Abstract
Whole-genome sequencing (WGS) is being applied increasingly to Bacillus cereus group species; however, misinterpretation of WGS results may have severe consequences. We report 3 cases, 1 of which was an outbreak, in which misinterpretation of B. cereus group WGS results hindered communication within public health and industrial laboratories.
Keywords: anthrax, Bacillus anthracis, Bacillus cereus, Bacillus paranthracis, Bacillus tropicus, bacteria, foodborne diseases, bioterrorism and preparedness, whole-genome sequencing, taxonomy, food safety
The Bacillus cereus group of bacteria is a complex of closely related species with varying pathogenic potential (1). Notable members, as defined in the US Food and Drug Administration’s Bacteriological Analytical Manual, include B. anthracis, an etiologic agent of anthrax, and B. cereus, which has been associated with numerous illnesses, including foodborne emetic intoxication, diarrheal toxicoinfection, anthrax-like illness, and nongastrointestinal infections (1–5).
Whole-genome sequencing (WGS) is used increasingly in clinical and industrial microbiology laboratories to characterize B. cereus group strains (6). However, interpreting WGS results from these organisms is challenging; insights derived from WGS may conflict with information provided by traditional microbiologic assays (6–8). Previously, we hypothesized that results from some WGS-based species classification methods can be easily misinterpreted when applied to the B. cereus group (7). Here, we show that this scenario is not hypothetical: we report 3 recent cases among public health and industrial laboratories in which misinterpretation of WGS results directly hindered public health and food safety investigations or responses.
The Study
We report 3 cases of WGS-based B. cereus group species assignment misinterpretations in 3 continents: Europe, North America, and Africa (Table 1; Appendix). Two cases (cases 2 and 3) occurred within the last 6 months and involved regional and national public health laboratories; 1 case (case 1) involved an industrial laboratory (Table 1). All cases involved strains known colloquially as group III B. cereus, a phylogenetic lineage within the B. cereus group that was identified using pantoate-β-alanine ligase gene (panC) sequencing (9).
Table 1. Summary of cases of laboratory misidentifications caused by taxonomic changes to Bacillus cereus group species, 2018–2022*.
| Case | Date | Location | Inquiring party | WGS-assigned species of inquiry | Case summary† |
|---|---|---|---|---|---|
| 1 |
November 2018 |
Europe |
Industrial laboratory |
B. anthracis
|
Two B. cereus strains isolated from a food processing facility were assigned to the B. anthracis species but were not closely related to the B. anthracis lineage most commonly responsible for anthrax illness and did not possess anthrax encoding genes or represent an anthrax threat. They would historically be referred to as B. cereus or group III B. cereus. |
| 2 |
October 2021 |
North America (USA) |
Government laboratory |
B. paranthracis
|
A B. cereus strain isolated from a food product responsible for a foodborne outbreak was assigned to the B. paranthracis species using WGS-based methods. B. paranthracis was historically referred to as B. cereus or group III B. cereus and encompasses B. cereus group strains capable of causing emetic and/or diarrheal foodborne illness. |
| 3 | January 2022 | Africa (South Africa) | Government laboratory | B. anthracis | Two B. cereus strains isolated during routine surveillance of meat products were classified using multiple WGS-based methods; they were assigned to the B. anthracis species but did not represent an anthrax threat. They would historically be referred to as B. cereus or group III B. cereus. |
*WGS, whole genome sequencing; ANI, average nucleotide identity; MLST, multilocus sequence typing; ST, sequence type; Group III, panC phylogenetic Group III; PubMLST, https://pubmlst.org; GTDB, Genome Taxonomy Database Releases R95 and R202, https://gtdb.ecogenomic.org. †B. cereus refers to the historical and/or colloquial species definition assigned using traditional microbiological methods, as outlined in the US Food and Drug Administration’s Bacteriological Analytical Manual (FDA BAM) (5).
Case 1 occurred in November 2018 at an industrial microbiology laboratory in Europe (Table 1). The inquiring party isolated B. cereus group strains from a food processing facility, then characterized them by WGS (protocols unknown). Each B. cereus group strain was assigned to a species by comparing its genome to the genomes of all B. cereus group species type strains and identifying the most similar species type strain by average nucleotide identity (7). Two strains were classified as B. anthracis using this approach (Table 1), and the inquiring party was concerned that the strains represented a potential anthrax threat (because of their B. anthracis label). However, subsequent investigation by M.W. and L.M.C. revealed that, although the strains most closely resembled B. anthracis, neither strain belonged to the historical, clonal B. anthracis lineage typically associated with anthrax toxin production (6), and neither strain possessed anthrax toxin- or capsule-encoding genes (Table 1; Figure 1). Thus, the strains were deemed incapable of causing anthrax, despite being assigned to B. anthracis by WGS. The authors noted that historically, these strains would be known colloquially as group III B. cereus, using microbiologic methods and panC phylogenetic group assignment (Figure 2, panel A).
Figure 1.
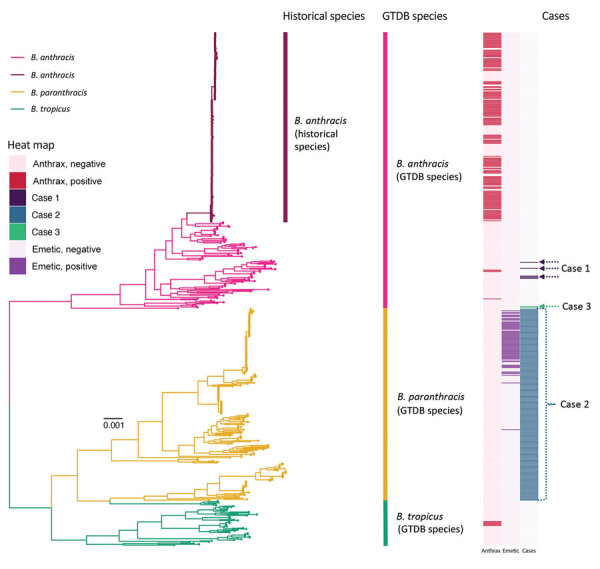
Maximum-likelihood phylogeny constructed using core genes detected among 605 genomes assigned to the GTDB B. anthracis, B. paranthracis, and B. tropicus species. Branch colors and clade labels denote GTDB species assignments or, for B. anthracis, historical species assignments. The heatmap to the right of the phylogeny shows whether a strain possessed anthrax toxin-encoding genes or not (anthrax); whether a strain possessed cereulide synthetase (emetic toxin)-encoding genes or not (emetic); and the strains or lineages discussed in the cases we detailed here (cases) (Table 1). For case 1, the actual genomes were not publicly available; thus, genomes assigned to the same sequence types (STs, via 7-gene multilocus sequence typing) are highlighted. For case 2, the only information provided to the authors was that the genome in question belonged to species B. paranthracis; thus, all genomes assigned to GTDB’s B. paranthracis species are highlighted. For case 3, the actual strain genomes associated with the case are highlighted. The phylogeny was rooted using panC Group II B. cereus group strain FSL W8-0169 as an outgroup (National Center for Biotechnology Information RefSeq Assembly accession no. GCF_001583695.1; omitted for readability). Branch lengths are reported in substitutions per site. GTDB, Genome Taxonomy Database.
Figure 2.
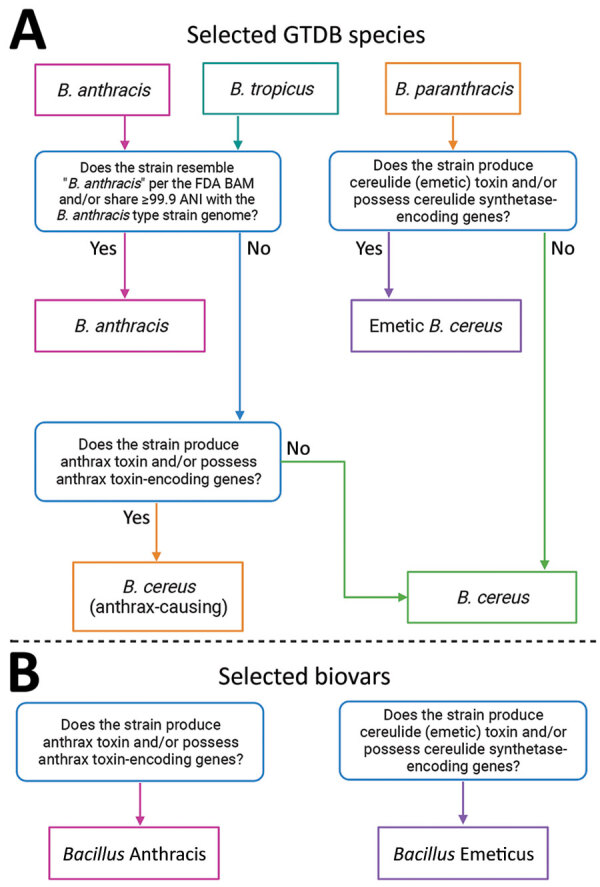
Flowcharts of Bacillus species and biovar name assignments. A) Flowchart depicting how 3 Bacillus species names assigned using GTDB releases R95 and R202 can be translated to historically important or colloquial names for B. cereus group species, as outlined in the US FDA’s BAM (5). B) Chart depicting how anthrax and cereulide (emetic) toxin-producing strains can be referred to using a previously proposed standardized collection of B. cereus group biovar terms (6). Figure was created using BioRender.com. ANI, average nucleotide identity; BAM, Bacteriological Analytical Manual; FDA, Food and Drug Administration; GTDB, Genome Taxonomy Database.
Case 2 occurred in October 2021 at a regional public health microbiology laboratory in the United States (Table 1). The inquiring party was responding to a foodborne outbreak that occurred at a correctional facility in Maryland in mid-September 2021. During the outbreak investigation, a B. cereus group strain was isolated from rehydrated dehydrated potatoes using standard protocols (5). The strain underwent WGS and was classified as B. paranthracis (protocols unknown) (Table 1). The inquiring party had never heard of B. paranthracis before and conducted a literature search, noting that the species was first described in 2017 (10); because of limited documented history of B. paranthracis, the inquiring party contacted M.W., J.K., and L.M.C. for assistance. We informed the inquiring party that B. paranthracis has historically been identified as group III B. cereus on the basis of microbiologic methods and panC phylogenetic group assignment. We also noted that B. paranthracis encompasses all strains known colloquially as emetic B. cereus (for their ability to produce cereulide, an emetic toxin) and some group III B. cereus strains capable of causing diarrheal foodborne illness (Figure 2, panel A) (11). We suggested that the inquiring party use multilocus sequence typing and virulence factor detection to determine if the strain belonged to a lineage previously associated with foodborne illness.
Case 3 occurred in January 2022 at a national veterinary public health microbiology laboratory in South Africa (Table 1). I.M. and collaborators isolated B. cereus group strains during routine surveillance of meat products (12). WGS was conducted on some strains (13). I.M. and L.M.C. assigned B. cereus group strains to species using multiple WGS-based methods (13); one method relied on the Genome Taxonomy Database (GTDB), a popular contemporary microbial species classification framework (14). GTDB releases R95 and R202 classified 2 strains as B. anthracis (Table 1); however, neither strain belonged to the historical, clonal B. anthracis lineage (6), and neither possessed anthrax toxin- or capsule-encoding genes (Table 1; Figure 1). Nevertheless, an inquiring party was concerned that the strains represented an anthrax threat because of the GTDB B. anthracis label (Table 1). We informed the inquiring party that neither possessed anthrax toxin-encoding genes. We noted that historically these strains would be known as group III B. cereus, using microbiologic methods and panC phylogenetic group assignment (Figure 2, panel A).
Conclusions
The growing popularity of WGS offers tremendous potential for improving B. cereus group surveillance, source tracking, and outbreak investigations. However, taxonomic issues in the B. cereus group have become more pronounced as researchers grapple with historical and WGS-based species definitions.
Here, we detailed 3 cases in which misinterpretation of B. cereus group WGS results directly hindered public health and food safety efforts. Two cases (cases 1 and 3) represented false-positive scenarios, in which group III B. cereus strains incapable of causing anthrax were incorrectly assumed to be anthrax-causing agents (Table 1). As noted previously, strains that lack anthrax toxin-encoding genes but are assigned to B. anthracis using WGS-based methods are not uncommon (Table 2); these strains have been isolated from diverse environments (e.g., meat, milk, spices, egg whites, baby wipes) on 6 continents and the International Space Station, and although some may cause illness, they cannot cause anthrax (6). One way of denoting that a B. cereus group strain may produce anthrax toxin is to append the term “biovar Anthracis” to the genus/species name (Figure 2, panel B) (2).
Table 2. Selected GTDB Bacillus species names and the clinically important strains they encompass*.
| GTDB species name | Encompasses strains which: |
Notes† | ||
|---|---|---|---|---|
| Can cause anthrax illness | Can cause emetic illness | Cannot cause anthrax or emetic illness | ||
|
B. anthracis
|
Yes |
No |
Yes |
Encompasses all anthrax-causing B. anthracis strains, some anthrax-causing B. cereus strains, and many B. cereus strains that are incapable of causing anthrax illness but are commonly isolated from environmental and food sources (6,7). |
|
B. paranthracis
|
No |
Yes |
Yes |
Encompasses all cereulide-producing B. cereus strains known colloquially as emetic B. cereus, including the high-risk ST26 lineage; also encompasses many environmental and food isolates that are incapable of causing emetic illness (7,11). |
| B. tropicus | Yes | No | Yes | Encompasses some anthrax-causing B. cereus strains, as well as B. cereus strains that are incapable of causing anthrax illness (6,7). |
*Obtained using GTDB Releases R95 and R202, but is applicable to any taxonomic framework, in which species names are assigned relative to B. cereus group species type strain genomes, e.g., by a species threshold of 95–96 average nucleotide identity or species threshold of 70% in silico DNA-DNA hybridization (7). GTDB, Genome Taxonomy Database; ST, sequence type assigned using the PubMLST 7-gene multilocus sequence typing scheme for B. cereus (https://pubmlst.org). †B. anthracis and B. cereus refer to historical and/or colloquial species definitions assigned using traditional microbiological methods, as outlined in the US Food and Drug Administration’s Bacteriological Analytical Manual (5).
The remaining case (case 2) represented a worst-case, false-negative scenario, in which a WGS-assigned species label with limited clinical interpretability or previous associations to foodborne illness (B. paranthracis) was assigned to an established pathogen (group III B. cereus) and directly hindered an outbreak investigation (Table 1). We anticipate that similar problems may arise with anthrax-causing B. cereus, because WGS-based methods assign some of these strains to B. tropicus, a species proposed in 2017 (Table 2) (7).We encourage readers to be mindful of this potential issue (Table 2). Overall, we hope that the cases we described can serve as cautionary tales for those who are transitioning to WGS for B. cereus group strain characterization.
Additional information about misidentifications resulting from taxonomic changes to Bacillus cereus group species, 2018–2022
Acknowledgments
This work was supported by United States Department of Agriculture National Institute of Food and Agriculture (USDA NIFA) Hatch Appropriations (project no. PEN04646; accession no. 1015787) and USDA NIFA grants (no. GRANT12686965 and 2019-67017-29591).
Biography
Dr. Carroll is a computational biologist at the European Molecular Biology Laboratory in Heidelberg, Germany. Her research focuses on developing and utilizing bioinformatic approaches to study the evolution and transmission dynamics of foodborne and zoonotic pathogens. She has specific expertise in the genomics and taxonomy of the Bacillus cereus group.
Footnotes
Suggested citation for this article: Carroll LM, Matle I, Kovac J, Cheng RA, Wiedmann M. Laboratory misidentifications resulting from taxonomic changes to Bacillus cereus group species, 2018–2022. Emerg Infect Dis. 2022 Sep [date cited]. https://doi.org/10.3201/eid2809.220293
References
- 1.Stenfors Arnesen LP, Fagerlund A, Granum PE. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev. 2008;32:579–606. 10.1111/j.1574-6976.2008.00112.x [DOI] [PubMed] [Google Scholar]
- 2.Baldwin VM. You can’t B. cereus—a review of Bacillus cereus strains that cause anthrax-like disease. Front Microbiol. 2020;11:1731. 10.3389/fmicb.2020.01731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottone EJ. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010;23:382–98. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jovanovic J, Ornelis VFM, Madder A, Rajkovic A. Bacillus cereus food intoxication and toxicoinfection. Compr Rev Food Sci Food Saf. 2021;20:3719–61. 10.1111/1541-4337.12785 [DOI] [PubMed] [Google Scholar]
- 5.Tallent SM, Knolhoff A, Rhodehamel EJ, Harmon SM, Bennett RW. Bacillus cereus. In: Bacteriological analytical manual (BAM). 8th ed. Silver Spring (MD): Food and Drug Administration; 2019. [Google Scholar]
- 6.Carroll LM, Wiedmann M, Kovac J. Proposal of a taxonomic nomenclature for the Bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. MBio. 2020;11:e00034–20. 10.1128/mBio.00034-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll LM, Cheng RA, Wiedmann M, Kovac J. Keeping up with the Bacillus cereus group: taxonomy through the genomics era and beyond. Crit Rev Food Sci Nutr. 2021;•••:1–26. 10.1080/10408398.2021.1916735 [DOI] [PubMed] [Google Scholar]
- 8.Balloux F, Brønstad Brynildsrud O, van Dorp L, Shaw LP, Chen H, Harris KA, et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26:1035–48. 10.1016/j.tim.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guinebretière MH, Velge P, Couvert O, Carlin F, Debuyser ML, Nguyen-The C. Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J Clin Microbiol. 2010;48:3388–91. 10.1128/JCM.00921-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Du J, Lai Q, Zeng R, Ye D, Xu J, et al. Proposal of nine novel species of the Bacillus cereus group. Int J Syst Evol Microbiol. 2017;67:2499–508. 10.1099/ijsem.0.001821 [DOI] [PubMed] [Google Scholar]
- 11.Carroll LM, Wiedmann M. Cereulide synthetase acquisition and loss events within the evolutionary history of group III Bacillus cereus sensu lato facilitate the transition between emetic and diarrheal foodborne pathogens. MBio. 2020;11:e01263–20. 10.1128/mBio.01263-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madoroba E, Magwedere K, Chaora NS, Matle I, Muchadeyi F, Mathole MA, et al. Microbial communities of meat and meat products: an exploratory analysis of the product quality and safety at selected enterprises in South Africa. Microorganisms. 2021;9:507. 10.3390/microorganisms9030507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll LM, Pierneef R, Mathole A, Atanda A, Matle I. Genomic surveillance of Bacillus cereus sensu lato strains isolated from meat and poultry products in South Africa enables inter- and intra-national surveillance and source tracking. Microbiol Spectr. 2022;10:e0070022. 10.1128/spectrum.00700-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;•••:btz848. 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information about misidentifications resulting from taxonomic changes to Bacillus cereus group species, 2018–2022


