumination conditions (white light, 50 μE m−2 s−1) (L) or after transfer of the culture to the dark for 0.25, 1, 2, 4, or 8 h. (B) Synechocystis sp. strain 6803 cells growing under illumination (lane 1) were transferred to the dark for 1 h (lane 2), and then the culture was divided into two fractions; one of them was subjected to an additional 30-min period of darkness (lane 3), while the other one was reilluminated for 5 min (lane 4) or 30 min (lane 5). Fifteen micrograms of total RNA was subjected to Northern blot analysis with internal cph1 or rcp1 probes (see Materials and Methods). The filters were stripped and rehybridized with an rnpB gene probe. Transcript size was estimated by comparison with 23S, 16S, and 5S rRNAs.
Redox- and glucose-dependent control of cph1 rcp1 mRNA expression.
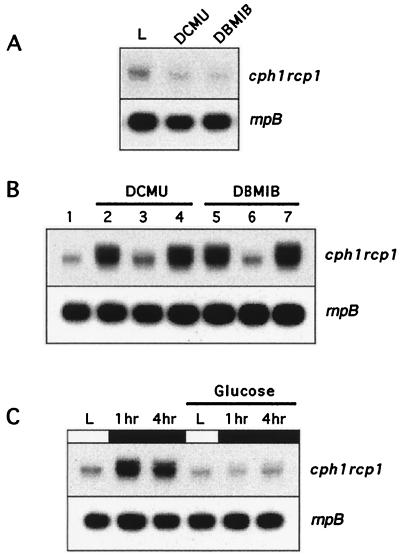
Activity of the photosynthetic apparatus is likely to exert a feedback regulatory role on the expression of photosynthetic genes. In fact, the expression of many cyanobacterial genes has been shown to be affected by redox signals (12, 22, 28). Figure 2A shows that inhibition of photosynthetic flow by DCMU (which blocks transfer of electrons between the PSII complex and the plastoquinone pool [35]) or DBMIB (which prevents the oxidation of plastoquinone by the cytochrome b6f complex [29]) did not upregulate cph1 rcp1 mRNA levels. These data suggest that it is the absence of light per se, and not the absence of photosynthetic flow, that mediates the dark-dependent increase in cph1 rcp1 mRNA levels. This was demonstrated by transferring DCMU- or DBMIB-treated cells to the dark. Under these conditions, cph1 rcp1 mRNA levels increased in a way similar to that in nontreated cells (Fig. 2B).
FIG. 2.
Effects of photosynthetic inhibitors and glucose on cph1 rcp1 mRNA levels. (A) Light-growing Synechocystis sp. strain 6803 cells were incubated either in the absence (L) or in the presence of 5 μM DCMU or DBMIB. RNA was isolated after 1 h and processed and hybridized as for Fig. 1. (B) Synechocystis sp. strain 6803 cells grown under illumination conditions (lane 1) were treated with DCMU (lane 2) or DBMIB (lane 5) and transferred to the dark for 1 h or maintained in the light for 1 h (lanes 3 and 6, respectively) and then subjected to darkness for 1 h (lanes 4 and 7, respectively). Northern blot hybridizations were performed with RNA from cells grown under the different conditions. (C) Total RNA was isolated from mid-log-phase Synechocystis sp. strain 6803 cells growing under illumination conditions (L) or after 1 or 4 h of darkness in the presence or absence of 10 mM glucose. RNA was subjected to Northern blot analysis with an internal cph1 probe.
Synechocystis sp. strain PCC 6803 is a heterotrophic facultative cyanobacterium that can utilize glucose as a source of energy, redox power, and carbon (30). Interestingly, when Synechocystis sp. strain PCC 6803 cells grown in the light and in the presence of glucose (mixotrophic growth) were transferred to the dark, the level of cph1 rcp1 mRNA remained constant (Fig. 2C). This suggests that not only the absence or the presence of light but also other metabolic signals are involved in the control of the expression of the cph1 rcp1 operon.
Determination of the cph1 rcp1 promoter region.
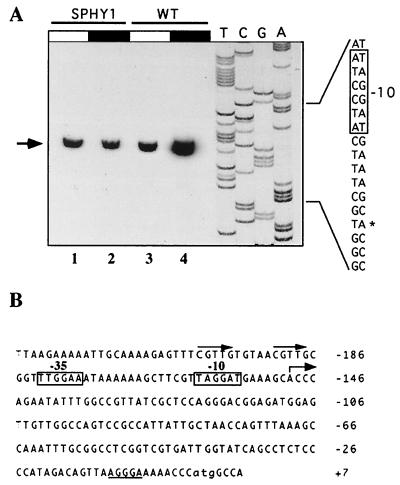
As a first step in the characterization of the cyanobacterial phytochrome promoter, we have determined the transcription start point of the cph1 rcp1 operon. Reverse primer extension of total RNA from light-grown cultures or from cultures subjected to 1 h of darkness was carried out. The transcription start point was localized at −150 bp with respect to the first translated nucleotide (Fig. 3A, lanes 3 and 4). −10 (TAGGAT) and −35 (TTGGAA) sequences with four of six sites matching the −10 (TATAAT) and −35 (TTGACA) boxes of the Escherichia coli ς70-like consensus promoters (14) were found upstream of the cph1 rcp1 first transcribed nucleotide (Fig. 3B). cDNA was more abundant when primer extension was carried out with RNA isolated from dark-treated wild-type Synechocystis sp. strain PCC 6803 cells than with RNA isolated from continuously illuminated cells (Fig. 3A). A direct nucleotide repetition in the form CGTTGN5CGTTG centered at position −45 was found (Fig. 3B). To determine whether the cph1 rcp1 5′-upstream region contains all of the cis-regulatory elements responsible for the observed light-dark regulation, we subcloned the −245 to +168 region of the cph1 gene into pFF11, a promoter-probe plasmid based on the cat reporter gene and constructed for testing promoters in Synechocystis sp. strain PCC 6803 (11). In vivo cph1 rcp1 promoter activity was monitored by determining CAT activity of the cyanobacterial reporter strain under normal illumination conditions or after 3 h of darkness. The −245 to +168 region of the cph1 gene displayed a significant promoter activity, and strong dark induction of the reporter gene activity was not observed. However, a 1.5- to 1.8-fold increase in CAT activity was consistently observed after 3 h of darkness (4.66 ± 1 mU/mg under normal illumination conditions, versus 8.3 ± 1.1 mU/mg in darkness). CAT activity levels of control strains harboring a promoterless cat gene or a cat gene driven by its own promoter were not affected by light-dark changes (data not shown).
FIG. 3.
Primer extension analysis of the cph1 rcp1 transcript. (A) Total RNA (20 μg) from illuminated (white segment) or 1-h-dark-incubated (black segment) Synechocystis sp. strain 6803 wild-type (WT) and SPHY1 mutant cells was annealed to the pht3 oligonucleotide and extended with reverse transcriptase as described previously (15). A sequencing ladder used with the same primer is also shown. An arrow indicates extension products. The transcription start nucleotide is marked with an asterisk on the sequence. These results were confirmed for three times with RNAs from three independent sets of cultures. (B) Sequence of the promoter region of the cph1 rcp1 operon. The transcription start point is indicated by a bent arrow. −10 and −35 sequences based on the transcriptional start site are boxed. The translation start codon is in lowercase. The putative Shine-Dalgarno sequence is underlined. The nucleotides are numbered with respect to the first nucleotide of the translation start codon. Direct repeat sequences are noted by arrows.
Light-decreased stability of cph1 rcp1 mRNA.
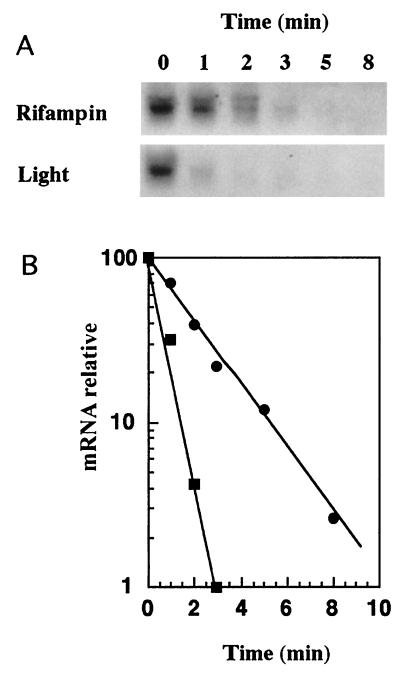
In order to investigate whether light affects cph1 rcp1 mRNA stability, we have determined the half-life of the cph1 rcp1 transcript under dark conditions or upon reillumination. For that purpose, rifampin (400 μg/ml) was added to 15-min-dark-treated cells, and cultures were kept in the dark. Aliquots were taken at 0, 1, 2, 3, 5, and 8 min after the addition of rifampin, and total RNA was isolated and subjected to Northern blotting. The data showed that, under these conditions, the half-life of cph1 rcp1 mRNA was close to 2 min (Fig. 4). However, when dark-treated cells were exposed to light in the absence of rifampin, a rapid decrease in transcript level was seen, with a half-life of about 30 s.
FIG. 4.
Light-decreased stability of cph1 rcp1 mRNA. (A) Rifampin (400 μg/ml) was added to 15-min-dark-treated cells, and cultures were kept in the dark. Aliquots were taken at the indicated times, and total RNA was isolated. Alternatively, 15-min-dark-treated cultures were reilluminated (light), and aliquots were taken at the indicated times for total RNA isolation. Fifteen micrograms of total RNA was subjected to Northern blot analysis with internal cph1 probes. (B) Band intensity was determined with an InstantImager, normalized with respect to the rnpB RNA level, and plotted against time. Values are the averages of two independent experiments. Symbols: ■, reilluminated cells; ●, rifampin-treated cells.
Effect of spectral quality on expression of the cph1 rcp1 operon.
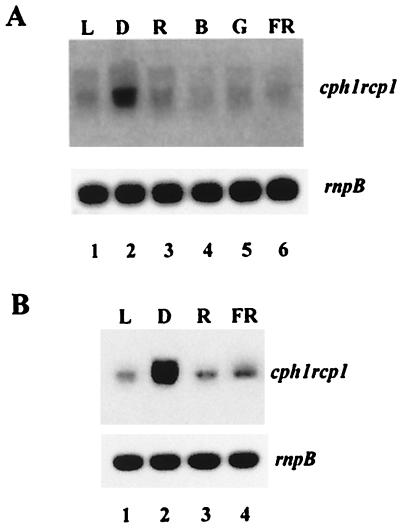
The results presented above suggest that redox signals from the photosynthetic apparatus are not involved in the upregulation of the expression of cph1 rcp1 operon, and therefore a photoreceptor might be involved in such regulation. As a first approach to identify this putative photoreceptor, the effect of spectral quality on expression of the cph1 rcp1 operon was analyzed. For that analysis, Synechocystis sp. strain PCC 6803 cells grown under normal illumination conditions were transferred to darkness for 15 min and then exposed to a 3-min pulse (20 μE m−2 s−1) of blue (λmax = 455 nm), green (λmax = 500 nm), red (λmax = 650 nm), or far-red (λmax = 725 nm) light. As previously shown, dark-treated cells displayed high cph1 rcp1 mRNA levels. Exposure of the cells to 20 μE of blue, green, red, or far-red filtered light m−2 s−1 resulted in a drastic decrease in the cph1 rcp1 transcript level (Fig. 5A). In a second set of experiments, exponentially growing Synechocystis sp. strain PCC 6803 cultures were transferred to darkness for 15 min, followed by one pulse of 3 min of red (λmax = 650 nm) light (20 μE m−2 s−1). The cells were then harvested or incubated for another 10 min in the presence of far-red (λmax = 725 nm) light (20 μE m−2 s−1). As shown above, 3 min of red light was enough to elicit a drastic reduction in the quantity of cph1 rcp1 mRNA. This effect was not reversed by far-red light (Fig. 5B), suggesting that cph1 rcp1 mRNA levels do not respond in a typical phytochrome-mediated way.
FIG. 5.
Effect of spectral quality on expression of cph1 rcp1 operon. (A) Northern blot hybridization of total RNA from Synechocystis sp. strain 6803 cultures grown under normal illumination conditions (lane 1 [L]), transferred to darkness for 15 min (lane 2 [D]), and then divided into four aliquots that were illuminated with a 3-min pulse of red (lane 3 [R]), blue (lane 4 [B]), green (lane 5 [G]), or far-red (lane 6 [FR]) light. (B) Northern blot hybridization of total RNA from Synechocystis sp. strain 6803 cultures grown under normal illumination conditions (lane 1), transferred to darkness for 15 min (lane 2), and then given a red light pulse of 3 min (lane 3), followed by a far-red light pulse of 10 min (lane 4).
Cph1 kinase activity is not involved in control of cph1 rcp1 mRNA levels.
In monocots, it has been extensively shown that transcription of the phyA gene is phytochrome dependent (3, 19). In order to investigate whether Cph1 is responsible for the light-dark-dependent regulation of the cph1 rcp1 operon expression, we constructed a cph1 mutant strain of Synechocystis sp. strain PCC 6803 (SPHY1) by interrupting the cph1 gene with a neomycin phosphotransferase (npt)-containing cassette (C.K1). The cassette was inserted into the HpaI site localized 712 bp downstream of the cph1 ATG codon (Fig. 6A). Complete segregation of the mutation was confirmed by Southern blot analysis (data not shown). Mutant cells were viable under normal illumination conditions, and growth rates were similar to those of the wild-type strain. The study of the expression of cph1 rcp1 operon in SPHY1 mutant cells was carried out by reverse primer extension of total RNA by using the oligonucleotide pht3, which is complementary to a region of the cph1 gene upstream of the cassette integration site. In the SPHY1 mutant cells, the cph1 transcription start point was localized in the same position as in the wild-type cells (Fig. 3A, lanes 1 and 2). Analysis of RNA from light- or dark-treated cells indicated that the cph1 transcript was not induced by darkness in the Cph1-deficient cells (Fig. 3A, lanes 1 and 2). Similar results were observed by Northern blotting with a DNA fragment upstream of the C.K1 insertion point as a probe (data not shown). These data might suggest that Cph1 is the photoreceptor responsible for the light-dark-dependent regulation of cph1 rcp1 mRNA levels. We have shown above that cph1 rcp1 transcript levels are regulated, at least in part, by controlling cph1 rcp1 mRNA stability. Since insertion of the C.K1 cassette into the cph1 rcp1 operon strongly modifies the structure of the cph1 rcp1 transcript, this mutation might dramatically change the stability of the transcript. Therefore, we have constructed, by site-directed mutagenesis, a Synechocystis strain with a more subtle inactivation of Cph1 that does not affect cph1 rcp1 mRNA structure. It has been shown previously that the His538 residue of Cph1 is essential for autophosphorylation and phosphotransfer to Rcp1 and, therefore, for transduction of the light signal (37). We have constructed an H538R mutant strain of Cph1 (Synechocystis sp. strain SPHY6) by replacing the endogenous wild-type locus by the mutated variant as described in Materials and Methods (Fig. 6B). Complete segregation of the mutation was confirmed by Southern blot analysis (data not shown). Mutant cells were viable under normal illumination conditions, and growth rates were similar to those of the wild-type strain. SPHY6 mutant cells displayed a wild-type phenotype with respect to the light-dark-dependent regulation of the cph1 rcp1 mRNA levels (Fig. 6C).
DISCUSSION
The sequencing of the complete genome of the cyanobacterium Synechocystis sp. strain PCC 6803 has uncovered the presence of a two-component regulatory system (Cph1-Rcp1) whose sensor component shows very significant amino acid identity to the plant phytochrome (20). Our data demonstrate that cph1 and rcp1 are cotranscribed and that cph1 rcp1 transcript expression is controlled by light.
Yeh et al. have demonstrated that only Cph1-Pr exhibits kinase activity, suggesting that Cph1-Pr is the active form that transduces the light signal by phosphate transfer to Rcp1 (37). Since Pfr is supposed to be the active form in plants (27), this is an important difference between the cyanobacterial system and the plant signal transduction system. Our results indicate that Cph1 and Rcp1 are expressed mostly under dark conditions. Since Cph1 is de novo synthesized in the form Pr and in the absence of light is not photoconverted to Cph1-Pfr (37), our current hypothesis is that, in the dark, Rcp1 would be phosphorylated. In contrast, in the light, the low level of Cph1-Pr synthesized is immediately converted into Pfr, and Rcp1 would be mostly unphosphorylated.
Absence of light seems to be the signal that triggers the accumulation of cph1 rcp1 transcript. Two obvious possible pathways for sensing the absence of light can be imagined: directly by a photoreceptor or indirectly through photosynthetic electron transport and the redox state of intermediate carriers. Our experiments with the photosynthetic inhibitors DCMU and DBMIB indicate that complete cessation of photosynthetic electron transport does not elicit the accumulation of cph1 rcp1 transcript. Furthermore, the presence of these inhibitors does not impair the dark effect. These data suggest that the redox state of photosynthetic electron carriers is not involved in the regulation of cph1 rcp1 expression and therefore support a direct photoreceptor-dependent-mediated mechanism. In order to investigate what kind of photoreceptor may be involved in the control of cph1 rcp1 expression, reillumination experiments were carried out with light of four different spectral qualities. Light that was red, far-red, blue, or green was able to downregulate the levels of cph1 rcp1 transcript, suggesting that more than one photoreceptor pathway could be involved in downregulation of cph1 rcp1 transcript levels. One obvious possibility is that Cph1 is able to autoregulate its own mRNA levels. In monocots, it has been shown extensively that transcription of the phyA gene is phytochrome dependent (3, 19). However, the results shown in Fig. 5 suggest that the pattern of cph1 rcp1 accumulation under red and far-red light does not follow a typical phytochrome-dependent response. Two Synechocystis cph1 mutant strains were generated in order to further investigate this possibility: a cph1::C.K1 insertion mutant (SPHY1) and an H538R point mutation (SPHY6) that produces a Cph1 protein unable to phosphorylate Rcp1 and, therefore, unable to transduce the light signal (37). SPHY1 cells showed uninducible levels of the 5′ region of the cph1 rcp1 transcript (Fig. 3A). In contrast, in SPHY6 cells, a normal dark-dependent induction of the cph1 rcp1 transcript was observed. While the results obtained with the SPHY1 mutant are consistent with an autoregulatory mechanism, the results obtained with the SPHY6 mutant exclude this hypothesis. We propose the following interpretation for our results. The cph1::C.K1 mutation (SPHY1) results in gross alterations of the cph1 mRNA. The short half-life of the cph1 rcp1 transcript upon reillumination suggests the existence of a specific degradation mechanism controlling cph1 rcp1 transcript levels in the light (Fig. 1B and Fig. 4). The molecular mechanism by which light controls cph1 rcp1 transcript stability remains to be elucidated. Thus, it is possible that the interruption of the cph1 mRNA by the C.K1 cassette affects elements within the cph1 mRNA coding region that confer stability to the message in darkness. Light-dependent control of mRNA stability determined by coding region elements has been reported recently for the psbAI and psbAII genes in the cyanobacterium Synechococcus sp. strain PCC 7942 (17). In the SPHY6 strain, we have introduced a point mutation that abolishes the kinase activity of Cph1 without affecting the structure of the cph1 rcp1 transcript. Since this strain shows a normal regulation of cph1 rcp1 transcript levels, we conclude that the Cph1 signal transduction pathway is not involved in the light-dark-mediated regulation of the cph1 rcp1 transcript levels.
In addition to the control of cph1 rcp1 transcript stability, we have investigated the possibility that cph1 rcp1 operon transcription is upregulated by absence of light. Transcriptional fusion experiments with a cat reporter gene driven by the promoter and leader regions of cph1 showed only a minor (1.8-fold in the best case) dark-dependent induction, in contrast to the 5-fold increase in the amount of cph1 rcp1 mRNA promoted by darkness. Furthermore, a transcriptional fusion of the −245 to +168 cph1 region with the green fluorescent protein gene was also used as a reporter mRNA. The amount of this chimeric mRNA was only slightly increased by darkness (1.3- to 1.7-fold, depending on the experiment) (data not shown). Therefore, our experiments suggest that dark-dependent transcriptional induction represents a minor contribution to the regulation of the cph1 rcp1 operon expression.
Our results demonstrate that cph1 rcp1 mRNA levels are also affected by other factors in addition to light. In fact, accumulation of cph1 rcp1 mRNA in dark-incubated Synechocystis sp. strain PCC 6803 cells is completely inhibited by the presence of exogenous glucose. Evidence for a specific interaction between plant phytochrome signaling and carbohydrate metabolism has also been reported in plants. For example, sucrose can inhibit PhyA-dependent far-red light-mediated inhibition of greening (1, 7). A number of anabolic Synechocystis sp. strain PCC 6803 genes have been shown to be switched off in the dark; however, glucose is able to abolish this dark-mediated inhibition of expression (12, 21, 22, 28). Since glucose is a source of energy, redox power, and carbon for Synechocystis sp. strain PCC 6803, it seems reasonable to imagine that the presence or absence of glucose could change the way that Synechocystis sp. strain PCC 6803 adapts to dark conditions.
The fact that cph1 rcp1 mRNA levels are upregulated in darkness suggests that Synechocystis sp. strain PCC 6803 phytochrome might be involved in the regulation of functions required for the adaptation from light to dark conditions and vice versa. The expression of many cyanobacterial genes has been shown to be dependent on the circadian rhythms (for a review, see reference 13). A role for the Cph1-Rcp1 system in setting the circadian clock might also be speculated. Finally, cyanobacterial phytochrome might be required only under specific stress conditions. In this regard, preliminary experiments indicate that cph1 rcp1 mRNA levels increase under conditions of nitrogen deficiency (data not shown). Characterization of cph1 and rcp1 mutants will be required in order to identify the biological functions controlled by the cyanobacterial phytochrome.
ACKNOWLEDGMENTS
We thank F. Chauvat for providing pFF11 plasmid and strains SFF16 and SFC57. We are grateful to J. Weitzman for critical reading of the manuscript.
This work was supported by grants from DGESID (PB97-0732) (Spain) and by Junta de Andalucía (CVI-0112). M.G.-D. was the recipient of a predoctoral fellowship from M.E.C. (Spain).
REFERENCES
- 1.Barnes S A, Nishizawa N K, Quaggio R B, Whitelam G C, Chua N H. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell. 1996;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Bruce W B, Quail P H. Cis-acting elements involved in photoregulation of an oat phytochrome promoter in rice. Plant Cell. 1990;2:1081–1089. doi: 10.1105/tpc.2.11.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauvat F, De Vries L, Van der Ende A, Van Arkel G A. A host vector system for gene cloning in the cyanobacterium Synechocystis PCC 6803. Mol Gen Genet. 1988;204:165–191. [Google Scholar]
- 5.Chauvat F, Labarre J, Ferino F. Development of gene transfer system for the cyanobacterium Synechocystis PCC 6803. Plant Physiol Biochem. 1988;26:629–637. [Google Scholar]
- 6.Clack T, Mathews S, Sharrock R A. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 7.Dijkwel P P, Huijser C, Weisbeek P J, Chua N H, Smeekens S C. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 9.Elich T D, Chory J. Phytochrome: if it looks and smells like a histidine kinase, is it a histidine kinase? Cell. 1997;91:713–716. doi: 10.1016/s0092-8674(00)80458-4. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- 11.Ferino F, Chauvat F. A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene. 1989;84:257–266. doi: 10.1016/0378-1119(89)90499-x. [DOI] [PubMed] [Google Scholar]
- 12.García-Domínguez M, Florencio F J. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 13.Golden S S, Ishiura M, Johnson C H, Kondo T. Cyanobacterial circadian rhythms. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- 14.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology Press; 1995. [Google Scholar]
- 16.Hughes J, Lamparter T, Mittmann F, Hartmann E, Gartner W, Wilde A, Borner T. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 17.Kulkarni R D, Golden S S. mRNA stability is regulated by a coding-region element and the unique 5′untranslated leader sequences of the three Synechococcus psbA transcripts. Mol Microbiol. 1997;24:1131–1142. doi: 10.1046/j.1365-2958.1997.4201768.x. [DOI] [PubMed] [Google Scholar]
- 18.Lamparter T, Mittmann F, Gartner W, Borner T, Hartmann E, Hughes J. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc Natl Acad Sci USA. 1997;94:11792–11797. doi: 10.1073/pnas.94.22.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lissemore J L, Quail P H. Rapid transcriptional regulation by phytochrome of the genes for phytochrome and chlorophyll a/b-binding protein in Avena sativa. Mol Cell Biol. 1988;8:4840–4850. doi: 10.1128/mcb.8.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno T, Kaneko T, Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–414. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- 21.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 22.Mohamed A, Jansson C. Photosynthetic electron transport controls degradation but not production of psbA transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1991;16:891–897. doi: 10.1007/BF00015080. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 24.Pepper A E. Molecular evolution: old branches on the phytochrome family tree. Curr Biol. 1998;8:R117–R120. doi: 10.1016/s0960-9822(98)70985-6. [DOI] [PubMed] [Google Scholar]
- 25.Quail P H. Photosensory perception and signal transduction in plants. Curr Opin Genet Dev. 1994;4:652–661. doi: 10.1016/0959-437x(94)90131-l. [DOI] [PubMed] [Google Scholar]
- 26.Quail P H. The phytochromes: a biochemical mechanism of signaling in sight? Bioessays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 27.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 28.Reyes J C, Florencio F J. Electron transport controls transcription of the glutamine synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1995;27:789–799. doi: 10.1007/BF00020231. [DOI] [PubMed] [Google Scholar]
- 29.Rich P R, Jeal A E, Madgwick S A, Moody A J. Inhibitor effects on redox-linked protonations of the b haems of the mitochondrial bc1 complex. Biochim Biophys Acta. 1990;1018:29–40. doi: 10.1016/0005-2728(90)90106-e. [DOI] [PubMed] [Google Scholar]
- 30.Rippka R, Deruelles J, Waterbury J B, Herman M, Stanier R Y. Genetics assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sharrock R A, Quail P H. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 33.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 34.Smith H. Phytochrome transgenics: functional, ecological and biotechnological applications. Semin Cell Biol. 1994;5:315–325. doi: 10.1006/scel.1994.1038. [DOI] [PubMed] [Google Scholar]
- 35.Trebst A. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- 36.Vioque A. Analysis of the gene encoding the RNA subunits of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992;20:6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh K C, Wu S H, Murphy J T, Lagarias J C. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]