Abstract
Regulation of toxin production in the gram-positive anaerobe Clostridium perfringens occurs at the level of transcription and involves a two-component signal transduction system. The sensor histidine kinase is encoded by the virS gene, while its cognate response regulator is encoded by the virR gene. We have constructed a VirR expression plasmid in Escherichia coli and purified the resultant His-tagged VirR protein. Gel mobility shift assays demonstrated that VirR binds to the region upstream of the pfoA gene, which encodes perfringolysin O, but not to regions located upstream of the VirR-regulated plc, colA, and pfoR genes, which encode alpha-toxin, collagenase, and a putative pfoA regulator, respectively. The VirR binding site was shown by DNase I footprinting to be a 52-bp core sequence situated immediately upstream of the pfoA promoter. When this region was deleted, VirR was no longer able to bind to the pfoA promoter. The binding site was further localized to two imperfect direct repeats (CCCAGTTNTNCAC) by site-directed mutagenesis. Binding and protection analysis of these mutants indicated that VirR had the ability to bind independently to the two repeated sequences. Based on these observations it is postulated that the VirR positively regulates the synthesis of perfringolysin O by binding directly to a region located immediately upstream of the pfoA promoter and activating transcription.
Bacteria use two-component signal transduction systems to respond to changes in environmental factors such as nutrient availability, temperature, osmolarity, pH, and oxygen tension (1). These regulatory networks usually consist of a membrane-associated sensor histidine kinase and its cognate response regulator, which communicate by a phosphorelay cascade that generally leads to the modulation of gene expression (56). The majority of two-component signal transduction systems have been identified in prokaryotes, but they have also been found in some eukaryotes (21). The phosphorelay process usually involves the detection of a specific environmental stimulus, which induces the autophosphorylation of the sensor kinase at a conserved histidine residue located in the cytoplasmic C-terminal region (38). This domain also contains conserved motifs that are involved in ATP binding and kinase activity (40). Once phosphorylated, it can act as phosphodonor for its cognate cytoplasmic response regulator. The N-terminal domain of the response regulator catalyzes the transfer of the phosphoryl group from the histidine residue to a conserved aspartate residue, which in essence activates the response regulator so that it is able to bind to its target DNA and subsequently modulate gene expression (38).
Clostridium perfringens is a gram positive anaerobic bacterium that is the causative agent of gas gangrene, or clostridial myonecrosis, and is characterized by its ability to produce many extracellular toxins and enzymes (44). Of these toxins, alpha-toxin (phospholipase C) and perfringolysin O (theta-toxin) have been implicated in gas gangrene (2, 13, 54). The production of these toxins and collagenase (kappa-toxin), protease, and sialidase has been shown to be regulated by a two-component signal transduction system that comprises the VirS sensor histidine kinase and the VirR response regulator, which are encoded by the virS and virR genes, respectively (27, 48). Mutation or inactivation of either virR or virS alters the ability to produce the various toxins (3, 27, 48). It has been proposed that when the transmembrane region of VirS detects an as yet unidentified environmental or growth phase stimulus, it autophosphorylates at His-255 and then acts as a phosphodonor for the phosphorylation of Asp-57 of VirR. The phosphorylated VirR protein positively regulates the transcription of its target genes either directly or by activating the transcription of other regulatory genes (3, 27, 43).
Comparison of the putative amino acid sequence of VirR with those of other response regulators revealed significant sequence similarity in the N-terminal region (27), in particular two conserved aspartate residues and conserved lysine and glutamate residues, all of which are proposed to form the catalytic domain where phosphorylation occurs (4, 53, 55, 58). Studies with other response regulators have shown that they often activate transcription by binding to a promoter region upstream of the target gene. However, unlike those of many response regulators, the C-terminal domain of VirR does not contain any DNA binding motifs such as a helix-turn-helix motif (38, 39) or a helix-loop-helix domain (29). Consensus binding sequences have been identified upstream of the genes regulated by other response regulators such as OmpR (15, 16, 41), PhoP (23, 24), and AlgR (35, 36). However, no common nucleotide sequences that could act as consensus binding sites were identified upstream of the VirR-regulated genes (3).
The objectives of this study were to determine if VirR could bind to the promoter regions of its target genes and if so to identify its precise binding sites. The initial target regions that were tested were located upstream of the VirR-regulated plc, colA, pfoR, and pfoA genes, which, respectively, encode alpha-toxin, collagenase, the putative regulatory protein PfoR, and perfringolysin O. The results show that VirR binds to two imperfect direct repeats located upstream of the pfoA gene.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
All bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultured at 37°C in 2× YT broth or agar medium or SOC broth (46) supplemented with ampicillin (100 μg ml−1).
TABLE 1.
Relevant strains and plasmid used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | Bethesda Research Laboratories |
| BL21(DE3)(pLysS) | F−ompT hsdSB(rB− mB−) gal dcm (DE3)(pLysS) | Novagen |
| Plasmids | ||
| pRSET A | pUC derived expression vector; N-terminal 6-His tag; Ampr; 2.9 kb | Invitrogen |
| pUC18 | Ampr cloning vector; 2.7 kb | 60 |
| pTox6 | pTZ18Ω(HindIII: C. perfringens, 2.7 kb)(plc+) | 45 |
| pTS302 | pUC19Ω(HindIII: C. perfringens, 4.3 kb)(pfoR+ pfoA+) | 49 |
| pKY3132 | pUC18Ω(XbaI/PstI: C. perfringens, 7.6 kb)(colA+) | 31 |
| pJIR870 | pJIR751Ω(HindIII:pJIR869, 2.7 kb)(virR+ virS+) | 27 |
| pJIR1342 | pRSET AΩ(BamHI/EcoRI: 2799/3155 PCR product, 0.711 kb)(virR+) | This study |
| pJIR1546 | pUC18Ω(SmaI: 4565/4566 PCR product, 0.278 kb) | This study |
| pJIR1781 | pUC18Ω(SmaI: 6926/5126 PCR product, 0.342 kb) (VirR binding site deleted) | This study |
| pJIR1804 | CCA→TAG change in DR1 | This study |
| pJIR1803 | CCA→TAG change in DR2 | This study |
| pJIR1821 | CCA→TAG change in DR1 and DR2 | This study |
Molecular techniques.
Plasmid DNA was routinely isolated by an alkaline lysis method (37). When used for sequencing, DNA was isolated by the modified mini-alkaline-lysis/polyethylene glycol precipitation procedure outlined in the PRISM Ready Reaction Dye Deoxy terminator cycle sequencing kit protocol (Applied Biosystems, Foster City, Calif.). Rubidium chloride-competent E. coli cells were prepared and transformed as previously described (14). Transformation of electrocompetent E. coli cells (52) was carried out with a Bio-Rad (Hercules, Calif.) Gene Pulser in 0.1-ml cuvettes under conditions outlined by the manufacturer.
PCR amplification was performed with Taq DNA polymerase (Boehringer GmbH, Mannheim, Germany) and a 0.5 μM concentration of each primer (Table 2) in a total volume of 100 μl. Reactions were carried out in a GeneAmp PCR System 2400 (Perkin-Elmer Corp., Foster City, Calif.), and the 94°C denaturation (1 min), 50°C annealing (2 min), and 72°C extension (3 min) steps were carried out for 30 cycles. The final cycle consisted of 2 min of annealing and 5 min of extension at the temperatures indicated above. All oligonucleotide primers used (Table 2) were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer.
TABLE 2.
Oligonucleotide primers
| Primer and use | Sequence (5′–3′) | Location, use, or reference |
|---|---|---|
| PCR | ||
| 2799 | AATAAGGATCCATGTTTAGTATTGCCTTAT | 5′ end of virR gene |
| 2798 | AATAAGAATTCTCATTAACATATTAAATCCCC | 3′ end of virR gene |
| 4826 | TTTGCCTTATAATTTATTTC | plc promoter region |
| 4824 | CTTTAGTTGATACCCCAGCC | plc promoter region |
| 4825 | CAAAAAATAAAAAATAATAGG | colA promoter region |
| 4823 | TAAAATAAATGCTGCTAAAG | colA promoter region |
| 3732 | AATATGAAGTGCTTAGAAAG | Upstream of pfoR |
| 2245 | TCCATGTGGTGCCTTATAACTA | Upstream of pfoR |
| 4567 | TTAAAGTTCAAAATAATAAG | Upstream of pfoA (inverted repeats) |
| 4568 | TTTGAGAAACTGAATACTGG | Upstream of pfoA (inverted repeats) |
| 4565 | GGAACTCATATTATAATTGG | pfoA promoter region |
| 4566 | TTTAAGTAAACATTTTCATC | pfoA promoter region |
| 5126 | CTCTAATTTTTTCTTTTCCC | Upstream of pfoA (promoter) |
| 5125 | GGGAAAAGAAAAAATTAGAG | Upstream of pfoA (direct repeats) |
| SOE PCR | ||
| 7352 | CTCATTCAATACTTTTGAGTTCCATTTATG | Delete VirR binding region |
| 6926 | CGCGAATTCAATGATTTAGAGGTAGAA | End of pfoR gene |
| 6927 | ATGGAACTCAAAAGTATTGAATGAGAT | Delete VirR binding region |
| 6928 | CGCGGATCCATCAGTTTTTACTTTAG | End of pfoA gene |
| DNase I footprinting | ||
| 6519 | TGGAATTGTGAGCGGATAAC | 55 bp upstream of pUC18 EcoRI site |
| Site-directed mutagenesis | ||
| 2927 | GTGCCACCTGAAGTCTAAGAAACC | 20 |
| 9781 | GTCGACTCTATAGGATCCCCG | Eliminates XbaI site of pUC18 |
| 8482 | TCGTGAATAACCTAGATTCCAATTAT | Introduces DR1 mutation |
| 8481 | TGTGCAGAACCTAGCTTTAATCG | Introduces DR2 mutation |
Nucleotide sequence analysis was carried out with a PRISM Big Dye terminator cycle sequencing Ready Reaction kit and AmpliTaq polymerase FS (Applied Biosystems) in accordance with the manufacturer's instructions. Sequencing samples were resolved and analyzed on a 373 DNA STRETCH sequencer (Applied Biosystems). Sequence analysis was performed with Sequencher 3.0 software (GeneCodes Corp., Ann Arbor, Mich.).
Construction of recombinant plasmids.
Plasmid DNA was digested with restriction endonucleases as specified by the manufacturers (Boehringer GmbH and New England Biolabs, Beverly, Mass.). Insert and vector DNA was isolated with the BRESA-CLEAN nucleic acid purification kit (Bresatec, Adelaide, Australia). When required, insert DNA and vector DNA were treated with T4 polynucleotide kinase (Promega Corp., Madison, Wis.) or alkaline phosphatase (Boehringer GmbH) as specified by the manufacturers. Vector and insert DNA was ligated with T4 DNA ligase (3 U μl−1; Promega Corp.) at 16°C overnight.
The plasmid pJIR1342 was constructed as follows to facilitate overexpression and purification of a His-tagged VirR protein. The 711-bp virR gene was amplified by PCR from pJIR870 (Table 1) with oligonucleotides 2799 and 2798 (Table 2). These primers introduced BamHI and EcoRI sites at the 5′ and 3′ ends of the PCR-generated virR gene, respectively, and enabled the amplified gene to be inserted into the BamHI and EcoRI sites of the expression vector pRSET A (Invitrogen, Carlsbad, Calif.) (Table 1) in the correct reading frame. Sequence analysis confirmed the in-frame insertion and confirmed that no mutations had been introduced by PCR.
To facilitate DNase I footprinting and site-directed mutagenesis, plasmid pJIR1546 was constructed by cloning a 278-bp PCR product containing the pfoA promoter region into the SmaI site of pUC18 (60). The PCR product was obtained by amplification with oligonucleotides 6565 and 6566 (Table 2). Similarly, pJIR1781 was constructed by cloning a 342-bp PCR product, from which the VirR binding site had been deleted, into the SmaI site of pUC18.
Expression and purification of His-tagged VirR.
E. coli BL21(DE3)(pLysS) cells (Novagen, Madison, Wis.) harboring pJIR1342 were cultured overnight at 37°C in 2× YT broth supplemented with ampicillin (100 μg ml−1) before being diluted 1 in 10 with the same medium. After incubation at 37°C for 1 h, the expression of His-tagged VirR was induced with 2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Progen, Darra Q, Australia) at 37°C for 30 min to 1 h. Cells were harvested by centrifugation at 16,300 × g for 10 min, resuspended in lysis buffer (20 mM Tris-HCl, 0.3 M NaCl, 10% glycerol, 5 mM imidazole, pH 7.9) and lysed by passage twice through a French press. Cellular debris was removed by centrifugation at 4°C as described above. The supernatant was applied to a Talon (Clontech, Palo Alto, Calif.) affinity column consisting of 1 ml of bed resin which had been previously equilibrated with lysis buffer. Proteins were allowed to bind for 1 h at 4°C while rotating, followed by three washes with 5 ml of lysis buffer. His-VirR was eluted with 5 ml of elution buffer (20 mM Tris-HCl, 0.3 M NaCl, 10% glycerol, pH 7.9) supplemented with 20, 60, 100, or 200 mM imidazole, and 1-ml fractions were collected. Samples of each fraction were mixed with gel loading buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22). The SDS–12% PAGE gels were stained with Coomassie brilliant blue essentially as described previously (46). Fractions containing highly purified VirR protein (the 20 and 60 mM imidazole fractions) were then pooled and dialyzed overnight in dialysis buffer (100 mM NaCl, 20 mM Tris [pH 7.5], 1 mM EDTA, 10% glycerol) at 4°C. The protein was concentrated by use of a Biomax-10 Ultrafree-15 centrifugal filter device (Millipore Corp., Bedford, Mass.) and stored at −70°C. Protein concentrations were determined by use of the BCA protein assay kit (Pierce, Rockford, Ill.).
Gel mobility shift assays.
PCR-generated target DNA fragments were labelled with digoxigenin-11-ddUTP (DIG) at their 3′ termini with the DIG gel shift kit (Boehringer GmbH) as described by the manufacturer. Binding reactions were carried out in a total volume of 20 μl, and reaction mixtures consisted of 4 μl of binding buffer (DIG gel shift kit), 1 μg of poly(dI-dC), 0.1 μg of poly-l-lysine, 15 fmol of DIG-labelled target DNA, 1 or 2 μg of purified VirR, and sterile distilled water. Binding reaction mixtures were incubated at room temperature for 15 min before the addition of gel loading buffer (supplied with the kit) that did not contain bromophenol blue. Reaction mixtures were then immediately loaded onto a preelectrophoresed 4% native 0.25× TBE (22.3 mM Tris, 22.3 mM boric acid, 0.5 mM EDTA, pH 8.0) polyacrylamide gel, alongside a control lane containing gel loading buffer with bromophenol blue. Samples were separated at 170 V and 4°C until the blue dye front was on the verge of running off the gel. The gel was then capillary transferred onto an Nylon+ membrane (Amersham Life Science, Buckinghamshire, United Kingdom) as described for Southern blots (46), with 0.25× TBE as the transfer buffer. Following overnight transfer at room temperature, the membrane was soaked in 10× SSC (1.5 M NaCl, 0.15 M sodium citrate, pH 7.0) and then cross-linked for 3 min at 312 nm with a Hybaid cross-linker (Integrated Sciences, Melbourne, Australia). Chemiluminescent detection of the bound probe was carried out as described by the manufacturer. The chemiluminescent signals were recorded by exposure to X-ray film (Fuji, Tokyo, Japan) at room temperature.
To quantitate the concentration dependence of VirR binding, the 183-bp pfoA target fragment was end-labelled with [α-32P]dATP by terminal transferase (Boehringer GmbH) in accordance with the manufacturer's instructions. Labelled DNA was used in gel mobility shift assays as described for DIG-labelled DNA, with the exception that the target fragment was incubated with various concentrations of VirR. Following electrophoresis, the acrylamide gel was vacuum dried and then exposed overnight to a phosphor screen (Molecular Dynamics, Sunnyvale, Calif.). Quantitative data were obtained with the STORM 690 PhosphorImager (Molecular Dynamics) with ImageQuant software (Molecular Dynamics).
DNase I footprinting.
The DNA probes were generated by PCR with oligonucleotide primers that had been end-labelled with [γ-32P]ATP by T4 polynucleotide kinase as described previously (19). For protection studies on the sense and antisense strands, PCR primers 6519 and 5126, respectively, were labelled. The amplified labelled products were isolated with the BRESA-CLEAN nucleic acid purification kit and quantitated in a Wallac 1410 scintillation counter (Wallac Oy, Turku, Finland).
Binding reactions were carried out in a total volume of 80 μl. The reaction mixtures consisted of the labelled DNA probe (25,000 cpm μl−1), 16 μl of binding buffer [100 mM HEPES (pH 7.6), 50 mM (NH4)2SO4, 5 mM 1,4-dithiothreitol, 1% (wt/vol) Tween 20, 150 mM KCl], 1 μg of poly(dI-dC), and the appropriate amount of purified VirR. Following incubation for 15 min at room temperature, MgCl2 was added to a final concentration of 4 mM prior to DNase I digestion. The target DNA in the no-VirR control was digested with 1 U of RQ1 RNase-free DNase (Promega); 2 U of RQ1 DNase was added to the reaction mixtures containing VirR. The samples were partially digested at room temperature for 1 min, and then the reaction was stopped by the addition of phenol (saturated with 1 mM Tris–0.1 mM EDTA, pH 8.0). The footprinting reaction products were then extracted and precipitated as described before (19), with the exception that the reaction products were resuspended in 3 μl of Stop solution from the T7 sequencing kit (Pharmacia Biotech, Uppsala, Sweden) and run on an 8% sequencing gel. To localize the DNase I footprint, sequencing reaction products generated by the T7 sequencing kit with primers 6519 or 5126 were run alongside the footprinting reaction products.
Deletion by SOE PCR.
Deletion of the VirR binding site in the pfoA promoter region was achieved by the splice overlap extension (SOE) PCR method (17), with a few modifications. Briefly, two separate PCR products were generated with pTS302 (Table 1) as the template and the primer pairs 6926 and 7352 and 6927 and 6928 (Table 2). These products, which contain complementary sequences, were isolated with the QIAquick gel extraction kit (Qiagen GmbH, Hilden, Germany). The purified products were then spliced together by PCR with primers 6926 and 6928; the 91°C denaturation (1 min), 37°C annealing (1 min), and 72°C extension (2 min) steps were carried out for 30 cycles. The final cycle consisted of 1 min of denaturation and annealing and 5 min of extension at the temperatures indicated above. The 2,080-bp SOE PCR product was then used as the template in a PCR with primers 6926 and 5126. The resultant 342-bp product was completely sequenced to ensure that the appropriate region had been deleted and to confirm that no other mutations had been introduced.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by the unique site elimination method (11) with a U.S.E. mutagenesis kit (Pharmacia Biotech) by a modification of the manufacturer's instructions. The second round of restriction enzyme selection was performed in a total volume of 20 μl, and the digested DNA was used to transform rubidium chloride-treated competent E. coli cells. These cells were heat shocked at 37°C for 2 min before inoculation into SOC broth. Plasmids were screened by restriction analysis, and the desired mutations were confirmed by nucleotide sequence analysis. All mutated DNA inserts were completely sequenced to confirm that no additional mutations had been introduced.
RESULTS
Overexpression and purification of His-tagged VirR.
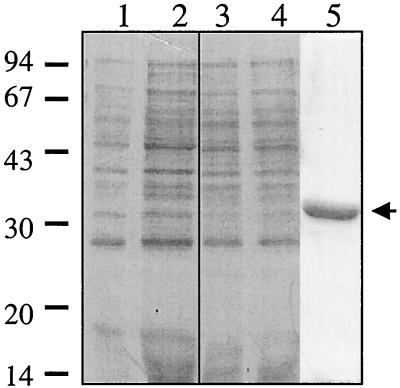
To overexpress the VirR protein, the virR gene was amplified by PCR and cloned into expression vector pRSET A. Sequence analysis of the resultant plasmid, pJIR1342, confirmed that no mutations had been introduced into the virR gene. IPTG induction in E. coli BL21(DE3)(pLysS)(pJIR1342) cells and Western blotting with an antibody specific for the fused vector-specific leader region showed that these cells produced an immunoreactive protein of the same size as the expected N-terminal fusion protein (His-VirR) (data not shown). However, even after 4 h of IPTG induction, the levels of expression were not sufficient to observe this protein in crude extracts separated by SDS-PAGE and stained with Coomassie blue (Fig. 1). Nonetheless, the presence of a six-His tag at the N terminus of the fusion protein facilitated the purification of the protein under native conditions by metal ion chelation chromatography with Talon affinity resin. Cells used in purification were induced for 30 min to 1 h, since a time course experiment demonstrated that a longer induction period resulted in significant protein degradation (data not shown). The purified protein was visualized on Coomassie-stained SDS–12% PAGE gel (Fig. 1) and was shown to migrate in accordance with its estimated molecular size of 33 kDa.
FIG. 1.
Purification of VirR. Low-molecular-weight standards (Pharmacia Biotech), in kilodaltons, are shown adjacent to the gel. Lanes 1 and 3, uninduced whole-cell extracts from cells harboring the vector pRSET A or pJIR1342, respectively; lanes 2 and 4, postinduction (4 h) whole-cell extracts from cells carrying the vector or pJIR1342, respectively; lane 5, His-VirR purified from cells induced for 30 to 60 min with IPTG (arrow).
Does VirR bind to the target gene regions?
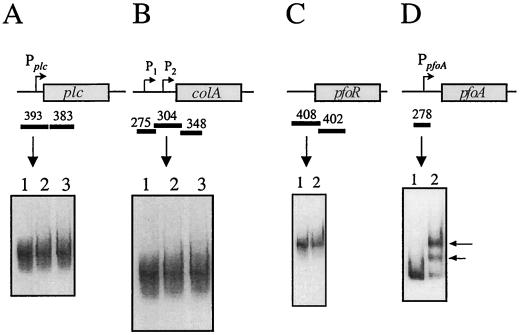
To identify possible VirR binding sites, purified VirR was used in gel mobility shift experiments with upstream promoter regions of the plc, colA, pfoA, and pfoR genes. In previous studies, the transcription of these genes was found to be affected by mutations in either virR or virS (3, 27, 49). The upstream regions were amplified by PCR (Table 2) and labelled with DIG. Each of the target fragments contained the VirR-dependent promoter of the respective gene (Fig. 2), with the exception of pfoR, since its promoter has not yet been identified.
FIG. 2.
Gel mobility shift analysis of target gene regions. Shown are the results of gel mobility shift assays using plc (A), colA (B), pfoR (C), and pfoA (D) gene regions. These regions are shown in the schematic above each gel shift result, with the rectangular boxes representing the respective genes. The direction of transcription is shown by arrows from each promoter (P). The DNA fragments used in the assays are shown by the solid bars, and their respective sizes (in base pairs) are shown above the bars. (A and B) Lane 1, no-VirR control; lanes 2 and 3, DIG-labelled target DNA incubated with 1 and 2 μg of VirR, respectively. (C) Lane 1, no-VirR control; lane 2, target DNA incubated with 1 μg of VirR. (D) Lanes 1 and 2, 278-bp fragment incubated with no protein or 1 μg of VirR, respectively. Shifted bands are indicated by the arrows.
A 393-bp fragment (from −252 to +142 with respect to the transcription start site) containing the single plc promoter (3), as well as a regulatory deoxyribosyladenine rich region that is involved in DNA bending (30, 57), was used as the target DNA in gel mobility shift experiments. The results showed that when this DNA target was incubated with different amounts of VirR, no shifts in DNA mobility were observed (Fig. 2A).
In contrast to what was found for the plc gene, two promoters have been identified upstream of the colA gene (3). Based on primer extension analysis, the promoter closer to the start of the gene, P2, was found to be virRS dependent, while the promoter located further upstream, P1, was found to be virRS independent (3). Therefore, initially a 304-bp fragment (−136 to +169) containing the P2 promoter was amplified, labelled, and used as the DNA target. Once more the results showed that VirR did not bind to this region, since no shifts in DNA mobility were observed when the target fragment was incubated with VirR (Fig. 2B).
The pfoR gene is located immediately upstream of pfoA (49). Since the pfoR promoter has not been identified, a 408-bp DNA fragment that encompassed the start of pfoR and 327 bp of sequence located upstream of the ATG start codon was used as the target (Fig. 2C). Like that of the regions upstream of the plc and colA genes, the mobility of the pfoR upstream regions did not shift in the presence of VirR (Fig. 2C). Based on these data it was concluded that VirR does not bind to the regions immediately upstream of the plc, colA, and pfoR genes. Note that with all of the target DNA fragments, the gel shift experiments were repeated in the presence of 50 mM acetyl phosphate. The same results were obtained in the presence of this low-molecular-weight phosphodonor (data not shown).
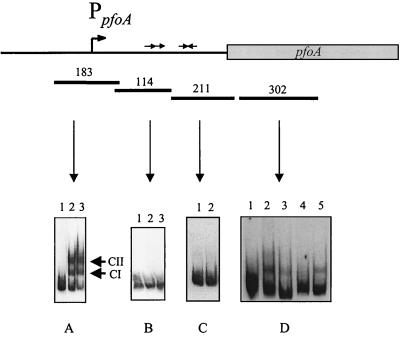
The final region of interest was upstream of the pfoA gene, which is transcribed from a major virRS-dependent promoter. This region was divided into two target fragments, a 211-bp fragment (+176 to +387) that contained two inverted repeats and a 278-bp region (−99 to +180) that contained a cluster of direct repeats as well as the pfoA promoter. When the 211-bp fragment was incubated with VirR, its mobility was found to be unaltered (Fig. 3C), providing evidence that VirR does not bind to the inverted repeats. However, when the 278-bp fragment was incubated with VirR, two bands of altered mobility were observed (Fig. 2D). This experiment provided the first evidence that VirR had the ability to bind to a region located directly upstream of one of its target genes.
FIG. 3.
Gel shift analysis of the pfoA gene region. The pfoA-derived DNA fragments used in the gel mobility shift assays (bars) and their respective sizes (in base pairs) are shown. The direct repeats in the 114-bp fragment, the promoter in the 183-bp fragment, and the inverted repeats in the 211-bp fragment, are shown as directly repeated arrows, a bent arrow (PpfoA), and inverted arrows, respectively. (A and B) Lane 1, no-VirR control; lanes 2 and 3, DIG-labelled DNA incubated with 1 and 2 μg of VirR, respectively. The VirR-DNA complexes CI and CII are indicated. (C) Lane 1, no-VirR control; lane 2, DIG-labelled fragment incubated with 1 μg of VirR. (D) Lane 1, no-VirR control; lanes 2 to 5, target DNA incubated with 1 μg of VirR. Reaction mixtures in lanes 3 to 5 also contained 15 pmol of the following unlabelled fragments: lane 3, 302-bp fragment (specific competitor); lane 4, 183-bp of unlabelled upstream pfoA fragment; lane 5, 408-bp upstream pfoR fragment (nonspecific competitor).
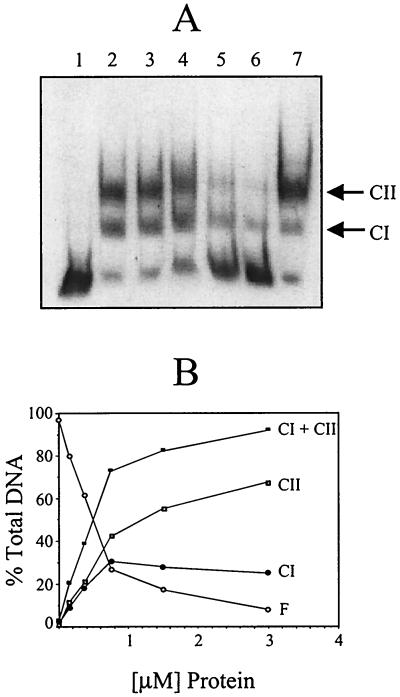
To further localize the VirR binding site and to determine whether the regulatory protein was bound to the cluster of direct repeats or to the region surrounding the promoter, the 278-bp fragment was divided into two separate, slightly overlapping target fragments, a 114-bp fragment (+66 to +180) that contained the direct repeats and a 183-bp fragment (−99 to +85) that encompassed the promoter region (Fig. 3). When these fragments were incubated with 1 or 2 μg of VirR, only the fragment containing the pfoA promoter had an altered electrophoretic mobility (Fig. 3A and B). As previously observed with the 278-bp fragment, two bands of altered mobility were observed. These bands were designated complex I (CI) and complex II (CII) (Fig. 3). These experiments demonstrated that VirR could bind to the promoter region of pfoA. Furthermore, this binding was shown to be specific, since the addition of various amounts of unlabelled 183-bp DNA fragment gradually reduced the amount of the CII band, and, to a lesser extent, the CI band (Fig. 4A). In addition, binding specificity was demonstrated by the lack of inhibition by a nonspecific competitor (the unlabelled 408-bp pfoR upstream region) when added at the same concentration as that of the specific competitor (Fig. 4A). Binding to the 183-bp fragment was dependent upon the concentration of VirR, since increasing the amount of VirR in the binding assay mixture progressively led to an increase in the amount of CI and CII, with a concomitant decrease in the amount of free unbound target DNA (Fig. 4B). As before, the addition of acetyl phosphate did not have any effect on the binding of the protein; that is, incubation in the presence of acetyl phosphate did not increase the VirR binding affinity.
FIG. 4.
Specificity and VirR dependence of DNA binding. (A) Competitive binding gel shift assay. The DIG-labelled 183-bp pfoA fragment was incubated with 1 μg of VirR and various amounts of unlabelled 183-bp DNA (specific competitor). Lane 1, no-VirR control; lanes 2 to 6, target DNA that was incubated with 1 μg of VirR. These incubation mixtures also contained 0, 3.0, 7.5, 15, and 30 pmol of specific competitor DNA, respectively. Lane 7, DIG-labelled DNA incubated with 1 μg of VirR and 30 pmol of nonspecific competitor (408-bp upstream pfoR fragment). (B) Concentration dependence of VirR binding. The [α-32P]dATP-labelled 183-bp pfoA fragment was incubated with various amounts of VirR and examined by gel shift analysis as described above except that the data were obtained and quantitated in a phosphorimager. The amount of labelled DNA in each band was calculated and plotted as shown. The amounts of free DNA (F), CI complex, CII complex, and total complex (CI+CII) are shown.
Binding of VirR was also detected with an internal 302-bp pfoA region (+436 to +737) (Fig. 3D, lane 2). However, the binding was weaker and was found to be nonspecific, since the addition of a specific competitor (the same unlabelled fragment) (Fig. 3D, lane 3) gave a binding profile similar to that obtained in the presence of the nonspecific competitor (the 408-bp upstream pfoR fragment) (Fig. 3D, lane 5). Furthermore, when the unlabelled 183-bp pfoA fragment was added at the same concentration, the shifted band was no longer observed (Fig. 3D, lane 4). Similar nonspecific binding was also found with the 383-bp internal plc gene region (+251 to +634), the 348-bp internal colA gene region (+150 to +397), the 402-bp internal pfoR gene region, and the 275-bp region (−389 to −117) encompassing the VirR-independent colA promoter, P1 (data not shown). The locations of these fragments are shown in Fig. 2. Once more, the addition of acetyl phosphate to these binding reaction mixtures did not have any effect on VirR binding.
Identification of the VirR binding site.
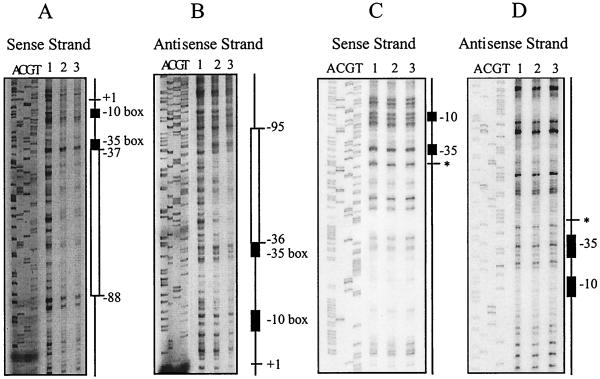
To identify the VirR binding site, DNase I footprinting was carried out on both sense and antisense strands. The 257-bp DNA target, which encompassed the pfoA promoter region, was obtained by PCR from pJIR1546 (Table 1) with primers 6519 and 5126. In separate PCR experiments, one primer was labelled with [γ-32P]ATP so that only one DNA strand was end-labelled. When these labelled DNA fragments were incubated separately with VirR and then partially digested with DNase I, a protected region was observed on both DNA strands (Fig. 5A and B). Comparison with a DNA sequencing ladder revealed that the region of protection on both strands overlapped and was located immediately upstream of the pfoA promoter (Fig. 6A). The region of overlap was termed the core binding region and was found to be approximately 52 bp in length.
FIG. 5.
DNase I footprinting analysis. Results of footprinting reactions where either the sense or antisense DNA strands were labelled with [γ-32P]ATP are shown. Sequencing reactions with the same oligonucleotide primers used to generate the PCR products are shown next to the footprinting reactions. The −10 and −35 boxes are represented by the black rectangles. (A and B) Identification of the VirR binding site. Regions protected from DNase I digestion are represented by the open rectangles. The transcription start point is shown as +1, and the positions of the regions of protection relative to +1 are as indicated. (C and D) Analysis of the VirR binding site deletion derivative. The site of deletion is indicated by the asterisk. In all panels, lane 1 contains no VirR and lanes 2 and 3 contain 1 and 2 μg of VirR, respectively. All footprinting reaction mixtures contained 25,000 cpm of labelled DNA.
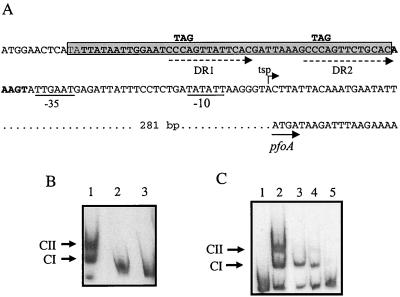
FIG. 6.
Sequence and mutations of the VirR binding site. (A) Sequence of the core binding region site (boldface). The imperfect direct repeats within this region are indicated by the dashed arrows. The nucleotide residues that were changed by site-directed mutagenesis are shown above the original nucleotide sequence. The −35 and −10 boxes of the pfoA promoter are underlined, the transcriptional start point (tsp) is indicated by the bent arrow, and the start of the pfoA gene is indicated by the solid arrow. The 49-bp region deleted by SOE PCR is shown as the gray rectangle. (B) Gel mobility shift assays carried out on the deletion derivatives. Lane 1, wild-type 183-bp fragment incubated with 1 μg of VirR; lanes 2 and 3, 134-bp deletion fragment incubated with either no VirR or 1 μg of VirR, respectively. (C) Gel mobility shift assay with direct repeat mutation derivatives. The imperfect direct repeats were altered by site-directed mutagenesis. Lanes 1 and 2, DNA fragment with intact direct repeats; lanes 3 to 5, DNA fragments with mutations in DR1, DR2, or DR1 and DR2, respectively. All binding reaction mixtures contained 1 μg of VirR, with the exception of the no-protein control in lane 1.
To confirm that this region was required for VirR binding, 49 bp of the 52-bp core binding region was deleted by SOE PCR. The resultant PCR product was then labelled with DIG and used as the target DNA in gel mobility shift experiments. The results showed that when this region was deleted, no DNA mobility shift was observed in the presence of purified VirR (Fig. 6B). These observations were confirmed by DNase I footprinting. When the core binding region was removed, the DNA footprint was no longer observed after incubation with VirR (Fig. 5C and D). These data provided clear evidence that the core binding region identified by DNase I footprinting was essential for the binding of VirR to the pfoA promoter region.
VirR binds to directly repeated sequences located immediately upstream of the pfoA promoter.
In other two-component signal transduction systems, the response regulators have been shown to bind to direct or inverted repeats located in the promoter region (16, 33). Previous nucleotide sequence analysis of the pfoA upstream region revealed the presence of two imperfect direct repeats (CCCAGTTNTNCAC) immediately upstream of the −35 box of the pfoA promoter (3). These repeats, which we have designated DR1 and DR2 (Fig. 6A), were located in the core binding region and are different from those in the 114-bp DNA fragment. Therefore, to determine whether DR1 or DR2 or both were directly involved in VirR binding, the repeats were altered by site-directed mutagenesis. In these experiments, the CCA residues of bases 2 to 4 of the direct repeats in the target plasmid, pJIR1546 (Table 1), were changed to TAG either individually to produce pJIR1804 (DR1 altered) and pJIR1803 (DR2 altered) or together to construct pJIR1821 (both DR1 and DR2 mutated) (Fig. 6A). Each altered insert was completely resequenced to confirm that no other mutations had been introduced. The 183-bp DNA fragments from pJIR1546 and its mutated derivatives were then amplified and analyzed for their abilities to bind VirR in gel mobility shift assays. The results showed that when either DR1 or DR2 was altered, VirR could bind to the target DNA to form CI but that very little CII was observed (Fig. 6C). However, when both DR1 and DR2 were mutated, VirR binding was almost eliminated, as no CII was observed and only a very faint CI band was evident (Fig. 6C). These results clearly demonstrated that the CCA residues in the direct repeats were required for VirR binding.
To determine if VirR bound to only the wild-type sites in the mutated regions or bound to both sites but was no longer able to form a second-stage complex, DNase I footprinting studies were carried out. For each plasmid derivative the sense DNA stand was end-labelled with [γ-32P]ATP, incubated with VirR, and then partially digested with DNase I. When both direct repeats were intact (pJIR1546 template), the normal footprint was observed (Fig. 7, lane 2). When DR1 was mutated, a region of DNA protection was observed in the DR2 region, but no footprint was observed in the DR1 region (Fig. 7, lane 4). The DNase I digestion profile in this area was the same as that for the corresponding region in the no-protein control (Fig. 7, lane 3). Similarly, when DR2 was mutated, a DNA footprint was observed in the DR1 region but no DR2 footprint was evident (Fig. 7, lane 6). Again the digestion profile was the same as that for the corresponding area in the no-VirR control (Fig. 7, lane 5). Finally, when both repeats were mutated, no footprints were detected in either region (Fig. 7, lane 8). Based on these data it was concluded that the direct repeats constituted two individual VirR binding sites. Furthermore, binding to these sites was not cooperative, since mutation of one site did not eliminate binding to the other site.
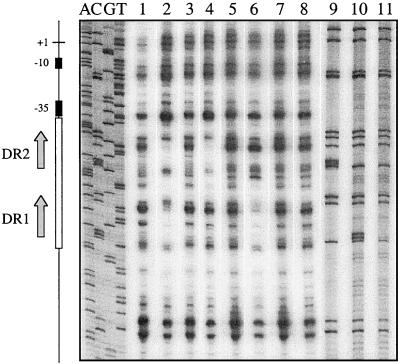
FIG. 7.
DNase I footprinting analysis of direct-repeat mutants. The labelled plasmid templates used in this experiment were the wild-type plasmid pJIR1546 (lanes 1 and 2) and the mutated derivatives pJIR1804 (DR1 mutant; lanes 3 and 4), pJIR1803 (DR2 mutant; lanes 5 and 6), and pJIR1821 (DR1-DR2 double mutant; lanes 7 and 8). Lanes 1, 3, 5, and 7, control reaction mixtures that were not preincubated with VirR. Lanes 2, 4, 6, and 8, test reaction mixtures that were incubated with 2 μg of VirR prior to partial digestion by DNase I and electrophoresis. The first four lanes (ACGT) show the sequencing reaction products from the wild-type pJIR1546 template. Lanes 9 to 11, C track sequencing reaction products from pJIR1804, pJIR1803, and pJIR1821, respectively. The positions of the DR1 and DR2 repeats are shown by the arrows, and the region of protection is represented by the open rectangle. The −10 and −35 boxes are depicted as black rectangles, and the transcription start point is shown as +1.
DISCUSSION
Most response regulators consist of a conserved N-terminal receiver domain and a unique C-terminal domain that interacts with the target genes. These proteins may be divided into different subclasses according to the nature of their C-terminal domains (1). Many response regulators contain a helix-turn-helix DNA binding motif in this region (1, 38), although others, such as OmpR, have a helix-loop-helix or winged-helix domain (29, 34). In C. perfringens, the VirR response regulator is responsible for the transcriptional activation of genes involved in toxin production. These genes include the plc, colA, and pfoA genes (27, 48). The N-terminal region of VirR has significant amino acid sequence similarity to those of other response regulators, in particular the conserved residues involved in phosphorylation (27). However, no helix-turn-helix motif has been identified in the C-terminal domain of VirR. In addition, upstream of the plc, colA, and pfoA genes, no common sequences which could act as conserved DNA binding motifs have been identified (3). Therefore, it was possible that VirR did not bind to a single consensus site but to various regions located upstream of the different target genes. Alternatively, VirR may regulate transcription of one or more of the target genes by activating the transcription of genes that encode other, as yet unidentified, regulatory proteins.
In this study, a His-tagged VirR protein was purified and subsequently tested in gel mobility shift assays for its ability to bind to the various promoter regions of the plc, colA, pfoR, and pfoA genes. VirR was found to only bind specifically to the pfoA promoter region, producing two complexes of altered electrophoretic mobilities. This result is consistent with the previously observed differential effects of mutation of the chromosomal virR (48) and virS (27) genes. These mutants produce reduced but detectible levels of alpha-toxin and collagenase. However, they do not have any detectible perfringolysin O activity, suggesting that the pfoA gene is regulated in a different manner from the other target genes (27, 48). Subsequent transcriptional analysis was in accordance with the observed toxin phenotypes, and it was suggested that expression of the plc, colA, and pfoR genes involves other regulatory genes in addition to the virS/virR system (3). Our results are in agreement with this hypothesis as they show that VirR does not bind to the promoter regions of these genes.
Previous studies indicated that the product of the pfoR gene, which is located approximately 500-bp upstream of pfoA, activated the expression of pfoA in E. coli (49). It was subsequently postulated that VirR may activate the transcription of pfoA indirectly by regulating the expression of the pfoR gene (3). In such circumstances VirR would not bind to the pfoA gene region. However, our results provide strong evidence that VirR acts directly on the pfoA promoter rather than through a regulatory cascade involving PfoR. These data do not rule out the possibility that PfoR may also regulate pfoA expression. Resolution of the role of PfoR awaits the construction and analysis of chromosomal pfoR mutants in C. perfringens.
DNase I footprinting and deletion analysis showed that VirR bound to a core 52-bp sequence located immediately upstream of the −35 region of the pfoA promoter. Many response regulators have been shown to bind in close proximity to the −35 boxes of their target gene promoters and to regulate transcription by modulating the binding of RNA polymerase to the promoter. Interaction between response regulators such as BvgA from Bordetella pertussis (6, 7), PhoP from Bacillus subtilis (42), and PhoB from E. coli (28) and their respective RNA polymerase has been demonstrated. We postulate that VirR activates pfoA transcription by either facilitating the binding of RNA polymerase to the promoter or by altering the conformation of the polymerase-promoter complex so that transcription can occur. Therefore, in the absence of VirR either RNA polymerase cannot bind to the promoter or it binds but cannot initiate transcription. In both situations no perfringolysin O is produced.
Sequence analysis of the VirR binding site revealed the presence of two CCCAGTTNTNCAC imperfect direct repeats (3). These repeats are not found in any of the other target gene regions. Many well-studied response regulators such as PhoP (12, 24, 25) and OmpR from E. coli (15, 41) have been shown to bind to short directly repeated sequences. Gel shift analysis of site-directed mutants of the pfoA repeats indicated that they were required for VirR binding. Mutation of either repeat almost eliminated the formation of the less-mobile CII VirR-DNA complex and significantly reduced the affinity of VirR for the DNA binding site, as observed previously with AlgR from Pseudomonas aeruginosa (36). Mutation of both repeats virtually eliminated VirR binding, providing direct evidence that VirR requires the CCA nucleotides of the repeat sequences for binding.
These results can be explained by postulating that the repeats represent two separate VirR binding sites, whereby CI represents a VirR-target DNA complex at either binding site and CII results from VirR binding at both sites. Alternatively, binding may have been cooperative, with both sites being required for VirR binding. However, DNase I footprinting of the site-directed mutants provided strong evidence that the direct repeats represented two separate VirR binding sites. When either repeat was mutated, only the footprint protecting the remaining wild-type repeat was observed. That is, the alteration of one repeat did not appear to affect the ability of VirR to bind to the other repeat. If binding was cooperative, mutation of one direct repeat would most likely reduce the ability of VirR to bind to the other binding site, as observed with NtrC from E. coli (9, 59), or prevent the binding of VirR to the wild-type site, as observed with PhoP (25).
In addition to the specific binding observed with the 183-bp pfoA fragment, several internal gene regions were also found to bind VirR, albeit nonspecifically. This interaction most likely represents genuine nonspecific binding. Sequence alignments of these regions did not show any regions of conservation, nor were there any similarities to the pfoA VirR binding region. The binding of response regulators to internal gene regions has recently been observed with genes regulated by the PhoP/PhoR system in B. subtilis (25). However, these regions were found to contain the consensus binding sequence. It is possible that although binding to the pfoA region is sequence specific, as demonstrated by the mutational analysis, the nonspecific binding to the internal gene regions may be due to secondary structure similarities. Alternatively, the nonspecific binding may be due to the absence of a binding cofactor such as RNA polymerase. The synergistic binding of RNA polymerase and the BvgA response regulator has been observed (6, 7). Important interactions between the α-subunit of RNA polymerase and OmpR have also been observed (47, 51). Future studies will need to involve an examination of the possible interactions between C. perfringens RNA polymerase and VirR.
With most response regulators, phosphorylation plays a key role in protein function. In most systems, phosphorylation can increase the binding affinity for the target DNA so that the regulatory protein can bind to secondary lower-affinity binding sites (6, 8, 16, 25), alter the DNA binding pattern (10), or initiate cooperative binding (9, 18, 59). However, some response regulators are able to bind when not phosphorylated, albeit with lower affinity (16, 23, 28, 42). The observation that the addition of acetyl phosphate did not alter the gel shift results suggested that DNA binding was not dependent on phosphorylation. One possible explanation for this result is that the VirR molecules may already be at least partially phosphorylated by sensor kinase cross talk or by low-molecular-weight phosphodonors present in the expression host cells. Nonspecific phosphorylation of response regulators does occur in other systems (40). However, it seems unlikely that the VirR protein phosphorylated by such mechanisms would remain in the active phosphorylated state during longer-term storage of the purified protein. Note that we cannot rule out the possibility that although acetyl phosphate did not affect VirR binding, other low-molecular-weight phosphodonors may have demonstrable effects.
While acetyl phosphate is often used as the phosphodonor for many response regulators, it is possible that this phosphodonor does not phosphorylate VirR or does so inefficiently. Either way, the addition of acetyl phosphate would have little or no observable effect. Different reactivities toward various phosphodonors have been previously demonstrated by several response regulators. CheY can be phosphorylated by acetyl phosphate, carbamoyl phosphate, and phosphoramidate (26), while phosphoramidate is used exclusively by CheB (26) and preferentially by NRI (or NtrC) (32). Therefore, further work is required to test for phosphorylation of VirR by other phosphodonors.
A more plausible explanation is that phosphorylation of the response regulator is not required for in vitro binding to the specific pfoA target site. Even though phosphorylation may be required in vivo for VirR binding, it may not be required at the VirR concentrations used in the in vitro binding experiments. BvgA (5) and PhoB (28) can still bind DNA when the N-terminal region, which is essential for phosphorylation, has been removed. However, phosphorylation is required for transcriptional activation (5, 28, 42). More specifically, it is the phosphorylated response regulator that interacts with RNA polymerase, which in turn leads to gene transcription. Therefore, it is also possible that phosphorylation of VirR is required for transcriptional activation but not for DNA binding.
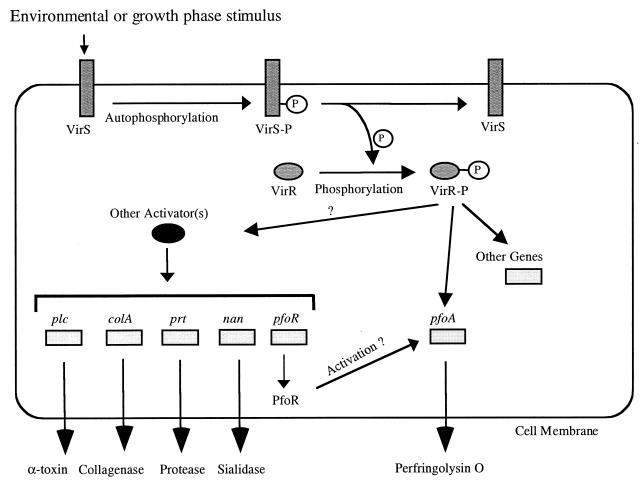
Based on the data presented in this paper we have modified the previous model (50) for the regulation of toxin production in C. perfringens (Fig. 8). We propose that when VirS detects an environmental or growth phase stimulus, it autophosphorylates at H255. Phosphorylated VirS is then able to donate the phosphoryl group to VirR, which is phosphorylated at D57. Once activated, VirR binds upstream of the pfoA promoter and activates the transcription of pfoA so that perfringolysin O is produced. Transcription of pfoA is VirR dependent, to the extent that no perfringolysin O is produced in the absence of a functional virRS operon. In addition, pfoA expression may also be controlled by another regulatory protein, PfoR (49). It also appears that VirR acts in conjunction with other regulatory factors to activate the transcription of the plc and colA genes so that increased levels of alpha-toxin and collagenase are produced (3). Although we have now shown that VirR binds directly to the pfoA promoter region there are still many aspects of the VirS/VirR two-component signal transduction system that are yet to be elucidated. These features include the nature of the external stimulus that activates VirS, the nature of the other putative regulatory genes that may be involved in the regulatory cascade, the extent of the regulatory network, and, most importantly, the effect of the mutated direct repeats on pfoA expression in vivo. However, it is hoped we are now at least one step closer to the elucidation of the overall mechanism by which toxin production and virulence are regulated in C. perfringens.
FIG. 8.
Proposed model of the VirS/VirR two-component signal transduction system. The genes postulated to be controlled by the VirS/VirR network are shown as light gray boxes. The dark gray rectangles represent the VirS sensor histidine kinase, while the VirR response regulator is depicted by the dark gray ovals. The presence of other activator(s) is symbolized by the black oval. The unknown protease and sialidase genes are represented by prt and nan, respectively.
ACKNOWLEDGMENTS
This research was supported by grants from the Australian National Health and Medical Research Council. J.K.C. was the recipient of an Australian Postgraduate Award.
We thank Rocco Iannello and Julia Young for helpful advice and discussions.
REFERENCES
- 1.Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 2.Awad M M, Bryant A E, Stevens D L, Rood J I. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 3.Ba-Thein W, Lyristis M, Ohtani K, Nisbet I T, Hayashi H, Rood J I, Shimizu T. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol. 1996;178:2514–2520. doi: 10.1128/jb.178.9.2514-2520.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellsolell L, Prieto J, Serrano L, Coll M. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J Mol Biol. 1994;238:489–495. doi: 10.1006/jmbi.1994.1308. [DOI] [PubMed] [Google Scholar]
- 5.Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators. Insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J Mol Biol. 1994;241:363–377. doi: 10.1006/jmbi.1994.1513. [DOI] [PubMed] [Google Scholar]
- 6.Boucher P E, Murakami K, Ishihama A, Stibitz S. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J Bacteriol. 1997;179:1755–1763. doi: 10.1128/jb.179.5.1755-1763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher P E, Stibitz S. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J Bacteriol. 1995;177:6486–6491. doi: 10.1128/jb.177.22.6486-6491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brissette R E, Tsung K, Inouye M. Mutations in a central highly conserved non-DNA-binding region of OmpR, an Escherichia coli transcriptional activator, influence its DNA-binding ability. J Bacteriol. 1992;174:4907–4912. doi: 10.1128/jb.174.15.4907-4912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Reitzer L J. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J Bacteriol. 1995;177:2490–2496. doi: 10.1128/jb.177.9.2490-2496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl J L, Wei B-Y, Kadner R J. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J Biol Chem. 1997;272:1910–1919. doi: 10.1074/jbc.272.3.1910. [DOI] [PubMed] [Google Scholar]
- 11.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 12.Eder S, Liu W, Hulett F M. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J Bacteriol. 1999;181:2017–2025. doi: 10.1128/jb.181.7.2017-2025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellemor D M, Baird R N, Awad M M, Boyd R L, Rood J I, Emmins J J. Use of genetically manipulated strains of Clostridium perfringens reveals that both alpha-toxin and theta-toxin are required for vascular leukostasis to occur in experimental gas gangrene. Infect Immun. 1999;67:4902–4907. doi: 10.1128/iai.67.9.4902-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 1. Oxford, England: IRL Press Ltd.; 1985. pp. 109–135. [Google Scholar]
- 15.Harlocker S L, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 16.Head C G, Tardy A, Kenney L J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 17.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 18.Huang K-J, Lan C-Y, Igo M M. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc Natl Acad Sci USA. 1997;94:2828–2832. doi: 10.1073/pnas.94.7.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iannello R C. DNase I footprinting using PCR-generated end-labeled DNA probes. In: Tymms M J, editor. In vitro transcription and translation protocols. Vol. 37. Totowa, N.J: Humana Press Inc.; 1995. pp. 379–391. [DOI] [PubMed] [Google Scholar]
- 20.Kennan R M, McMurray L M, Levy S B, Rood J I. Glutamate residues located within putative transmembrane helices are essential for TetA(P)-mediated tetracycline efflux. J Bacteriol. 1997;179:7011–7015. doi: 10.1128/jb.179.22.7011-7015.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennelly P J, Potts M. Fancy meeting you here! A fresh look at “prokaryotic” protein phosphorylation. J Bacteriol. 1996;178:4759–4764. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Hulett F M. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, Hulett F M. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology. 1998;144:1443–1450. doi: 10.1099/00221287-144-5-1443. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Qi Y, Hulett F M. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol Microbiol. 1998;28:119–130. doi: 10.1046/j.1365-2958.1998.00779.x. [DOI] [PubMed] [Google Scholar]
- 26.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyristis M, Bryant A E, Sloan J, Awad M M, Nisbet I T, Stevens D L, Rood J I. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 28.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol. 1996;259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita C, Matsushita O, Katayama S, Minami J, Takai K, Okabe A. An upstream activating sequence containing curved DNA involved in activation of the Clostridium perfringens plc promoter. Microbiology. 1996;142:2561–2566. doi: 10.1099/00221287-142-9-2561. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita O, Yoshihara K, Katayama S, Minami J, Okabe A. Purification and characterization of a Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J Bacteriol. 1994;176:149–156. doi: 10.1128/jb.176.1.149-156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCleary W R, Stock J B, Ninfa A J. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills S D, Lim C-K, Cooksey D A. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet. 1994;244:341–351. doi: 10.1007/BF00286685. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno T, Tanaka I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol Microbiol. 1997;24:665–667. doi: 10.1046/j.1365-2958.1997.3571723.x. [DOI] [PubMed] [Google Scholar]
- 35.Mohr C D, Hibler N S, Deretic V. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J Bacteriol. 1991;173:5136–5143. doi: 10.1128/jb.173.16.5136-5143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr C D, Leveau J H J, Krieg D P, Hibler N S, Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morelle G. A plasmid extraction procedure on a miniprep scale. Focus. 1989;11:7–8. [Google Scholar]
- 38.Msadek T, Kunst F, Rapoport G. Two-component regulatory systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other Gram-positive bacteria. Biochemistry, physiology and molecular genetics. Washington, D.C.: American Society for Microbiology; 1996. pp. 729–745. [Google Scholar]
- 39.Nelson H C M. Structure and function of DNA-binding proteins. Curr Opin Genet Dev. 1995;5:180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 41.Pratt L A, Silhavy T J. Identification of base pairs important for OmpR-DNA interaction. Mol Microbiol. 1995;17:565–573. doi: 10.1111/j.1365-2958.1995.mmi_17030565.x. [DOI] [PubMed] [Google Scholar]
- 42.Qi Y, Hulett F M. PhoP∼P and RNA polymerase ςA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol. 1998;28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 43.Rood J I. Virulence genes of Clostridium perfringens. Annu Rev Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 44.Rood J I, Cole S T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saint-Joanis B, Garnier T, Cole S T. Gene cloning shows the alpha-toxin of Clostridium perfringens to contain both sphingomyelinase and lecithinase activities. Mol Gen Genet. 1989;219:453–460. doi: 10.1007/BF00259619. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sharif T R, Igo M M. Mutations in the alpha subunit of RNA polymerase that affect the regulation of porin gene transcription in Escherichia coli K-12. J Bacteriol. 1993;175:5460–5468. doi: 10.1128/jb.175.17.5460-5468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J Bacteriol. 1994;176:1616–1623. doi: 10.1128/jb.176.6.1616-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu T, Okabe A, Minami J, Hayashi H. An upstream regulatory sequence stimulates expression of the perfringolysin O gene of Clostridium perfringens. Infect Immun. 1991;59:137–142. doi: 10.1128/iai.59.1.137-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T, Okabe A, Rood J I. Regulation of toxin production in C. perfringens. In: Rood J I, McClane B A, Songer J G, Titball R W, editors. The clostridia: molecular biology and pathogenesis. London, United Kingdom: Academic Press; 1997. pp. 451–470. [Google Scholar]
- 51.Slauch J M, Russo F D, Silhavy T J. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the α subunit of RNA polymerase. J Bacteriol. 1991;173:7501–7510. doi: 10.1128/jb.173.23.7501-7510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith M, Jessee J, Landers T, Jordan J. High efficiency bacterial electroporation: 1 × 1010E. coli transformants/mg. Focus. 1990;12:38–40. [Google Scholar]
- 53.Sola M, Gomis-Ruth F X, Serrano L, Gonzalez A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- 54.Stevens D L, Tweten R K, Awad M M, Rood J I, Bryant A E. Clostridial gas gangrene: evidence that α and θ toxins differentially modulate the immune response and induce acute tissue necrosis. J Infect Dis. 1997;176:189–195. doi: 10.1086/514022. [DOI] [PubMed] [Google Scholar]
- 55.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 56.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 57.Toyonaga T, Matsushita O, Katayama S, Minami J, Okabe A. Role of the upstream region containing an intrinsic DNA curvature in the negative regulation of the phospholipase C gene of Clostridium perfringens. Microbiol Immunol. 1992;36:603–613. doi: 10.1111/j.1348-0421.1992.tb02060.x. [DOI] [PubMed] [Google Scholar]
- 58.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 59.Weiss V, Claverie-Martin F, Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci USA. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]