Abstract
Shewanella putrefaciens MR-1 possesses a complex electron transport system which facilitates its ability to use a diverse array of compounds as terminal electron acceptors for anaerobic respiration. A previous report described a mutant strain (CMTn-1) deficient in CymA, a tetraheme cytochrome c. However, the interpretation of the electron transport role of CymA was complicated by the fact that CMTn-1 was also markedly deficient in menaquinones. This report demonstrates that the depressed menaquinone levels were the result of the rifampin resistance phenotype of the parent of CMTn-1 and not the interruption of the cymA gene. This is the first report of rifampin resistance leading to decreased menaquinone levels, indicating that rifampin-resistant strains should be used with caution when analyzing electron transport processes. A site-directed gene replacement approach was used to isolate a cymA knockout strain (MR1-CYMA) directly from MR-1. While MR1-CYMA retained menaquinone levels comparable to those of MR-1, it lost the ability to reduce iron(III), manganese(IV), and nitrate and to grow by using fumarate as an electron acceptor. All of these functions were restored to wild-type efficacy, and the presence of the cymA transcript and CymA protein was also restored, by complementation of MR1-CYMA with the cymA gene. The requirement for CymA in anaerobic electron transport to iron(III), fumarate, nitrate, and manganese(IV) is therefore not dependent on the levels of menaquinone in these cells. This represents the first successful use of a suicide vector for directed gene replacement in MR-1.
Shewanella putrefaciens MR-1 is a gram-negative facultative anaerobe with remarkable respiratory plasticity that can couple its anaerobic growth, and link respiratory proton translocation, to the reduction of a variety of compounds including manganese(IV) oxides, iron(III) oxides, fumarate, nitrate, trimethylamine N-oxide (TMAO), and many others (30, 32–34, 38, 40). Previous studies suggested that this bacterium contains a potentially complex multi-component branched electron transport system that includes cytochromes, quinones, dehydrogenases, and iron-sulfur proteins (30–36, 38, 40, 41). Some electron transport components are common to the use of several electron acceptors (32, 34), while others are unique to specific electron acceptors (35).
The mutant CMTn-1, which was previously generated by transposon mutagenesis of the rifampin-resistant derivative MR-1A (32), cannot use nitrate, Fe(III), or fumarate as electron acceptors, and its ability to reduce Mn(IV) is markedly compromised; it retains the ability to use TMAO and O2 as electron acceptors (32, 34). This is the same electron acceptor phenotype as that of the menaquinone-deficient mutant CMA-1 (32). In fact, compared to MR-1, CMTn-1 is markedly deficient in menaquinones, although unlike CMA-1 it cannot be restored to a wild-type phenotype by supplementation with menaquinone or menaquinone precursors (32). Subsequent characterization of CMTn-1 demonstrated that the transposon interrupted the cymA gene, which encodes a 21-kDa tetraheme cytochrome c (34). The wild-type cymA gene complements CMTn-1, restoring the CymA cytochrome and the cells' ability to utilize nitrate, Fe(III), Mn(IV), and fumarate (34); the complemented mutant, however, still has very low levels of menaquinone. The phenotype of CMTn-1 is therefore apparently due to the loss of the CymA cytochrome and not to a deficiency in menaquinones. However, two questions regarding the mutant CMTn-1 remain: (i) what is the reason for the menaquinone deficiency in CMTn-1? and (ii) would the phenotype of a cymA knockout be the same if menaquinone levels were normal? The latter is particularly important because the putative role of CymA orthologs in other bacteria (e.g., NapC of Thiosphaera pantotropha) is to accept electrons from membrane-bound quinones and transfer them to downstream electron transport components (e.g., nitrate reductase) (5).
This report demonstrates that menaquinone levels are markedly depressed in the spontaneous rifampin-resistant mutant MR-1A, which served as the parent of CMTn-1. This suggests that the rifampin resistance, and not the cymA knockout, is responsible for the depressed menaquinone levels in CMTn-1. This report also describes the isolation of a cymA knockout directly from MR-1 by a site-directed gene replacement approach. This cymA knockout (MR1-CYMA) has the same electron transport phenotype as CMTn-1 but has menaquinone levels comparable to those of the wild-type MR-1. The electron transport phenotype of MR1-CYMA can be restored to that of the wild-type by complementation with the cymA gene. The requirement for CymA in anaerobic electron transport to Fe(III), fumarate, nitrate, and Mn(IV) is therefore not dependent on the levels of menaquinone in these cells.
MATERIALS AND METHODS
Materials.
All materials were from sources previously described (34) with the following exceptions. The Expand High Fidelity PCR system and Expand Long Template PCR system were from Boehringer-Mannheim (Indianapolis, Ind.). Restriction enzymes and biotinylated RNA markers were from New England BioLabs (Beverly, Mass.). Custom oligonucleotide primers were synthesized by Operon Technologies (Alameda, Calif.) or by Genemed Biotechnologies (South San Francisco, Calif.). Vitamin K2 (menaquinone-4, MK-4) and coenzyme Q6 (ubiquinone-6) and Q10 (ubiquinone-10) standards were obtained from Sigma Chemical Co. (St. Louis, Mo.), and 1,4-dihydroxy-2-naphthoic acid (DHNA) was obtained from Aldrich Chemical (Milwaukee, Wis.).
Bacterial strains, plasmids, media, and growth conditions.
A list of the bacteria and plasmids used in this study is presented in Table 1. For molecular biology purposes, S. putrefaciens and Escherichia coli were grown aerobically on Luria-Bertani (LB) medium (49) supplemented, when required, with antibiotics at the following concentrations: ampicillin, 50 μg ml−1; kanamycin, 50 μg ml−1; chloramphenicol, 34 μg ml−1; rifampin, 50 μg ml−1. E. coli was grown at 37°C unless indicated otherwise. S. putrefaciens was grown at room temperature (23 to 25°C), except where indicated otherwise, under either aerobic or anaerobic conditions as previously described (30) in defined medium (40) supplemented with 15 mM lactate. For anaerobic growth or for testing electron acceptor use, the medium was also supplemented with vitamin-free Casamino Acids (0.1 g liter−1) and with one of the following electron acceptors: 20 mM TMAO, 20 mM fumarate, 10 mM ferric citrate, 2 mM nitrate, or 5 mM MnO2. For growth on TMAO, the medium was also supplemented with 30 mM HEPES to buffer against alkalinization by the product trimethylamine. Antibiotics were included when needed at the concentrations listed above.
TABLE 1.
Bacteria and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference(s) | Source |
|---|---|---|---|
| S. putrefaciens | |||
| MR-1 | Manganese-reducing strain from Lake Oneida, N.Y., sediments | 38 | Previous study |
| MR-1A | Spontaneous Rfr mutant of MR-1 | 32 | Previous study |
| MR-1B | Spontaneous Rfr mutant of MR-1 | This work | |
| CMTn-1 | Transposon mutant of MR-1A that is unable to use Fe(III), nitrate, and fumarate as terminal electron acceptors; cymA::TnphoA | 32, 34 | Previous study |
| MR1-CYMA | cymA mutant derived from MR-1; cymA::Kmr | This work | |
| E. coli | |||
| JM109 | recA1 F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14−(McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17 (rK− mK+) relA1 supE44 | 55 | Promega |
| S17-1 | C600::RP-4 2-Tc::Mu-Km::Tn7 hsdR hsdM+ recA thi pro; donor strain to mate pVK100-derived plasmids into MR-1 | 50 | M. L. P. Collins |
| S17-1λ pir | λ(pir) hsdR pro thi; chromosomally integrated RP4-2 Tc::Mu Km::Tn7; donor strain to mate pEP185.2-derived plasmids into MR-1 | 50 | V. L. Miller |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen | |
| Plasmids | |||
| pVK100 | 23-kb broad-host-range cosmid; Tcr Kmr Tra+ | 19 | ATCC 37156 |
| pGEM-7Zf(+) | 3.00-kb cloning/sequencing vector; Apr | Promega | |
| pCR2.1-TOPO | 3.9-kb vector for cloning PCR products | Invitrogen | |
| pCMTN1-VK | pVK100 with the MR-1 cymA open reading frame plus associated 5′ and 3′ regions | 34 | Previous study |
| pUT/mini-Tn5Km | Apr; Tn5-based delivery plasmid; used as source of Kmr | 10 | D. Frank |
| pTOPO-cymA | pCR2.1-TOPO with the MR-1 cymA open reading frame plus associated 5′ and 3′ regions; 4.9 kb | This work | |
| pTOPO/cymA:Km | pTOPO-cymA with the Kmr gene replacing bases 932–1214 of cymA; 6.7 kb | This work | |
| pEP185.2 | 4.28-kb mobilizable suicide vector derived from pEP184; oriR6K mobRP4 Cmr | 18 | V. L. Miller |
| pDSEPcymAII | Kmr gene-interrupted cymA gene cloned into the SmaI site of pEP185.2; Cmr Kmr; 7.1 kb; used for gene replacement to generate strain MR1-CYMA | This work |
To examine the effect of DHNA and MK-4 on the fumarate-dependent anaerobic growth of strains MR-1A and MR-1B, the defined medium was supplemented with either DHNA or MK-4 (10 μM [each] final concentration). Stock solutions of 1.5 mM DHNA and MK-4 were prepared in dimethylformamide. Control cultures were supplemented with the equivalent amount of dimethylformamide.
DNA manipulations.
Restriction digests and mapping, cloning, subcloning, and DNA electrophoresis were done according to standard techniques (49) following manufacturers' recommendations as appropriate. DNA ligations were done with Fast-Link DNA ligase (Epicentre Technologies, Madison, Wis.) or T4 DNA ligase (Life Technologies, Rockville, Md.). Isolation of plasmid and cosmid DNA was accomplished with the QIAprep Spin Plasmid kit (Qiagen, Chatsworth, Calif.). The sizes of DNA fragments, RNA, and proteins were estimated based on their relative electrophoretic mobilities to known standards by using a computer program kindly provided by G. Raghava (46). Colony PCR (51) with primers specific to cymA was utilized to screen transconjugants for gene replacement events.
Aerobically grown mid-logarithmic-phase E. coli cells were prepared for electroporation as suggested by Bio-Rad; the cells were stored in 10% glycerol at −80°C until needed. Plasmids and cosmids were introduced into E. coli by electroporation as previously described (34).
To verify the DNA sequences of the ends of cloned fragments, thermal cycle DNA sequencing was done as previously described (34), except that the SequiTherm EXCEL II DNA sequencing system (Epicentre Technologies) was used. Computer-assisted sequence analysis and comparisons were done with MacVector software (IBI, New Haven, Conn.).
Northern blotting and RT-PCR.
Total RNA was purified from anaerobically grown cells by a hot phenol method, followed by treatment with RNase-free DNase as previously described (34). All standard precautions to prevent RNase contamination were followed (49). Electrophoresis of RNA and Northern blot analysis were done as previously described (34) with the following exceptions. Immediately after electrophoresis, the gel was sequentially soaked (20 min each) in 50 mM NaOH–150 mM NaCl and 150 mM NaCl–100 mM Tris-HCl (pH 7.5). The RNA was transferred to a Magna (MSI, Westboro, Mass.) nylon membrane (0.45 μm pore size) by using 10× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) and the Posiblot pressure blotter (Stratagene, La Jolla, Calif.) at 70 to 75 mm Hg pressure for 1 h. The RNA was fixed to the membrane by UV cross-linking (33 mJ cm−2). The membrane was prehybridized, hybridized, and washed as previously described (34) prior to development with the Phototope Star detection system (New England BioLabs). Biotinylated DNA probes were generated with a PCR-based labeling system (25).
Reverse transcription-PCR (RT-PCR) was done with the Titan One Tube RT-PCR system (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. Total RNA (2 μg) from each strain served as the template, and sense (5′-GGCTATTTTGCAACTCAGCAGAC-3′) and antisense (5′-CGAGTATGGCAGTGTTGACAGTTT-3′) primers were based on the sequence of cymA from MR-1 (34).
Quinone analysis.
Quinones were extracted from the cells essentially as described previously (21). The cells were harvested by centrifugation, and the cell pellets were washed in cold 20 mM MgCl2–0.1 M triethanolamine–HCl (pH 7.2); the washed pellets were resuspended in this buffer (1.0 ml of buffer per 0.6 g wet cell weight). Per 0.6 g of cells, 6.0 ml of methanol-acetone (1/1 [vol/vol]) was added to the cell suspension, followed by vigorous agitation and incubation at room temperature for 30 min. Petroleum ether (2.0 ml) was added, followed by vigorous agitation. Following a 1-min centrifugation at 2,800 × g at 15°C to break the phases, the upper layer was removed; the lower layer was reextracted with another 2.0 ml of petroleum ether, followed by centrifugation and recovery of the upper layer. The upper layers were pooled and evaporated under a stream of nitrogen at 37°C. The dried residue was dissolved in chloroform-methanol (2/1 [vol/vol]; 50 μl per 0.6 g original wet cell weight).
Quinones were resolved by thin-layer chromatography (TLC) essentially as described previously (14) on Merck Kieselgel 60 F254 plates (20 by 20 cm; 0.25 mm thick) developed with petroleum ether-diethyl ether (9/1 [vol/vol]). The plates were examined under reflective UV light (254 nm). To record the image, the plates were photographed with Kodak T-MAX film (ASA100), and the photograph was scanned into a computer to label the image. Alternatively, the image was captured directly with a video camera and the FOTO/Analyst Archiver electronic documentation system (Fotodyne, Hartland, Wis.).
High-pressure liquid chromatography (HPLC) analysis of quinones was conducted with a Hewlett-Packard 1090 series II/L HPLC system equipped with a diode array detector. Samples (10 μl) were separated on a Nucleosil 5-μm, 120-Å C18 column (inside diameter, 200 by 2.0 mm; Phenomenex, Torrance, Calif.) including a 30-mm C18 guard column. A gradient mobile phase at a constant flow rate (0.25 ml min−1) was utilized: the initial mobile phase was 100% methanol and was changed in a linear fashion over time to methanol-isopropanol (60/40) at 15 min; this was held for 25 min, after which the mobile phase was returned in a linear fashion to 100% methanol over a 15-min interval to prepare the column for the next sample. Detection was monitored at 270 and 275 nm, which are λmax values for menaquinones and ubiquinones, respectively (42). The detector also continuously monitored the spectra between 220 and 370 nm. Data were collected and analyzed with the ChemStation software provided with the HPLC system. Peaks were quantified based on peak area relative to external standards of ubiquinone-6 and menaquinone-4.
Miscellaneous procedures.
Growth was assessed by measuring culture turbidity at 500 nm in a Beckman DU-64 spectrophotometer. Nitrate (9) and nitrite (52) levels were determined colorimetrically in cell-free filtrates. The Fe(II) level was determined by a ferrozine extraction procedure (23, 39). The Mn(II) level was determined in filtrates by a formaldoxime method (3, 7), and MnO2 was prepared as described previously (38).
Cytoplasmic membrane, intermediate membrane, outer membrane, and soluble fractions (periplasm plus cytoplasm) were purified from cells by an EDTA-lysozyme-Brij protocol as previously described (30). The intermediate membrane is a hybrid of the cytoplasmic and outer membranes (30). The separation and purity of these subcellular fractions were assessed by examining spectral cytochrome content (30), membrane buoyant density (30), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (22, 28) stained for protein with Pro-Blue (Owl Separation Systems, Woburn, Mass.) or for heme as previously described (30). Protein was determined by a modified Lowry method, with bovine serum albumin as the standard (34).
RESULTS
Construction of cymA insertion mutation.
While an early report suggested that pACYC184 (which contains the p15A origin of replication) could be used as a suicide vector in MR-1 (48), this was subsequently shown not to be the case (37). For these studies we utilized the mobilizable vector pEP185.2; it contains the R6K origin of replication, which requires the pir gene product for replication (20); pir is carried on the chromosome of the E. coli donor strain S17-1λ pir. In several matings of E. coli S17-1λ pir/pEP185.2 with MR-1, we were unable to obtain any MR-1 cells which survived chloramphenicol selection because of resistance encoded by the cat gene of pEP185.2. These results suggested that pEP185.2 is not maintained in MR-1 and could therefore be useful as a suicide vector for gene replacement strategies.
To construct a plasmid suitable for gene replacement of cymA, the following strategy was used. A 984-kb DNA fragment containing the entire cymA gene plus 5′ and 3′ flanking sequences was generated by PCR of MR-1 genomic DNA with primers bracketing bases 604 to 1588 of the published cymA sequence (34). This PCR product was cloned into pCR2.1-TOPO generating pTOPO/cymA. Inverse PCR (43) was done with pTOPO/cymA as the template and specific cymA primers (with additional ClaI restriction sites at the 5′ ends) designed to generate TOPO/cymA(Δ932–1214), a 4.6-kb fragment that contains all of pCR2.1-TOPO and the 5′ and 3′ ends of cymA but that is missing bases 932 to 1214 of the published cymA sequence. The 2.1-kb Kmr gene from pUT/mini-Tn5Km was generated by PCR with primers that included ClaI restriction sites at the 5′ ends. Following digestion with ClaI, the Kmr gene was ligated to the TOPO/cymA(Δ932–1214) fragment, generating pTOPO/cymA:Km. A 2.8-kb DNA fragment containing the Kmr gene-interrupted cymA gene was generated by PCR with pTOPO/cymA:Km as the template and primers bracketing bases 604 to 1588 of the published cymA sequence; this fragment was blunt-ended and ligated into the SmaI site of the suicide vector pEP185.2, generating pDSEPcymAII, which was electroporated into the donor strain E. coli S17-1λ pir. At each step during the construction process, appropriate analyses (restriction mapping, PCR, DNA sequence analysis) were done to assure that the expected construct was obtained.
E. coli S17-1λ pir/pDSEPcymAII was mated with MR-1 and MR-1 exconjugants were selected with kanamycin under aerobic conditions on defined medium with 15 mM lactate as the electron donor. Colonies were screened by colony PCR (51) with primers bracketing bases 20 to 1873 of the published cymA sequence; those lacking the expected wild-type PCR product of 1.9 kb in two independent PCR analyses were pursued as putative insertional mutants. One strain, designated MR1-CYMA met all criteria expected for a cymA knockout: (i) it was found to lack the expected wild-type PCR product of 1.9 kb in several independent PCR analyses with primers bracketing bases 20 to 1873 of the published cymA sequence; (ii) it was found to be negative for the expected PCR product by using primers specific to the cat gene of pEP185.2; (iii) it could not grow in the presence of chloramphenicol but did grow in the presence of kanamycin; (iv) it was found to be positive for the expected PCR product by using primers specific to the Kmr gene used to interrupt cymA in pDSEPcymAII; and (v) it remained negative for Fe(III) reduction even after 1 week of incubation under anaerobic conditions. The lack of the cat gene and the sensitivity to chloramphenicol are consistent with a double-crossover gene replacement, as a single-crossover insertion into the genome should retain the cat gene of the suicide vector. Strain MR1-CYMA was fully characterized as described below.
Characterization and complementation of the cymA insertion mutant.
A cymA probe detected RNA bands of approximately 900 and 1,200 bases in MR-1 cells grown anaerobically with TMAO as the electron acceptor (Fig. 1A). These two bands were previously detected in Northern blot analysis of total RNA from MR-1 cells grown anaerobically with fumarate or TMAO as the electron acceptor (34); the smaller band was predominant in fumarate-grown cells, and the larger band could represent a cymA transcript generated from a different transcriptional start site, or it could represent a larger mRNA precursor (34). Even though the cymA transcript is more prominent in fumarate-grown cells (34), we used TMAO-grown cells to provide for direct comparisons because MR1-CYMA will not grow on fumarate (see below). These two cymA RNA bands were also detected in MR-1 cells containing the control vector pVK100, but they were absent in the gene replacement mutant MR1-CYMA (Fig. 1A). Plasmid pCMTN1-VK (34) is pVK100 containing an insert consisting of the 561-bp cymA gene and 262 and 166 bp of the associated 5′ and 3′ DNA from MR-1, respectively (i.e., the insert spans bases 604 to 1592 of the published sequence [34]). Plasmid pCMTN1-VK restored the 900-base cymA transcript to MR1-CYMA (Fig. 1A). Since the levels of the cymA transcript are low in TMAO-grown cells, RT-PCR analysis of total RNA from these strains was done with primers bracketing bases 950 to 1290 of the published sequence (34). RT-PCR analysis confirmed the presence of the expected 341-bp RT-PCR product in MR-1 and MR-1/pVK100, as well as its absence in MR1-CYMA (Fig. 1B); plasmid pCMTN1-VK restored this 341-bp RT-PCR product, derived from the cymA transcript, to MR1-CYMA (Fig. 1B).
FIG. 1.
Expression of cymA in S. putrefaciens cells grown anaerobically with TMAO as the electron acceptor as determined by Northern blotting (A) and RT-PCR (B). The lanes contain samples from the following strains: lanes 1, MR1-CYMA(pCMTN1-VK); lanes 2, MR1-CYMA(pVK100); lanes 3, MR-1(pVK100); lanes 4, MR-1. (A) Northern blot of 100 μg of total RNA from each strain hybridized with a biotin-labelled cymA probe (representing bases 604 to 1588 of the published sequence [34]). No bands were seen in lane 2 with extended exposure times. Lane M, 3 μl of biotinylated RNA markers (New England BioLabs), with their sizes in bases indicated at the right; the position of each marker was assessed from a short exposure. The major cymA RNA is indicated by the arrow at the left. (B) RT-PCR of 2 μg of total RNA from each strain with primers specific for cymA. The sizes of the DNA markers (lane M) in kilobases are indicated at the right. The cymA product from RT-PCR is indicated by the arrow at the left.
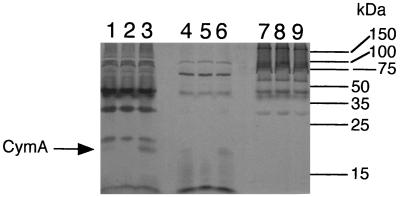
It was previously shown that cymA encodes a 21-kDa tetraheme c-type cytochrome, which is most prominent in the cytoplasmic membrane, with lesser amounts in the soluble fraction (34). Subcellular fractions of the strains used in this study were prepared and examined for CymA content. While CymA is more prominent in fumarate-grown cells (34), TMAO-grown cells were used to allow for a direct comparison with the mutant MR1-CYMA. Analysis of these subcellular fractions for membrane buoyant density, cytochrome content, and SDS-PAGE patterns (see below) confirmed a prominent separation of the various fractions and demonstrated that they were comparable to the analogous fractions from previous experiments (29–31, 34, 36, 41). The 21-kDa heme-positive band corresponding to CymA is present in MR-1 but is absent in the gene replacement mutant MR1-CYMA (Fig. 2). Complementing the mutant with pCMTN1-VK restores the 21-kDa heme-positive CymA band (Fig. 2). A small amount of CymA is also seen in the soluble fractions of MR-1 and the complemented mutant, but CymA is absent from MR1-CYMA (Fig. 2). Assuming it is similar to its orthologs in other bacteria, CymA is likely associated with the cytoplasmic membrane; the subcellular fractionation process could cause the release of a small amount of CymA into the soluble fraction. Certain other c-type cytochromes are loosely associated with the cytoplasmic membrane and are readily removed by changes in ionic strength (2, 8).
FIG. 2.
Heme-stained SDS-PAGE profiles of subcellular fractions prepared from S. putrefaciens cells grown anaerobically with TMAO as the electron acceptor. The lanes were loaded with 50 μg of protein of each of the following subcellular fractions: cytoplasmic membrane (lanes 1 to 3), soluble fraction (lanes 4 to 6), and outer membrane (lanes 7 to 9). Fractions were prepared from MR-1/pVK100 (lanes 1, 4, and 7), MR1-CYMA/pVK100 (lanes 2, 5, and 8), and MR1-CYMA/pCMTN1-VK (lanes 3, 6, and 9). The bars and numbers at the right indicate the migration and masses of the protein standards.
No other differences in heme stain profiles between the various strains were apparent (Fig. 2), and protein stains of these same fractions did not reveal any other differences. The sum total of evidence supports the absence of the CymA cytochrome and its corresponding RNA in MR1-CYMA, both of which are restored by complementing this mutant with the cymA gene.
In contrast to the wild-type, MR1-CYMA could not reduce nitrate, Fe(III), or Mn(IV), and it could not grow with fumarate as a terminal electron acceptor (Table 2). As was the case for CMTn-1, the block was nearly complete for nitrate, iron, and fumarate (≤2% of the wild-type value). The ability of MR1-CYMA to reduce Mn(IV) was also severely compromised (6.5% of the activity of MR-1) (Table 2). In the previous report, strain CMTn-1 was reported to have 27% of the Mn(IV)-reducing activity of MR-1 (34); however, this rate was not corrected for the small amount of Mn(IV) reduction that occurred in sterile controls. The data in Table 2 have been corrected for this background and hence more accurately reflect the prominent role of CymA in Mn(IV) reduction. If the phenotype of MR1-CYMA is solely due to the loss of the CymA cytochrome, then complementation with the wild-type cymA gene should restore the electron transport phenotype to that of MR-1. This is in fact the case; relative to the pVK100 control, pCMTN1-VK fully restored the ability of MR1-CYMA to use nitrate, fumarate, and Fe(III), and Mn(IV) reduction was restored to 85% that of the wild type (Table 2). MR1-CYMA also could not grow on nitrate, Fe(III), or Mn(IV), and pCMTN1-VK restored the ability to grow on these electron acceptors (not shown). As was the case for CMTn-1, MR1-CYMA retained the ability to grow with O2 or TMAO as electron acceptors (Table 2).
TABLE 2.
Use of electron acceptors by plasmid-bearing strains of S. putrefaciensa
| Strain (plasmid) | Reduction of:
|
Growthd on:
|
Growth, no electron acceptord | ||||
|---|---|---|---|---|---|---|---|
| Fe(III)b | Mn(IV)b | Nitratec | Fumarate | TMAO | Oxygen | ||
| MR-1 | 53.7 ± 2.65 | 0.770 ± 0.036 | 29.5 ± 0.481 | 0.150 ± 0.010 | 0.168 ± 0.009 | 0.568 ± 0.014 | 0.004 ± 0.004 |
| MR-1(pVK100) | 83.1 ± 1.14 | 0.742 ± 0.009 | 28.7 ± 0.351 | 0.139 ± 0.001 | 0.141 ± 0.005 | 0.519 ± 0.006 | 0.001 ± 0.001 |
| MR1-CYMA(pVK100) | 0.286 ± 0.052 | 0.048 ± 0.005 | 0.182 ± 0.182 | 0.003 ± 0.000 | 0.155 ± 0.006 | 0.478 ± 0.018 | 0.005 ± 0.001 |
| MR1-CYMA(pCMTN1-VK) | 85.1 ± 0.143 | 0.627 ± 0.003 | 28.3 ± 0.202 | 0.142 ± 0.008 | 0.135 ± 0.003 | 0.296 ± 0.024 | 0.001 ± 0.001 |
All values represent means plus or minus the differences between the means and the high and low values for two parallel but independent experiments.
Rate of formation of Mn(II) or Fe(II) (in micromoles per day in a 13-ml culture) to the point of maximum reduction observed over a period of 4 days.
Rate of nitrate loss (in micromoles per day in a 15-ml culture) to the point of maximum loss observed over a period of 4 days.
Maximum increase in culture turbidity (optical density at 500 nm) observed over a period of 4 days.
Quinone analysis of various strains.
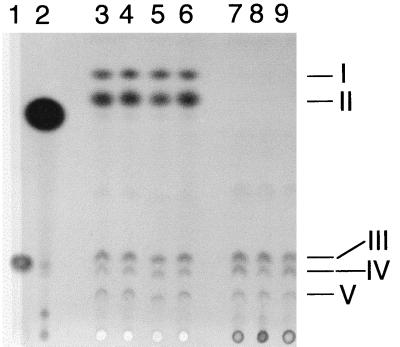
TLC analysis of quinones from the various strains (MR-1, MR-1/pVK100, MR1-CYMA/pVK100, MR1-CYMA/pCMTN1-VK) demonstrated that there were no apparent significant differences in ubiquinone or menaquinone content (Fig. 3, lanes 3 to 6). Based on HPLC analysis of extracts pooled from two or more cultures, levels of menaquinone (21 to 23 nmol per g of wet cells), methylmenaquinone (4.3 to 5.0 nmol per g of wet cells), and total ubiquinones (5.9 to 7.2 nmol per g of wet cells) were similar for all four strains. The spectra of the quinones in spots I and II were typical for menaquinones, whereas those in spots III to V were typical for ubiquinones (Fig. 4). Therefore the knockout of cymA in MR1-CYMA is solely responsible for the electron transport phenotype, as this mutant retains wild-type levels of menaquinone. This is in contrast to the marked deficiency of menaquinones in the previous mutant, CMTn-1, and its parent, MR-1A (Fig. 3, lanes 7 and 8). Based on HPLC analysis, MR-1A and its derived strains contained similar levels of total ubiquinones (4.0 to 7.2 nmol per g of wet cells), whereas menaquinone and methylmenaquinone were not detected. This suggests that the spontaneous rifampin resistance in MR-1A is somehow responsible for the menaquinone deficiency in CMTn-1 and that it is not related to the knockout of cymA in CMTn-1. Even though the introduction of the wild-type cymA gene into CMTn-1 did restore its ability to reduce Mn(IV), Fe(III), nitrate, and fumarate (34), the levels of menaquinones remained markedly depressed in CMTn-1/pCMTN1-VK (Fig. 3, lane 9). It is clear that the presence or absence of CymA does not influence the levels of menaquinones. Conversely, the levels of menaquinones do not affect the overall electron acceptor phenotype of a cymA knockout.
FIG. 3.
Thin-layer chromatogram of quinone standards and of quinones isolated from S. putrefaciens cells grown anaerobically with TMAO as the electron acceptor. Lanes 1 and 2 were loaded with ubiquinone-6 and menaquinone-4 standards, respectively. The other lanes were loaded with quinone extracts (equivalent to 0.10 g of wet cells) isolated from cells of the following strains: lane 3, MR-1; lane 4, MR-1/pVK100; lane 5, MR1-CYMA/pVK100; lane 6, MR1-CYMA/pCMTN1-VK; lane 7, MR-1A; lane 8, CMTn-1/pVK100; lane 9, CMTn-1/pCMTN1-VK. The samples were loaded at the bottom, and migration was upward. The quinone standards did not exactly migrate with the quinones from S. putrefaciens because the length and composition of the isoprenyl side chain affect the migration. In a previous report (32), spots I and II were identified as methylmenaquinone and menaquinone, respectively, because of their migration relative to the identified quinones in S. putrefaciens ATCC 8071 (14). Spots III, IV, and V were identified as ubiquinones by a similar comparison.
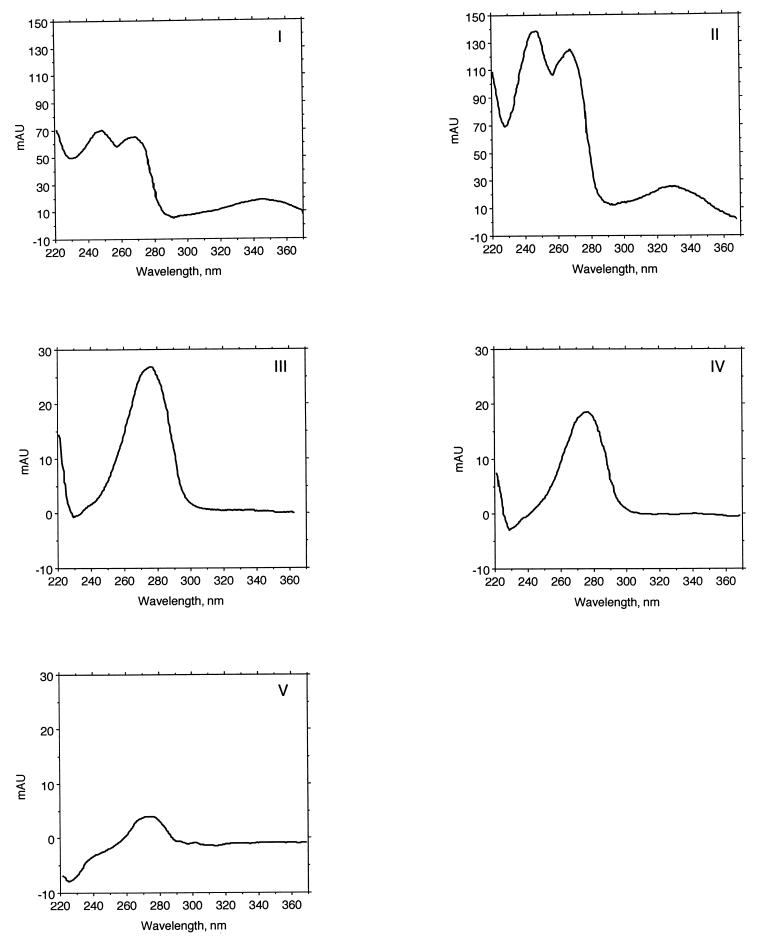
FIG. 4.
UV absorption spectra for quinone peaks. The quinone spots were recovered from the TLC plate (Fig. 3) and were subjected to HPLC analysis with diode array detection. Each quinone eluted as a single peak. The spectra for the quinones in spots I and II are typical menaquinone spectra, with λmax at ∼245, 270, and 330 to 340 nm (42), while those of spots III, IV, and V are consistent with ubiquinones with a λmax of ∼275 nm.
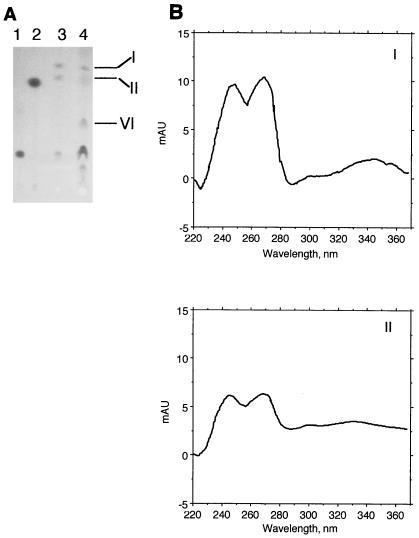
The TLC quinone data in Fig. 3 still do not address a puzzling issue for MR-1A, i.e., it appears to be menaquinone deficient but it retains the ability to grow anaerobically on fumarate and to reduce nitrate, Mn(IV), and Fe(III) (34). This conflicts with the data for strain CMA-1, a menaquinone-deficient strain isolated directly from MR-1, in which menaquinones were required for growth on fumarate and for reduction of nitrate, Mn(IV), and Fe(III) (32). It is possible that MR-1A is not totally devoid of menaquinones but that levels are so low as to be undetectable by TLC analysis or by analyzing unconcentrated samples by HPLC. Consistent with this hypothesis is the observation that MR-1A is significantly slower than MR-1 in its ability to utilize fumarate (see below), nitrate, Fe(III), and Mn(IV) under anaerobic conditions. We therefore reexamined the quinone content of MR-1A by using larger cell amounts to enhance quinone detection. While still not visible by TLC, two menaquinones (λmax at 245, 270, and 330 to 340 nm) were seen in HPLC analysis of quinone extracts of MR-1A grown on TMAO, although levels were approximately 5% of those observed for MR-1. Since menaquinone is not required for growth on TMAO, fumarate-grown cells were also examined because menaquinone is required for growth on fumarate (32). When cell amounts equivalent to those used in Fig. 3 were used, two menaquinones were faintly visible by TLC analysis of fumarate-grown MR-1A (not shown); they were readily apparent by HPLC analysis (λmax at 245, 270, and 330 to 340 nm), and levels in independent cultures varied from 9 to 30% of those seen in MR-1 cells when normalized to equal wet cell weights. When significantly larger amounts of MR-1A cells were analyzed, these two menaquinones were visible by TLC analysis and they comigrated with those seen in MR-1 (Fig. 5A). The spectra of these spots were consistent with those of menaquinones (Fig. 5B).
FIG. 5.
(A) Thin-layer chromatogram of quinone standards and of quinones isolated from S. putrefaciens cells grown anaerobically with fumarate as the electron acceptor. Lanes 1 and 2 were loaded with ubiquinone-6 and menaquinone-4 standards, respectively. The other lanes were loaded with quinone extracts isolated from cells of the following strains: lane 3, MR-1 (equivalent to 0.18 g of wet cells); lane 4, MR-1A (equivalent to 0.72 g of wet cells). The samples were loaded at the bottom, and migration was upward. The numbered quinone spots were recovered from the TLC plate and subjected to HPLC analysis using diode array detection. Each quinone eluted as a single predominant peak, and the UV absorption spectra of the quinone peaks from MR-1A are shown (B). The spectra for the quinones in spots I and II are typical menaquinone spectra, with λmax at ∼245, 270, and 330 to 340 nm (42). The spectrum for spot VI (not shown) was consistent with ubiquinone with a single λmax of 275 nm (42).
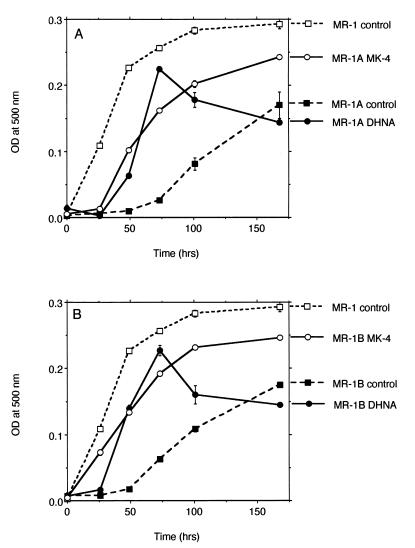
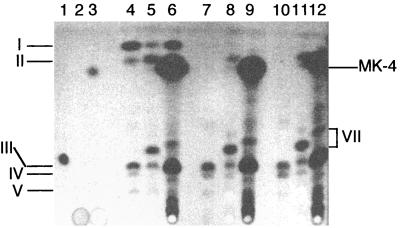
Therefore, while the Rfr phenotype in MR-1A is associated with depressed menaquinone levels (e.g., 9 to 30% of those of MR-1 for anaerobic growth on fumarate), this is apparently enough menaquinone to support MR-1A's ability to reduce nitrate, Fe(III), Mn(IV), and fumarate. However, MR-1A is much slower than MR-1 in its use of these electron acceptors. For example, it takes MR-1A 5 days to achieve pronounced growth on fumarate, and even then it is less turbid than is MR-1 after only 2 days on fumarate (Fig. 6A). Supplementation of the medium with either a menaquinone analog (MK-4) or a precursor in the menaquinone synthesis pathway (DHNA) significantly stimulated the rate of growth of MR-1A on fumarate, although it was still not as fast as MR-1 (Fig. 6A). The results were similar for MR-1B (Fig. 6B), another spontaneous Rfr MR-1 mutant, which was isolated independently from MR-1A. Similar to MR-1A, MR-1B also has markedly depressed levels of menaquinones (Fig. 7). Some menaquinone was synthesized by MR-1A and MR-1B cells that were grown in the presence of DHNA (Fig. 7). While less than levels seen in MR-1, this is presumably enough menaquinone to enhance their anaerobic growth on fumarate (Fig. 6). For cells grown in the presence of MK-4, the levels of MK-4 are so high as to interfere with the detection of cell-synthesized menaquinone (Fig. 7). Nonetheless, cells can likely utilize MK-4 directly, although it may not be as efficiently anchored in membranes because its isoprenyl side chain is approximately 15 carbons shorter than that of menaquinone-7 typically found in S. putrefaciens (1, 27, 42). Together, the data suggest that the depressed menaquinone levels in MR-1A contribute significantly to its slower anaerobic growth, but additional unknown effects associated with the Rfr phenotype cannot be discounted at this point.
FIG. 6.
Anaerobic growth of MR-1 versus MR-1A (A) or MR-1B (B) cells on fumarate. The medium was supplemented with vehicle (control; dimethylformamide only), MK-4 (10 μM), or DHNA (10 μM). Growth was measured by increases in optical density (OD) measured at 500 nm with a Beckman DU-64 spectrophotometer. Points represent the means for n = 2, and bars represent the range of high and low values; for points lacking apparent range bars, the bars were smaller than the points as shown.
FIG. 7.
Thin-layer chromatogram of quinone standards and of quinones isolated from S. putrefaciens cells grown anaerobically with fumarate as the electron acceptor and supplemented with either DHNA or MK-4. Standards were loaded in lanes 1 to 3 as follows: lane 1, ubiquinone-10; lane 2, DHNA; lane 3, menaquinone-4. The other lanes were loaded with quinone extracts (equivalent to 0.18 g of wet cells) isolated from cells of the following strains: lanes 4 to 6, MR-1; lanes 7 to 9, MR-1A; lanes 10 to 12, MR-1B. During cell growth, the medium was supplemented with 10 μM DHNA (lanes 5, 8, and 11), 10 μM MK-4 (lanes 6, 9, and 12), or the solvent for these quinones as a control (lanes 4, 7, and 10). The samples were loaded at the bottom, and migration was upward. As described above for MR-1, spots I and II are methylmenaquinone and menaquinone, respectively, and spots III, IV, and V are ubiquinones. The prominent presence of MK-4 in cells grown in the presence of MK-4 is indicated at the right. The DHNA standard (15 nmol; lane 2) was not visually detectable under these conditions. The spectra for the spots indicated by VII (not shown) were consistent with ubiquinone.
DISCUSSION
The interpretation of previous studies with the cymA mutant CMTn-1 (34) were complicated by its low levels of menaquinones (32), which could have contributed to its phenotype. This issue required clarification because the putative role of CymA orthologs in other bacteria is to accept electrons from membrane-bound quinones and transfer them to downstream electron transport components (5, 17, 24). The results in this study clarify the role of CymA in anaerobic electron transport in MR-1. The menaquinone levels in the mutant MR1-CYMA are the same as those in the wild type, and yet the knockout of cymA renders the cells unable to reduce nitrate, Fe(III), and Mn(IV) and unable to grow anaerobically with fumarate as the electron acceptor. Complementing MR1-CYMA with wild-type cymA restored its phenotype to that of the wild type. It seems likely that CymA is a common component in the electron transport chains for at least these four anaerobic electron acceptors and that these chains diverge to separate components somewhere after CymA. CymA is not required for aerobic growth or for anaerobic growth coupled to TMAO reduction. Since the CymA-deficient mutant of MR-1 bears the same electron transport deficiencies as the menaquinone-deficient mutant CMA-1 (which is restored to a wild-type phenotype by DHNA, a precursor in the menaquinone synthesis pathway) (32), it is possible that CymA is somehow involved in the transfer of electrons from menaquinone to other electron transport components. One possible role for CymA is to transfer electrons to components which are localized in the periplasm and/or outer membrane. For example, the majority of Fe(III) reductase activity in MR-1 is localized in the outer membrane (31). MR-1 localizes a majority of its membrane-bound cytochromes to the outer membrane when grown under anaerobic conditions (30), and one or more of these outer membrane cytochromes could be part of the electron transport chain for the reduction of Fe(III) and/or Mn(IV) (36). CymA is also necessary for fumarate reduction, which is catalyzed by a periplasmic enzyme in S. putrefaciens (26, 29, 44). However, CymA does not likely serve as a terminal reductase for these various oxidants, e.g., a mutant lacking the 65-kDa fumarate reductase is only deficient in anaerobic growth on fumarate (35).
Prior to this report, a successful suicide vector approach had not been described for MR-1. One report indicated that pACYC184 could not replicate in MR-1 (48), but this was subsequently disproven (37). Another report described the use of pJQ200KS to generate a gene replacement knockout of the fumarate reductase of S. putrefaciens (now S. frigidimarina [47]) NCIMB 400 (13). This use cannot be extrapolated to MR-1 because MR-1 maintains pJQ200KS as a free replicon (C. Myers, unpublished data); this is not surprising because it contains the p15A origin of replication from pACYC184 (45). The use of pEP185.2 was successful in the present study, and the plasmid should prove useful as a suicide vector for the directed replacement of other genes in MR-1.
Quinone analyses of several other strains of S. putrefaciens, including the type strain, ATCC 8071, have been reported. Most strains synthesize two or three ubiquinones (Q-6, Q-7, Q-8) (1, 27, 42), consistent with the three ubiquinones seen in our TLC analysis of MR-1 (Fig. 3). In our previous studies (32), these same three ubiquinones were present and were identified based on relative migration in TLC; the HPLC spectral analysis (λmax at 275 nm) is consistent with their identity as ubiquinones (42). Other strains of S. putrefaciens synthesize two predominant menaquinones, MK-7 and methylmenaquinone-7 (MMK-7), while a few strains also produce small amounts of MK-8 (1, 27, 42). Our TLC analysis demonstrated two prominent menaquinones in MR-1, which were previously designated MK and MMK based on relative migration in TLC (32); the HPLC spectral analysis (λmax at 245, 270, and 330 to 340 nm) is consistent with their identity as MK and MMK (42).
The exact reason for the depressed menaquinone levels in the spontaneous Rfr strain MR-1A is not known. However, they are not due to the presence of rifampin itself as MR-1A cells grown in the absence of rifampin also have markedly depressed menaquinone levels (not shown). The decreased menaquinone levels could be due to changes in the RNA polymerase. Rifampin binds the β (RpoB) subunit of RNA polymerase, and several mutations in rpoB are associated with rifampin resistance (6, 15). In other species, mutations in rpoB leading to rifampin resistance can have pleiotropic effects, including effects on promoter clearance, RNA elongation, transcriptional termination, paused transcription complexes, and DNA supercoiling (11, 12, 16). While the core RNA polymerase (α2ββ′) carries out elongation and termination of RNA synthesis, the addition of a ς subunit to make the holopolymerase (α2ββ′ς) is required for initiation of RNA synthesis (54). The ς subunit of the holoenzyme plays a prominent role in the recognition of promoters (54), and most eubacteria synthesize several different ς factors which direct RNA polymerase to distinct promoters (54). The effect of rifampin on gene expression can vary significantly depending on the ς factor which initiates transcription from that gene (53). Some mutations in rpoB can lead to decreased transcription of certain ς factors, thereby affecting the expression of several genes (56). While the exact rpoB mutation responsible for the Rfr phenotype of MR-1A is unknown at this time, it is plausible that it could be responsible for decreased transcription of one or more of the genes required for menaquinone synthesis. The addition of MK-4 or DHNA to the medium significantly improved the rate of growth of MR-1A on fumarate as well as the menaquinone content of these cells (Fig. 6 and 7).
To the best of our knowledge, this is the first report of a spontaneous rifampin-resistant mutant that leads to markedly depressed levels of menaquinones. This marked effect on menaquinone levels is of concern because spontaneous Rfr mutants are frequently used as recipients in mating protocols designed to generate mutants. Even though the reduced levels of menaquinones in the Rfr strains (MR-1A and CMTn-1) did not affect the ultimate electron acceptor phenotype of a cymA knockout, effects related to the Rfr phenotype must be considered possible contributing factors to the phenotypes of other Shewanella mutants, e.g., those lacking fumarate reductase (13, 35) or MtrB (a putative outer membrane protein required for Fe(III) and Mn(IV) reduction) (4). Even if menaquinone levels are not affected by other rpoB mutations, other as yet uncharacterized effects must be considered. Caution in interpreting the phenotypes of Shewanella mutants derived from Rfr strains is therefore warranted. Given the potential for rpoB mutations to affect paused transcriptional complexes and transcriptional termination, particular caution is advised in interpreting the phenotypes resulting from mutations of genes arranged in operons, e.g., mtrAB of MR-1 (4).
The interpretation of results for the previously described mutant CMA-1 are not complicated by Rfr effects, as it was isolated directly from MR-1 by acridine orange mutagenesis (32). CMA-1 is MK deficient and cannot reduce Fe(III), Mn(IV), or nitrate or grow anaerobically with fumarate. Supplementation of the medium with either MK-4 or DHNA restores the presence of MK in CMA-1 and its ability to utilize nitrate, Fe(III), Mn(IV), and fumarate (32). A recent menC mutant isolated directly from MR-1 confirmed this role for MK (D. K. Newman and R. G. Kolter, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. K-17, p. 404, 1999), i.e., it is deficient in o-succinylbenzoic acid synthase, an enzyme involved in menaquinone synthesis; similar to CMA-1, it could not reduce fumarate, Fe(III), or Mn(IV) and was restored to the wild-type phenotype by supplementation of the medium with DHNA (Newman and Kolter, Abstr. 99th Gen. Meet. Am. Soc. Microbiol.).
In summary, by using a site-directed gene replacement approach, a cymA knockout strain (MR1-CYMA), which was derived directly from MR-1, lost the ability to reduce Fe(III), Mn(IV), and nitrate and to grow with fumarate as an electron acceptor. All of these functions, were restored to wild-type efficacy, and the presence of the cymA transcript and CymA protein was also restored, by complementation with the cymA gene. However, unlike the previous mutant CMTn-1, MR1-CYMA retained menaquinone levels comparable to those of the wild type, thereby providing a definitive demonstration of the requirement for CymA in anaerobic electron transport associated with at least four different electron acceptors. This also represents the first successful use of a suicide vector for directed gene replacement in MR-1. Furthermore, the depressed menaquinone levels in CMTn-1 were the result of the Rfr phenotype of the parent strain, MR-1A, and were unrelated to the interruption of the cymA gene. Rifampin-resistant strains of Shewanella should be used with caution, particularly when analyzing electron transport processes.
ACKNOWLEDGMENTS
This work was supported by National Institute of Health grant R01GM50786 to C.R.M.
We are grateful to V. L. Miller for graciously providing pEP185.2, to D. Frank for providing pUT/mini-Tn5Km, and to K. Nithipatikom for providing assistance with the HPLC analyses.
REFERENCES
- 1.Akagawa-Matsushita M, Itoh T, Katayama Y, Kuraishi H, Yamasato K. Isoprenoid quinone composition of some marine Alteromonas, Marinomonas, Deleya, Pseudomonas, and Shewanella species. J Gen Microbiol. 1992;138:2275–2281. [Google Scholar]
- 2.Alefounder P R, Ferguson S J. The location of dissimilatory nitrite reductase and the control of dissimilatory nitrite reductase by oxygen in Paracoccus denitrificans. Biochem J. 1980;192:231–240. doi: 10.1042/bj1920231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong P B, Lyons W B, Gaudette H E. Application of formaldoxime colorimetric method for the determination of manganese in the pore water of anoxic estuarine sediments. Estuaries. 1979;2:198–201. [Google Scholar]
- 4.Beliaev A S, Saffarini D A. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol. 1998;180:6292–6297. doi: 10.1128/jb.180.23.6292-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berks B C, Richardson D J, Reilly A, Willis A C, Ferguson S J. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem J. 1995;309:983–992. doi: 10.1042/bj3090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodmer T, Zürcher G, Imboden P, Telenti A. Mutation position and type of substitution in the β-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 1995;35:345–348. doi: 10.1093/jac/35.2.345. [DOI] [PubMed] [Google Scholar]
- 7.Brewer P G, Spencer D W. Colorimetric determination of manganese in anoxic waters. Limnol Oceanogr. 1971;16:107–110. [Google Scholar]
- 8.Carver M A, Jones C W. The detection of cytochrome patterns in bacteria. In: Goodfellow M, Minnikin D E, editors. Chemical methods in bacterial systematics. New York, N.Y: Academic Press; 1985. pp. 383–399. [Google Scholar]
- 9.Cataldo D A, Haroon M, Schrader L E, Youngs V L. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal. 1975;6:71–80. [Google Scholar]
- 10.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drlica K, Franco R J, Steck T R. Rifampin and rpoB mutations can alter DNA supercoiling in Escherichia coli. J Bacteriol. 1988;170:4983–4985. doi: 10.1128/jb.170.10.4983-4985.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher R F, Yanofsky C. Mutations of the beta subunit of RNA polymerase alter both transcriptional pausing and transcription termination in the trp operon leader region. J Biol Chem. 1983;258:8146–8150. [PubMed] [Google Scholar]
- 13.Gordon E H J, Pealing S L, Chapman S K, Ward F B, Reid G A. Physiological function and regulation of flavocytochrome c3, the soluble fumarate reductase from Shewanella putrefaciens NCIMB 400. Microbiology. 1998;144:937–945. doi: 10.1099/00221287-144-4-937. [DOI] [PubMed] [Google Scholar]
- 14.Itoh T, Funabashi H, Katayama-Fujimura Y, Iwasaki S, Kuraishi H. Structure of methylmenaquinone-7 isolated from Alteromonas putrefaciens IAM 12079. Biochim Biophys Acta. 1985;840:51–55. [Google Scholar]
- 15.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 16.Jin D J, Zhou Y N. Mutational analysis of structure-function relationship of RNA polymerase in Escherichia coli. Methods Enzymol. 1996;273:300–319. doi: 10.1016/s0076-6879(96)73027-6. [DOI] [PubMed] [Google Scholar]
- 17.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 18.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 19.Knauf V C, Nester E W. Wide host range cloning vectors: cosmid clone bank of Agrobacterium Ti plasmids. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 20.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 21.Kröger A, Dadák V. On the role of quinones in bacterial electron transport; the respiratory system of Bacillus megaterium. Eur J Biochem. 1969;11:328–340. doi: 10.1111/j.1432-1033.1969.tb00776.x. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lovley D R, Phillips E J P. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol. 1986;51:683–689. doi: 10.1128/aem.51.4.683-689.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M-C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 25.Mertz L M, Westfall B, Rashtchian A. PCR nonradioactive labelling system for synthesis of biotinylated DNA probes. FOCUS (Life Technologies, Inc) 1994;16.2:49–51. [Google Scholar]
- 26.Morris C J, Black A C, Pealing S L, Manson F D C, Chapman S K, Reid G A, Gibson D M, Ward B F. Purification and properties of a novel cytochrome: flavocytochrome c from Shewanella putrefaciens. Biochem J. 1994;302:587–593. doi: 10.1042/bj3020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moule A L, Wilkinson S G. Polar lipids, fatty acids, and isoprenoid quinones of Alteromonas putrefaciens (Shewanella putrefaciens) Syst Appl Microbiol. 1987;9:192–198. [Google Scholar]
- 28.Myers C R, Collins M L P. Cell-cycle-specific oscillation in the composition of chromatophore membrane in Rhodospirillum rubrum. J Bacteriol. 1986;166:818–823. doi: 10.1128/jb.166.3.818-823.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers C R, Myers J M. Fumarate reductase is a soluble enzyme in anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1992;98:13–20. [Google Scholar]
- 30.Myers C R, Myers J M. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J Bacteriol. 1992;174:3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers C R, Myers J M. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1993;108:15–22. [Google Scholar]
- 32.Myers C R, Myers J M. Role of menaquinone in the reduction of fumarate, nitrate, iron(III) and manganese(IV) by Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1993;114:215–222. [Google Scholar]
- 33.Myers C R, Myers J M. Ferric iron reduction-linked growth yields of Shewanella putrefaciens MR-1. J Appl Bacteriol. 1994;76:253–258. doi: 10.1111/j.1365-2672.1994.tb01624.x. [DOI] [PubMed] [Google Scholar]
- 34.Myers C R, Myers J M. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers C R, Myers J M. Isolation and characterization of a transposon mutant of Shewanella putrefaciens MR-1 deficient in fumarate reductase. Lett Appl Microbiol. 1997;25:162–168. doi: 10.1046/j.1472-765x.1997.00196.x. [DOI] [PubMed] [Google Scholar]
- 36.Myers C R, Myers J M. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim Biophys Acta. 1997;1326:307–318. doi: 10.1016/s0005-2736(97)00034-5. [DOI] [PubMed] [Google Scholar]
- 37.Myers C R, Myers J M. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett Appl Microbiol. 1997;24:221–225. doi: 10.1046/j.1472-765x.1997.00389.x. [DOI] [PubMed] [Google Scholar]
- 38.Myers C R, Nealson K H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 39.Myers C R, Nealson K H. Microbial reduction of manganese oxides: interactions with iron and sulfur. Geochim Cosmochim Acta. 1988;52:2727–2732. [Google Scholar]
- 40.Myers C R, Nealson K H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol. 1990;172:6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers J M, Myers C R. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim Biophys Acta. 1998;1373:237–251. doi: 10.1016/s0005-2736(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 42.Nishijima M, Araki-Sakai M, Sano H. Identification of isoprenoid quinones by frit-FAB liquid chromatography—mass spectrometry for the chemotaxonomy of microorganisms. J Microbiol Methods. 1997;28:113–122. [Google Scholar]
- 43.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pealing S L, Black A C, Manson F D C, Ward F B, Chapman S K, Reid G A. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens: a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry. 1992;31:12132–12140. doi: 10.1021/bi00163a023. [DOI] [PubMed] [Google Scholar]
- 45.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 46.Raghava G P S. Improved estimation of DNA fragment length from gel electrophoresis data using a graphical method. BioTechniques. 1994;17:100–104. [PubMed] [Google Scholar]
- 47.Reid G A, Gordon E H. Phylogeny of marine and freshwater Shewanella: reclassification of Shewanella putrefaciens NCIMB 400 as Shewanella frigidimarina. Int J Syst Bacteriol. 1999;49:189–191. doi: 10.1099/00207713-49-1-189. [DOI] [PubMed] [Google Scholar]
- 48.Saffarini D A, Nealson K H. Sequence and genetic characterization of etrA, an fnr analog that regulates anaerobic respiration in Shewanella putrefaciens MR-1. J Bacteriol. 1993;175:7938–7944. doi: 10.1128/jb.175.24.7938-7944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1–3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 51.Townsend K M, Frost A J, Lee C W, Papadimitriou J M, Dawkins H J S. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van'T Riet J, Stouthamer A H, Planta R J. Regulation of nitrate assimilation and nitrate respiration in Aerobacter aerogenes. J Bacteriol. 1968;96:1455–1464. doi: 10.1128/jb.96.5.1455-1464.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wegrzyn A, Szalewska-Palasz A, Blaszczak A, Liberek K, Wegrzyn G. Differential inhibition of transcription from ς70- and ς32-dependent promoters by rifampicin. FEBS Lett. 1998;440:172–174. doi: 10.1016/s0014-5793(98)01449-5. [DOI] [PubMed] [Google Scholar]
- 54.Wösten M M S M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Y N, Jin D J. RNA polymerase β mutations have reduced ς70 synthesis leading to a hyper-temperature-sensitive phenotype of a ς70 mutant. J Bacteriol. 1997;179:4292–4298. doi: 10.1128/jb.179.13.4292-4298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]