Abstract
The SARS-CoV-2 pandemic has highlighted the need for improved technologies to help control the spread of contagious pathogens. While rapid point-of-need testing plays a key role in strategies to rapidly identify and isolate infectious patients, current test approaches have significant shortcomings related to assay limitations and sample type. Direct quantification of viral shedding in exhaled particles may offer a better rapid testing approach, since SARS-CoV-2 is believed to spread mainly by aerosols. It assesses contagiousness directly, the sample is easy and comfortable to obtain, sampling can be standardized, and the limited sample volume lends itself to a fast and sensitive analysis. In view of these benefits, we developed and tested an approach where exhaled particles are efficiently sampled using inertial impaction in a micromachined silicon chip, followed by an RT-qPCR molecular assay to detect SARS-CoV-2 shedding. Our portable, silicon impactor allowed for the efficient capture (>85%) of respiratory particles down to 300 nm without the need for additional equipment. We demonstrate using both conventional off-chip and in-situ PCR directly on the silicon chip that sampling subjects’ breath in less than a minute yields sufficient viral RNA to detect infections as early as standard sampling methods. A longitudinal study revealed clear differences in the temporal dynamics of viral load for nasopharyngeal swab, saliva, breath, and antigen tests. Overall, after an infection, the breath-based test remains positive during the first week but is the first to consistently report a negative result, putatively signalling the end of contagiousness and further emphasizing the potential of this tool to help manage the spread of airborne respiratory infections.
Keywords: SARS-CoV-2, Aerosols, Breath, Impactor, Diagnostics, Lab-on-a-chip
1. Introduction
Person-to-person transmission facilitated by respiratory droplets plays an important role in the spreading of infectious diseases. For SARS-CoV-2, airborne transmission is thought to occur over both short and long distances by a continuum of exhaled particle sizes. While large droplets settle quickly, smaller particles can remain aloft for hours and travel long distances (Greenhalgh et al., 2021; Vuorinen et al., 2020). Especially in poorly ventilated spaces where people congregate, exhaled particles potentially containing infectious virus particles can accumulate, leading to a significantly increased infection risk (Li et al., 2021). While transmission through aerosols is difficult to demonstrate directly, multiple studies have identified airborne SARS-CoV-2 to be viable (Lednicky et al., 2020; Tang et al., 2021) and reports of super-spreader events point to exhaled particles playing a key role in viral spreading (Lemieux et al., 2021). Rapid diagnosis and contact tracing together with quarantining of potentially infected persons has proven to be a cornerstone of public health measures deployed by many countries to contain spread in the absence of immunity (Raymenants et al., 2022). While nasopharyngeal swab tests are the most common method of sampling (Kevadiya et al., 2021), it is perceived as unpleasant by most subjects (Takeuchi et al., 2021) and only indicates if a person has been infected recently. Given the clear role of exhaled particles in the transmission of SARS-CoV-2, there is a need for techniques to analyze a person's contagiousness at the point-of-need. Various techniques have been used to collect airborne viral particles, including large liquid impingers, solid impactors and electrostatic precipitators (Verreault et al., 2008). These approaches are, however, limited due to their excessive size, long sampling times, large pressure drop that is required for efficient particle collections (Coleman et al., 2021), and the use of harsh conditions that limit the amount of detectable material. Portable solutions to collect exhaled viral particles, relying on sampling the exhaled breath condensate (Daniels et al., 2021; Nwanochie and Linnes, 2022; Ryan et al., 2021) or using filter papers (Malik et al., 2021), allow little design flexibility and need significant sample volumes for analysis which impacts sensitivity negatively. A recent study that used face mask filters yielded a subpar sensitivity (Smolinska et al., 2021). To overcome these limitations, we have developed a portable breath sampler capable of collecting respiratory particles in combination with a standard molecular test as a non-invasive method for routine sampling and to yield insight into a person's contagiousness (see Fig. 1 ).
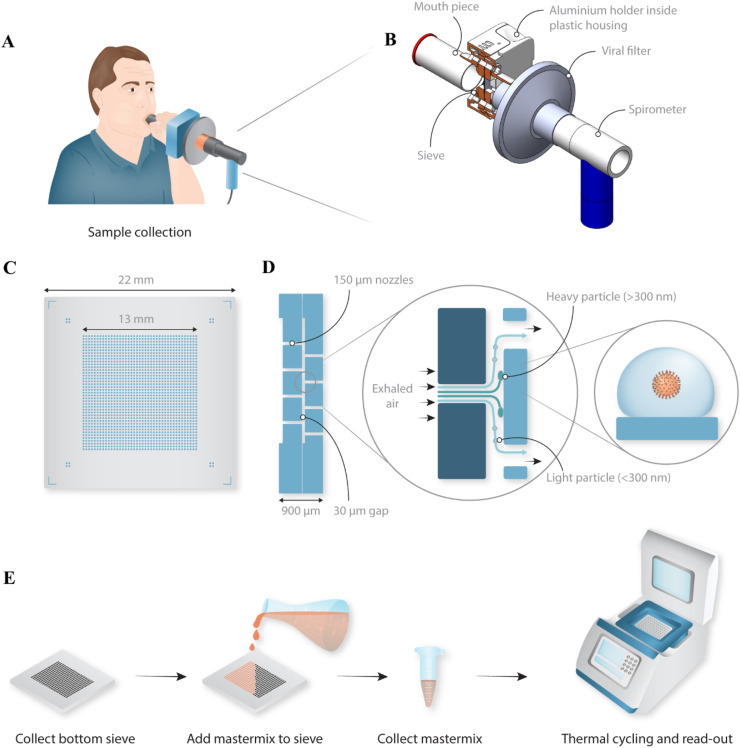
Fig. 1.
Schematic overview of the portable device to sample exhaled particles. (A) Schematic representation of a person breathing into the sampling device. (B) Design of the disposable sampling device with the position of the silicon sieve indicated and kept in place by a holder consisting of an aluminium pre-heated block with O-rings for sealing. A mouthpiece is used in front, and a viral filter in between the silicon impactor and a spirometer (indicated in blue) for measuring flow rate during sampling. (C) Schematic top-view of the final sieve, 22 × 22 mm2 in size, consisting of an array of 1600 nozzles with a diameter of 150 μm. (D) The non-integrated, non-monolithic impactor consists of two sieves stacked on top of each other, creating a gap of 30 μm between the two arrays of holes (the single piece, monolithic impactor is described in Fig. 4). Exhaled particles, some containing virus, are collected on the bottom sieve by inertial impaction, while air and very small particles (<300 nm) are directed to the outlet nozzles and exit without impacting. (E) Schematic overview of the used protocol for this non-monolithic version of the impactor. The bottom sieve is removed from the sample device and master mix is pipetted on top followed by a brief spin to collect the sample. The sample is transferred to a 96-well plate and an RT-qPCR is conducted using a commercial thermal cycler.
2. Materials and methods
2.1. Impactor design and simulations
The finite volume method and 3-D simulations of the impactor chip were performed using ANSYS Fluent commercial software. The discrete phase model (DPM) was used to track the particles’ motion in the Lagrangian domain while the Eulerian formulation is simultaneously used for the continuous phase (Zahari et al., 2018). The pressure and velocity fields were calculated by solving the steady state laminar Navier-Stokes equation. The collection efficiency for a specific particle diameter was estimated by tracking the particles injected at the inlet of the nozzle using the DPM. The collection efficiency is the fraction of particles that are trapped on the impaction surface over the total number of particles injected. Because of the small size of tracked particles (<1 μm), the Cunningham correction factor had to be applied to the solution using a user defined function (UDF) (Gussman, 1969).
2.2. Fabrication of the silicon impactor
To produce non-monolithic impactors, sieves were fabricated from 200 mm Si wafers using standard lithographic techniques. In short, after a standard clean to remove any particles, the Si wafers were patterned followed by a Bosch dry etch step (200 mm DSi-Rapier etcher, SPTS Technologies Ltd), resulting in a front side 30 μm deep shallow cavity. The cavity surface was then protected by a 200 nm thin thermal oxide layer. A temporary carrier was used to shield the front side cavity prior to grinding the wafers to 450 μm. The nozzles were then processed using back-side litho and etch landing on the shallow cavity. Afterward, the temporary carrier was removed using laser debonding. Extensive wet cleans followed by an ozone clean were applied to remove all the residues. The oxide to protect the Si surface was then removed by dipping the wafers in a diluted (10% v/v) HF solution. A new 150 nm thin high quality oxide layer was formed by thermal oxidation at 1050 °C prior to standard dicing. It is expected that exhaled particles attach to any surface with which they come into contact, hence, no coating other than the silicon oxide finishing during processing was applied. A shown in Fig. 1, two sieve samples were mounted on top of another to create a fully functional impactor with a 30 μm gap.
Monolithic impactors were fabricated on Si–Si fusion bonded 200 mm wafers. The bottom wafers were first processed using deep reactive ion etching (DRIE) to form the 30 μm shallow cavity. After a thermal oxidation step to have a 200 nm thin oxide at the surface, these wafers were oxide-oxide fusion bonded with another blank Si wafer called top wafer. The top Si wafer was then grinded, resulting in a 100 μm thick membrane on top of the shallow cavity. Next, the top nozzles as well as the fluidic inlet/outlet were formed by a standard sequence of lithographic patterning followed by a dry etch step as described for the non-monolithic sieves. Next, a temporary carrier was used again to protect the holes at the top wafers, enabling backside grinding to 450 μm total Si–Si thickness and etching to form the backside nozzles. Special consideration was given to the timing of the backside etch to limit any over-etching. After removal of the temporary carriers, a sequence of wet clean steps followed, including a mesitylene and sulphuric acid-ozonclean, to remove all residues from temporary bonding process as well as passivation polymers from the Bosch dry etch step. Finally, a new 150 nm thin high-quality oxide was formed by thermal oxidation and the wafers were diced to the final impactors (2 × 1.8 cm) used for testing. The monolithic impactors were used for the studies using the in-situ PCR, while the non-monolithic impactors were used for all other clinical studies. A comparison of the designs of the impactors is shown in Fig. S1, while more performance attributes of the monolithic impactors are shown in Fig. S2. Both impactors were heated prior to use to avoid water condensation (see Fig. S5C).
2.3. Impactor capture efficiency tests
To determine the capture efficiency of the breath sampler (see Fig. 2C and Fig. S2), a nebulizer set-up was used to generate aerosols in a controlled manner. Aerosols were generated with an AGK2000 particle generator (Palas, Germany) from a 1.25% KCl solution at 1 bar. The concentration of particles was adjusted by venting and diluting with air. Pneumatic switch valves enabled us to choose the flow path, and either went through an empty tube (control) or through our silicon chip. The aerosolized particle size distribution and concentration that left the control line or the chip was measured by a spectrometer Promo 2300 (Palas, Germany). The average concentration of particles arriving to the spectrometer during control Cnaerosol and during collection Cncollection was calculated. Collection efficiency was calculated as follows (Cnaerosol - Cncollection)/Cnaerosol. The flow was regulated using an additional vacuum pump (RZ6 Vacuubrand), a flow meter and an adjustable flow restrictor allowing measurements from 5 L/min to 35 L/min.
Fig. 2.
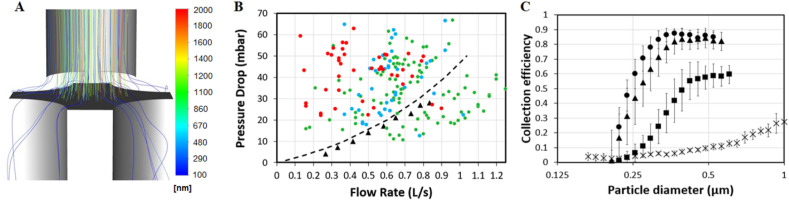
Non-monolithic silicon impactor characteristics. (A) Fluidic simulation for the designed sieve visualizing particle trajectories of different sizes, coloured by particle diameter, generated using Ansys Fluent software for a nozzle with a diameter of 150 μm at a flow rate of 0.6 L/s. (B) Experimentally (triangles) measured and simulated (dotted line) pressure drop of the sieve versus flow rate. The rated comfort levels of a test panel for different flow rates and pressure drops are indicated as well with green dots being perceived as a comfortable, blue as a neutral and red as an uncomfortable user experience. (C) Normalized capture efficiency of the impactor as a function of particle diameter for different flow rates (crosses: 0.08 L/s, rectangles: 0.25 L/s, triangles: 0.42 L/s, and circles: 0.6 L/s). The error bars correspond to the standard deviation over 4 different impactor chips.
2.4. Impactor acceptance measurements
A group of participants (N = 32) was recruited to perform a series of breathing exercises in a lab setup created to determine the acceptable pressure drop for the sieve (see Fig. 2B). Participants were selected based on a short survey, taking into account differences in age, fitness and health conditions. Next to a group of healthy controls (N = 12), the recruitment focused on profiles expected to have difficulties with performing the resistance tests, namely (1) elderly (N = 8), (2) sedentary people (N = 4) and (3) persons with respiratory difficulties (N = 8). The lab setup (see Fig. S3) included a spirometer and an adjustable valve to modify the pressure drop. Conditions with varying pressure drop were presented to participants in randomized order. Based on the spirometer data, a custom-built software program provided feedback on their flow rate (target 0.6 L/s) and progress (target of 20 L exhaled air). Following each exercise, participants rated subjective experience on a Likert scale ranging between 1 (very comfortable) to 7 (very uncomfortable), and perceived effort on a Borg scale ranging from 1 (very light activity) to 10 (max effort activity). To visualize the Likert data, a rating of 1–3 was recoded as comfortable, 4 as neutral, and 5–7 as uncomfortable. To adhere to hygienic and safety measures, a strict protocol was followed, including disinfection steps and disposable HEPA filters and mouthpieces. Participants received a small monetary reward in return.
2.5. RT-qPCR protocols
For the nasopharyngeal and saliva samples, the standard validated protocol at University Hospitals Leuven was used. More specifically, the nasopharyngeal samples were collected in 1.5 mL zymo-medium (Zymo Research) and saliva samples (target 1.3 mL) were collected in fertipro kits containing 2 mL InactivBlue transport medium (InActiv Blue™). Sample transfer was performed using Tecan Evo200, Air liha. Extraction was performed using the KingFisher extraction robot. RT-qPCR was performed using the Taqpath 2019-ncov assay kit v2 (ThermoFisher) on 384-well plates using a Quantstudio 5 thermocycler. Analysis was performed using FastFinder analysis v4.x.
For the different impactors (see Fig. S1), slightly different direct (i.e. single step) RT-qPCR methods were used. The sequences of primers and probes are listed in Table S1. For all protocols, a reference curve was obtained using synthetic RNA (Twist Biosciences) or genomic RNA (Vircell) (see Fig. S4B). More specifically.
-
(1)
for the non-monolithic impactor, a direct RT-qPCR was performed on the collected, exhaled particles. The RT-qPCR mix contained both primers (500 nM 2019-nCoV_N2–F and 2019-nCoV_N2-R) and Taqman N2 probe (125 nM) ordered from IDT (IDT, Belgium) targeting the SARS-CoV-2 N2 gene, and feline infectious peritonitis virus (FIPV) primers (FcoV1128f 400 nM, FcoV1129r 900 nM) and probes (250 nM) and feline coronavirus extracted RNA template (1e2 RNA copies per reaction) used as internal control.
The mix further contained ready to use RT-qPCR buffer (TaqPath™ 1-Step RT-qPCR Master Mix, ThermoFisher) supplemented with 0.1% Triton X100 for virus lysis (see Supplementary Materials) and nuclease free water to a total volume of 50 μl. This mix was then added to the surface of the bottom silicon sieve, on which the exhaled particles impacted. After a short 2′ incubation, the RT-qPCR mix was collected by briefly spinning the sieve using a 50 mL plastic tube as a holder. The as such collected RT-qPCR mix was transferred to PCR strips and loaded into a benchtop thermal cycler (LC96, Roche) for RT-qPCR (50 °C for 15′ followed by 3’ at 95 °C and 50 cycles of 15″ at 90 °C and 60” at 60 °C). The Cq values were determined using the LightCycler application software.
-
(2)
for the monolithic impactor and when using the custom instrument (see Fig. S6), a similar, somewhat faster protocol (30′ qPCR) with minor modifications was used. More specifically, the RT-qPCR protocol used similar N2 primers and probes, but at an elevated concentration compared to the off-sieve PCR. The master mix contained 5 μM of both 2019-nCoV_N2–F and 2019-nCoV_N2-R, 400 nM N2 probe, 0.1U/μl KAPA2G HS fast polymerase and TaqPath™ 1-Step RT-qPCR Master Mix supplemented with 1% Tergitol 15-S-9 for virus lysis. After an RT step of 15′ at 50C, the mix was held at 95 °C for 3′, followed by 45 PCR cycles (10″ 60 °C, 1″ 95 °C with a ramp rate of 10 °C/s). The data analysis is detailed in the Supplementary Materials.
-
(3)
for demonstrating an ultra-fast, below 5′ RT-qPCR on chip, a further modified protocol was developed as detailed in the Supplementary Materials and results shown in Figs. S4C–D.
2.6. Trial design and participants: exhaled particles sampling in hospitalized patients
The trial complied with the Declaration of Helsinki, the International Conference on Harmonization Guidelines for Good Clinical Practice, and applicable local regulations. The protocol was reviewed and approved by the ethics committee, and all subjects provided written informed consent before study entry. Subjects were recruited at the low-care covid ward at University Hospital Leuven (Belgium). For inclusion of SARS-CoV-2 positive subjects, patients hospitalized at the ward, either for COVID-19-related symptoms or for other health issues, were approached to participate in the study. All patients were aged 42 years or older, and tested positive for SARS-CoV-2 infection within 3 days of study inclusion. Both symptomatic and asymptomatic subjects were included. Specific symptoms were not systematically recorded.Patients with significant breathing problems were excluded from the study. A nasopharyngeal swab test was taken on the first day of inclusion in the study, and analyzed with the reference RT-PCR test at the University Hospital to confirm the SARS-CoV-2 infection status. Healthy volunteers were recruited amongst the hospital staff at the low-care covid ward. At the time of this study, no subjects had received a vaccine or had confirmed previous SARS-CoV-2 infection.
After enrolment, study participants were asked to breathe into the breath sampler. A breathing test was defined as 20 tidal exhalations through the breath sampler, while the air flow was measured with a spirometer. During the breathing test, all subjects were instructed to stay within their comfort levels, and thus the air flow was variable in this study. In all cases, the subjects were instructed to exhale into the mouthpiece of the breath sampler and inhale away from the device or through the nose. Subjects were asked to repeat the breathing test typically 2 times. A number of patients were tested again up to 4 days later.
2.7. Trial design and participants: exhaled particles sampling in student testing center
The trial complied with the Declaration of Helsinki, the International Conference on Harmonization Guidelines for Good Clinical Practice, and applicable local regulations. The protocol was reviewed and approved by the ethics committee, and participants provided written informed consent before study entry.In order to be included in the study, the subjects had to be tertiary education students residing in Leuven, Belgium, who wanted to be tested following a high-risk contact, symptoms typical of a SARS-CoV-2 infection, or after returning from a high-risk country. For the selection of subjects, a combination of risk assessment (questionnaire) and pre-selection through rapid tests (Abbott Panbio Covid-19 Ag test kit) was used. As the students were sampled using a nasopharyngeal swab test, the result of this test was used as reference. It should be noted that even subjects selected as a “negative subject” were still at risk of being positive. The clinical study contained 3 parts: (1) a comparison of different breathing techniques, (2) a longitudinal study, following subjects over multiple days with multiple tests daily and (3) a comparison between on-sieve and off-sieve RT-qPCR methods.
-
(1)
For comparison of breathing techniques, volunteering students were asked to breathe through the sieve using different breathing methods (4 min tidal breathing; 2 min tidal breathing; 1 min exhaling accompanied by the sound “e”; 10 deep exhalations into the device). In all cases, the subjects were instructed to alternatingly exhale into the mouthpiece and inhale away from the device or via the nose. Subjects were asked never to go beyond their comfort zone and could stop at any moment.
-
(2)
For the longitudinal study, subjects were screened for recruitment by the KU Leuven contact tracing team focusing on contact tracing of the Leuven tertiary education student population (Raymenants et al., 2021). Subjects needed to be recently exposed to a confirmed COVID-19 case. Preferentially, this source case had a high viral load and had likely caused a secondary infection already. Exclusion criteria were: previous vaccination or COVID-19 infection (based on previous positive RT-qPCR test or antibody test at inclusion), exposure more than 7 days prior to assessment for inclusion, physical inability to attend the testing center or inability to provide informed consent. Participants were preferentially recruited if they were thought to be in an early phase of infection based on initial diagnostic tests, symptom onset and exposure history. Saliva (saliva N) and breath RT-qPCR (breath test N) were performed once to twice daily while a nasopharyngeal RT-qPCR (NP Swab N) and antigen (Abbott AG) test were performed once daily. Participants remaining negative were excluded 5–7 days after exposure. Participants who tested positive during follow-up were initially asked to provide breath and saliva samples twice daily. However, intermediate data analysis revealed limited variation in same day Ct values, prompting a switch to once daily sampling of subjects for all diagnostic tests. Symptoms were not systematically recorded. When participants remained positive on at least one of the performed tests until day 10, follow up was prolonged until subjects were deemed unlikely to still be infectious by a medical doctor based on Ct values and symptom resolution. Due to the availability of the subject or test taker, no sample was taken on a limited number of days. The selection procedure and inclusion flowchart are visualized in Fig. S8.
-
(3)
For the comparison between on-sieve and off-sieve RT-qPCR, SARS-CoV-2 positive subjects were asked to breathe through the 2 different systems, using the vocalization protocol. Each method was repeated twice, resulting in a total of 4 datapoints per subject.
3. Results
3.1. Impactor design and particle capture efficiency
To efficiently capture exhaled particles from breath while minimizing perceived effort and resulting sample volume, a silicon impactor was designed (see Fig. 1C-D). The dimensions and performance attributes of the impactor chip were determined by numerical simulations (see Fig. 2A-B). As the relationship between flow rate, pressure drop, and collection efficiency needs to be balanced carefully; the design requirement was to efficiently capture particles as small as 300 nm-diameter at a flow rate of 0.6 L/s and with a pressure drop of less than 30 mbar, while keeping the functional chip area to a minimum. The pressure drop was chosen to ensure most people felt comfortable while exhaling through the sieve, which was confirmed based on results of a mixed test panel (see Fig. 2B). Results of the same test panel showed that a higher pressure drop – especially in combination with a low achieved flow rate – resulted in a less comfortable user experience (see Fig. 2B). The measured pressure drop and the collection efficiency of the silicon impactor matched simulation results (see Fig. 2B–C). Consistent capture efficiencies over 85% were measured for particles larger than 300 nm diameter at a flow rate of 0.6 L/s. While larger particles are more easily diverted from the air flow, the collection of smaller particles requires a higher flow rate with an associated pressure drop through the nozzles.
3.2. SARS-CoV-2 detection in exhaled particles
To demonstrate the efficient detection of SARS-CoV-2 in exhaled breath using the silicon impactors, a first clinical study was performed focusing on sampling hospitalized patients. Both SARS-CoV-2 positive (considered positive based on an earlier nasopharyngeal swab-sampled laboratory RT-qPCR test) and healthy volunteers (considered negative) were asked to perform 20 tidal exhalations into the portable breath sampler (Fig. 1B). A nasopharyngeal swab sample was taken contemporaneously. Next, the impactor chips were retrieved from the sampler and rinsed with master mix to perform a direct RT-qPCR test (see Fig. 1E). The swabs were analyzed with a reference RT-qPCR assay. Of the 55 subjects tested, 23 were negative using both nasopharyngeal swab and breath tests. Of the 32 patients determined positive using the nasopharyngeal swab, the breath sampling method confirmed presence of SARS-CoV-2 in 24 patients and 8 yielded a negative result (i.e. 75 % positive agreement and 100% negative agreement, see Supplementary Materials). This apparent lower sensitivity of breath testing is considered to result from the phase of infection in which sampling took place (see Longitudinal study and Discussion section). The threshold cycle (Ct) value of the breath samples tends to be higher than that of the nasopharyngeal samples, indicating that significantly less viral RNA is captured using breath sampling. However, for 4 subjects, three of which were asymptomatic, the breath sample had a lower Ct value compared to the nasopharyngeal swab (see Source Data S1). Three of these patients were retested 3 or 4 days later and consistently showed a positive breath test. These results corroborate earlier reports describing high viral loads in persons without symptoms and stressing the importance of asymptomatic and pre-symptomatic transmission (Kenyon, 2020).
To assess the impact of different breathing protocols on the sensitivity of the breath-based RT-qPCR test, a second clinical study was conducted in ambulatory patients comparing 2 and 4 min of tidal breathing, 10 forced exhalations, and 1 min of vocalizing whilst exhaling. In this trial, 56 subjects visiting a student COVID-19 test center (Raymenants et al., 2021) were sampled with all four breathing protocols. A rapid antigen test was used to pre-screen these participants. Of the 56 participants, 33 tested negative and 23 tested positive on a nasopharyngeal swab laboratory RT-qPCR test which was used as a reference. Breath sampling using vocalization was the most sensitive breath sampling protocol, yielding a positive result in 17 out of 23 positive participants (i.e. 74 % positive agreement and 100% negative agreement, see Supplementary Materials). The better performance of vocalization can be explained by a higher rate of emission of particles upon vocalizing as opposed to tidal breathing (Asadi et al., 2019; Chen et al., 2021a). The forced exhalation approach appeared much less sensitive (i.e. 22% positive agreement) in this student group (average ∼22 years old, ambulatory, upright position during sampling) as compared to the first study on a cohort of hospitalized patients (average ∼62 years old, in-patient, recumbent position during sampling), suggesting a possible effect of age, sampling position, a lower respiratory tract infection prompting admission, or another covariate not assessed within this study (Chen et al., 2021b; Edwards et al., 2021). The observed difference in positive percentage agreement between breathing techniques (22% vs 74%) is striking and warrants further research. In both studies, the observed Ct values were reproducible (SD 0.94) all above 23 (below ∼104 viral copies/sieve) and mostly above 27 (below ∼103 copies/sieve), in the same order of magnitude as earlier empirical data assessing concentrations of viral pathogens in exhaled breath during 30 min of tidal breathing (Coleman et al., 2021; Leung et al., 2020; Yan et al., 2018). All Ct values and associated viral copy numbers are listed in Source Data File S2.
3.3. Longitudinal study
A longitudinal study was set up to assess how RT-qPCR on exhaled particles collected while vocalizing corresponds to RT-qPCR results using nasopharyngeal swabs and saliva samples, and to a rapid antigen test performed on nasopharyngeal swabs, during the course of a COVID-19 infection. High risk contacts of confirmed SARS-CoV-2 positive subjects were followed up prospectively to study infections from an early phase onwards (see Methods and Fig. S8). Of the 58 high-risk contacts included in the study, 11 developed an infection.
In contrast to the moderate 74% positive agreement between the breath and nasopharyngeal tests in our in-patient study, the individual graphs of study participants in this ambulatory longitudinal study show very similar trends between the different RT-qPCR based tests (Fig. 3 ). The first day subjects became positive using any of the four tests, they were detected using breath in 9 out of 11 subjects, using nasopharyngeal swabs in 6 out of 8 subjects, using saliva in 10 out of 11 subjects, and using rapid antigen tests in 5 out of 9 subjects (see Source data S3). In other words, we find that RT-qPCR on exhaled particles turned positive as early as RT-qPCR on nasopharyngeal and saliva samples while the antigen test appeared less sensitive at the start of an infection.
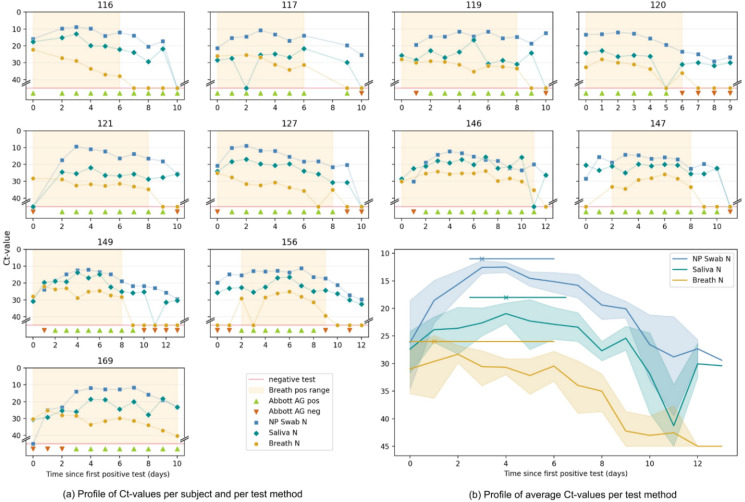
Fig. 3.
Longitudinal study. The findings of the longitudinal study are summarized comparing Ct values of the N gene in breath (breath test N), nasopharyngeal swab (NP Swab N) and saliva (saliva N) to a rapid antigen test on nasopharyngeal swab (Abbott AG). (A) Individual graphs of the 11 participants followed up over the course of their infection, day 0 being the first day any diagnostic test turned positive. The period in which the breath test is positive is shaded. As shown, 3/11 participants were positive on all tests concurrently, while 8/11 had discrepant results on the first day of testing positive. In the latter cases, the breath test turned positive before NP swab on two occasions (subject 121 and 169) and after in two others (subject 147 and 156). The Abbott AG test turned positive 0–3 days after a PCR test (mean 1.4 days). (B) A summed graph in which the Ct values of all tests performed in the 11 participants on a particular day are averaged for one particular sample type. A trend towards an earlier peak in the breath test N in comparison to NP Swab N and saliva N is shown on top. Median and 95%CI were calculated using the bootstrap method. Lastly, as shown and more clearly visible in Fig. S7, the breath test turns negative before RT-qPCR on other respiratory samples.
Quickly after the first positive test (day 2 onwards), a 100% positive agreement is observed between the nasopharyngeal and breath RT-qPCR up to day 6. From day 7 onwards, the agreement drops as less exhaled virus particles were detected and more breath tests turned negative (see Fig. S7). Exhaled particle-based viral loads appear to have an early peak value followed by a monotonic decline, in contrast to the other sample types (Fig. 3B). In general, we observe that the breath-based RT-qPCR becomes negative before the other molecular tests. As the Ct values also rapidly increase to high Ct values, we have not applied a cut-off value as often done for nasopharyngeal based tests.
3.4. In-situ rapid on-chip RT-qPCR
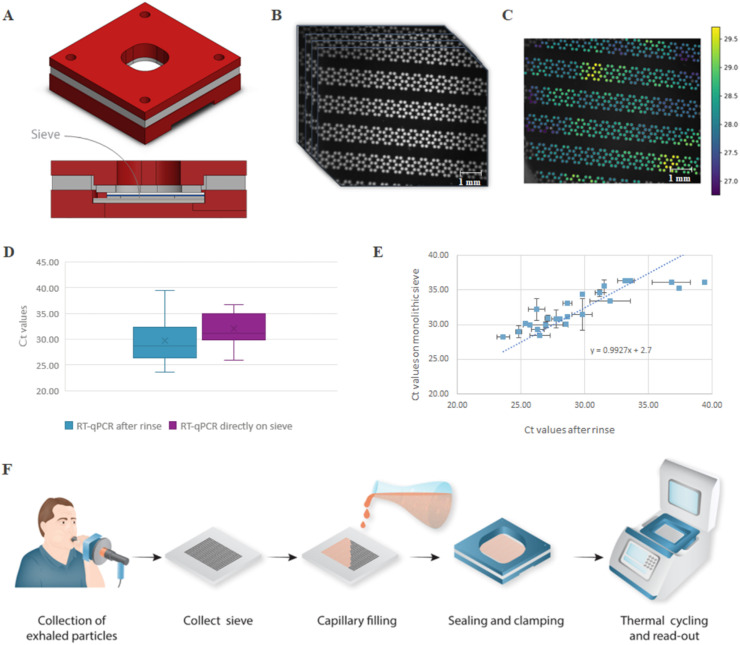
The minute volume of the exhaled particle samples, in combination with the thermal properties of the silicon impactor chips, is ideally suited for conducting rapid and direct molecular tests at the point-of-need. To demonstrate this, an integrated workflow was designed, eliminating the need for a rinsing step after sampling (see Fig. 4 ). After sample collection, reagents were pipetted directly into the silicon impactor that filled by capillary actuation. The impactor was then sealed using a custom clamp (Fig. 4A), followed by a direct, in situ- RT-qPCR using a dedicated thermal cycler (see Fig. 4F). Results of the monolithic sieve were very similar to the non-monolithic approach using rinsing and off-sieve amplification (see Fig. 4D and E). Of the 40 clinical samples tested, 28 were positive and 5 were negative for both methods performed in duplicate. Of the remaining 7 samples, 1 sample was only positive on the non-monolithic sieve for currently unknown reasons. The other 6 samples showed non-identical duplicates, high Ct values, or a non-uniform amplification (i.e. discrete positive regions) on the monolithic sieve, all indications of the presence of very few viral copies and results being Poisson limited (see Table S4).
Fig. 4.
In-situ RT-qPCR using an integrated, monolithic silicon chip. (A) A 3D CAD image and cross-section of the clamped sieve. The poly-methyl-methacrylate (PMMA) housing has an opening on top for optical access and at the bottom for thermal access. The monolithic sieve itself is clamped in between a top glass substrate with a clear silicone sheet and a bottom silicon substrate with a Li2000 thermal tape for good thermal contact. More details of the housing and clamp are shown in Fig. S5 (B). The optical signal of the sieve is captured for each cycle during thermal cycling using an in-house developed RT-qPCR set-up resulting in a series of images. (C) Resulting heat-map of the fluorescent signals. An R-script is used to generate a Ct value for every nozzle/well. In the example shown, the median Ct value is 28.2 and the mean Ct value is 29.3, calculated over a total of 701 nozzles. (D) Ct values obtained from the positive clinical samples comparing the rinsing method (mean 28.6) with the in-situ RT-qPCR method (mean 31.1). Note that the shift in Ct value was also apparent in the reference curves (see Fig. S4B). (E) Scatterplot of the individual clinical samples that were positive for both RT-qPCR performed on the non-monolithic sieve with rinsing (x-axis) and monolithic sieve (y-axis) showing a linear relationship. The error bars represent the standard deviation from 2 samples gathered from the same subject at the same time point. (F) Schematic overview of the used protocol for the monolithic version of the impactor. The monolithic sieve is removed from the sample device, followed by adding the master mix. The impactor fills by capillary fluidic movement after which both sides of the sieve are sealed using a PMMA clamp. The sieve with clamp is positioned in the custom thermal cycler for direct, in-situ RT-qPCR. More details of the set-up are shown in Fig. S6.
4. Discussion
Molecular tests on nasopharyngeal swabs have been the reference method for SARS-CoV-2 testing because of their purported sensitivity and specificity. Key disadvantages of this testing approach are that nasopharyngeal swab sampling is usually experienced as uncomfortable and requires a healthcare worker. Furthermore, it is prone to detecting viral particles that are no longer acutely infective. The superior sensitivity of molecular assays means they are better at detecting infections early as compared to rapid antigen tests (Dinnes et al., 2020), but also risk over-diagnosing active SARS-CoV-2 replication at the tail end of an infection (Mancuso et al., 2020). Rapid antigen tests are preferred for their ease-of-use, but do not have the performance benefits of a molecular test. Most importantly, no existing test approach targets air-borne transmission. Since SARS-CoV-2 is believed to spread mostly through exhaled particles, the exhaled viral load may allow for a more accurate measurement of the actual transmission window. Some efforts have focused on analyzing exhaled volatile organic compounds borne from the host response (Chen et al., 2021c; Ibrahim et al., 2021), providing indirect evidence of an infection.
Using a novel sampling device, we have demonstrated a rapid, integrated workflow for the sensitive capture and molecular detection of virus particles in breath. While the detected viral concentrations show significant subject-to-subject variability, good reproducibility is obtained for a given subject over multiple days or when tested in duplicates. Vocalizing while exhaling appeared most sensitive amongst different breathing protocols tested, consistent with reports on the effect of vocalization on exhaled aerosol production (Asadi et al., 2019; Gregson et al., 2021) and anecdotal evidence of the importance of vocalization in superspreading events (Hamner et al., 2020). Using our breath sampling method, we demonstrated in a longitudinal study a similar accuracy and sensitivity compared to the nasopharyngeal swab during the first week of an infection (see Fig. S7). The rapid decline of detectable viral particles after ∼1 week also clarifies the apparent lower positive agreement (75% PPA) observed in the earlier clinical studies in which subjects throughout the entire course of an infection were included. None of the tests was able to detect at the start (day 0) all 11 of the 58 subjects that turned positive. All RT-qPCR based tests showed a similar detection rate, but especially the rapid antigen tests performed less (56% at day 0; 50% at day 1) confirming the better reliability of a molecular test early on. Viral load assessed using the breath sample tended to peak earlier compared to and became undetectable before other sample types. These results suggest that the exhaled viral load decreases gradually over time after a peak early in the course of an infection. This is consistent with contact tracing studies showing that SARS-CoV-2 transmission peaks early on in the infection, already starting few days prior to symptom onset, and declines rapidly in the first week thereafter (Cheng et al., 2020; Hu et al., 2021a). These results also correspond to the rapid disappearance of infectious virus one week after it can be first detected (Bullard et al., 2020; Perera et al., 2020; Wölfel et al., 2020). If these preliminary findings revealing high sensitivity in the initial infection phase, early peak value in viral concentration measured and more rapid return to negativity as opposed to other respiratory samples are confirmed, breath RT-qPCR is closer to a contagiousness test than current state-of-the-art tests are.
The described test using a non-monolithic sieve can already be used in a clinical setting by shipping the collected breath sample, similar as for a nasopharyngeal swab, to a centralized testing center. The true value of a portable breath test, however, is to employ the integrated workflow in a decentralized setting. A rapid molecular test based on breath would be of great value for containing the spread of pandemic respiratory virus (Giovannini et al., 2021). This would require that the following improvements are implemented. Firstly, the current test does not yet include an internal control. As the captured volume is very small and largely diluted, we do not expect nor have observed any PCR inhibition. Still, an internal control is good standard practice and, especially if an exhaled human control can be identified, it would be a further means to standardize results. While large differences in both flow rate and total volume were observed between subjects, we have not identified a direct correlation between these parameters and the recorded Ct values. Further normalization of the test may help to further compare data on population level. Besides an internal control, the option to move to a multiplex assay to include additional targets may be wanted to detect different targets or variants of concern. Secondly, the entire assembly is still labor-intensive and a next design will be needed to focus on more user-friendly, integrated cartridges. The fact that our workflows still need manual intervention as well as the fact that the impactors need to be heated to avoid condensation will require further work to increase user-friendliness. Thirdly, the described test with current hardware still takes close to 1 h to perform. We already have used faster hardware to show that an ultrafast, on-chip PCR can be performed on clinical samples with equal sensitivity (Cai et al., 2019). Similarly, a sub-5 minutes RT-qPCR test was realized on a silicon chip for the detection of SARS-CoV-2 (See Fig. S4) indicating the possibility of a rapid screening test for exhaled particles. Further development of the proposed platform for rapid testing will facilitate on-site testing and help to contain outbreaks as exemplified by the success of rapid antigen tests, with the added benefit of improved sensitivity and specificity of molecular testing (Hu et al., 2021b).
5. Conclusion
Conventional sampling for SARS-CoV-2 using nasopharyngeal swabs or saliva samples can reliably indicate whether a person has been infected but reveals very little information whether a person is contagious. To assess this more important parameter, one needs to focus on exhaled aerosols. We here present a compact, point-of-care method -leveraging silicon micromachining-to sample subjects’ exhaled breath particles efficiently in a miniature volume without requiring additional equipment. Performed clinical studies demonstrate that the sensitivity and specificity of exhaled breath-based testing for SARS-CoV-2 is comparable to that of swab-based testing during the first week of infection. The longitudinal data suggest that RT-qPCR on breath appears to constitute a respiratory sample associated with viral kinetics which are distinct from other frequently used sample types proving its potential impact for early diagnostics and transmission control. As a result, breath sampling might lead to an improved understanding of the dynamics of pathogen transmission by exhaled particles with implications for the effective monitoring and control of airborne infectious diseases even beyond SARS-CoV-2 (Giovannini et al., 2021).
Ethics
The clinical studies were set up via the Ethical Commission of UZ Leuven: OMAKA pilot study: In-vitro Validation of imec Aerosol Sieve for Capturing SARS-CoV-2 Virus (study reference S64765); YAS Open-label PoC study: IMEC Breath Sampler for Capturing and Detecting SARS-CoV-2 Virus in Aerosols and Droplets of Exhaled Breath” (study reference S65005).
CRediT authorship contribution statement
Tim Stakenborg: Conceptualization, Methodology, Writing - Original Draft, Supervision, Visualization, Project administration. Joren Raymenants: Methodology, Formal analysis, Writing - Original Draft, Supervision. Ahmed Taher: Conceptualization, Formal analysis, Writing - Original Draft, Visualization. Elisabeth Marchal: Validation, Formal analysis, Investigation, Writing - Review & Editing, Supervision. Bert Verbruggen: Methodology, Formal analysis, Supervision, Project administration. Sophie Roth: Formal analysis, Investigation, Writing - Original Draft, Supervision, Visualization. Ben Jones: Conceptualization, Formal analysis, Writing - Review & Editing. Abdul Yurt: Conceptualization, Methodology, Writing - Original Draft, Supervision, Visualization, Project administration. Wout Duthoo: Methodology, Formal analysis, Investigation. Klaas Bombeke: Methodology. Maarten Fauvart: Validation, Formal analysis, Investigation, Writing - Review & Editing. Julien Verplanken: Software, Formal analysis, Visualization. Rodrigo S. Wiederkehr: Validation, Investigation, Writing - Review & Editing. Aurelie Humbert: Conceptualization, Validation, Supervision. Chi Dang: Validation, Writing - Original Draft, Supervision. Evi Vlassaks: Methodology, Supervision. Alejandra L. Jáuregui Uribe: Software, Investigation, Writing - Original Draft, Visualization. Zhenxiang Luo: Methodology, Software, Formal analysis. Chengxun Liu: Conceptualization, Investigation. Kirill Zinoviev: Investigation. Riet Labie: Conceptualization, Supervision. Aduen Darriba Frederiks: Software. Jelle Saldien: Resources, Supervision. Kris Covens: Validation, Formal analysis, Investigation, Writing - Review & Editing. Pieter Berden: Validation, Formal analysis, Investigation, Writing - Review & Editing. Bert Schreurs: Software, Formal analysis. Joost Van Duppen: Validation. Rabea Hanifa: Validation. Megane Beuscart: Validation. Van Pham: Methodology, Validation. Erik Emmen: Investigation, Visualization. Annelien Dewagtere: Investigation. Ziduo Lin: Investigation. Marco Peca: Methodology, Validation. Youssef El Jerrari: Validation. Chinmay Nawghane: Investigation. Chad Arnett: Investigation. Andy Lambrechts: Resources, Supervision, Project administration. Paru Deshpande: Resources, Supervision. Katrien Lagrou: Resources, Supervision. Paul De Munter: Resources, Supervision. Emmanuel André: Resources, Writing - Review & Editing, Supervision. Nik Van den Wijngaert: Resources, Writing - Review & Editing, Supervision, Project administration, Funding acquisition. Peter Peumans; Conceptualization, Writing - Review & Editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We want to thank Hanne Lenaerts, Chris D'haemer, Sabine Clabots, Hannelore De Mulder, Zakia Madour, Soumia El Mahmoudi, Mehdi Humbert, and Pieter-Jan Eelen for help during the clinical tests and breath sampler assembly; Karen Van Keer for facilitating lab tests; Younjae Choe, Antonio Pappaterra, Comate Engineering & Design as well as EXD Excogitate design for help in designing the breath sampler housing and clamp; Somersault1824 for help in art work; and Virovet Livestock Solutions for help with feline virus tests.
The work was partially funded by a project grant of the Flemish Government. Pieter Berden acknowledges support of the Research Foundation Flanders.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114663.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
The authors do not have permission to share data.
References
- Asadi S., Wexler A.S., Cappa C.D., Barreda S., Bouvier N.M., Ristenpart W.D. Aerosol emission and superemission during human speech increase with voice loudness. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-38808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., van Caeseele P.G., Poliquin G. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Fauvart M., Wiederkehr R.S., Jones B., Cools P., Goos P., Vaneechoutte M., Stakenborg T. Ultra-fast, sensitive and quantitative on-chip detection of group B streptococci in clinical samples. Talanta. 2019:192. doi: 10.1016/j.talanta.2018.09.041. [DOI] [PubMed] [Google Scholar]
- Chen P.Z., Bobrovitz N., Premji Z., Koopmans M., Fisman D.N., Gu F.X. Heterogeneity in transmissibility and shedding SARS-CoV-2 via droplets and aerosols. eLife. 2021;10:1–32. doi: 10.7554/ELIFE.65774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Z., Bobrovitz N., Premji Z.A., Koopmans M., Fisman D.N., Gu F.X. SARS-CoV-2 shedding dynamics across the respiratory tract, sex, and disease severity for adult and pediatric COVID-19. eLife. 2021;10 doi: 10.7554/eLife.70458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Qi X., Zhang L., Li X., Ma J., Zhang C., Feng H., Yao M. COVID-19 screening using breath-borne volatile organic compounds. J. Breath Res. 2021;15(4):047104. doi: 10.1088/1752-7163/ac2e57. [DOI] [PubMed] [Google Scholar]
- Cheng H.Y., Jian S.W., Liu D.P., Ng T.C., Huang W.T., Lin H.H. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern. Med. 2020;180:1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K.K., Tay D.J.W., Sen Tan K., Ong S.W.X., Son T.T., Koh M.H., Chin Y.Q., Nasir H., Mak T.M., Chu J.J.H., Milton D.K., Chow V.T.K., Tambyah P.A., Chen M., Wai T.K. Viral load of SARS-CoV-2 in respiratory aerosols emitted by COVID-19 patients while breathing, talking, and singing. Clin. Infect. Dis. 2021;74(10):1722–1728. doi: 10.1093/cid/ciab691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J., Wadekar S., DeCubellis K., Jackson G.W., Chiu A.S., Pagneux Q., Saada H., Engelmann I., Ogiez J., Loze-Warot D., Boukherroub R., Szunerits S. A mask-based diagnostic platform for point-of-care screening of Covid-19. Biosens. Bioelectron. 2021;192 doi: 10.1016/j.bios.2021.113486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., Beese S., Dretzke J., Ferrante di Ruffano L., Harris I.M., Price M.J., Taylor-Phillips S., Hooft L., Leeflang M.M.G., Spijker R., Van den Bruel A. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8(8):CD013705. doi: 10.1002/14651858.CD013705. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.A., Ausiello D., Salzman J., Devlin T., Langer R. 2021. Exhaled Aerosol Increases with COVID-19 Infection, Age, and Obesity 118; pp. 1–7. DCSupplemental.Published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini G., Haick H., Garoli D. Detecting COVID-19 from breath: a game changer for a big challenge. ACS Sens. 2021;6:1408–1417. doi: 10.1021/acssensors.1c00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T., Jimenez J.L., Prather K.A., Tufekci Z., Fisman D., Schooley R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet. 2021;397:1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson F.K.A., Watson N.A., Orton C.M., Haddrell A.E., McCarthy L.P., Finnie T.J.R., Gent N., Donaldson G.C., Shah P.L., Calder J.D., Bzdek B.R., Costello D., Reid J.P. Comparing aerosol concentrations and particle size distributions generated by singing, speaking and breathing. Aerosol Sci. Technol. 2021;55:681–691. doi: 10.1080/02786826.2021.1883544. [DOI] [Google Scholar]
- Gussman R.A. On the aerosol particle slip correction factor. J. Appl. Meteorol. Climatol. 1969;8:999–1001. doi: 10.1175/1520-0450(1969)008<0999:OTAPSC>2.0.CO;2. [DOI] [Google Scholar]
- Hamner L., Dubbel P., Capron I., Ross A., Jordan A., Lee J., Lynn J., Ball A., Narwal S., Russell S., Patrick D., Leibrand H. High SARS-CoV-2 attack rate following exposure at a choir practice - Skagit County, Washington, March 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:606–610. doi: 10.15585/mmwr.mm6919e6. [DOI] [PubMed] [Google Scholar]
- Hu S., Wang W., Wang Y., Litvinova M., Luo K., Ren L., Sun Q., Chen Xinghui, Zeng G., Li J., Liang L., Deng Z., Zheng W., Li M., Yang H., Guo J., Wang K., Chen Xinhua, Liu Z., Yan H., Shi H., Chen Z., Zhou Y., Sun K., Vespignani A., Viboud C., Gao L., Ajelli M., Yu H. Infectivity, susceptibility, and risk factors associated with SARS-CoV-2 transmission under intensive contact tracing in Hunan, China. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-21710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Guo J., Li G., Lu X., Li X., Zhang Y. Role of efficient testing and contact tracing in mitigating the COVID-19 pandemic : a network modelling study. BMJ Open. 2021 doi: 10.1136/bmjopen-2020-045886. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim W., Cordell R.L., Wilde M.J., Richardson M., Carr L., Dasi A.S.D., Hargadon B., Free R.C., Monks P.S., Brightling C.E., Greening N.J., Siddiqui S. Diagnosis of covid-19 by exhaled breath analysis using gas chromatography–mass spectrometry. ERJ Open Res. 2021;7 doi: 10.1183/23120541.00139-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The prominence of asymptomatic superspreaders in transmission mean universal face masking should be part of COVID-19 de-escalation strategies. Int. J. Infect. Dis. 2020;97:21–22. doi: 10.1016/j.ijid.2020.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J.E., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Lauzard M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., Usmani M., Shankar S.N., Mohamed K., Eiguren-Fernandez A., Stephenson C.J., Alam M.M., Elbadry M.A., Loeb J.C., Subramaniam K., Waltzek T.B., Cherabuddi K., Morris J.G., Wu C.Y. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J.E., Siddle K.J., Shaw B.M., Loreth C., Schaffner S.F., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., DeRuff K.C., Rudy M., Bauer M.R., Lagerborg K.A., Normandin E., Chapman S.B., Reilly S.K., Anahtar M.N., Lin A.E., Carter A., Myhrvold C., Kemball M.E., Chaluvadi S., Cusick C., Flowers K., Neumann A., Cerrato F., Farhat M., Slater D., Harris J.B., Branda J.A., Hooper D., Gaeta J.M., Baggett T.P., O'Connell J., Gnirke A., Lieberman T.D., Philippakis A., Burns M., Brown C.M., Luban J., Ryan E.T., Turbett S.E., LaRocque R.C., Hanage W.P., Gallagher G.R., Madoff L.C., Smole S., Pierce V.M., Rosenberg E., Sabeti P.C., Park D.J., MacInnis B.L. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021;371 doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.H., McDevitt J.J., Hau B.J.P., Yen H.L., Li Y., Ip D.K.M., Peiris J.S.M., Seto W.H., Leung G.M., Milton D.K., Cowling B.J. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Qan H., Hang J., Chen X., Cheng P., Ling H., Wang S., Liang P., Li J., Xiao S., Wei J., Liu L., Cowling B.J., Kang M. Probable airborne transmission of SARS-CoV-2 in a poorly ventilated restaurant. Build. Environ. 2021;196:107788. doi: 10.1016/j.buildenv.2021.107788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M., Kunze A.C., Bahmer T., Herget-Rosenthal S., Kunze T. SARS-CoV-2: viral loads of exhaled breath and oronasopharyngeal specimens in hospitalized patients with COVID-19. Int. J. Infect. Dis. 2021;110:105–110. doi: 10.1016/j.ijid.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso P., Venturelli F., Vicentini M., Perilli C., Larosa E., Bisaccia E., Bedeschi E., Zerbini A., Rossi P.G. Temporal profile and determinants of viral shedding and of viral clearance confirmation on nasopharyngeal swabs from SARS-CoV-2-positive subjects: a population-based prospective cohort study in Reggio Emilia. Italy. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanochie E., Linnes J.C. Review of non-invasive detection of SARS-CoV-2 and other respiratory pathogens in exhaled breath condensate. J. Breath Res. 2022;16 doi: 10.1088/1752-7163/ac59c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A.P.M., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y., Chin A.W.H., Chu D.K.W., Cheng S.M.S., Poon L.L.M., Chuang V.W.M., Peiris M. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymenants J., Geenen C., Gorissen S., Andre E. Empirical evidence on the efficiency of backward contact tracing in COVID-19. Nat. Commun. 2022 Aug 13;13(1):4750. doi: 10.1038/s41467-022-32531-6. PMID: 35963872; PMCID: PMC9375086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymenants Joren, Geenen Caspar, Thibaut Jonathan, et al. Integrated PCR testing and extended window contact tracing system for COVID-19 to improve comprehensiveness and speed. 16 November 2021. PROTOCOL (Version 2) available at Protocol Exchange. [DOI]
- Ryan D.J., Toomey S., Madden S.F., Casey M., Breathnach O.S., Morris P.G., Grogan L., Branagan P., Costello R.W., De Barra E., Hurley K., Gunaratnam C., Mcelvaney N.G., Obrien M.E., Sulaiman I., Morgan R.K., Hennessy B.T. Use of exhaled breath condensate (EBC) in the diagnosis of SARS-COV-2 (COVID-19) Thorax. 2021;76:86–88. doi: 10.1136/thoraxjnl-2020-215705. [DOI] [PubMed] [Google Scholar]
- Smolinska A., Jessop D.S., Pappan K.L., Saedeleer A. De, Kang A., Martin A.L., Allsworth M., Tyson C., Bos M.P., Clancy M., Morel M., Cooke T., Dymond T., Harris C., Galloway J., Bresser P., Dijkstra N., Jagesar V., Savelkoul P.H.M., Beuken E.V.H., Nix W.H.V., Louis R., Delvaux M., Calmes D., Ernst B. The SARS-CoV-2 viral load in COVID-19 patients is lower on face mask filters than on nasopharyngeal swabs. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., Notake S., Nakamura K., Ishikawa H., Suzuki H. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study. Sci. Rep. 2021;11:1–8. doi: 10.1038/s41598-021-90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Marr L.C., Li Y., Dancer S.J. Covid-19 has redefined airborne transmission. BMJ. 2021;373:1–2. doi: 10.1136/bmj.n913. [DOI] [PubMed] [Google Scholar]
- Verreault D., Moineau S., Duchaine C. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 2008;72:413–444. doi: 10.1128/mmbr.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorinen V., Aarnio M., Alava M., Alopaeus V., Atanasova N., Auvinen M., Balasubramanian N., Bordbar H., Erästö P., Grande R., Hayward N., Hellsten A., Hostikka S., Hokkanen J., Kaario O., Karvinen A., Kivistö I., Korhonen M., Kosonen R., Kuusela J., Lestinen S., Laurila E., Nieminen H.J., Peltonen P., Pokki J., Puisto A., Råback P., Salmenjoki H., Sironen T., Österberg M. Modelling aerosol transport and virus exposure with numerical simulations in relation to SARS-CoV-2 transmission by inhalation indoors. Saf. Sci. 2020;130 doi: 10.1016/j.ssci.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yan J., Grantham M., Pantelic J., De Mesquita P.J.B., Albert B., Liu F., Ehrman S., Milton D.K. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Natl. Acad. Sci. U.S.A. 2018;115:1081–1086. doi: 10.1073/pnas.1716561115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahari N.M., Zawawi M.H., Sidek L.M., Mohamad D., Itam Z., Ramli M.Z., Syamsir A., Abas A., Rashid M. Introduction of discrete phase model (DPM) in fluid flow: a review. AIP Conf. Proc. 2018;2030:020234. doi: 10.1063/1.5066875. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors do not have permission to share data.