Abstract
Cysteine is the major source of fixed sulfur for the synthesis of sulfur-containing compounds in organisms of the Bacteria and Eucarya domains. Though pathways for cysteine biosynthesis have been established for both of these domains, it is unknown how the Archaea fix sulfur or synthesize cysteine. None of the four archaeal genomes sequenced to date contain open reading frames with identities to either O-acetyl-l-serine sulfhydrylase (OASS) or homocysteine synthase, the only sulfur-fixing enzymes known in nature. We report the purification and characterization of OASS from acetate-grown Methanosarcina thermophila, a moderately thermophilic methanoarchaeon. The purified OASS contained pyridoxal 5′-phosphate and catalyzed the formation of l-cysteine and acetate from O-acetyl-l-serine and sulfide. The N-terminal amino acid sequence has high sequence similarity with other known OASS enzymes from the Eucarya and Bacteria domains. The purified OASS had a specific activity of 129 μmol of cysteine/min/mg, with a Km of 500 ± 80 μM for sulfide, and exhibited positive cooperativity and substrate inhibition with O-acetyl-l-serine. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a single band at 36 kDa, and native gel filtration chromatography indicated a molecular mass of 93 kDa, suggesting that the purified OASS is either a homodimer or a homotrimer. The optimum temperature for activity was between 40 and 60°C, consistent with the optimum growth temperature for M. thermophila. The results of this study provide the first evidence for a sulfur-fixing enzyme in the Archaea domain. The results also provide the first biochemical evidence for an enzyme with the potential for involvement in cysteine biosynthesis in the Archaea.
The serine and homoserine pathways are the two major routes for cysteine biosynthesis in nature. Serine transacetylase and O-acetylserine sulfhydrylase (OASS) catalyze steps in the serine pathway (reactions 1 and 2) (28):
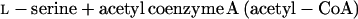 |
1 |
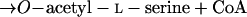 |
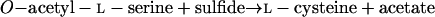 |
2 |
Homoserine transacetylase, homocysteine synthase, cystathionine β-synthase, and γ-cystathionase catalyze steps of the homoserine pathway (reactions 3 to 6) (28):
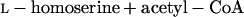 |
3 |
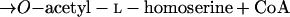 |
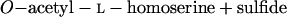 |
4 |
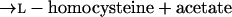 |
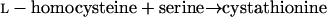 |
5 |
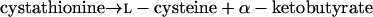 |
6 |
Cysteine is the major source of fixed sulfur for the synthesis of sulfur-containing compounds in organisms from the Bacteria and Eucarya domains; thus, the sulfur-fixing enzymes catalyzing reactions 2 and 4 are key enzymes in sulfur metabolism (13). Plants and members of the Bacteria domain synthesize cysteine and fix sulfur via the serine pathway. Plants and procaryotes from the Bacteria domain also fix sulfur by synthesizing homocysteine; however, they cannot utilize homocysteine for cysteine biosynthesis (21). Fungi fix sulfide and synthesize cysteine by using both pathways (26). Although in yeast the homocysteine synthase also has OASS activity (28), the homoserine pathway appears to be the major route for cysteine biosynthesis (21). Many members of the Archaea are autotrophic and do not require cysteine or other forms of fixed sulfur for growth; however, it is unknown how the Archaea fix sulfur or synthesize cysteine.
The genomes of four members of Archaea have been sequenced. The Methanococcus jannaschii (6) and Archaeoglobus fulgidus (20) genomes contain no open reading frames having a deduced sequence with significant identity to any enzymes known to be involved in the fixation of sulfur or in cysteine biosynthesis. The methanoarchaeal genomes are also void of any open reading frames with deduced sequence identity to any known cysteinyl-tRNA synthetases. The genome of Pyrococcus horikoshii (19) contains an open reading frame with sequence similarity to γ-cystathionase, and the genome of Methanobacterium thermoautotrophicum (44) contains an open reading frame with sequence similarity to homoserine transacetylase. However, these putative genes have not been expressed, and it is not known whether the gene products have the expected enzyme activities. It is also possible that O-acetyl-l-homoserine is an intermediate only for the biosynthesis of methionine and not for the biosynthesis of l-cysteine (3). Furthermore, there are no open reading frames in either the P. horikoshii or M. thermoautotrophicum genome with a deduced sequence having significant identity to other enzymes of the homoserine pathway or any enzymes in the serine pathway for cysteine biosynthesis. Remarkably, none of the four archaeal genomes sequenced to date contain open reading frames with deduced sequence identities to either OASS or homocysteine synthase, the only sulfur-fixing enzymes known in nature. We present here the first purification from an archaeon of an enzyme catalyzing sulfur fixation and cysteine biosynthesis, the pyridoxal 5′-phosphate-dependent OASS from the methanoarchaeon Methanosarcina thermophila.
MATERIALS AND METHODS
Cell material.
M. thermophila TM-1 was grown on acetate as described previously (46). The medium contained the following constituents in demineralized water at the indicated final percent (wt/nt/volume) concentrations: NH4Cl, 0.14; K2HPO4, 0.13; KH2PO4, 0.133; NaCl, 0.05; MgSO4, 0.05; Na2S · 9H2O, 0.027; CaCl2 · 2H2O, 0.006; Fe(NH4)2(SO4)2, 0.001; cysteine-HCl · H2O, 0.027; yeast extract (Difco, Detroit, Mich.), 0.01; Trypticase (BBL, Cockeysville, Md.), 0.01; and sodium acetate, 0.41. In addition, the medium contained 1% (vol/vol) each vitamin and trace mineral solutions as described elsewhere (46). The cells were harvested at the end of exponential growth and stored in liquid nitrogen until use.
Purification of OASS.
OASS was purified by monitoring cysteine production from O-acetyl-l-serine and sulfide in reaction mixtures following each purification step. All procedures were done aerobically and at 21°C unless otherwise indicated. Protein concentrations were determined by the method of Bradford (5), using Bio-Rad dye reagents and bovine serum albumin (Pierce) as a standard.
(i) Preparation of cell extract.
Thawed cell paste (20 g [wet weight]) was suspended in 25 ml of buffer A [50 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-KOH (pH 6.8)] containing 10% (vol/vol) glycerol. DNase I (0.25 mg) was added to the suspension and then passed twice through a chilled French pressure cell at 20,000 lb/in2 (1 lb/in2 = 6.9 kPa). The cell lysate was centrifuged at 2,000 × g for 20 min, and the supernatant was recentrifuged at 78,400 × g for 2 h.
(ii) Q-Sepharose chromatography.
The supernatant from step i was applied to a Q-Sepharose HP (Pharmacia Biotechnology) column (bed volume = 150 ml) equilibrated with buffer A. The column was washed with 300 ml of buffer A, and a 1-liter linear 0 to 1 M KCl gradient was applied at 7.0 ml/min. Peak fractions containing OASS activity, which eluted between 0.28 and 0.35 M KCl, were pooled and stored at −20°C. The procedure was repeated twice. The pooled peak fractions were combined and concentrated in dialysis tubing (3.5-kDa cutoff; Spectrum) embedded in dry polyethylene glycol (Mr = 8,000; Sigma) at 4°C.
(iii) Phenyl-Sepharose chromatography.
The concentrated protein solution from step ii was raised to a final concentration of 2 M NaCl by addition of 5 M NaCl in 50 mM Tris-Cl (pH 7.5) and loaded onto a phenyl-Sepharose FF HS (Pharmacia Biotechnology) column (bed volume = 160 ml) equilibrated with buffer B (50 mM Tris-Cl [pH 7.5] containing 2 M NaCl). After a 320-ml wash, the column was developed with a 1.1-liter decreasing linear gradient of 2.0 to 0 M NaCl at 2 ml/min. The peak of activity, which eluted between 0.1 and 0.9 M NaCl, was pooled and concentrated as described in step ii.
(iv) Mono Q chromatography I.
The pooled samples were dialyzed against 4 liters of buffer C (50 mM Tris-Cl [pH 8.0]) containing 1 mM pyridoxal 5′-phosphate and loaded onto a Mono Q HR 10/10 anion-exchange column (Pharmacia) equilibrated with buffer C. After a 20-ml wash, the column was developed with a 200-ml linear gradient from 0 to 1 M NaCl applied at 2.0 ml/min. The enzyme eluted between 0.3 and 0.4 M NaCl. Fractions with highest specific activity were pooled and stored at −20°C.
(v) Mono Q chromatography II.
Step iv was repeated except that buffer A was used, and the column was developed from 0 to 1 M KCl. The purified enzyme, which eluted between 0.4 and 0.5 M KCl, was stored at −20°C.
Enzyme assay.
OASS activity was measured as described previously (11) except that reactions were performed at 40°C and were allowed to proceed for 1 min. Solutions were made anaerobic and stored under N2 for sulfide Km determination to avoid oxidation of the substrate.
Kinetic data analysis.
The kinetic data were fitted to the indicated rate equations by using the Levenberg-Marquardt algorithm with KaleidaGraph for Windows version 3.09 (Abelbeck Software) and a Pentium Dell XPS M200s computer. The Hill equation (42) is v = Vmax[S]n/(Kh + [S]n), where v is the velocity of the reaction (rate of product formation), Vmax is the maximum velocity, Kh is the product of the two kinetic dissociation constants, and n is the Hill cooperativity constant. At S0.5, v = Vmax/2 and thus S0.5 = Kh1/n.
A modified Hill equation incorporating an ordinate intercept (b) was proposed by Morgan et al. (27) and called MMF: v = (bk + Vmax [S]n)/(K + [S]n). At S0.5, v = (Vmax + b)/2, and thus S0.5 = K1/n.
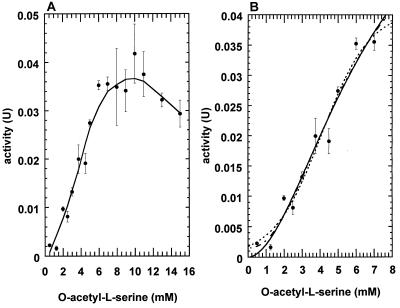
The logistic equation (34, 35) is v = Vmax/(1 + exp (β − γ[S]). At S0.5, v = Vmax/2 and thus S0.5 = β/γ. Saturation curves included data points from 0 to 7 mM O-acetyl-l-serine to preclude the inhibition portion of the curve (see Fig. 5).
FIG. 5.
(A) Dependence of activity of purified OASS from M. thermophila on O-acetyl-l-serine concentration. Standard assays were used except that O-acetyl-l-serine was varied as indicated, and 0.1 μg of enzyme was used per assay. (B) At low O-acetyl-l-serine concentrations, the data were fit by using the Hill equation (—), the logistic equation (---), and the MMF equation (- · - · -).
N-terminal analysis.
Purified enzyme was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto an Immobilon-P polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) at 23V for 14 to 16 h at 4°C with 10 mM (3-[cyclohexylamino]-1-propanesulfonic acid) (CAPS)-Na (pH 11) containing 10% (vol/vol) methanol. The N-terminal sequence was determined with a PE Biosystems 477A sequencer coupled to a 120A analyzer (PE Biosystems, Foster City, Calif.). The BLAST blastp program (1) was used to search the nonredundant sequence databases at the National Center for Biotechnology Information (Bethesda, Md.) for enzymes with similar N termini and later to search for OASS sequences. Representative OASS enzymes from the eucaryotic group (spinach CSaseA [38], CSase B [39], and CSase C [40], Citrullus vulgaris CSase A [32], wheat O-acetylserine lyase [49], Arabidopsis thaliana CSase A and CSase B [14], Capsicum annuum CSase B [36], and Entamoeba histolytica isozymes [33]) and the procaryotic group (Escherichia coli CysK [8] and CysM [43], Salmonella enterica serovar Typhimurium CysK [8], Synechococcus sp. strain PCC 7942 CysK [31], Flavobacterium strain K3-15 CysK [29], Campylobacter jejuni CysM [12], and Aspergillus nidulans CysM [47]) were aligned by using Pattern-Induced (local) Multiple Alignment 1.4 from the Baylor College of Medicine Search Launcher (45).
Molecular mass determination.
SDS-PAGE was performed as described previously (24). The molecular mass of the native enzyme was determined with a Superose 12 HP (Pharmacia) gel filtration column. After equilibration with buffer D (50 mM Tris-Cl [pH 7.5], 100 mM KCl), 0.2 ml of sample was injected, and the column was developed at a flow rate of 0.3 ml/min. The column was calibrated with a molecular weight kit (Sigma) containing cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), and β-amylase (200 kDa).
UV-visible absorption spectroscopy.
The UV-visible absorption spectrum was recorded at 21°C with a Beckman DU640 apparatus. The concentration of O-acetyl-l-serine in solution was increased stepwise by addition of 2 to 5 μl from 0 to final concentrations of 0.5, 1.0, 2.0, 5.0, 7.5, 10, and 15 μM. Spectra were taken at each concentration. Sodium sulfide was added to a final concentration of 1 mM, and a final spectrum was recorded.
Isoelectric focusing.
Isoelectric focusing was performed in a Rotofor system (Bio-Rad).
Materials.
All chemicals were of reagent grade from Sigma or Fisher. Molecular mass standards were from Sigma.
RESULTS
Purification.
The assay of OASS activity in the soluble and membrane fractions obtained by centrifugation of cell extract through a sucrose gradient revealed that 95% of the activity resides in the soluble fraction. A typical purification of OASS is summarized in Table 1. The level of enzyme activity in cell extract was found to be within the range reported for procaryotes from the Bacteria domain (0.03 to 50 U/mg) (9, 15). The M. thermophila enzyme could be stored at −20°C for up to 7 days between each purification step with no significant loss of activity. OASS was purified to apparent homogeneity as indicated by SDS-PAGE (Fig. 1). The purified enzyme was stable to several freeze-thaw cycles, but activity was completely absent after 2 months of storage at −20°C. However, approximately 10% of the activity could be recovered by incubating the enzyme at 21°C for 30 min in Tris-Cl buffer (pH 8.0) containing 50 mM dithiothreitol and 80 μM pyridoxal 5′-phosphate.
TABLE 1.
Scheme for purification of OASS from M. thermophila
| Step | Vol (ml) | Protein (mg) | Total activity (Ua) | Sp act (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cell extract | 96 | 2,891 | 979 | 0.339 | 100 | 1 |
| Q-Sepharose | 60 | 105 | 660 | 6.30 | 67 | 19 |
| Phenyl-Sepharose | 220 | 36 | 525 | 14.5 | 54 | 43 |
| Mono Q chromatography | ||||||
| I | 12 | 6.7 | 413 | 61.0 | 42 | 180 |
| II | 7 | 3.5 | 445 | 129 | 45 | 381 |
Defined as micromoles of Cys produced per minute.
FIG. 1.
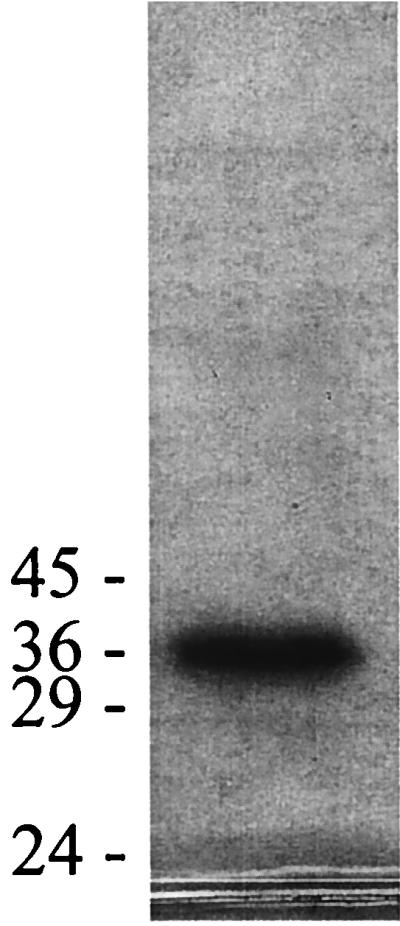
SDS-PAGE of OASS purified from M. thermophila. The 12% gel was loaded with 9 μg of purified OASS. The positions to which the molecular mass markers migrated are shown in kilodaltons at the left.
Physical properties.
A plot of activity versus the assay temperature revealed a broad optimum between 40 and 60°C (data not shown). Significant activity was detected up to 80°C. These results are compatible with the optimum growth temperature of M. thermophila (55°C). The N-terminal sequence of the purified OASS from M. thermophila (Fig. 2) shows significant identity and similarity to OASS enzymes from a variety of plants and procaryotes from the Bacteria domain. The identity extends to residues that are perfectly conserved among all these enzymes.
FIG. 2.
N-terminal amino acid sequence alignment of select OASS enzymes. Completely conserved amino acids are indicated in boldface, partially conserved amino acids with respect to the M. thermophila sequence are indicated by gray shading, and gaps introduced by sequence alignment are indicated by dashes. Numbers on the right refer to positions of the right-most residues shown with respect to the entire sequence.
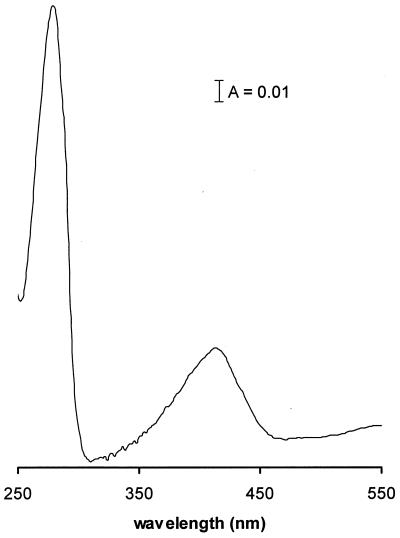
The UV-visible spectrum (Fig. 3) contained two major peaks with absorbance maxima at 278 and 413 nm, the former due to aromatic amino acids. The absorbance at 413 nm is a property of all OASS enzymes studied in plants and in procaryotes from the Bacteria domain. The absorbance is attributed to the aldoxime form of pyridoxal 5′-phosphate (10), produced when a Schiff base is formed between the cofactor and an ɛ-amino group of a lysine residue in the protein. The OASS purified from M. thermophila has an A280/A413 ratio of 4.0, which compares favorably to the ratio found for OASS from Salmonella serovar Typhimurium of 3.5 (2). Incubation of OASS with a 1:10 molar ratio of enzyme to pyridoxal 5′-phosphate did not significantly increase activity (data not shown), suggesting that the purified enzyme had a full complement of this cofactor.
FIG. 3.
UV-visible absorption spectrum of OASS purified from M. thermophila. The enzyme (0.13 mg/ml) was in 50 mM Tris-KOH buffer (pH 6.8) containing 350 mM KCl.
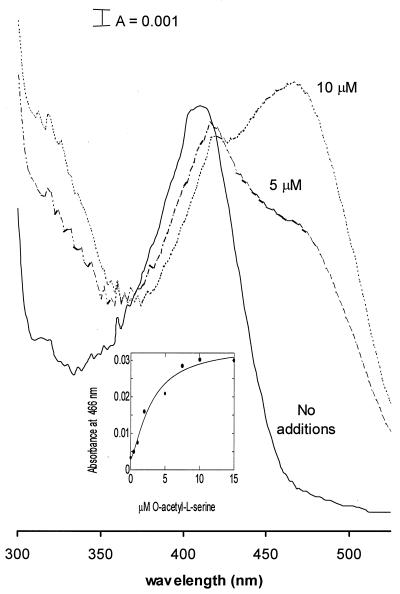
A marked change in the spectrum, attributed to pyridoxal 5′-phosphate, occurs upon addition of O-acetyl-l-serine. Titration with increasing amounts of O-acetyl-l-serine produced a shift in absorbance from 413 to 466 nm (Fig. 4) and formation of a broad shoulder at 330 nm (Fig. 4). The increase in absorbance at 466 nm was concentration dependent and was saturated at 10 μM O-acetyl-l-serine (Fig. 4, inset). A similar shift in absorbance was observed by Schnackerz et al. (41) upon binding of d-serine to d-serine dehydratase. The species responsible for this shift in absorbance is the α-aminoacrylic acid in the Schiff base with pyridoxal 5′-phosphate. The formation of this α-aminoacrylate intermediate in the OASS from M. thermophila was reversible upon addition of sulfide. The original spectrum with its absorption at 413 nm was regenerated upon addition of 1 mM sodium sulfide, consistent with reaction of this second substrate with the intermediate and transfer of the acetyl group of O-acetyl-l-serine to sulfide, forming l-cysteine (10). The observed spectral changes upon addition of O-acetyl-l-serine and reversion upon addition of sulfide demonstrate an active role of pyridoxal 5′-phosphate in the reaction mechanism of the OASS from M. thermophila.
FIG. 4.
Spectroscopic titration of OASS purified from M. thermophila with O-acetyl-l-serine. Protein concentration was 0.10 mg/ml in 50 mM Tris-KOH buffer (pH 6.8), containing 350 mM KCl. O-Acetyl-l-serine additions were made in 2 to 5-μl increments in a total volume of 1.0 ml. The amount of O-acetyl-l-serine for each spectrum is shown. (Inset) Absorption of enzyme at 466 nm versus amount of O-acetyl-l-serine added.
Isoelectric focusing determined that the pI of M. thermophila OASS is 5.0 ± 0.5, similar to the calculated pI values of the spinach OASS isoenzymes, which range from 5.0 to 6.0 (40). No data are available for any of the procaryotic enzymes. SDS-PAGE analysis of the purified M. thermophila OASS revealed a single band at 36 kDa (Fig. 1). The native molecular mass of OASS was determined to be 93 kDa by gel filtration chromatography (data not shown). This is the largest native molecular mass reported for any OASS, the next closest being an OASS from the plant Datura innoxia, with a native molecular mass of 86 kDa (23). The SDS-PAGE and native gel chromatography results suggest that the OASS purified from M. thermophila is either a homodimer or a homotrimer. All other OASS enzymes purified to date are homodimeric with native molecular masses ranging from 52 to 72 kDa (2, 4, 11, 12, 16, 17, 22, 25, 29, 30, 33, 36, 38, 43, 48, 49), the only exception being a heterodimeric OASS from D. innoxia (23).
Kinetic analyses.
Plots of increasing concentrations of O-acetyl-l-serine versus the initial velocity of the reaction were sigmoidal (Fig. 5B), a result suggesting the OASS purified from M. thermophila displays positive kinetic cooperativity at low concentrations of this substrate. Similar positive cooperativity has been noted in some plant enzymes (23, 37) and in Salmonella serovar Typhimurium when OASS type B (CysM) is bound to serine transacetylase the first enzyme in the serine pathway (22). However, in Salmonella serovar Typhimurium no positive cooperativity is observed when the enzyme is not associated with serine transacetylase. The activity of the purified M. thermophila OASS was inhibited at concentrations of O-acetyl-l-serine above 10 mM (Fig. 5A). Inhibition of activity has also been reported for the OASS from Salmonella serovar Typhimurium at concentrations above 7.5 mM O-acetyl-l-serine (10) and in Phaseolus OASS at concentrations above 10 mM (4). Analyses of the data in Fig. 5B by using three different equations all gave similar fits, indicated by the similar chi square error and coefficients of determination (R2), the best fit being provided by the MMF model (Table 2). The MMF model is a generalized form of the Hill equation which allows for a nonzero intercept. No data were available close to zero; thus, it might be an error to assume that the curve is strictly sigmoidal from zero to the first data point. The Vmax and S0.5 values obtained by the logistic equation, describing a very general growth curve, were slightly lower than those obtained by the other two methods. Both models allowing for a nonzero intercept had better fits than the Hill equation. The S0.5 values obtained were similar to those found for the D. innoxia enzymes (23). The variations in Vmax are probably due to the restricted data set used to preclude portions of the curve affected by the substrate inhibition.
TABLE 2.
Kinetic constants from substrate saturation experiments with OASS purified from M. thermophila
| Model | Vmax | S0.5 (mM) | n | Chi square (10−5) | R2 |
|---|---|---|---|---|---|
| Hill | 0.0711 | 6.667 | 1.847 | 4.94 | 0.9647 |
| MMF | 0.0587 | 5.597 | 2.214 | 4.75 | 0.9661 |
| Logistic | 0.0408 | 4.111 | NAa | 4.76 | 0.9660 |
NA, not applicable.
SDS-PAGE and native gel chromatography of the OASS purified from M. thermophila suggested that the enzyme is either a homodimer or a homotrimer. Consistent with all OASS enzymes studied, it is anticipated that each subunit of the M. thermophila OASS contains an active site with one pyridoxal 5′-phosphate. The values of n determined by the Hill equation and the MMF model are also consistent with two or more active sites. Generally the Hill equation underestimates the amount of cooperativity (42), while the MMF model tends to overestimate it (23). The Hill model revealed a Kh value of 33 ± 12 mM O-acetyl-l-serine. The Kh value is the product of the two kinetic dissociation constants; however, substrate inhibition precluded an accurate determination of the individual constants.
Plots of increasing concentrations of sodium sulfide versus the initial velocity of the reaction exhibited normal Michaelis-Menten kinetics (data not shown). The Km for sulfide was 500 ± 80 μM, a value within the range of Kms reported for sulfide for other OASS enzymes that have been purified (20 to 2,700 μM) (7, 15, 36).
DISCUSSION
The genomic sequences of four phylogenetically and metabolically diverse members of the Archaea provide little insight into mechanisms of archaeal sulfur fixation. The results presented here provide the first documentation of a sulfur-fixing enzyme, OASS, in the Archaea. The M. thermophila OASS enzyme was identified through its sulfur-fixing activity, the presence of a pyridoxal 5′-phosphate cofactor, and high sequence similarity between the N terminus and those of other known OASS enzymes.
Although OASS is essential for the serine pathway of cysteine biosynthesis in plants and Bacteria, additional functions for OASS enzymes have been proposed. In Eucarya and Bacteria, OASS is involved in the recycling of released sulfur during sulfur starvation (14) and sequestering of sulfide into cysteine to prevent toxic levels in the cell (49). In the Eucarya domain, OASS enzymes can also catalyze the formation of heterocyclic β-substituted alanines from O-acetyl-l-serine and heterocyclic compounds (18). Thus, it is possible that M. thermophila OASS has additional functions, or a function unrelated to the serine pathway in the cell, and that another pathway exists for cysteine biosynthesis in the Archaea.
M. thermophila was cultured with acetate as the carbon and energy source in 10-liter fermentors to obtain the large amounts of cell material necessary for purification of OASS. The fermentors require continuous gassing; thus, in addition to volatile sulfide, the presence of cysteine is essential to maintain the reduction potential necessary for growth. The level of OASS activity in cell extracts of M. thermophila was at the lower end of the range reported for procaryotic OASS enzymes. In E. coli and Salmonella serovar Typhimurium, expression of OASS and other enzymes required for cysteine biosynthesis is maximally repressed in the presence of cysteine (21). If a similar regulation was effective in M. thermophila, then cysteine present in the growth medium would repress the synthesis of OASS to constitutive levels; however, this hypothesis could not be tested without the ability to grow M. thermophila in the absence of cysteine. An increase in the levels of OASS when cells are grown in the absence of cysteine would support a role for OASS in cysteine biosynthesis. Positive cooperativity in response to O-acetyl-l-serine may be important if OASS functions in the serine pathway and transcription of the genes encoding enzymes of this pathway are regulated by the same mechanism as proposed for E. coli and Salmonella serovar Typhimurium. In the proposed mechanism, the levels of O-acetyl-l-serine in the cell influence the expression of serine transacetylase and OASS. Inactivity of the M. thermophila OASS at low concentrations of O-acetyl-l-serine may be necessary to allow accumulation of O-acetyl-l-serine to levels required for transcription of serine transacetylase and OASS. Clearly, more research is necessary to provide evidence in support of this hypothesis and a role for OASS in cysteine biosynthesis.
The presence of open reading frames in the genomes of M. thermoautotrophicum and P. horikoshii with deduced sequences having identity to homoserine transacetylase and γ-cystathionase sequences is consistent with the homocysteine pathway for cysteine biosynthesis in these Archaea. However, M. thermoautotrophicum and P. horikoshii are classified in different taxonomic orders than the methanosarcina; thus, M. thermophila may have inherited enzymes of the serine pathway or acquired them by horizontal gene transfer from the Bacteria domain. The complete gene sequence may help to determine the evolutionary relationship to other OASS enzymes and the origin of OASS in the Archaea.
ACKNOWLEDGMENTS
This work was funded by NASA Ames Cooperative Agreement NCC21059 and Department of Energy Basic Science Grant DE-FG02-95ER20198.
We thank J. Martin Bollinger for help with the kinetic data analysis, Kevin Gutshall for help with the N-terminal sequence determination, Jonna Coombs for help with the pI determination, and Jonna Coombs, Robert Barber, and Ubolsree Leartsakuplanich for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Becker M A, Kredich N M, Tomkins G M. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J Biol Chem. 1969;244:2418–2427. [PubMed] [Google Scholar]
- 3.Bender D A. Amino acid metabolism. 2nd ed. London, England: John Wiley & Sons Ltd.; 1985. [Google Scholar]
- 4.Bertagnolli B L, Wedding R T. Purification and initial kinetic characterization of different forms of O-acetylserine sulfhydrylase from seedlings of two species of Phaseolus. Plant Physiol. 1977;60:115–121. doi: 10.1104/pp.60.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschi. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Burnell J N, Whatley F R. Sulphur metabolism in Paracoccus denitrificans purification, properties and regulation of serine transacetylase, O-acetylserine sulphydrylase and β-cystathionase. Biochim Biophys Acta. 1977;481:246–265. doi: 10.1016/0005-2744(77)90157-7. [DOI] [PubMed] [Google Scholar]
- 8.Byrne C R, Monroe R S, Ward K A, Kredich N M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J Bacteriol. 1988;170:3150–3157. doi: 10.1128/jb.170.7.3150-3157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers L A, Trudiger P A. Cysteine and S-sulfocysteine biosynthesis in bacteria. Arch Mikrobiol. 1971;77:165–184. doi: 10.1007/BF00408609. [DOI] [PubMed] [Google Scholar]
- 10.Cook P F, Wedding R T. A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J Biol Chem. 1976;251:2023–2029. [PubMed] [Google Scholar]
- 11.Droux M, Martin J, Sajus P, Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch Biochem Biophys. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- 12.Garvis S G, Tipton S L, Konkel M E. Identification of a functional homolog of the Escherichia coli and Salmonella typhimurium cysM gene encoding O-acetylserine sulfhydrylase B in Campylobacter jejuni. Gene. 1997;185:63–67. doi: 10.1016/s0378-1119(96)00631-2. [DOI] [PubMed] [Google Scholar]
- 13.Giovanelli J, Mudd H S, Dakto A H. Sulfur amino acids in plants. In: Miflin B J, editor. The biochemistry of plants. Vol. 5. New York, N.Y: Academic Press; 1980. pp. 453–505. [Google Scholar]
- 14.Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R. Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Lett. 1994;351:257–262. doi: 10.1016/0014-5793(94)00872-8. [DOI] [PubMed] [Google Scholar]
- 15.Hensel G, Trueper H G. Cysteine and S-sulfocysteine biosynthesis in phototrophic bacteria. Arch Microbiol. 1976;109:101–103. doi: 10.1007/BF00425119. [DOI] [PubMed] [Google Scholar]
- 16.Ikegami F, Kaneko M, Kamiyama H, Murakoshi I. Purification and characterization of cysteine synthase from Citrullus vulgaris. Phytochemistry. 1988;27:697–701. [Google Scholar]
- 17.Ikegami F, Kaneko M, Lambein F, Kuo Y, Murakoshi I. Difference between uracilylalanine synthases and cysteine synthases in Pisum sativum. Phytochemistry. 1987;26:2699–2704. [Google Scholar]
- 18.Ikegami F, Murakoshi I. Enzymic synthesis of non-protein β-substituted alanines and some higher homologues in plants. Phytochemistry. 1994;35:1089–1104. [Google Scholar]
- 19.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 20.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 21.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Ingraham J C, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 419–428. [Google Scholar]
- 22.Kredich N M, Becker M A, Tomkins G M. Purification and characterization of cysteine synthase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- 23.Kuske C R, Ticknor L O, Guzman E, Gurley L R, Valdez J G, Thompson M E, Jackson P J. Purification and characterization of O-acetylserine sulfhydrylase isoenzymes from Datura innoxia. J Biol Chem. 1994;269:6233–6232. [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Leon J, Romero L C, Galvan F, Vega J M. Purification and physicochemical characterization of O-acetyl-l-serine sulfhydrylase from Chlamydomonas reinhardtii. Plant Sci. 1987;53:93–99. [Google Scholar]
- 26.Marzluf G A. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- 27.Morgan P H, Mercer L P, Flodin N W. General model for nutritional responses of higher organisms. Proc Natl Acad Sci USA. 1975;72:4327–4331. doi: 10.1073/pnas.72.11.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morzycka E, Paszewski A. Two pathways of cysteine biosynthesis in Saccharomycopsis lipolytica. FEBS Lett. 1979;101:97–100. doi: 10.1016/0014-5793(79)81302-2. [DOI] [PubMed] [Google Scholar]
- 29.Mueller R, Kuttler E, Lanz C, Drewke C, Schmidt K. Isolation of a gene encoding cysteine synthase from Flavobacterium K3-15. FEMS Microbiol Lett. 1996;136:305–308. doi: 10.1111/j.1574-6968.1996.tb08065.x. [DOI] [PubMed] [Google Scholar]
- 30.Murakoshi I, Ikegami F, Kaneko M. Purification and properties of cysteine synthase from Spinacia oleracea. Phytochemistry. 1985;24:1907–1911. [Google Scholar]
- 31.Nicholson M L, Gaasenbeek M, Laudenbach D E. Two enzymes together capable of cysteine biosynthesis are encoded on a cyanobacterial plasmid. Mol Gen Genet. 1995;247:623–632. doi: 10.1007/BF00290354. [DOI] [PubMed] [Google Scholar]
- 32.Noji M, Murakoshi I, Saito K. Molecular cloning of a cysteine synthase cDNA from Citrullus vulgaris (watermelon) by genetic complementation in an Escherichia coli Cys− auxotroph. Mol Gen Genet. 1994;244:57–66. doi: 10.1007/BF00280187. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki T, Asai T, Kobayashi S, Ikegami F, Noji M, Saito K, Takeuchi T. Molecular cloning and characterization of the genes encoding two isoforms of cysteine synthase in the enteric protozoan parasite Entamoeba histolytica. Mol Biochem Parasitol. 1998;97:33–44. doi: 10.1016/s0166-6851(98)00129-7. [DOI] [PubMed] [Google Scholar]
- 34.Ratkowsky D A. Non-linear regression modeling. New York, N.Y: Marcel Dekker, Inc.; 1983. [Google Scholar]
- 35.Ratkowsky D A. A suitable parameterization of the Michaelis-Menten enzyme reaction. Biochem J. 1986;240:357–360. doi: 10.1042/bj2400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roemer S, d'Harlingue A, Camara B, Schantz R, Kuntz M. Cysteine synthase from Capsicum annuum chromoplasts. J Biol Chem. 1992;267:17966–17970. [PubMed] [Google Scholar]
- 37.Rolland N, Ruffet M L, Job D, Douce R, Droux M. Spinach chloroplast O-acetylserine (thiol)-lyase exhibits two catalytically non-equivalent pyridoxal-5′-phosphate-containing active sites. Eur J Biochem. 1996;236:272–282. doi: 10.1111/j.1432-1033.1996.00272.x. [DOI] [PubMed] [Google Scholar]
- 38.Saito K, Miura N, Yamazaki M, Hirano H, Murakoshi I. Molecular cloning and bacterial expression of cDNA encoding a plant cysteine synthase. Proc Natl Acad Sci USA. 1992;89:8078–8082. doi: 10.1073/pnas.89.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito K, Tatsuguchi K, Murakoshi I, Hirano H. cDNA cloning and expression of cysteine synthase B localized in chloroplasts of Spinacia oleracea. FEBS Lett. 1993;324:247–252. doi: 10.1016/0014-5793(93)80127-g. [DOI] [PubMed] [Google Scholar]
- 40.Saito K, Tatsuguchi K, Takagi Y, Murakoshi I. Isolation and characterization of cDNA that encodes a putative mitochondrion-localizing isoform of cysteine synthase (O-acetylserine (thiol)-lyase) from Spinacia oleracea. J Biol Chem. 1994;269:28187–28192. [PubMed] [Google Scholar]
- 41.Schnackerz K D, Ehrlich J H, Giesemann W, Reed T A. Mechanism of action of d-serine dehydratase. Identification of a transient intermediate. Biochemistry. 1979;18:3557–3563. doi: 10.1021/bi00583a019. [DOI] [PubMed] [Google Scholar]
- 42.Segel I H. Enzyme kinetics, behavior and analysis of rapid equilibrium and steady-state systems. New York, N.Y: John Wiley & Sons; 1975. [Google Scholar]
- 43.Sirko A, Hryniewicz M, Hulanicka D, Boeck A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of cysTWAM gene cluster. J Bacteriol. 1990;172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 46.Sowers K R, Nelson M J, Ferry J G. Growth of acetotrophic, methane-producing bacteria in a pH auxostat. Curr Microbiol. 1984;11:227–230. [Google Scholar]
- 47.Topczewski J, Sienko M, Paszewski A. Cloning and characterization of the Aspergillus nidulans cysB gene encoding cysteine synthase. Curr Genet. 1997;31:348–356. doi: 10.1007/s002940050215. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi T, Zhu X, Masada M. Purification and characterization of a novel cysteine synthase isozyme from spinach hydrated seeds. Biosci Biotechnol Biochem. 1998;62:501–507. doi: 10.1271/bbb.62.501. [DOI] [PubMed] [Google Scholar]
- 49.Youssefian S, Nakamura M, Sano H. Tobacco plants transformed with the O-acetylserine (thiol) lyase gene of wheat are resistant to toxic levels of hydrogen sulfide gas. Plant J. 1993;4:759–769. doi: 10.1046/j.1365-313x.1993.04050759.x. [DOI] [PubMed] [Google Scholar]