Abstract
Alzheimer’s disease involves many neurobiological alterations from molecular to macroscopic spatial scales, but we currently lack integrative, mechanistic brain models characterizing how factors across different biological scales interact to cause clinical deterioration in a way that is subject-specific or personalized. As important signalling molecules and mediators of many neurobiological interactions, neurotransmitter receptors are promising candidates for identifying molecular mechanisms and drug targets in Alzheimer's disease.
We present a neurotransmitter receptor-enriched multifactorial brain model, which integrates spatial distribution patterns of 15 neurotransmitter receptors from post-mortem autoradiography with multiple in vivo neuroimaging modalities (tau, amyloid-β and glucose PET, and structural, functional and arterial spin labelling MRI) in a personalized, generative, whole-brain formulation.
In a heterogeneous aged population (n = 423, ADNI data), models with personalized receptor-neuroimaging interactions showed a significant improvement over neuroimaging-only models, explaining about 70% (±20%) of the variance in longitudinal changes to the six neuroimaging modalities. In Alzheimer's disease patients (n = 25, ADNI data), receptor-imaging interactions explained up to 39.7% (P < 0.003, family-wise error-rate-corrected) of inter-individual variability in cognitive deterioration, via an axis primarily affecting executive function. Notably, based on their contribution to the clinical severity in Alzheimer’s disease, we found significant functional alterations to glutamatergic interactions affecting tau accumulation and neural activity dysfunction and GABAergic interactions concurrently affecting neural activity dysfunction, amyloid and tau distributions, as well as significant cholinergic receptor effects on tau accumulation. Overall, GABAergic alterations had the largest effect on cognitive impairment (particularly executive function) in our Alzheimer’s disease cohort (n = 25). Furthermore, we demonstrate the clinical applicability of this approach by characterizing subjects based on individualized ‘fingerprints’ of receptor alterations.
This study introduces the first robust, data-driven framework for integrating several neurotransmitter receptors, multimodal neuroimaging and clinical data in a flexible and interpretable brain model. It enables further understanding of the mechanistic neuropathological basis of neurodegenerative progression and heterogeneity, and constitutes a promising step towards implementing personalized, neurotransmitter-based treatments.
Keywords: neurotransmitter receptors, multimodal neuroimaging, Alzheimer’s disease, whole-brain computational model, personalized medicine
Many neurobiological processes altered in Alzheimer's disease are regulated by neurotransmitter receptors. Khan et al. use a personalized, multimodal computational model of the brain to identify the role of receptor-mediated pathways in mediating interactions between processes affecting cognitive deterioration.
Introduction
Alzheimer’s disease involves degenerative changes to several neurobiological processes spanning molecular to macroscopic scales, including proteinopathies, modified gene expression, synaptic alterations, vascular dysregulation, hypometabolism and structural atrophy.1 In Alzheimer’s disease, these processes begin decades before the manifestation of cognitive deterioration,2 with vast inter-patient heterogeneity in age of disease onset, spatial distribution of neuropathologies, progression patterns and clinical presentation.3 Currently, there are no effective disease-modifying treatments for Alzheimer’s disease, despite many expensive attempts.2,3 These failures may be attributed to: (i) the use of a generalized medicine approach to treatment without considering the pathophysiological and clinical heterogeneity of the disease4-6; (ii) the focus on single disease factors (e.g. tau and amyloid) whereas most biological mechanisms in Alzheimer’s disease are multifactorial7; and, importantly, (iii) an incomplete multi-scale understanding of how molecular and macroscopic factors interact to cause disease progression.8
Recently, multimodal neuroimaging models9,10 have unravelled the temporal ordering of macroscopic structural, functional, vascular and proteinopathy changes in Alzheimer’s disease. Furthermore, personalized models of longitudinal neuroimaging data have been used to identify subject-specific alterations of neurobiological processes including tau and amyloid accumulation, blood flow and neural activity at rest.11 Nevertheless, such neuroimaging models lack a mechanistic basis in molecular and cellular processes. While these modalities may involve molecular imaging, such as amyloid or tau PET, their spatial resolution is limited in practice.12 Identifying important pathways between truly microscopic-scale variables and observable macroscopic neuroimaging (i.e. molecular PET and MRI) in Alzheimer’s disease would both advance the understanding of the underlying biology and improve the selection of therapeutic targets tailored to an individual’s particular disease subtype or presentation.
One particularly relevant class of molecules is neurotransmitter receptors, which regulate a variety of biological processes known to be dysfunctional in neurodegeneration. As neurotransmitter receptors are mediators of many relevant neurobiological factors, studying them is critical for a complete mechanistic understanding and the potential treatment of abnormal brain conditions such as neurodegeneration.1 For example, dopamine receptors expressed by the cerebral microvasculature and glial cells appear to modulate the coupling between neural activity and vascular response,13 which is altered in Alzheimer’s disease.14 As an organ, the brain consumes energy disproportionately to its mass.15 A significant fraction of this energy expenditure is attributed to synaptic signalling and molecular synthesis, with ∼37% of this associated with postsynaptic receptors and housekeeping processes.16 The production and degradation of neurotransmitter receptors is a complex, dynamic process that is regulated in response to changes in many variables, such as receptor activation, gene expression and external stimuli.17 Since these processes are energy-intensive, changes to their concentrations are likely to indicate relevant biological alterations, making them a potential therapeutic target. Although it is not primarily considered a neurotransmitter disease, Alzheimer’s disease is associated with dysfunction in several important neurotransmitter receptor systems. Particularly, acetylcholine and glutamate receptors are implicated in essential stages of a pathological neurodegenerative cascade, including cholinergic hydrolysis and glutamatergic excitotoxicity.1 Neurotransmitter receptor alterations are also suspected of being a mechanistic pathway in healthy ageing.18 Thus, integrating neurotransmitter receptors with macroscopic neuroimaging data has the potential to uncover molecular pathways important to ageing and disease progression. However, in vivo neurotransmitter receptor imaging is difficult, due to the lack of specific in vivo radiolabels.19 Typically, receptor mapping has involved either post-mortem histology, or expensive PET imaging for a limited set of molecules with available radionuclides. As such, large longitudinal in vivo datasets for several receptors would be extremely expensive or technologically infeasible to collect. Consequently, alterations to neurotransmitter systems during disease progression are not well characterized.20
Motivated by these concerns, we propose a whole-brain generative formulation integrating high resolution in vitro neurotransmitter receptor density maps and in vivo multimodal neuroimaging. For the first time, this model allows a quantitative comparison of the causal role of different neurotransmitter receptors and neuroimaging modalities in healthy ageing and neurodegeneration. Specifically, we fit subject-specific generative models of neuroimaging data in an ageing population covering the Alzheimer’s disease spectrum (n = 423, ADNI data), augmented with 15 whole-brain neurotransmitter receptor distribution patterns. We then treat the parameters of these personalized models as subject-specific measures representing latent receptor-neuroimaging interactions, and identify multi-scale interactions that explain mechanistic variability and cognitive heterogeneity between patients with Alzheimer’s disease. We find that receptor density maps and their interactions with neuroimaging significantly improve the fit of neuroimaging models, providing a valid proxy for true, longitudinal in vivo receptor imaging. Examining model parameters in Alzheimer’s disease patients, we found an axis of variability between receptor–imaging interactions and cognitive decline, primarily affecting executive function. Specifically, this axis is influenced by predictors of tau distribution and resting-state neural activity, concordant with recent reports in late-onset Alzheimer’s disease.21,22 Via this axis, mechanisms of glutamatergic, cholinergic and GABAergic receptor interactions correlated significantly with cognitive decline in Alzheimer’s disease. In contrast, while receptor–imaging interactions in healthy individuals did not vary significantly with cognitive status, mechanisms affecting cerebral blood flow (CBF) changes and grey matter atrophy accounted for most of the inter-individual heterogeneity. This work represents the earliest attempt to integrate several neurotransmitter receptors and multimodal neuroimaging data in a universal formulation, representing a notable advance towards implementing individually tailored neurotransmitter-based diagnosis and treatment in neurodegeneration.
Materials and methods
Ethics statement
The study was conducted according to Good Clinical Practice guidelines, the Declaration of Helsinki, US 21CFR Part 50–Protection of Human Subjects and Part 56–Institutional Review Boards, and pursuant to state and federal HIPAA regulations (http://adni.loni.usc.edu). Study subjects and/or authorized representatives gave written informed consent at the time of enrolment for sample collection and completed questionnaires approved by each participating site Institutional Review Board. The authors obtained approval from the ADNI Data Sharing and Publications Committee for data use and publication, see documents http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Data_Use_Agreement.pdf and http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Manuscript_Citations.pdf, respectively.
Data description and processing
Study participants
This study used longitudinal data from n = 423 participants [149 healthy, 151 early mild cognitive impairment (MCI), 103 late MCI and 20 Alzheimer’s disease-diagnosed subjects at baseline] from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu). Demographic information is summarized in Supplementary Table 1. At least three different imaging modalities were acquired for each included subject (i.e. structural MRI, fluorodeoxyglucose PET, resting functional MRI, arterial spin labelling and/or amyloid-β PET). The ADNI was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early Alzheimer’s disease.
Structural MRI acquisition/processing
Brain structural T1-weighted 3D images were acquired for all n = 423 subjects. For a detailed description of acquisition details, see http://adni.loni.usc.edu/methods/documents/mri-protocols/. All images underwent non-uniformity correction using the N3 algorithm.23 Next, they were segmented into grey matter, white matter and CSF probabilistic maps, using SPM12 (http://fil.ion.ucl.ac.uk/spm). Grey matter segmentations were standardized to MNI space24 using the DARTEL tool.25 Each map was modulated to preserve the total amount of signal/tissue. Mean grey matter density and determinant of the Jacobian (DJ)25 values were calculated for the regions described in the section ‘Receptor densities and brain parcellation’. For each region, obtained grey matter density and DJ values were statistically controlled for differences in acquisition protocols. Both measurements provided equivalent modelling results. All the results/figures presented in this study correspond to the DJ, which constitutes a robust local measure of structural atrophy.
Fluorodeoxyglucose PET acquisition/processing
A 185-MBq (5 ± 0.5 mCi) injection of 18F-FDG was administered to each participant (n = 418) and brain PET imaging data were acquired ∼20 min post-injection. All images were corrected using measured attenuation. Also, images were preprocessed according to four main steps26: (i) dynamic coregistration (separate frames were coregistered to one another lessening the effects of patient motion); (ii) across time averaging; (iii) resampling and reorientation from native space to a standard voxel image grid space (‘AC-PC’ space); (iv) spatial filtering to produce images of a uniform isotropic resolution of 8 mm full-width at half-maximum (FWHM); and (v) affine registration to the participant’s structural T1 image. Next, using the registration parameters obtained for the structural T1 image with nearest acquisition date, all FDG-PET images were spatially normalized to the MNI space.24 Regional standardized uptake value ratio (SUVR) values for the regions considered were calculated using the cerebellum as thereference region.
Resting functional MRI acquisition/processing
Resting-state functional images were obtained using an echo-planar imaging sequence on a 3.0-T Philips MRI scanner for n = 127 subjects. Acquisition parameters were: 140 time points, repetition time (TR) = 3000 ms, echo time (TE) = 30 ms, flip angle = 80°, number of slices = 48, slice thickness = 3.3 mm, in-plane resolution = 3 mm and in-plane matrix = 64 × 64. Preprocessing steps included: (i) motion correction; (ii) slice timing correction; (iii) alignment to the structural T1 image; (iv) spatial normalization to MNI space using the registration parameters obtained for the structural T1 image with the nearest acquisition date; and (v) signal filtering to keep only low-frequency fluctuations (0.01–0.08 Hz).27 For each brain region, our model requires a local (i.e. intra-regional, non-network) measure of functional activity, to maintain mechanistic interpretability and to prevent data leakage of network information into local model terms (described further in the section ‘Receptor-enriched multifactorial causal model’). Due to its high correlation with glucose metabolism28 and validation as an Alzheimer’s disease-sensitive metric,29,30 we calculated regional fractional amplitude of low-frequency fluctuation (fALFF)31 as a measure of functional integrity.
Furthermore, while our model uses structural connectivity as the network along which inter-region propagation occurs, we also calculated and used a functional connectome, as the average of the absolute Pearson correlation matrices across all healthy subjects with functional MRI data (n = 42). Based on this, we compared model performance using structural and functional connectivity, characterizing the choice of connectivity metrics (see ‘Multi-scale interactions involving neurotransmitter receptors are important to explaining multifactorial brain reorganization’ section and Supplementary Fig. 8).
Arterial spin labelling acquisition/processing
Resting arterial spin labelling (ASL) data were acquired using the Siemens product PICORE sequence for n = 195 subjects. Acquisition parameters were: TR/TE = 3400/12 ms, TI1/TI2 = 700/1900 ms, field of view = 256 mm, 24 sequential 4-mm thick slices with a 25% gap between the adjacent slices, partial Fourier factor = 6/8, bandwidth = 2368 Hz/pixel and imaging matrix = 64 × 64. For preprocessing details see ‘UCSF ASL Perfusion Processing Methods’ in http://adni.loni.usc.edu. In summary, main preprocessing steps included: (i) motion correction; (ii) perfusion-weighted images computation; (iii) intensity scaling; (iv) CBF images calculation; (v) alignment to the structural T1 image; (vi) spatial normalization to MNI space24 using the registration parameters obtained for the structural T1 image with the nearest acquisition date; and (vii) mean CBF calculation for each considered brain region.
Amyloid-β PET acquisition/processing
A 370-MBq (10 mCi ± 10%) bolus injection of AV-45 was administered to each participant (n = 422), and 20 min continuous brain PET imaging scans were acquired ∼50 min post-injection. The images were reconstructed immediately after the 20-min scan, and when motion artefact was detected another 20-min continuous scan was acquired. For each individual PET acquisition, images were initially preprocessed according to four main steps26: (i) dynamic coregistration (separate frames were coregistered to one another lessening the effects of patient motion); (ii) across time averaging; (iii) resampling and reorientation from native space to a standard voxel image grid space (‘AC-PC’ space); (iv) spatial filtering to produce images of a uniform isotropic resolution of 8 mm FWHM; and (v) affine registration to the participant’s structural T1 image. Next, using the registration parameters obtained for the structural T1 image with the nearest acquisition date, all amyloid images were spatially normalized to the MNI space.24 Considering the cerebellum as an amyloid-β non-specific binding reference, SUVR values for the regions were calculated.
Tau PET acquisition/processing
A 370-MBq/kg bolus injection of tau specific ligand 18F-AV-1451 ([F-18] T807) was administered to each participant (n = 238), and 30 min (6 × 5 min frames) brain PET imaging scans were acquired starting at 75 min post-injection (n = 200). Images were preprocessed according to four main steps26: (i) dynamic coregistration (separate frames were coregistered to one another lessening the effects of patient motion); (ii) across time averaging; (iii) resampling and reorientation from native space to a standard voxel image grid space (‘AC-PC’ space); (iv) spatial filtering to produce images of a uniform isotropic resolution of 8 mm FWHM; and (v) affine registration to the participant’s structural T1 image. Next, using the registration parameters obtained for the structural T1 image with the nearest acquisition date, all tau images were spatially normalized to the MNI space.24 Considering the cerebellum as a non-specific binding reference, SUVR values for the grey matter regions considered were calculated.
Receptor densities and brain parcellation
In vitro quantitative receptor autoradiography was applied to measure the densities of 15 receptors in 44 cytoarchitectonically defined cortical areas spread throughout the brain.32 These receptors span major neurotransmitter systems, and show significant regional variability across the brain. Brains were obtained through the body donor programme of the University of Düsseldorf. Donors (three male and one female; between 67 and 77 years of age) had no history of neurological or psychiatric diseases, or long-term drug treatments. Causes of death were non-neurological in each case. Each hemisphere was sliced into 3-cm slabs, shock frozen at −40°C and stored at −80°C.
Receptors for the neurotransmitters glutamate (AMPA, NMDA, kainate), GABA (GABAA, GABAA-associated benzodiazepine binding sites, GABAB), acetylcholine (muscarinic M1, M2, M3, nicotinic α4β2), noradrenaline (α1, α2), serotonin (5-HT1A, 5-HT2) and dopamine (D1) were labelled according to previously published binding protocols consisting of pre-incubation, main incubation and rinsing steps.32 The ligands used are summarized in Supplementary Table 3. Receptor densities were quantified by densitometric analysis of the ensuing autoradiographs, and areas were identified by cytoarchitectonic analysis in sections neighbouring those processed for receptor autoradiography, and which had been used for the visualization of cell bodies.33
A brain parcellation was then defined with the aid of the Anatomy Toolbox34 using 44 regions of interest for which receptor densities were available.35 This parcellation was based on areas identified by cortical cytoarchitecture, as well as other cyto- and receptor-architectonically defined regions with receptor measurements (regions are summarized in Supplementary Table 4). These 44 regions were mirrored across left and right hemispheres for a total of 88 brain regions in our parcellation. For each receptor, regional densities were normalized using the mean and standard deviation across all 88 brain regions.
The structural T1 images of the Jülich34 and Brodmann36 brain parcellations were registered to the MNI ICBM152 T1 template using FSL 5.0’s FLIRT affine registration tool,37 and the obtained transformations were used to project the corresponding parcellations to the MNI ICBM152 space (using nearest neighbour interpolation to conserve original parcellation values). In the MNI ICBM152 space, voxels corresponding to the cytoarchitectonically defined regions from Zilles and Palomero-Gallagher35 were identified from the regions in the Anatomy Toolbox, with the remaining Brodmann regions (Supplementary Table 4) filled in using the Brodmann brain atlas. The resulting parcellation of 88 brain regions in the common template space was then used to extract whole-brain multimodal neuroimaging data and estimate the diffusion-based connectivity matrix, as described in the next section.
Anatomical connectivity estimation
The connectivity matrix was constructed using DSI Studio (http://dsi-studio.labsolver.org). A group average template was constructed from a total of 1065 subjects.38 A multishell diffusion scheme was used, and the b-values were 990, 1985 and 2980 s/mm2. The numbers of diffusion sampling directions were 90, 90 and 90, respectively. The in-plane resolution was 1.25 mm. The slice thickness was 1.25 mm. The diffusion data were reconstructed in the MNI space using q-space diffeomorphic reconstruction39 to obtain the spin distribution function.40 A diffusion sampling length ratio of 2.5 was used, and the output resolution was 1 mm. The restricted diffusion was quantified using restricted diffusion imaging.41 A deterministic fibre tracking algorithm42 was used. A seeding region was placed at the whole brain. The QA threshold was 0.159581. The angular threshold was randomly selected from 15° to 90°. The step size was randomly selected from 0.5 to 1.5 voxels. The fibre trajectories were smoothed by averaging the propagation direction with a percentage of the previous direction. The percentage was randomly selected from 0 to 95%. Tracks with length shorter than 30 or longer than 300 mm were discarded. A total of 100 000 tracts were calculated. A custom brain atlas based on cytoarchitectonic regions with neurotransmitter receptor data35 was used as the brain parcellation, as described in the section ‘Receptor densities and brain parcellation’, and the connectivity matrix was calculated by using count of the connecting tracks.
Multimodal neuroimaging data
After preprocessing ADNI neuroimaging data for all six modalities and extracting it for the cytoarchitectonically defined atlas described in the section ‘Receptor densities and brain parcellation’, subjects lacking sufficient longitudinal or multimodal data were discarded. The disqualification criteria were (i) fewer than four imaging modalities with data; or (ii) fewer than three longitudinal samples for all modalities. For the remaining subjects, missing neuroimaging modalities at each time point with actual individual data were imputed using trimmed scores regression with internal principal component analysis (PCA).43 Imputation accuracy was validated using 10-fold cross-validation, showing a strong capacity to recover the real data (correlation values: rCBF = 0.44, ramyloid = 0.60, rneural activity = 0.95, rgrey matter = 0.80, rmetabolism = 0.81, rtau = 0.71; all P < 10−6). Finally, a total of 423 subjects were left with all six neuroimaging modalities with an average of 4.75 (±2.71) time points. We used the mean and variance of each neuroimaging modality across all regions and healthy subjects to calculate z-scores of neuroimaging data across all (healthy, MCI and AD) subjects. See Supplementary Tables 1 and 2 for demographic characteristics, and the ‘Multimodal neuroimaging data’ section and Supplementary Fig. 1 for a detailed flow chart of the selection and analysis of the participants.
Cognitive scores
We used multiple composite scores derived from the ADNI neuropsychological battery. Protocols for deriving each score are described in the respective ADNI protocols documentation or relevant publication for executive function,44 memory,44 language,45 visuospatial functioning,45 Mini-Mental State Examination (MMSE)46 and the Alzheimer’s Disease Assessment Scale (ADAS11/13) 46. With an average of 7.27 ± (2.55) evaluations per subject in our cohort (n = 423), we calculated cognitive decline as the linear best fit rate of change of each cognitive score with respect to examination date. Thus, for each patient, cognitive decline was represented by a set of seven rates of change.
Receptor-enriched multifactorial causal model
Under the framework of the multifactorial causal model (MCM) introduced in Iturria-Medina et al.11 we consider the brain as a dynamical system of anatomically connected regions defined by interacting, neuroimaging-derived biological factors. These biological factors are tissue structure, neuronal activity, blood flow, metabolism and the accumulation of misfolded proteins (amyloid, tau), quantified by structural MRI, functional MRI, ASL MRI, FDG-PET, amyloid PET and tau PET, respectively. Each biological factor at a particular brain region is represented by a single variable , whose rate of change is a function of (i) local states of other factors; and (ii) the propagation of the same factor across anatomically connected regions. Thus, in our model, pathological factors can propagate throughout the brain, but any direct interactions between factors must occur locally within a region.
In this study, for a given subject, and at each of the nROI = 88 brain regions, the system is defined by nfac = 6 state variables or factors. Each factor Sm,i represents the mth neuroimaging modality at the brain region. Factor dynamics can be decomposed into local effects due to factor–factor interactions and network propagation of the factor. In general, the differential equation describing this coupled system for a given subject is:
| (1) |
where f and g and are functions that determine the effects of local multimodal interactions and propagation, respectively, and is the net connectivity of region . Here, we extend the basic MCM formulation [Equation (1)] to include the local effects of neurotransmitter receptors. With being a matrix of spatial maps, composed of local densities of a receptor at a region , and being a vector of all receptor densities in region , we define the general form of the receptor-enriched MCM (re-MCM) as:
| (2) |
The first term represents the local component, which is the interaction between the factor and all other factors in region , mediated by the local densities of receptors in that region. The second term represents the contribution due to network propagation of the factor , mediated by the net anatomical connectivity of the region . The functions and in Equation (2) define the global imaging factor dynamics, which are valid for all brain regions. Thus, regional differences are due to different imaging factor states, receptor distributions and anatomical connectivity, but the mechanisms of their interactions, represented by and , are consistent across the whole brain.
Given the decades-long temporal scale of neurodegeneration compared to the relatively short few months between neuroimaging samples, we assume a locally linear, time-invariant dynamical system:
| (3) |
where is the directed anatomical connectivity from region to , and was defined by the local rate of change of neuroimaging data for successive longitudinal samples at times and :
| (4) |
In this work, we expand the local effect term to include (i) direct factor–factor effects; (ii) interaction terms mediated by nrec = 15 receptor types; and (iii) direct receptor effects [Equation (3)] on the neuroimaging factor rate of change . The local factor effects term in Equation (3) is now expanded:
| (5) |
Although the receptor maps are constant templates with spatial but no temporal variation, their interaction terms add a dynamic element, as they imply a regional heterogeneity to neuroimaging predictors that is not directly explained by the direct receptor term in Equation (3). For instance, we might notice that (hypothetically) the interaction between a glutamatergic receptor and functional activity is a significant predictor of grey matter atrophy. Whether or not functional activity or the glutamatergic receptor map are significant predictors on their own, the significance of the interaction term would imply that the spatial distribution template of the glutamatergic receptor is informative when combined with functional activity.
Additionally, for propagation, we consider only symmetric connectivity between regions and , using a template connectivity matrix for all subjects, as described in the ‘Anatomical connectivity estimation’ section, to give the propagation term
| (6) |
This reduces the net propagation of a factor to a region to a single propagation term. A more complete treatment may consider vascular connectivity as well,4,11 as this measure may be more relevant for different processes (such as functional activity, CBF and metabolism, respectively).
| (7) |
Formulated in this way, each model contains a set of parameters for subject and factor (or 678 total parameters per subject). Apart from the propagation term, which is specific to the imaging modality output of the model, all predictors are identical for the six neuroimaging modalities. That is, a common set of receptor maps, multimodal neuroimaging states and pseudo-personalized receptor–imaging interactions are used as predictors. However, based on their respective effects on each output modality, we obtain 678 distinct biological parameters per subject, each with a distinct mechanistic interpretation (e.g. the effect of neural activity on metabolism or the effect of neural activity on CBF). We then perform linear regression, using the terms in Equation (7) as predictors with longitudinal ADNI neuroimaging samples and receptor maps , to estimate subject- and modality-specific parameters for each subject and modality . Separate regression models were built for (i) each of the n = 423 qualifying subjects; and (ii) each of the six neuroimaging factors. These subjects were drawn from the ADNI dataset with at least four recorded neuroimaging modalities, and at least three longitudinal samples for at least one modality.
To evaluate model fit, we calculate the coefficient of determination ( for each subject. With the data vector with elements , and model predictions with , the coefficient of determination is
| (8) |
where is the mean of neuroimaging data for a particular modality across all brain regions and longitudinal samples.
Statistical analysis
Model fit
Personalized model fit quantified by the coefficient of determination (R2) was evaluated for each subject and neuroimaging modality. F-tests were used to compare receptor-neuroimaging (113 parameters per modality) and neuroimaging-only (eight parameters per modality) to fitting neuroimaging data in each subject (F-test with P < 0.05). The model fit (R2) was evaluated for each subjects’ neuroimaging models using 1000 iterations of randomly permuted receptor maps (with receptor densities shuffled across regions independently for each receptor type), and we calculated the P-value of the true receptor data model R2 compared to this distribution.
Biological parameters and relationship with cognition
We aimed to further clarify how the cognitive decline observed in Alzheimer’s disease progression is modulated by specific neurotransmitter receptor systems and their causal interactions with macroscopic biological factors (i.e. amyloid, tau, CBF, neural activity, glucose metabolism and grey matter density). As changes in several receptor densities are difficult to image in vivo, we analysed the receptor terms from our personalized re-MCM approach as a proxy for the importance of each particular receptor’s distribution or interactions in predicting multi-domain cognitive deterioration in Alzheimer’s disease. To consider the inter-subject variability in the diseased population, we used a combination of cognitive assessment scores as disease severity descriptors (i.e. executive function, memory, language, visuospatial functioning, MMSE, ADAS11 and ADAS13; see ‘Cognitive scores’).
We aimed to robustly identify significant and relevant re-MCM parameters that represent molecular-neuroimaging interactions associated with cognitive decline, using a data-driven multivariate cross-correlation analysis in combination with a randomized permutation test to ensure the statistical stability of our results. By concurrently analysing the multivariate changes across all re-MCM parameters, this multidimensional analysis searched for large clusters of functionally related receptor-neuroimaging interaction mechanisms statistically associated with Alzheimer’s disease-associated cognitive changes. In other words, the singular value decomposition (SVD) method used here (and its associated permutation test) identified the specific set of receptors and/or imaging features that were maximally related to cognitive decline. To this end, we selected a clinical subgroup of interest (either n = 112 cognitively healthy subjects or n = 25 Alzheimer’s disease patients from the n = 423 total subjects with sufficient multimodal neuroimaging data), and performed the following procedure on the original set of 678 re-MCM parameters and seven rates of cognitive decline per subject (executive function, memory, language, visuospatial functioning, MMSE and ADAS11/13):
To identify correlated axes of variation, we performed PCA on all 678 biological parameters separately on the healthy and Alzheimer’s disease participants, and ranked parameters based on the variance explained in the first principal component (PC).
- To relate biological parameters to cognition, we performed SVD on the cross-covariance matrix between significant parameters and rates of cognitive decline for Alzheimer’s disease patients, after adjusting for covariates (baseline age, education and gender). SVD allows us to simultaneously reduce the dimensionality of the seven cognitive assessments and to rank parameters by their variation with cognition. Where is a matrix of z-scores of each re-MCM parameter for this clinical subgroup and is a matrix of the corresponding z-scores of the rates of clinical decline, the cross-covariance matrix is decomposed to:
where and are orthonormal matrices of spatial loadings for the coefficients and cognitive scores, respectively, and is a (diagonal) matrix of singular values .(9) To evaluate the significance of SVD components, we performed permutation tests by shuffling the mapping between subjects’ re-MCM parameters and cognitive scores, and repeating SVD. To compare permuted iterations, we performed a Procrustes transformation to align the axes of singular components. We kept only those singular components that are significant P < 0.05) compared to 1000 permutation iterations of SVD components.
We performed 1000 iterations of bootstrapping on the parameters , and discarded the parameters with non-significant 95% confidence intervals.
- For the remaining significant re-MCM parameters and SVD components, we computed the variance explained per parameter. We then summed the contribution of each significant parameter to each significant SVD component , weighted by the fraction of total variance explained by the ith component
(10)
Inter-subject mechanistic variability
To explore the potential clinical utility of our approach at the personalized level, we performed a quantitative comparison between diseased participants in terms of their inter-subject variability across different receptor systems. To this end, we defined individual-specific ‘fingerprints’ of the alterations in receptor-modulated synergistic interactions. Specifically, for each participant i and receptor system r, we calculated the Mahalanobis distance of re-MCM parameters associated with cognitive decline in our Alzheimer’s disease cohort. This distance is calculated between subject’s parameters , and the distribution of healthy subjects’ parameters for receptor r, with means and a covariate matrix
| (11) |
To quantify the relationship between this summary metric of receptor alterations and specific cognitive domains, we performed multivariate linear regression on rates of cognitive decline (adjusted by age, gender, education level and APOE4 status; n = 25) using the z-scores of the Mahalanobis distances for the six receptor systems. We also estimated the explanatory importance of each receptor system, as the percentage improvement in model fit (R2) by including a particular receptor Mahalanobis distance.
Data and code availability
The three datasets used in this study are available from the ADNI database (neuroimaging and cognitive evaluations; http://www.adni.loni.usc.edu), the HCP database (tractography template for connectivity estimation; http://www.humanconnectomeproject.org/) and receptor density data published in Zilles and Palomero-Gallagher.35 We anticipate that the re-MCM method will be released soon as part of our available and open-access, user-friendly software (https://www.neuropm-lab.com/neuropm-box.html).47
Results
Capturing receptor-mediated multifactorial brain reorganization
Here, we aimed to develop a multi-scale generative brain model linking regional receptor densities (for 15 neurotransmitter receptors) and multimodal neuroimaging-based factors (for six biological variables) in a flexible, unified formulation. We aimed to use this mathematical framework to infer receptor alterations associated with the long-term physiological changes of complex brain reorganization processes (namely ageing and neurodegeneration) and their cognitive impact. Because changes in receptor concentrations are difficult to measure in vivo, our receptor density maps were composed of group-averaged templates, with spatial distributions of receptors but no inter-individual variability or intra-individual longitudinal progression. Consequently, we use the predictive importance of receptor distributions in generative models of abnormal neuroimaging-derived biological variables as a proxy for alterations in either receptor density or mechanistic interactions with other imaging-derived variables.
We proceeded to characterize the multifactorial brain dynamics of each participant using the developed neurotransmitter re-MCM (Fig. 1) and the quality-controlled, multimodal longitudinal neuroimaging data (described in the ‘Data description and processing’ section). For each participant with sufficient longitudinal and multimodal data (n = 423), the re-MCM was fit for all six neuroimaging modalities, to obtain receptor-imaging biological parameters reflecting local factor–factor interactions mediated by neurotransmitter receptor distributions (e.g. amyloid-tau interactions modulated by NMDA receptors) and the spreading of effects via anatomical networks (e.g. amyloid and tau propagation along white matter connections).
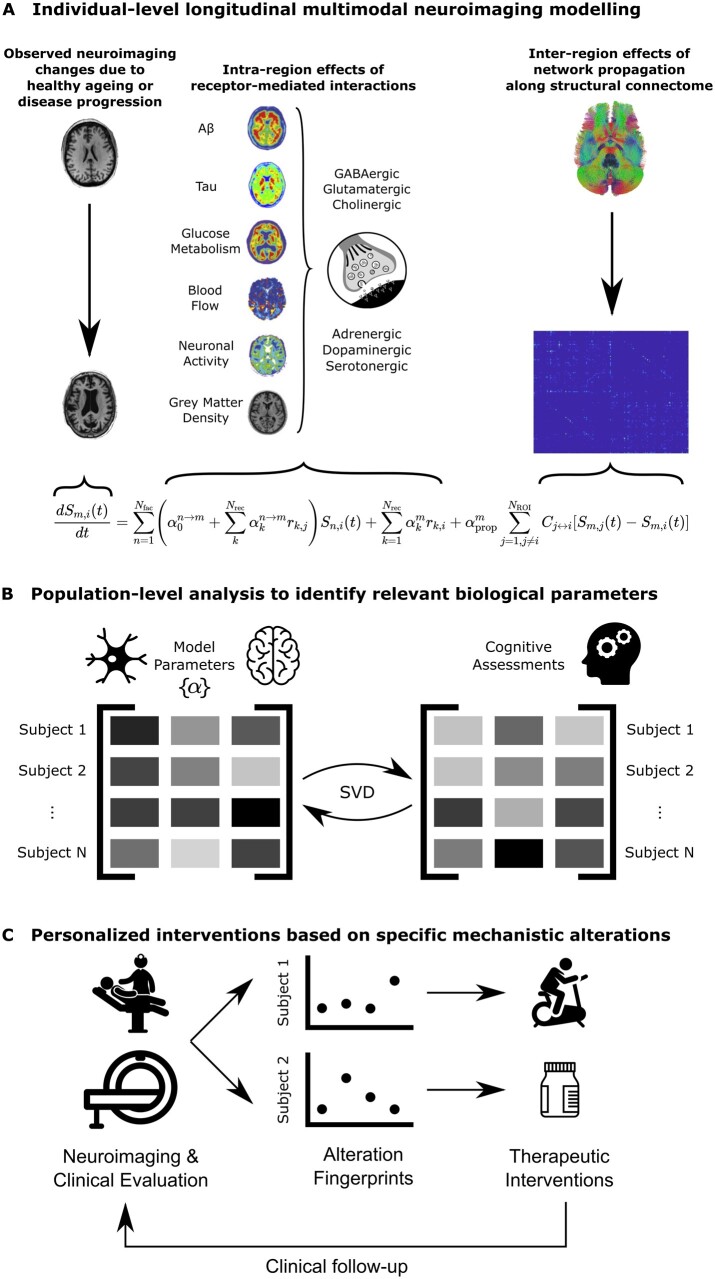
Figure 1.
Neurotransmitter receptor-enriched multifactorial causal modelling. (A) For each subject with longitudinal neuroimaging data, changes between subsequent samples in each neuroimaging modality are decomposed into local synergistic effects due to (i) the direct influence of all neuroimaging-quantified biological factors; (ii) receptor density distributions; and (iii) multi-scale receptor–imaging interactions; and (iv) global network-mediated intra-brain propagation. Combining this data across (nROI = 88) brain regions and multiple neuroimaging samples results in a multivariate regression problem to identify the subject-specific parameters {α}. (B) At a group level, these personalized model parameters are then compared to subjects’ cognitive assessments (specifically, the rates of decline for seven composite cognitive scores described in the ‘Cognitive scores’ section) using a singular value decomposition (SVD) procedure on the cross-covariance matrix, to identify multi-scale receptor-neuroimaging interactions that are robustly correlated with the severity of cognitive symptoms in Alzheimer’s disease (outlined in the ‘Biological parameters and relationship with cognition’ section). (C) In the context of personalized applications, inter-subject variability in receptor–imaging interactions can be used as clinical ‘fingerprints’ of molecular alterations representing different disease mechanisms. Patients can then receive individually tailored treatment plans to address their underlying aetiology, based on their specific fingerprints. For example, patients with greater vascular alterations may benefit more from lifestyle interventions such as physical exercise, whereas patients with greater receptor alterations may require neurotransmitter-based medication (depending on the most affected receptor). Furthermore, treatment plans can be continually adjusted with follow-up visits.
Multi-scale interactions involving neurotransmitter receptors are important to explaining multifactorial brain reorganization
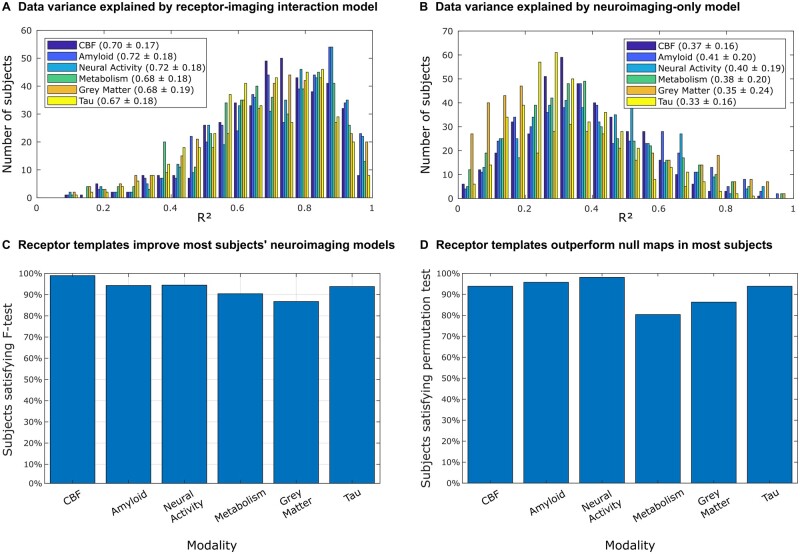
Firstly, we proceeded to evaluate the ability of the re-MCM approach to fit longitudinal neuroimaging data with and without receptor maps and multi-scale receptor–imaging interactions (Fig. 2A and B). For each of the six neuroimaging modalities per subject, we calculated the coefficient of determination (R2) as a measure of model accuracy for explaining the real imaging-specific longitudinal changes. While model accuracy varied by imaging modality, we observed that the personalized models including receptor-neuroimaging interactions explained ∼70% (±20%) of observed variance in all modalities (Fig. 2A).
Figure 2.
Receptor density templates and multi-scale receptor-neuroimaging interactions significantly improve individual longitudinal neuroimaging models. The improvement in neuroimaging modelling was evaluated in terms of (i) including direct receptor terms and receptor-neuroimaging interactions in the model; and (ii) using true receptor density maps compared to randomized, spatially permuted maps. The histograms in A and B show the distribution of the coefficient of determination (R2) of n = 423 individual models of neuroimaging changes including (A) and excluding (B) receptor predictors. Subject-specific linear models fit neuroimaging changes reasonably well, with a significant improvement by including receptor terms. This is confirmed by the F-test between subject models with and without receptor densities and receptor–imaging interactions (113 and 8 parameters, respectively). The proportion of subjects for whom the F-statistic is above the critical threshold is shown in (C). This critical threshold corresponds to a statistically significant (P < 0.05) improvement due to the receptor terms in the re-MCM model, accounting for the increase in adjustable model parameters. Furthermore, to validate the benefit of the receptor templates over randomized null maps, re-MCM models were fit with 1000 spatially shuffled receptor maps for each subject. The P-value of the model fit (R2) using true receptor templates compared to the distribution of R2 of models using randomized templates was calculated for each subject. The proportion of subjects for whom the true receptor maps resulted in a statistically significant improvement in model fit (P < 0.05) is shown in D. The results of these two analyses in C and D validate the use of averaged receptor templates in personalized neuroimaging models.
Inter-region propagation in our model occurs along structural connectivity. While functional connectivity can be a better predictor of functional MRI data, structural connectivity is a better measure of the actual physical substrate connecting brain regions. Nevertheless, to explore the effects of alternate connectivity measures, we repeated our modelling steps using functional connectivity in place of the structural connectivity derived from diffusion-MRI tractography. While the connectivity matrices differed, we found almost no change in model fit or parameters across subjects, with a high correlation r > 0.99 of model R2 (P < 0.001) across all modalities (Supplementary Fig. 8). We attribute this to the dominance of intra-regional effects in our model, with many interacting local receptor and neuroimaging predictors, and also to the shared information in structural and functional connectivity.48
Next, to evaluate the relevance of receptor densities and receptor-mediated interactions between biological factors quantified by imaging (e.g. amyloid-tau interaction modulated by GABA), we compared the model fit of full re-MCM models (incorporating receptor–factor interactions as previously described) with restricted models (using only neuroimaging predictors and network propagation). The models including receptor maps and receptor–imaging interactions explained, on average, more than twice as much of the variance in longitudinal neuroimaging changes (Supplementary Table 7; P < 0.001 with a two-sample t-test). To account for the greater explanatory power of a larger model with more parameters, we quantified the improvement in individual neuroimaging modelling due to the receptor terms, we conducted F-tests between the full re-MCM formulation (Fig. 2A) and the restricted model (Fig. 2B). As hypothesized, we observed that the inclusion of receptor maps and multi-scale (receptor–imaging) interaction terms significantly improved (P < 0.05) the model accuracy for 86.8–99.0% of the subjects (Fig. 2C) while accounting for the additional degrees of freedom in the model with receptors. While the inclusion of receptors and receptor–imaging interactions improved model performance for all subjects and modalities, this improvement was not always significant, most notably in 13.2% of grey matter atrophy models (Fig. 2C). We attribute this to the use of a shared, group-averaged set of neurotransmitter receptors templates (further tested next).
Having established that receptor maps and receptor-neuroimaging interactions do significantly improve personalized neuroimaging models, we then performed a permutation analysis on the receptor maps to test the informativeness compared model performance using averaged receptor templates to a set of null receptor maps. For each subject, the model fitting procedure was repeated using 1000 random permutations of the spatial receptor maps. Receptor densities were shuffled across regions of interest, independently for each receptor. We then compared the distribution of model fit (R2) using these randomly permuted data with the R2 obtained for the models using the true receptor templates. We observed that the significance of the improvement in model fitting over randomized receptor maps varied by imaging modality, for example, being lower for metabolism than for neural activity (Fig. 2D). Nevertheless, the true receptor templates perform significantly better in ∼80–98% of all subjects, depending on the modality. The gain in model performance by imaging modality is presented in Supplementary Table 8, and generally fell between 15.6% ± 13.3% (P < 0.0417) for glucose metabolism to 22.3% ± 15.0% (P < 0.003) for neural activity. Notably, the modalities for which true receptor data was the least informative (metabolism and grey matter atrophy), were also the ones for which augmenting the model with receptor data provided the least significant improvements across all subjects. Furthermore, we compared the proportion of subjects with significant improvements over null maps across diagnoses, shown in Supplementary Fig. 7. On average across modalities, 96.2% of healthy subjects’ models were significantly improved, whereas this was progressively lower for MCI subjects (89.4% for early MCI and 89.8% for late MCI) and Alzheimer’s disease patients (78.3%).
We hypothesize that accentuated ageing processes and neurodegeneration may alter receptor densities or interaction mechanisms in each individual, requiring the biological parameters in our personalized models to compensate. Identifying these specific alterations is the subject of the remaining subsections.
Characterizing receptor–imaging interaction variability in healthy ageing and Alzheimer’s disease
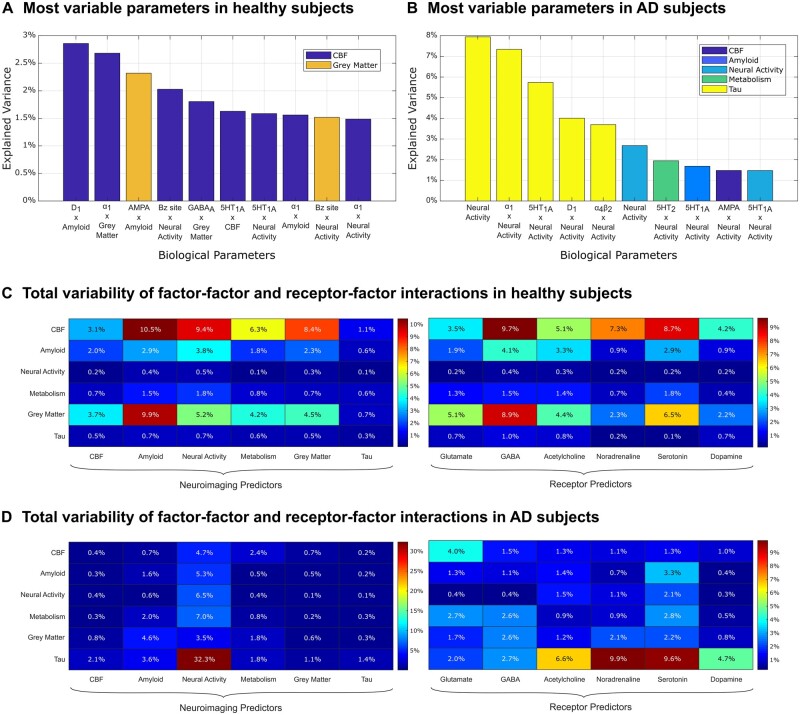
We aimed to characterize the variability in receptor-mediated brain reorganization in the studied healthy ageing (n = 112) and Alzheimer’s disease subpopulations (n = 25). In the healthy population, we performed a PCA on all re-MCM biological parameters (678 in total) across the six neuroimaging modalities, finding that the first principal component (PC1) is able to explain 97.3% of the group’s variance. The most variable parameters contributing to PC1 belonged to CBF and grey matter models (Fig. 3A). That is, if current CBF in a region becomes less important (relative to other re-MCM predictors) to predicting its future change, grey matter density also becomes less important to predicting future atrophy, whereas the current level of amyloid becomes more important to predicting future accumulation. These results indicate that, in the absence of an influential disease process (e.g. neurodegeneration), inter-individual differences in the long-term brain reorganization are mechanistically driven by receptor-mediated processes affecting CBF and grey matter density. Most prominently, these include the CBF effects due to interactions between the dopaminergic D1 receptor and amyloid distribution (2.9%), the adrenergic α1 receptor and grey matter density (2.7%), the GABAA benzodiazepine site and neural activity (GABAA/BZ; 2.0%), and the GABAA receptor and grey matter density (1.8%). Additionally, the interaction between the glutamatergic AMPA receptor and amyloid distribution as a predictor of grey matter atrophy (2.3%) are also notably variable.
Figure 3.
Variability of biological parameters across healthy and Alzheimer’s disease participants. (A and B) PCA-based sources of variability in the 678 re-MCM parameters across healthy subjects (n = 112) and Alzheimer’s disease patients (n = 25), respectively. The first principal component (PC1) captured 97.3% of the variance across parameters in healthy subjects, and 26.2% in Alzheimer’s disease patients. The top 10 biological parameters and their contributions to PC1 are plotted (with their target neuroimaging models in the legend), highlighting the receptor–imaging interactions that characterize the main axis of variability in each clinical subgroup. In healthy subjects, a multifactorial combination of receptor–imaging interactions affecting atrophy and CBF changes were the most variable parameters along PC1. Notably, for Alzheimer’s disease patients, the top parameters were direct or receptor-mediated effects of neural activity on various (but especially tau) imaging models. (C and D) To evaluate the relative importance of receptor– and factor–factor interactions, we then aggregated the importance of all direct or interaction terms involving a given predictor class (factor or receptor type) along PC1, for (C) healthy subjects and (D) for Alzheimer’s disease patients, respectively. Note that the percentage variation across all parameters is shown. As such, there is an overlap in terms between the two heat maps (receptor–factor interaction terms contribute to both) and they should be interpreted separately.
In the Alzheimer’s disease group (n = 25), with the presence of a neurodegenerative condition, the first PC of the re-MCM biological parameters only explained 26.2% of the population variability (with subsequent PCs explaining <10% each). Along this main axis of variability, inter-individual differences are primarily due the effects of neural activity as a direct or receptor-mediated predictor of tau accumulation (Fig. 3B; 7.9% of PC1 via the direct term, 7.3% via adrenergic α1 receptors, 5.7% via serotonergic 5HT1A receptors, 4.0% via dopaminergic D1 receptors and 3.7% via cholinergic α4β2 receptors). The next subsection covers a deeper analysis of the Alzheimer’s disease group.
Interestingly, in the healthy subpopulation, when the individually small contributions of all receptor terms for each target neuroimaging modality were summed (Fig. 3C), we observed that the receptor mechanisms that affect CBF changes, grey matter atrophy and amyloid accumulation were the most variable, with GABAergic and serotonergic mechanisms dominating. For example, combined variability due to GABAergic (9.7% of PC1), serotonergic (8.7% of PC1) and adrenergic (primarily α1 receptors; 7.3% of PC1) interactions predicting CBF changes accounted for approximately a quarter of variability across all six neuroimaging modalities and 678 total parameters (25.7% of PC1). As seen in Fig. 3B, the main sources of biological parameter variability in Alzheimer’s disease (Fig. 3D) involved neural activity predictors of tau accumulation. Predictors of tau accumulation involving adrenergic (9.9% of PC1), serotonergic (9.6%), cholinergic (6.6%) and dopaminergic (4.7%) interactions were the most variable.
Receptor-imaging alterations underlying cognitive deterioration in Alzheimer’s disease
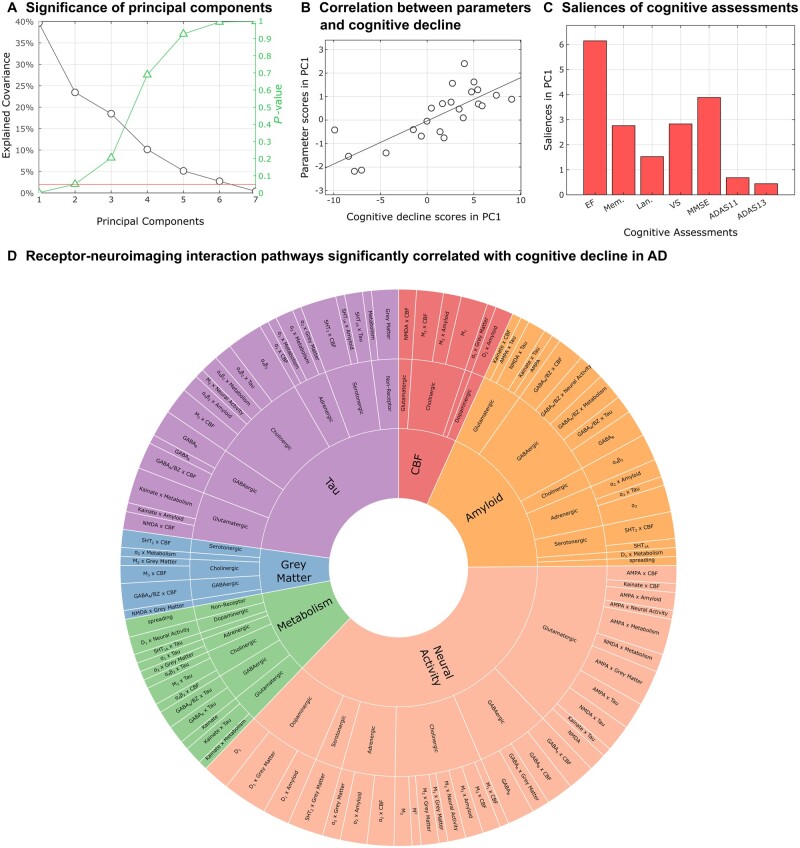
To determine the receptor-neuroimaging alterations underlying multiple cognitive variations in Alzheimer’s disease, we performed a multivariate cross-correlation analysis between the rate of changes of the selected cognitive descriptors and the biological parameters across all Alzheimer’s disease participants (see the section ‘Biological parameters and relationship with cognition’). Notably, we found that just the first component of the identified biological parameters can explain up to 39.7% [P < 0.004, family-wise error-rate (FWE)-corrected] of the inter-individual variability in Alzheimer’s disease cognitive deterioration (Fig. 4A). Furthermore, we identified the specific cognitive domains that are correlated with receptor-neuroimaging alterations (Fig. 4C), with executive dysfunction being the most salient cognitive feature with respect to receptor-neuroimaging parameters. Finally, Fig. 4D presents a detailed pathway of 95 receptor–imaging interactions significantly associated with cognitive decline based on feature bootstrapping, and their associated neuroimaging modalities mediating Alzheimer’s disease-related symptom severity. These results show that a multifactorial set of molecular alterations are relevant to cognitive decline in Alzheimer’s disease. Cumulative effects of different neuroimaging interactions and receptor subtypes from the same family are summarized in Fig. 5, quantified by the total cognitive variance explained by all parameters of the relevant category via the significant SVD component.
Figure 4.
Significant neurotransmitter receptor–imaging interactions underlying Alzheimer’s disease clinical severity. (A) The latent cross-correlation components are ranked by the fraction of cognitive decline variance explained by re-MCM biological parameters (along with the reported P-values based on the permutation analysis; see the ‘Biological parameters and relationship with cognition’ section). In this case, only a single latent component was significant (39.7% variance explained, P < 0.004, FWE-corrected). (B) A notable correlation (r = 0.80; P < 10−8) between the projections of statistically stable re-MCM parameters and rates of cognitive decline in the principal component space was observed, with the removal of an outlier subject more than three median absolute deviations from the median. (C) Saliences of cognitive decline to this first latent component, providing a relative ranking of cognitive domains. These saliences are proportional to the contribution of each term relative to every other term, for example showing that executive dysfunction is most correlated with alterations to receptor–imaging interactions in Alzheimer's disease. (D) Receptor-imaging pathways that are significantly correlated with cognitive decline, arranged by neuroimaging model and receptor type (Supplementary Table 5). The angle of each sector is proportional to the contribution of the corresponding parameter to explaining the variance in the rates of cognitive decline. The inner sectors represent the six neuroimaging modalities that together comprise each personalized re-MCM model. Within each modality, the intermediate sectors represent the neurotransmitter system involved, while the outer sector consists of the specific two-way receptor-neuroimaging interactions or direct predictor terms in the model. Notably, while receptors appear only as predictors in the outer sector, neuroimaging modalities appear both as predictors and as model outputs in the inner sectors. Thus, the relative importance of each neuroimaging modality to explaining cognitive differences is not fully represented by the angle of each inner sector.
Figure 5.
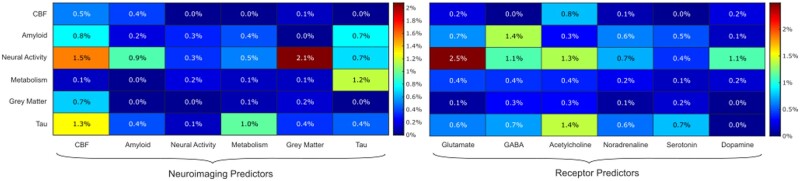
Contributions of mechanistic pathways to the severity of cognitive decline in Alzheimer’s disease. To better visualize the importance of neuroimaging factors and neurotransmitter receptor systems, heatmaps of the cumulative cognitive variance explained by each predictor category in each neuroimaging model are shown. These variances are the percentages of total cognitive variance that are explained by significant biological parameters of each category via the first significant SVD component. As such, the rows of the heat map on the left replicate the inner sector of Fig. 4D, while the columns show the importance of each imaging modality or receptor family as predictors, with CBF and tau predictors explaining the most variance in cognitive decline.
Grey matter density (2.1%) and CBF (1.5%) changes as predictors of neural activity dysfunction, and CBF (1.3%) and glucose metabolism (1.0%) as predictors of tau distribution were the most cognitively significant pathways between imaging modalities, although tau as a predictor of amyloid distribution (0.7%), neural activity dysfunction (0.7%) and glucose metabolism (1.2%) was also significant. Overall, as predictors, biological parameters involving CBF, tau and grey matter density were the most significant in relation to the cognitive severity of Alzheimer’s disease. The neuroimaging models of neural activity dysfunction and tau accumulation were the major sources of cognitively significant biological parameters.
In terms of receptor systems, glutamatergic, GABAergic and cholinergic alterations were significant to cognitive decline, as summarized in Supplementary Table 6. Alterations to glutamatergic predictors of resting-state functional activity (2.5%), GABAergic predictors of amyloid deposition (1.4%) and cholinergic predictors of tau distribution (1.4%) were the dominant receptor effects.
Furthermore, while the second component was borderline non-significant (P < 0.051), it explained 23.4% of the variance between model parameters and cognitive decline (r = 0.89, P < 10−8; Supplementary Fig. 10). In this axis, receptor-imaging parameters predicting neural activity were less prominent, with CBF and metabolism model parameters contributing more. Cognitively, this second component corresponded to non-executive function domains, primarily memory, language and visuospatial function.
As a control case, we performed an equivalent cross-correlation analysis in the healthy population, notably finding the first PC relating re-MCM parameter with rates of cognitive decline in health to be non-significant (Supplementary Fig. 3), although the second PC explaining a small amount of cognitive variance was significant (15.5% variance explained, P < 0.02; Supplementary Fig. 4). Furthermore, we found no significant component in amyloid-negative healthy subjects (P > 0.2 for all components). We attribute this effect to the lack of consistent cognitive decline in the analysed healthy population, in contrast to the large variability observed for Alzheimer’s disease.
To test the sensitivity of our findings to genetic covariates, we repeated our analyses both with and without APOE ε4 allele status and a polygenic hazard score (PHS)49 as covariates in the SVD analysis, in addition to age, gender and education in both cases. To overcome the low number of Alzheimer’s disease participants, we expanded our criteria to include MCI and Alzheimer’s disease participants (n = 177 for APOE status, n = 161 for PHS). Importantly, we confirmed that the previously identified Alzheimer’s disease-related significant latent variables and parameters are robust to the inclusion of APOE status and PHS (Supplementary Fig 5 and 6). Finally, to further restrict our analysis to subjects on the amyloid-mediated Alzheimer’s disease spectrum, we repeated the SVD analysis In amyloid positive subjects with MCI and Alzheimer’s disease (n = 52). As was the case in the initial Alzheimer’s disease group, we found one significant PC (44.3% variance explained, P < 0.003) with a high correlation between model parameters and cognitive decline (mainly executive function; r = 0.76, P < 0.001). The main receptor–imaging interactions along this axis were analogous to those in the Alzheimer’s disease group, namely cholinergic predictors of tau accumulation, although parameters of the neural activity model were less prominent in favour of predictors of metabolism (particularly for adrenergic and cholinergic systems; Supplementary Fig. 9).
Clinically-similar subjects have different underlying receptor alterations
Finally, for each participant and receptor family, we defined a summary metric quantifying how much receptor-based mechanisms differ from clinically healthy subjects (see the ‘Inter-subject mechanistic variability’ section). For example, a given subject’s glutamatergic Mahalanobis distance is a combined measure of the ‘unhealthiness’ of receptor-based interactions and spatial distributions involving NMDA, AMPA and kainate, while accounting for the variation of these mechanisms in healthy subjects.
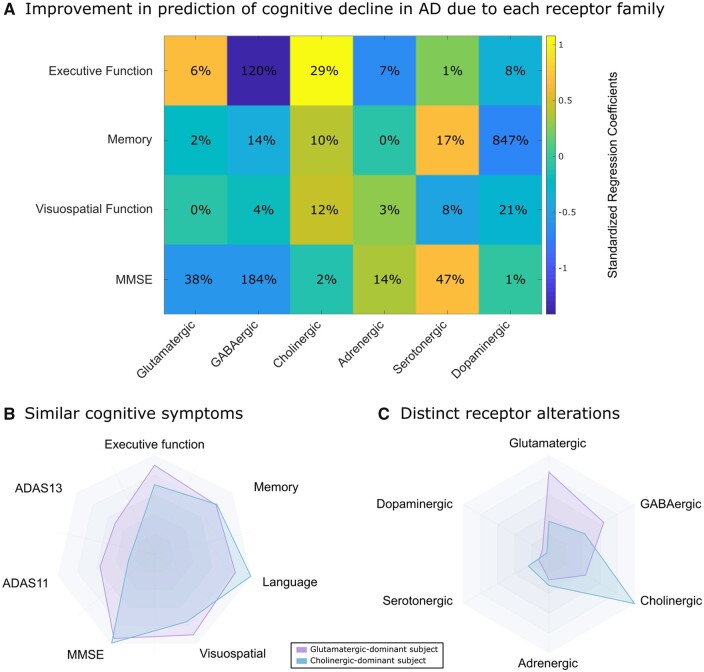
Although a simplified summary metric, the receptor Mahalanobis distances explained a large proportion of cognitive variance in the Alzheimer’s disease population, with 71.4% for executive function (P < 0.0004), 43.3% for memory (P < 0.08), 18.7% for language (P < 0.66), 40.1% for visuospatial function (P < 0.10), 43.8% for MMSE (P < 0.08) and 33.8% for ADAS11 (P < 0.22). Figure 6A shows the effects of each receptor family on cognitive domains, as well as the percentage improvement in explaining cognitive variance due to each receptor family. We note the large negative effects of GABAergic alterations on executive function and the MMSE, and dopaminergic alterations on memory. Interestingly, cholinergic alterations showed a moderate positive effect and explanatory importance towards executive function.
Figure 6.
Receptor alterations underlying inter-individual disease heterogeneity. (A) In Alzheimer’s disease patients (n = 25), we quantified the relative effect sizes of standardized Mahalanobis distances of receptor mechanisms on different cognitive domains. We also standardized the regression coefficients within each cognitive domain before visualizing to facilitate comparison across cognitive domains, and the percentage improvement in model fit (R2) due to each receptor system is also shown. For example, the explanation of inter-subject variability in executive function decline by glutamatergic, cholinergic, adrenergic, serotonergic and dopaminergic Mahalanobis distances is improved by 120% (i.e. more than doubled) by the inclusion of GABAergic Mahalanobis distance as well. (B and C) We show two Alzheimer’s disease participants, with similar symptoms across a variety of cognitive domains. For these participants, we calculated the Mahalanobis distance to the distribution of all healthy subjects (n = 112), along mechanisms involving each receptor family. The subjects show distinct receptor alterations based on their longitudinal neuroimaging changes, despite their shared designation as Alzheimer’s disease patients and similar cognitive profiles.
In Fig. 6B and C we illustrate how two Alzheimer’s disease patients with similar cognitive symptoms present distinct receptor alteration fingerprints, with primarily glutamatergic and cholinergic mechanisms respectively. Importantly, this result indicates that even subjects with identical clinical diagnoses present distinctive underlying spatiotemporal molecular alterations, and supports the use of whole-brain generative models to uncover patient-specific receptor and potential disease mechanisms to target clinically.
Discussion
In this work, we have presented a personalized, whole-brain and generative multimodal neuroimaging model incorporating receptor-neuroimaging interactions using in vivo data. Subsequent analyses on the resulting models have allowed, for the first time, the identification of (i) variability in receptor-neuroimaging interactions in healthy subjects and Alzheimer’s disease patients; and (ii) specific pathways of receptor-neuroimaging interactions that are important to cognitive decline in Alzheimer’s disease patients. This exploratory analysis provides a bridge between molecular-level mechanisms and observable macroscopic neuroimaging biomarkers of healthy ageing and Alzheimer’s disease, revealing which neurotransmitter receptor systems mediate dysfunctional interactions between neurobiological processes such as CBF, amyloid and tau deposition, grey matter atrophy, neural activity and metabolism.
Due to the difficulty of comprehensive, personalized in vivo receptor imaging for a large cohort, receptor maps were not specific to each subject, but instead the averaged templates of four post-mortem brain samples. Post-mortem in vitro autoradiography allowed the imaging of a large number of receptor types, even those without in vivo radioligands. First, our work demonstrates that (i) multi-scale interaction terms involving the spatial distributions of neurotransmitter receptors are highly informative to models of neuroimaging progression; and (ii) even group-averaged receptor map templates can significantly improve the personalized model fit in nearly all subjects when combined with personalized neuroimaging predictors. Specifically, incorporating receptor maps and multi-scale receptor–imaging interactions to personalized models with multimodal neuroimaging predictors improves the average data variance explained from ∼40 to 70% (Fig. 2A and B). This improvement is statistically significant (F-test with P < 0.05) in almost all subjects (Fig. 2C), even after accounting for the additional predictive power of the larger, multi-scale models. Including only receptor maps without receptor–imaging interactions also resulted in a more modest yet significant improvement in most subjects across all imaging modalities (Supplementary Fig. 2). This is a particularly strong result, validating the use of a group-averaged receptor template, given the large improvement and the stringent criterion accounting for additional model parameters.
Additionally, models using the true receptor templates perform significantly better (P < 0.05 of ) than models using randomly permuted, null receptor maps in almost all subjects (Fig 2D; 80.4–98.1%, depending on the modality), although this improvement was less evident with disease progression (Supplementary Fig. 7). These results, along with the consistency of regional receptor densities across the four (aged but healthy) brains used to produce the templates compared to inter-region variability,35 support the applicability of receptor templates to a wider population. Receptor mapping studies across more diverse clinical groups of patients would help validate or augment our modelling approach. Nevertheless, given the difficulty of acquiring a wide variety of in vivo molecular data, due to a limited number of appropriate radioligands, and the high cost of longitudinal molecular imaging, these results on model accuracy are a promising validation for the combination of other molecular templates (such as gene expression atlases) with personalized neuroimaging predictors. These ‘pseudo-personalized’ molecular-imaging predictors can then be incorporated into neuroimaging models and used to infer mechanistic alterations in a group of subjects. If these personalized models are sufficiently accurate, as in this work, the weights of their biological parameters then serve as proxies for individual-specific alterations to receptor-mediated mechanisms.
While interpreting these parameters, it is important to distinguish between the types of biological mechanism they represent, which include (for each neuroimaging model): (i) direct neuroimaging effects; (ii) direct receptor density effects; (iii) receptor–imaging interactions; (iv) network propagation; and (v) offset terms representing an intrinsic rate of change for the neuroimaging modality. We hypothesize that ageing and neurodegeneration alter the spatial distributions of and functional interactions involving neurotransmitter receptors, which would lead to subject-specific model parameters to compensate in the absence of inter-subject variability in receptor data. Thus, model parameters are a proxy for alterations to spatial maps of receptors or their interactions with neurobiological processes (represented by direct model receptor density terms and receptor–imaging interaction terms in the model, respectively). In our parameter analyses in the section ‘Receptor-imaging alterations underlying cognitive deterioration in Alzheimer’s disease’, direct receptor density terms represent alterations to the spatial distribution of a particular receptor. Each interaction biological parameter value can be interpreted as the effect of the corresponding receptor or imaging factor on the brain reorganization process, as measured by neuroimaging changes, given ‘normal’ (i.e. spatial mean) values of all related predictors involving the same receptor or imaging term, respectively. For example, we consider the case where the interaction term between a glutamatergic receptor and amyloid in the CBF model is significantly related to cognitive decline. This implies that, under normal levels of amyloid and the glutamatergic receptor individually, a functional alteration in this mechanism (quantified by the re-MCM parameter weight) is correlated with faster cognitive deterioration.
Biological parameters were evaluated for principal axes of variability in Fig. 3 and the cognitively relevant variability in Fig. 4. The former method was used to identify linear combinations of biological parameters that accounted for inter-individual differences in receptor and/or neuroimaging interaction strengths in healthy participants and Alzheimer’s disease patients. On the other hand, the goal of the latter analysis was to identify biological parameters that were robustly correlated with multivariate measures of cognitive decline in Alzheimer’s disease. The purpose of these analyses was not to compare effects sizes between predictors, but rather to explore inter-subject differences in receptor–imaging interactions in relation to cognitive decline. For example, if regional amyloid accumulation strongly predicts changes in functional activity, but this biological parameter is consistent across subjects with different clinical and cognitive states, it would not be significant to our analysis. Rather than using clinical diagnosis, which is subject to large variability due to patient presentation and clinician bias, we used a combination of cognitive test scores. Ultimately, cognitive performance is the phenotype of interest in neurodegeneration. Our SVD analysis allows us to identify parameters associated with cognitive scores, rather than simply those with a large variability between individuals due to other causes.
Sources of variability in healthy and Alzheimer’s disease participants (Fig. 3) reflect alterations to mechanisms of receptor–imaging interaction that predict the same or another imaging modality. Here, we observed that a single PCA component explains 97.3% of the inter-individual variability in healthy subjects. Along this axis, a multi-faceted combination of receptor–imaging interaction predictors of CBF alterations (e.g. the interaction between dopaminergic D1 receptors and amyloid) and grey matter atrophy (e.g. the interaction between glutamatergic AMPA receptors and amyloid) account for most of the variability (Fig. 3A and C). Interestingly, there is relatively low variability in the biological parameters of receptor influence on neural activity, glucose metabolism and tau distribution in healthy individuals (Fig. 3C). In healthy subjects, the receptor-imaging mechanisms affecting these factors are comparatively consistent, whereas the mechanisms behind atrophy, CBF regulation and amyloid accumulation display more inter-subject heterogeneity.
In contrast, the first PC of Alzheimer’s disease patients’ biological parameters explained only 26.2% of the total variance, but this was dominated by neural activity as a (receptor-modulated) predictor of tau accumulation (as well as other neuroimaging modalities; Fig. 3B and D). Receptor mechanism variability was largely explained by adrenergic and serotonergic predictors, for example the interactions of α1 and 5HT1A receptors with neural activity to predict tau accumulation. As tau is primarily present in axonal microtubules, the exacerbation of tau pathology has been linked to enhanced neural activity.50 Conversely, tau is also believed to suppress and silence neural activity.22 Thus, the PC of variability in Alzheimer’s disease participants may represent variability in an activity-dependent tau accumulation via adrenergic α1, serotonergic 5HT1A, dopaminergic D1 and cholinergic α4β2 receptors. This would be consistent with the observed mediation of tau hyperphosphorylation by adrenergic and serotonergic receptors in animal models.51,52
From the inner sectors of Fig. 4D, inter-individual differences in cognitive decline are most correlated with biological parameters of the neural activity, tau and amyloid models, and least correlated with biological parameters of the CBF, grey matter density and glucose metabolism models. In other words, differences in receptor–imaging interactions affecting CBF changes are less relevant to cognitive symptom severity in Alzheimer’s disease than those affecting resting-state functional activity. While neural activity is not a cognitively important predictor of other neuroimaging modalities, many predictors of neural activity dysfunction are correlated with cognitive severity in Alzheimer’s disease (Fig. 5). Conversely, predictors of CBF do not vary significantly with cognition, whereas CBF itself is an important predictor of many other neuroimaging modalities. This may imply a causal ordering, with CBF alterations preceding dysfunctional activity.
The glutamatergic system is implicated in cognitive decline via its role as the major excitatory mediator of neural activity.53–55 In Alzheimer’s disease, the glutamatergic system is involved in excitotoxicity due to calcium ion influx via NMDA receptors,53 resulting in synaptic loss and neuronal cell death.54 Tau and amyloid are involved via an overactivation of NMDA receptors.56 The synaptic activation of NMDA receptors is linked to specific neurophysiological conditions, particularly activity-dependent synaptic plasticity, as well as behavioural symptoms of multiple brain disorders including Alzheimer’s disease.57 In addition, AMPA receptors are involved in synaptic scaling, and consequently learning and memory. Reductions in AMPA receptor levels have been observed in mouse models of Alzheimer’s disease,58 as well as in the entorhinal cortices59 of Alzheimer’s disease patients, with a differential preservation of certain subunits in the hippocampus,60 and AMPA receptor endocytosis has been linked to the phosphorylated tau signalling cascade.61 Thus, the established Alzheimer’s disease-related alterations and cognitive roles of the glutamatergic NMDA and AMPA receptors would be consistent with their significant modulation of resting-state functional activity in relation to cognitive decline in Alzheimer’s disease via interactions with CBF, glucose metabolism and tau.
From the columns of Fig. 5, CBF changes are the largest neuroimaging driver of cognitively relevant dysfunction in other modalities, consistent with its precedence among Alzheimer’s disease imaging biomarkers.10 Closely coupled to neural activity, CBF is mediated by several neuronal factors, including vasodilatory neurotransmitters and vascular dysregulation is implicated in the pathogenesis of Alzheimer’s disease.14 CBF interactions with a multitude of receptors were correlated to cognitive severity via amyloid, neural activity, grey matter density and tau models. This is consistent with the amyloid-dependent relationship of CBF to memory performance,62 and the link to tau pathology via gene expression alterations in Alzheimer’s disease.63
Furthermore, inter-individual differences in the effects of tau on other imaging modalities are also major contributors to Alzheimer’s disease-associated cognitive decline, as seen in Fig. 5. These include glutamatergic interactions affecting neural activity and amyloid accumulation, and a multifactorial set of receptor interactions affecting metabolism. Cognitive decline in Alzheimer’s disease is accompanied by changes in the role of regional tau concentration as a predictor of amyloid distribution, suggesting synergistic or mediation effects such as the tau axis hypothesis.64 Tau is believed to mediate amyloid toxicity,64 which may explain the significant role of tau as a predictor of amyloid accumulation (Fig. 5). Multimodal PET imaging has shown a region-dependent relationship between tau burden and hypometabolism in Alzheimer’s disease.65 Furthermore, alterations to glucose metabolism in mice brains were found to lead to abnormal tau hyperphosphorylation.66 Along with the established neural activity dysfunction due to tau accumulation,67 these mechanisms are consistent with the cognitively significant role of tau as a predictor of other neuroimaging modalities.
Along with NMDA, acetylcholine is the neurotransmitter system most associated with Alzheimer’s disease and its clinical treatment.68 Based on the cholinergic hypothesis, dysfunction in acetylcholine-producing basal forebrain regions would eventually lead to synaptic de-afferentiation in the cortical regions to which they project,69 with cognitive implications.70 This is consistent with the significant role of cholinergic predictors of tau distribution, which appear to be more correlated with cognitive severity of Alzheimer’s disease than amyloid distribution (Fig. 5).
Although GABAergic receptors were not initially linked to Alzheimer’s disease, recent evidence has uncovered disease-related alterations, contributions to pathogenesis and a potential therapeutic role in Alzheimer’s disease.71 The disruption of the excitatory/inhibitory balance maintained by GABAergic signalling has been implicated in the cognitive symptoms of Alzheimer’s disease, such as an increase in epileptic seizures.72 Electrophysiological activity has found a functional remodelling of GABAergic neurons in Alzheimer’s disease, showing reduced currents and a faster rate of desensitization.73 The presence of amyloid was also found to affect the expression of the α6 subunit of the GABAA receptor.71 Furthermore, a role for tau has been proposed in the regulation of GABAergic function and synaptic plasticity to maintain normal cognition.72 Additionally, it has recently been found that the administration of benzodiazepine in mouse brains leads to tau hyperphosphorylation.74 Such drugs potentiate GABAergic neurotransmission by binding to the benzodiazepine binding site of GABAA receptors. As such, this may indicate a mechanistic pathway for the induction of tau pathology involving GABAergic receptors, based on the tau and grey matter sectors of Fig. 4D. From Fig. 4D, the GABAA-associated benzodiazepine site is particularly involved in cognitively significant interactions affecting amyloid accumulation. GABAA and GABAB receptors play a notable cognitive role by affecting neural activity dysfunction, and all three GABAergic targets included in this work are involved via tau accumulation.
Finally, we introduced a summary metric of alterations to receptor-mediated interactions with reference to their normal variation in healthy ageing. Particularly, we found that GABAergic alterations had the largest effect on cognitive impairment in Alzheimer’s disease patients, significantly affecting executive function and the MMSE (Fig. 6A). Furthermore, we showed that subjects with identical clinical diagnosis and similar cognitive symptoms can have distinct underlying dynamics and receptor alteration fingerprints (Fig. 6B and C). These results highlight the clinical utility of our dynamical modelling approach. By fusing in vitro receptor templates with longitudinal neuroimaging data and modelling the underlying dynamics of receptor-mediated neurobiological interactions, we are able to infer subject-specific mechanistic alterations despite the lack of subject-specific receptor data. As a demonstrative example, we summarized subject-specific alterations at the scale of receptor families. However, in a clinical context, subject-specific alterations at the broad scale of receptor families, the finer scale of specific receptors, or even specific receptor-neurobiological interactions (e.g. NMDA × CBF interactions) can be used to design personalized, precision treatments, which will be a topic of future work.
The whole-brain re-MCM models used cyto-architectonically identified cortical regions of interest, neglecting subcortical structures for which no receptor distribution data was available. Many neurotransmitter alterations relevant to neurodegeneration occur in these regions, notably dopamine deficiencies in the basal ganglia in Parkinson’s disease and early cholinergic neuronal death in Alzheimer’s disease. As such, including subcortical regions may better characterize important molecular pathways. Nevertheless, some effects of these phenomena are captured via projections to cortical neurons that are covered by our regions of interest. Additionally, future work will aim to integrate CSF into the model.
In this work, we used fALFF31 as the regional measure of functional integrity. Low-frequency (0.01–0.08 Hz) oscillations in the blood oxygenation level-dependent (BOLD) signal reflect the intensity of spontaneous activity in the resting brain, primarily in the default mode network.31 When the amplitude of low-frequency fluctuations (ALFF) is normalized by the overall power spectrum of the BOLD signal to calculate fALFF, the effects of physiological noise are suppressed.31 However, compared to ALFF, fALFF significantly amplifies the signal from some non-default mode network regions (namely temporal-parietal regions and the precentral gyrus),31 reducing its desired specificity to resting-state activity. Nevertheless, fALFF shows high temporal stability over the course of functional MRI scans,75 long-term (i.e. about 6 month) test-retest reliability,76 and high sensitivity to Alzheimer’s disease progression.29,30 Alternative functional MRI-based metrics include regional homogeneity (ReHo)77 or graph theoretic metrics such as functional connectivity degree.78 Comparing fALFF, ReHo and graph-based metrics using simultaneous resting-state functional MRI/PET scans, Aiello et al. found functional connectivity degree to be the least correlated to glucose uptake, while the difference in correlation to glucose uptake between fALFF and ReHo was not significant.28 Furthermore, as an intentional consideration to maintain model interpretability, our modelling framework avoids graph theoretic functional MRI metrics to separate local, intra-region effects from inter-region effects due to network propagation. Although graph theoretic features can have biophysical interpretation, such as weighted degree representing transneural propagation or regional participation coefficients reflecting metabolic demands, they integrate information from multiple regions, which causes a leakage of network information into the intra-regional component of our model. Thus, as fALFF is a local functional MRI measure that has been found to be at least as informative as ReHo in reflecting metabolic activity and validated as a measure sensitive to Alzheimer’s disease progression by multiple studies,29,30 all the analyses and results presented in this study correspond to fALFF as the measure of resting-state functional integrity. It is, however, important to note that all available functional MRI metrics have limitations in reflecting actual neuronal activity or integrity. Here, our choice of metric is aligned with the ‘neurocentric’ resting-state functional MRI model,79 which assumes that the spontaneous fluctuations in BOLD signal reflect ongoing neuronal processes. Multiple limitations to this model have been pointed out, including the lack of clear neurophysiological interpretability,79 suggesting that interpretations of resting-state functional MRI-based findings (including ours) should be taken with caution.
In addition to intra-region effects, our model considered network propagation along the tractography-derived white matter structural connectome. However, functional, metabolic80 and vascular connectivity define complementary biophysical networks that may also contribute to the propagation of neurodegenerative pathology. For simplicity and to focus on local, receptor-mediated interactions, we restricted our model to structural connectivity. The structural connectome is the physical substrate for the axonal propagation of pathology, and the scaffolding for the more abstract functional network. However, to estimate the effect of our choice of connectivity, we repeated our model fitting with functional connectivity, finding no significant change in model fit (see the section ‘Multi-scale interactions involving neurotransmitter receptors are important to explaining multifactorial brain reorganization’ and Supplementary Fig. 8). We attribute this to (i) the dominance of intra-regional effects in our model, with a relatively low contribution due to propagation effects; and (ii) the shared information between anatomical and functional connectivity.48 While this work has focused on local interactions between biological processes, dynamical interactions also occur at a network level. For example, structural connectivity81 and the vascular network82 are two of the factors that shape functional connectivity, and modelling the dynamic interactions between these networks may be a potential direction for future work.
The dynamical system modelling approach in this work relies on longitudinal and multimodal neuroimaging data to fit personalized models. Consequently, our results would benefit from larger cohorts with more longitudinal samples of multimodal data. Additionally, receptor map templates of patients at different stages of ageing and disease progression would improve the characterization of salient alterations. While we have attempted to uncover causal molecular-macroscopic mechanisms, due to the lack of personalized, longitudinal receptor maps, some identified biological model parameters may in fact reflect a molecular alteration (i.e. a change in either spatial distribution or functional alteration of a receptor) in response to a change in a macroscopic biological variable. As such, the exploratory interpretation of our results in relation to cognitive decline in Alzheimer’s disease should account for both possible causal directions between a given biological parameter and its target modality. For example, α1 receptors interacting with tau to predict functional activity represents a three-way interaction, which may in fact reflect a causal direction from functional activity to adrenergic alteration. Furthermore, we have assumed a direct relationship between each imaging modality and an underlying neurobiological process. For instance, CBF in our model was derived from ASL MRI, the temporal resolution of which is limited by the relaxation time of blood. However, recent work on venous blood flow using BOLD perfusion lag-mapping has shown significant age-related changes outside the temporal resolution of ASL MRI.83 As new or improved imaging biomarkers are developed for Alzheimer’s disease, their future inclusion in the re-MCM framework would improve the coverage of potential disease-related mechanisms.
Nevertheless, these results offer interpretable results via molecular targets and mechanisms of action. We find that receptor distributions mediate interactions between macroscopic biological factors that significantly affect cognitive decline in Alzheimer’s disease. Specifically, inter-individual differences in cognitive deterioration correlate with the modulation of neural activity dysfunction primarily by glutamatergic receptors, amyloid accumulation by GABAergic receptors and tau build-up by glutamatergic, GABAergic and cholinergic receptors. Traditionally, the accumulation of misfolded proteins, namely amyloid and tau, has been implicated in the pathogenesis of Alzheimer’s disease. However, our results indicate a multifactorial, and heterogeneous set of mechanisms involved in disease. Furthermore, our personalized, data-driven approach allows us to account for inter-subject heterogeneity in biological pathology and clinical presentation.
A growing body of evidence supports the critical role of neurotransmitter receptors in Alzheimer’s disease symptoms severity and their subsequent potential as therapeutic targets.84,85 Neurotransmitter-based drugs such as the acetylcholinesterase inhibitor donepezil and the NMDA antagonist memantine have long been proposed as potential treatments for Alzheimer’s disease patients. However, these drugs have shown limited efficacy and adverse side-effects.20,56 We propose that using personalized and multi-scale modelling can identify patient-specific alterations and therapeutic needs, by stratifying patients based on the biological parameter weights corresponding to the underlying, cognitively significant mechanisms (Figs 1C and 6). This information can then be used to design individually tailored multifactorial therapies to slow the process of cognitive decline in both diseased and normally ageing individuals.
Supplementary Material
Acknowledgements
Data used in preparation of this article were partly obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding
This research was undertaken thanks in part to funding from: the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives Initiative, the Canada Research Chair tier-2, Fonds de la recherche en santé du Québec (FRQS) Junior 1 Scholarship and Weston Brain Institute (Rapid Response AD program 2018) awards to Y.I.M., the Brain Canada Foundation and Health Canada support to the McConnell Brain Imaging Center at the Montreal Neurological Institute, and the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreements 785907 (Human Brain Project SGA2) and 945539 (Human Brain Project SGA3) awarded to N.P.G. and K.Z. Multimodal imaging and clinical data collection and sharing for this project was funded by ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd ; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Abbreviations
- ADAS
Alzheimer’s Disease Assessment Scale
- CBF
cerebral blood flow
- fALFF
fractional amplitude of low-frequency fluctuation
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
- re-MCM
receptor-enriched multifactorial causal model
- SVD
singular value decomposition
Contributor Information
Ahmed Faraz Khan, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal H3A 2B4, Canada; McConnell Brain Imaging Center, Montreal Neurological Institute, Montreal H3A 2B4, Canada; Ludmer Centre for Neuroinformatics and Mental Health, Montreal H3A 2B4, Canada.
Quadri Adewale, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal H3A 2B4, Canada; McConnell Brain Imaging Center, Montreal Neurological Institute, Montreal H3A 2B4, Canada; Ludmer Centre for Neuroinformatics and Mental Health, Montreal H3A 2B4, Canada.
Tobias R Baumeister, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal H3A 2B4, Canada; McConnell Brain Imaging Center, Montreal Neurological Institute, Montreal H3A 2B4, Canada; Ludmer Centre for Neuroinformatics and Mental Health, Montreal H3A 2B4, Canada.
Felix Carbonell, Biospective Inc ., Montreal H3B 2T9, Canada.
Karl Zilles, Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich, 52425 Jülich, Germany.
Nicola Palomero-Gallagher, Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich, 52425 Jülich, Germany; Cécile and Oskar Vogt Institute of Brain Research, Medical Faculty, Heinrich-Heine University, 40225 Düsseldorf, Germany; Department of Psychiatry, Psychotherapy, and Psychosomatics, Medical Faculty, RWTH Aachen, 52074 Aachen, Germany; JARA, Translational Brain Medicine, 52074 Aachen, Germany.
Yasser Iturria-Medina, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal H3A 2B4, Canada; McConnell Brain Imaging Center, Montreal Neurological Institute, Montreal H3A 2B4, Canada; Ludmer Centre for Neuroinformatics and Mental Health, Montreal H3A 2B4, Canada.
References
- 1. Francis PT, Ramírez MJ, Lai MK.. Neurochemical basis for symptomatic treatment of Alzheimer’s disease. Neuropharmacology. 2010;59(4-5):221–229. [DOI] [PubMed] [Google Scholar]
- 2. Jack CR Jr, Knopman DS, Jagust WJ.. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9(1):119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lam B, Masellis M, Freedman M, Stuss DT, Black SE.. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimer’s Res Ther. 2013;5(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iturria-Medina Y, Carbonell FM, Evans AC.. Multimodal imaging-based therapeutic fingerprints for optimizing personalized interventions: Application to neurodegeneration. NeuroImage. 2018;179:40–50. [DOI] [PubMed] [Google Scholar]
- 5. Kosik KS. Personalized medicine for effective Alzheimer disease treatment. JAMA Neurol. 2015;72(5):497–498. [DOI] [PubMed] [Google Scholar]
- 6. Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520(7549):609–611. [DOI] [PubMed] [Google Scholar]
- 7. Prakash A, Kalra J, Mani V, Ramasamy K, Majeed ABA.. Pharmacological approaches for Alzheimer’s disease: Neurotransmitter as drug targets. Expert Rev Neurother. 2015;15(1):53–71. [DOI] [PubMed] [Google Scholar]
- 8. Roy K. Computational modeling of drugs against Alzheimer’s disease. Springer; 2018. [Google Scholar]
- 9. Jack CR Jr, Knopman DS, Jagust WJ, et al. . Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-Pérez JM, Evans AC.. Initiative tADN. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun. 2016;7(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iturria-Medina Y, M.Carbonell F, Sotero RC, Chouinard-Decorte F, C.Evans A.. Multifactorial causal model of brain (dis)organization and therapeutic intervention: Application to Alzheimer’s disease. NeuroImage. 2017;152:60–77. [DOI] [PubMed] [Google Scholar]
- 12. Moses WW. Fundamental limits of spatial resolution in PET. Nucl Instrum Methods Phys Res Sect A. 2011;648:S236–S240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi JK, Chen YI, Hamel E, Jenkins BG.. Brain hemodynamic changes mediated by dopamine receptors: Role of the cerebral microvasculature in dopamine-mediated neurovascular coupling. Neuroimage. 2006;30(3):700–712. [DOI] [PubMed] [Google Scholar]
- 14. Iadecola C. Neurovascular regulation in the normal brain and in Alzheimer's disease. Nat Rev Neurosci. 2004;5(5):347–360. [DOI] [PubMed] [Google Scholar]
- 15. Mink JW, Blumenschine RJ, Adams DB.. Ratio of central nervous system to body metabolism in vertebrates: Its constancy and functional basis. Am J Physiol. 1981;231(3):R203–R212. [DOI] [PubMed] [Google Scholar]
- 16. Harris JJ, Jolivet R, Attwell D.. Synaptic energy use and supply. Neuron. 2012;75(5):762–777. [DOI] [PubMed] [Google Scholar]
- 17. McCann CM, Tapia JC, Kim H, Coggan JS, Lichtman JW.. Rapid and modifiable neurotransmitter receptor dynamics at a neuronal synapse in vivo. Nat Neurosci. 2008;11(7):807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mora F, Segovia G, del Arco A.. Aging, plasticity and environmental enrichment: Structural changes and neurotransmitter dynamics in several areas of the brain. Brain Res Rev. 2007;55(1):78–88. [DOI] [PubMed] [Google Scholar]
- 19. Heiss WD, Herholz K.. Brain receptor imaging. J Nucl Med. 2006;47(2):302–312. [PubMed] [Google Scholar]
- 20. Kandimalla R, Reddy PH.. Therapeutics of neurotransmitters in Alzheimer’s disease. J Alzheimer's Dis. 2017;57(4):1049–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bejanin A, Schonhaut DR, La Joie R, et al. . Tau pathology and neurodegeneration contribute to cognitive impairment in Alzheimer’s disease. Brain. 2017;140(12):3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Busche MA, Wegmann S, Dujardin S, et al. . Tau impairs neural circuits, dominating amyloid-β effects, in Alzheimer models in vivo. Nat Neurosci. 2019;22(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sled JG, Zijdenbos AP, Evans AC.. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- 24. Evans AC, Kamber M, Collins D, MacDonald D.. An MRI-based probabilistic atlas of neuroanatomy. In: Shorvon, SD, Fish DR, Andermann F, Bydder GM, Stefan H., eds. Magnetic resonance scanning and epilepsy. NATO ASI Series, vol 264. Springer; 1994:263–274. [Google Scholar]
- 25. Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38(1):95–113. [DOI] [PubMed] [Google Scholar]
- 26. Jagust WJ, Bandy D, Chen K, et al. . The Alzheimer's Disease Neuroimaging Initiative positron emission tomography core. Alzheimer's Dement. 2010;6(3):221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan C, Zang Y.. DPARSF: A MATLAB toolbox for ‘pipeline‘ data analysis of resting-state fMRI. Front Syst Neurosci. 2010;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aiello M, Salvatore E, Cachia A, et al. . Relationship between simultaneously acquired resting-state regional cerebral glucose metabolism and functional MRI: A PET/MR hybrid scanner study. Neuroimage. 2015;113:111–121. [DOI] [PubMed] [Google Scholar]
- 29. Yang L, Yan Y, Wang Y, et al. . Gradual disturbances of the amplitude of low-frequency fluctuations (ALFF) and fractional ALFF in Alzheimer spectrum. Front Neurosci. 2018;12:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Vos F, Koini M, Schouten TM, et al. . A comprehensive analysis of resting state fMRI measures to classify individual patients with Alzheimer's disease. Neuroimage. 2018;15(167):62–72. [DOI] [PubMed] [Google Scholar]
- 31. Zou QH, Zhu CZ, Yang Y, et al. . An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J Neurosci Methods. 2008;172(1):137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palomero-Gallagher N, Zilles K.. Cyto-and receptor architectonic mapping of the human brain. Handbook Clin Neurol. 2018;150:355–387. [DOI] [PubMed] [Google Scholar]
- 33. Merker B. Silver staining of cell bodies by means of physical development. J Neurosci Methods. 1983;9(3):235–241. [DOI] [PubMed] [Google Scholar]
- 34. Eickhoff SB, Paus T, Caspers S, et al. . Assignment of functional activations to probabilistic cytoarchitectonic areas revisited. NeuroImage. 2007;36(3):511–521. [DOI] [PubMed] [Google Scholar]
- 35. Zilles K, Palomero-Gallagher N.. Multiple transmitter receptors in regions and layers of the human cerebral cortex. Front Neuroanatomy. 2017;11:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Barth JA; 1909. [Google Scholar]
- 37. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM.. FSL. Neuroimage 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 38. Yeh FC, Panesar S, Fernandes D, et al. . Population-averaged atlas of the macroscale human structural connectome and its network topology. NeuroImage. 2018;178:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeh FC, Tseng WYI.. NTU-90: A high angular resolution brain atlas constructed by q-space diffeomorphic reconstruction. NeuroImage. 2011;58(1):91–99. [DOI] [PubMed] [Google Scholar]
- 40. Yeh FC, Wedeen VJ, Tseng WYI.. Generalized q-sampling imaging. IEEE Trans Med Imaging. 2010;29(9):1626–1635. [DOI] [PubMed] [Google Scholar]
- 41. Yeh FC, Liu L, Hitchens TK, Wu YL.. Mapping immune cell infiltration using restricted diffusion MRI. Magn Reson Med. 2017;77(2):603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng WYI.. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS ONE. 2013;8(11):e80713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Folch-Fortuny A, Arteaga F, Ferrer A.. Missing data imputation toolbox for MATLAB. Chemom Intell Lab Syst. 2016;154:93–100. [Google Scholar]
- 44. Gibbons LE, Carle AC, Scott-Mackin R, et al. Composite measures of executive function and memory: ADNI_EF and ADNI_Mem. October 23, 2015. Accessed May 2021. https://adni.bitbucket.io/reference/docs/UWNPSYCHSUM/ADNI_Methods_UWNPSYCHSUM.pdf
- 45. Choi SE, Mukherjee S, Gibbons LE, et al. . Development and validation of language and visuospatial composite scores in ADNI. Alzheimer's Dement. 2020;6(1):e12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alzheimer's Disease Neuroimaging Initiative . ADNI2 Procedures Manual. July 2008. Accessed May 2021. https://adni.loni.usc.edu/wp-content/uploads/2008/07/adni2-procedures-manual.pdf
- 47. Iturria-Medina Y, Carbonell FM, Assadi A, et al. . NeuroPM toolbox: integrating molecular, neuroimaging and clinical data for characterizing neuropathological progression and individual therapeutic needs. medRvix. [Preprint] 10.1101/2020.09.24.20200964 [DOI] [Google Scholar]
- 48. Honey CJ, Sporns O, Cammoun L, et al. . Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 2009;106(6):2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Desikan R, Fan C, Wang Y, et al. . Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14(3):e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu JW, Hussaini SA, Bastille IM, et al. . Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 2016;19(8):1085–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang F, Gannon M, Chen Y, et al. . β-amyloid redirects norepinephrine signaling to activate the pathogenic GSK3β/tau cascade. Sci Transl Med. 2020;12(526):eaay6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang YJ, Ren QG, Gong WG, et al. . Escitalopram attenuates β-amyloid-induced tau hyperphosphorylation in primary hippocampal neurons through the 5-HT1A receptor mediated Akt/GSK-3β pathway. Oncotarget. 2016;7(12):13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang R, Reddy PH.. Role of glutamate and NMDA receptors in Alzheimer’s disease. J Alzheimer’s Dis. 2017;57(4):1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hynd MR, Scott HL, Dodd PR.. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer’s disease. Neurochem Int. 2004;45(5):583–595. [DOI] [PubMed] [Google Scholar]
- 55. Butterfield DA, Pocernich CB.. The glutamatergic system and Alzheimer’s disease. CNS Drugs. 2003;17(9):641–652. [DOI] [PubMed] [Google Scholar]
- 56. Xu Y, Yan J, Zhou P, et al. . Neurotransmitter receptors and cognitive dysfunction in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol. 2012;97(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lau CG, Zukin RS.. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8(6):413–426. [DOI] [PubMed] [Google Scholar]
- 58. Chang EH, Savage MJ, Flood DG, et al. . AMPA receptor downscaling at the onset of Alzheimer’s disease pathology in double knockin mice. Proc Natl Acad Sci USA. 2006;103(9):3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yasuda RP, Ikonomovic MD, Sheffield R, Rubin RT, Wolfe BB, Armstrong DM.. Reduction of AMPA-selective glutamate receptor subunits in the entorhinal cortex of patients with Alzheimer's disease pathology: A biochemical study. Brain Res. 1995;678(1-2):161–167. [DOI] [PubMed] [Google Scholar]
- 60. Carter TL, Rissman RA, Mishizen-Eberz AJ, et al. . Differential preservation of AMPA receptor subunits in the hippocampi of Alzheimer's disease patients according to Braak stage. Exp Neurol. 2004;187(2):299–309. [DOI] [PubMed] [Google Scholar]
- 61. Miller EC, Teravskis PJ, Dummer BW, Zhao X, Huganir RL, Liao D.. Tau phosphorylation and tau mislocalization mediate soluble Aβ oligomer‐induced AMPA glutamate receptor signaling deficits. Eur J Neurosci. 2014;39(7):1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bangen KJ, Clark AL, Edmonds EC, et al. . Cerebral blood flow and amyloid-β interact to affect memory performance in cognitively normal older adults. Front Aging Neurosci. 2017;9:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bryant AG, Hu M, Carlyle BC, et al. . Cerebrovascular senescence is associated with tau pathology in Alzheimer's disease. Front Neurol. 2020;11:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ittner LM, Götz J.. Amyloid-β and tau—a toxic pas de deux in Alzheimer’s disease. Nat Rev Neurosci. 2011;12(2):67–72. [DOI] [PubMed] [Google Scholar]
- 65. Bischof GN, Jessen F, Fliessbach K, et al. . Impact of tau and amyloid burden on glucose metabolism in Alzheimer's disease. Ann Clin Transl Neurol. 2016;3(12):934–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Planel E, Miyasaka T, Launey T, et al. . Alterations in glucose metabolism induce hypothermia leading to tau hyperphosphorylation through differential inhibition of kinase and phosphatase activities: Implications for Alzheimer's disease. J Neurosci. 2004;24(10):2401–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Menkes-Caspi N, Yamin HG, Kellner V, Spires-Jones TL, Cohen D, Stern EA.. Pathological tau disrupts ongoing network activity. Neuron. 2015;85(5):959–966. [DOI] [PubMed] [Google Scholar]
- 68. Verma S, Kumar A, Tripathi T, Kumar A.. Muscarinic and nicotinic acetylcholine receptor agonists: Current scenario in Alzheimer's disease therapy. J Pharm Pharmacol. 2018;70(8):985–993. [DOI] [PubMed] [Google Scholar]
- 69. Hampel H, M-M M, Cuello AC, et al. . Revisiting the cholinergic hypothesis in Alzheimer’s disease: Emerging evidence from translational and clinical research. Alzheimer's Dement. 2017;6(1):2–15. [DOI] [PubMed] [Google Scholar]
- 70. Grothe M, Zaborszky L, Atienza M, et al. . Reduction of basal forebrain cholinergic system parallels cognitive impairment in patients at high risk of developing Alzheimer's disease. Cerebral Cortex. 2010;20(7):1685–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li Y, Sun H, Chen Z, Xu H, Bu G, Zheng H.. Implications of GABAergic neurotransmission in Alzheimer’s disease. Front Aging Neurosci. 2016;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Levenga J, Krishnamurthy P, Rajamohamedsait H, et al. . Tau pathology induces loss of GABAergic interneurons leading to altered synaptic plasticity and behavioral impairments. Acta Neuropathol Commun. 2013;1(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Limon A, Reyes-Ruiz JM, Miledi R.. Loss of functional GABAA receptors in the Alzheimer diseased brain. Proc Natl Acad Sci USA. 2012;109(25):10071–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Whittington RA, Virág L, Gratuze M, et al. . Administration of the benzodiazepine midazolam increases tau phosphorylation in the mouse brain. Neurobiol Aging. 2019;75:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Küblböck M, Woletz M, Höflich A, et al. . Stability of low-frequency fluctuation amplitudes in prolonged resting-state fMRI. Neuroimage. 2014;1(103):249–257. [DOI] [PubMed] [Google Scholar]
- 76. Zuo XN, Xing XX.. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: A systems neuroscience perspective. Neurosci Biobehav Rev. 2014;45:100–118. [DOI] [PubMed] [Google Scholar]
- 77. Zang Y, Jiang T, Lu Y, He Y, Tian L.. Regional homogeneity approach to fMRI data. NeuroImage. 2004;22(1):394–400. [DOI] [PubMed] [Google Scholar]
- 78. Heuvel MPVD, Pol HEH.. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20(8):519–534. [DOI] [PubMed] [Google Scholar]
- 79. Lu H, Jaime S, Yang Y.. Origins of the resting-state functional MRI signal: Potential limitations of the ‘neurocentric’ model. Front Neurosci. 2019;13:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Riedl V, Utz L, Castrillon G, et al. . Metabolic connectivity mapping reveals effective connectivity in the resting human brain. Proc Natl Acad Sci USA. 2016;113(2):428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deco G, Ponce-Alvarez A, Mantini D, Romani GL, Hagmann P, Corbetta M.. Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. J Neurosci. 2013;33(27):11239–11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bright MG, Whittaker JR, Driver ID, Murphy K.. Vascular physiology drives functional brain networks. NeuroImage. 2020;217:116907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Aso T, Sugihara G, Murai T, et al. . A venous mechanism of ventriculomegaly shared between traumatic brain injury and normal ageing. Brain. 2020;143(6):1843–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaur S, DasGupta G, Singh S.. Altered neurochemistry in Alzheimer’s disease: Targeting neurotransmitter receptor mechanisms and therapeutic strategy. Neurophysiology. 2019;51(4):1–17.31017844 [Google Scholar]
- 85. Whitehouse PJ, Au KS.. Neurotransmitter receptor alterations in Alzheimer's disease. In: Traber J, Gispen WH, eds. Senile dementia of the Alzheimer type. Advances in Applied Neurological Sciences, vol 2. Springer; 1985:175–182. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three datasets used in this study are available from the ADNI database (neuroimaging and cognitive evaluations; http://www.adni.loni.usc.edu), the HCP database (tractography template for connectivity estimation; http://www.humanconnectomeproject.org/) and receptor density data published in Zilles and Palomero-Gallagher.35 We anticipate that the re-MCM method will be released soon as part of our available and open-access, user-friendly software (https://www.neuropm-lab.com/neuropm-box.html).47