Abstract
Background
Broadened use of predictive molecular and phenotypic profiling amongst oncologists has facilitated optimal integration of targeted- and immuno-therapeutics into clinical care. However, the use of predictive immunomarkers in ovarian cancer (OC) has not consistently translated into clinical benefit. Vigil (gemogenovatucel-T) is a novel plasmid engineered autologous tumor cell immunotherapy designed to knock down the tumor suppressor cytokines, TGFβ1 and TGFβ2, augment local immune function via increased GMCSF expression and enhance presentation of clonal neoantigen epitopes.
Methods
All patients enrolled in the VITAL trial (NCT02346747) of maintenance Vigil vs. placebo as front-line therapy with homologous recombination proficient (HRP) stage IIIB-IV newly diagnosed ovarian cancer underwent NanoString gene expression analysis. Tissue was obtained from surgically resected ovarian tumor tissue following surgical debulking. A statistical algorithm was used to analyze the NanoString gene expression data.
Results
Using the NanoString Statistical Algorithm (NSA), we identify high expression of ENTPD1/CD39 (which functions as the rate-limiting step in the production of the immune suppressor adenosine from ATP to ADP) as a presumptive predictor of response to Vigil versus placebo regardless of HRP status on the basis of relapse free survival (median not achieved vs 8.1 months, p = 0.00007) and overall survival (median not achieved vs 41.4 months, p = 0.013) extension.
Conclusion
NSA should be considered for application to investigational targeted therapies in order to identify populations most likely to benefit from treatment, in preparation for efficacy conclusive trials.
Subject terms: Cancer, Biomarkers
Plain Language Summary
Treatment options are limited in patients with advanced stage ovarian cancer. Treatments that stimulate the immune system to target the cancer are sometimes effective, however determining which patients will have benefit has been difficult. It is therefore important to develop new markers to predict which patients will respond to therapy. In this study, we looked at the levels of a large number of genes in tumors from patients treated with Vigil (gemogenovatucel-T), a treatment that modifies patient’s own tumor cells to activate the immune system. We demonstrate that high expression of a gene named ENTPD1/CD39 predicts a positive response to Vigil therapy. This finding could help clinicians to determine which patients might benefit from Vigil treatment and therefore might guide decisions on who should receive this treatment.
Rocconi et al. analyse gene expression data from patients with ovarian cancer receiving the autologous tumor cell immunotherapy gemogenovatucel-T (Vigil) as part of the VITAL study. The authors identify ENTPD1/CD39 expression as a predictor of relapse-free and overall survival benefit.
Introduction
Vigil (gemogenovatucel-T) is a novel autologous tumor cell immunotherapy, which is constructed from harvested malignant tissue1–3. It incorporates a multigenic plasmid encoding the human immune-stimulatory GMCSF gene and a bifunctional short-hairpin RNA construct, which specifically knocks down the proprotein convertase furin and its downstream targets TGFβ1 and TGFβ21,3,4. It is also designed to facilitate both cancer-associated antigen and neoantigen expression, upregulate MHC-II and enhance bone-marrow derived dendritic cell maturation, thereby augmenting the afferent immune response and generating a systemic antitumor effect. The VITAL study (NCT02346747) was a Phase IIb double-blind, placebo-controlled trial involving women 18 years and older with Stage III/IV high-grade serous, endometroid or clear cell ovarian cancer (OC) in clinical complete response (CCR) following carboplatin and paclitaxel induction chemotherapy5,6. Results recently published in a subset of 67 patients with BRCA1/2-wildtype (wt) OC showed improved relapse free survival (RFS; HR = 0.51, p = 0.02) and overall survival (OS; HR = 0.49, p = 0.049) compared to placebo5. Moreover, ad hoc analysis of a subset of 45 patients with homologous repair proficient (HRP) tumors by Myriad MyChoice CDx (Myriad Genetics, Salt Lake City, UT) also showed improvement in RFS and OS (HR = 0.39, p = 0.007 and HR = 0.34, p = 0.019, respectively)6. Long term follow-up confirmed a durable survival effect7. Three-year survival proportion from time of procurement was 83% for Vigil and 40% for placebo (p = 0.0006)7. A correlation of systemic immune response to Vigil clinical benefit was noted using ELISPOT assay3,8.
Contemporary clinical management of oncology patients is increasingly being guided by predictive molecular and phenotypic profiling in order to optimize the use of targeted- and immuno-therapeutics9, e.g., tumor mutational burden (TMB), MMR, PD-1, and PD-L110. However, the use of predictive biomarkers for immunotherapy in OC has not consistently translated into clinical benefit11–16 despite documented responses in some patients17. Although genomically unstable, OC is not mutationally driven, thus the clinical efficacy of immunotherapy in this disease has been dismal (<10% which generally correlates with high TMB, a presumptive marker of neoantigen content), represented by several failed phase III clinical trials11–15,18.
Nevertheless, we have studied patient subpopulations most sensitive to Vigil therapy based on molecular profile using NanoString assessment, and demonstrated that TIShigh score (tumor inflammation score) and MHC-II expression correlated with ELISPOT reactivity and clinically to OS and RFS19. Likewise, using NanoString technology to assess OS and RFS in patients enrolled in the VITAL study20, we showed marked benefit in patients with BRCA1/2-wt and HRP profiles and improved outcomes in patients whose tumors had mutant TP53 (p = 0.0013). The current study explores the relationship of mRNA expression via NanoString analysis in harvested baseline tumor to RFS and OS in Vigil treated patients from the VITAL study. ENTPD1/CD39 demonstrated clinical significance as a presumptive predictor of Vigil response versus placebo regardless of HRP status.
Methods
Study design and Vigil construction
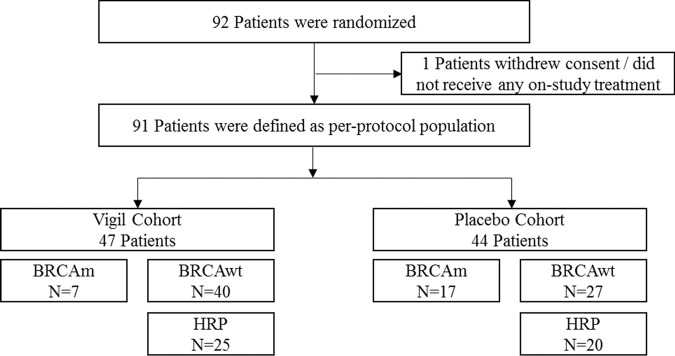
All patients provided written informed consent prior to study enrollment in the VITAL study. Briefly, the VITAL study (NCT02346747) was a phase 2b randomized, double-blind, placebo controlled trial involving women 18 years and older with stage III or IV high-grade serous, endometroid or clear cell ovarian cancer in clinical complete response. As specified in the approved clinical protocol (Mary Crowley IRB), patients provided consent for excess tissue to be used for additional immunotherapy research. Specimens were obtained from excess tissue harvested at the time of procurement for vaccine manufacture. Tissue is dissociated into cell suspension and cells are frozen at a concentration of 1.33 million cells/ml in freeze media (10% DMSO v/v in 1% HSA/plasma-Lyte A solution and stored long term in vapor phase nitrogen. Homologous recombination status [homologous recombination deficient (HRD) or HRP] was determined for all patients using the Myriad MyChoice CDx assay as previously described6,7. Patient demographics and CONSORT diagram are presented in Table 1 and Fig. 1, respectively.
Table 1.
Demographics summary of all patients by ENTPD1/CD39 status.
| ENTPD1/CD39 Status, No. (%) | ||
|---|---|---|
| Characteristic | ENTPD1/CD39 Low | ENTPD1/CD39 High |
| No. of patients | 45 | 46 |
| Frontline chemotherapy | ||
| Neoadjuvant | 6 (13.3%) | 9 (19.6%) |
| Adjuvant | 39 (86.7%) | 37 (80.4%) |
| Stage | ||
| III | 38 (84.4%) | 39 (84.8%) |
| IV | 7 (15.6%) | 7 (15.2%) |
| Age (years) | ||
| Median (IQR) | 62.0 (56–70) | 63.5 (55–68) |
| Range | 38–79 | 42–84 |
| <65 | 27 (60%) | 26 (56.5%) |
| >= 65 | 18 (40%) | 20 (43.5%) |
| ECOG | ||
| 0 | 31 (68.9) | 30 (65.2) |
| 1 | 14 (31.1) | 16 (34.8) |
| Residual disease post-surgery | ||
| Macroscopic | 13 (28.9%) | 14 (30.4%) |
| Microscopic/NED | 32 (71.1%) | 32 (69.6%) |
Fig. 1. CONSORT Diagram.
Flow of patients through the VITAL trial.
Vigil plasmid construction and cGMP manufacturing have been previously described5,6. Following VITAL study protocol guidelines, ovarian tumor tissue was excised at the time of initial tumor cytoreduction surgery and shipped to Gradalis, Inc. (Dallas, TX) for tissue processing, transfection, and vaccine manufacture.
RNA isolation and gene expression analysis
RNA expression was determined from total RNA isolated using RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). NanoString PanCancer Immuno-Oncology 360TM CodeSet using the nCounter SPRINT platform (NanoString Technologies, Seattle, WA, USA), which includes 750 cancer expression pathway genes, was used to analyze gene expression per manufacturer protocol.
Statistics
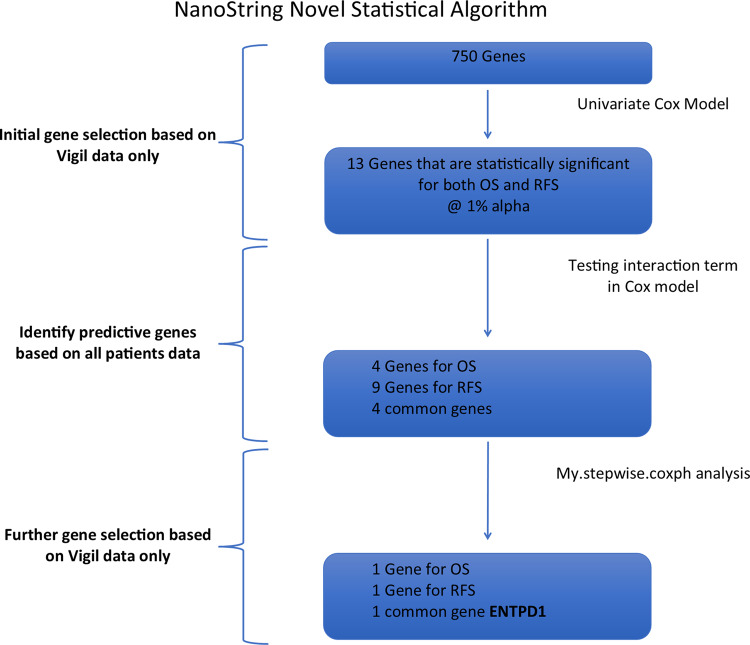
For all 750 genes a NanoString statistical algorithm (NSA) was defined prior to gene analysis (Fig. 2) to assess the correlation of NanoString gene expression results with clinical benefit as measured by both RFS and OS advantage effect with Vigil to specific mRNA expression. First, a univariate Cox model was used with gene Z-scores as a continuous variable and run for both OS and RFS in Vigil treated patients. From this data, the two-sided p-value, HR and corresponding 95% confidence interval (CI) were extracted. Genes that were significant for both OS and RFS advantage at the 1% significance level were identified. The more stringent variable selection criterion of 1% significance level was used due to the relatively small number of OS/RFS events compared to the large number of genes assessed. Next, Cox proportional hazards model with interaction term for each gene identified in the univariate Cox model was used to identify genes that were predictive of response to Vigil by analyzing data of both Vigil and placebo patients. A Cox proportional hazards model was used to determine if the interaction term between gene and treatment group was significant. The Cox model included the treatment group, gene and treatment-by-gene interaction term. The gene was considered predictive if the interaction term was significant (p ≤ 0.05). The model was run using the gene as a continuous variable or using binary high or low gene assignment. When using binary gene assignment, the median gene value for all 91 patients was calculated for each of the 750 cancer expression pathway genes. Patients were dichotomized into high or low gene expression groups if their value was either above or below the median. Kaplan–Meier (KM) curves were generated for genes identified as predictive for both OS and RFS. Since the identified predictive genes may not be independent, further model selection was performed using a multivariate Cox model in Vigil treated patients to further refine identification of relevant genes. We used the my.stepwise.coxph function in R (open source, R Core Team), which employs both forward selection and backward elimination methodology to further select genes that were significantly associated with the time-to-event data (OS or RFS) in Vigil treated patients21. The significance level for variable entry and for stay in the model was set at 0.01 and variable stay we set at 0.01 to account for potential multiplicity in the model selection process.
Fig. 2. NanoString Statistical Algorithm.
Flow chart of all patients’ analysis. Analyzed both with genes as raw continuous data and with genes dichotomized. Genes were selected if the interaction term was significant in both analyses. 5% alpha was used unless noted.
Biomarker vigil benefit over placebo
Previous analyses of Vigil relationship to BRCA1/2-wt, HRP and TP53 mutation (p53-mu) subpopulations revealed correlation to clinical benefit5–7,19,20. These subpopulations were explored via KM analysis to assess the effect of combination biomarkers BRCA1/2-wt, HRP, p53-mu and genes identified as significant following multivariate analysis in this study on Vigil and placebo treatment effects as measured by OS and RFS.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Results
Univariate analysis vigil patients only
First, a univariate Cox model was performed with the gene Z-score as a continuous variable to obtain the two-sided p-value, HR and 95% CI in Vigil treated patients only (n = 47). This analysis identified 13 genes that were statistically significant at the 1% significance level for both OS and RFS (Supplementary Table 1). All of these genes are associated with critical immunologic modulation function as per NanoString Pan Cancer Immuno-Oncology 360™ Code set (NanoString Technologies, Seattle, WA, USA).
Predictive genes using all patients data
While the previous analysis was able to identify genes of interest, they were not able to specify if genes were predictive. To determine genes predictive of Vigil treatment efficacy, Cox proportional hazards model with interaction term was used to analyze data from both Vigil and placebo patients (n = 91). The Cox model included the treatment group, gene and treatment-by gene interaction term.
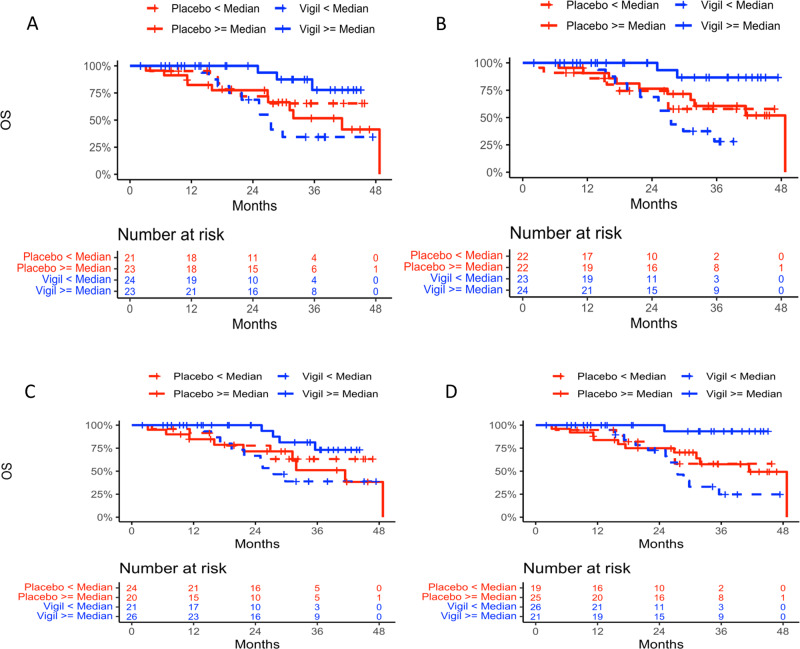
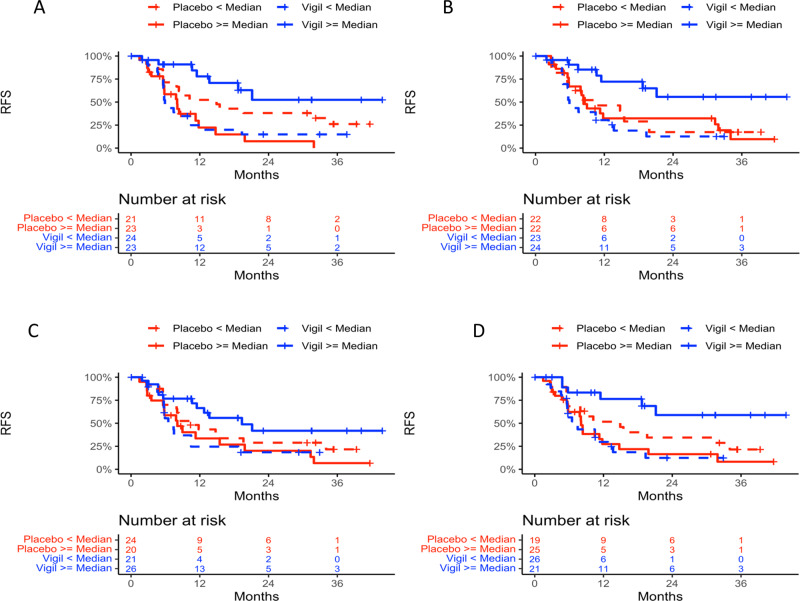
Demographics between Vigil and placebo were previously shown to not impact clinical benefit results5,6. Four genes were identified as predictive in both Cox models using continuous and binary data for both OS and RFS (CD79B, CCL13, ENTPD1/CD39 and MRC1). Four separate KM curves were generated for each gene in: (1) Vigil patients with gene expression < median and ≥ median; (2) placebo patients with gene expression < median and ≥ median; (3) Vigil patients with gene expression < median and placebo patients < median; and (4) Vigil patients with gene expression ≥ median and placebo patients ≥ median. KM curves for OS (Fig. 3) and RFS (Fig. 4) for placebo vs. Vigil with < or ≥ median expression from these 4 genes demonstrate benefit correlation with ≥ median expression. For patients with ≥ median ENTPD1/CD39 expression, OS was not achieved compared to placebo OS of 41.4 months (p = 0.013) and median RFS was not achieved in Vigil treated compared to 8.1 months with placebo (p = 0.00007). Patients with ≥ median expression levels of CCL13, CD79B and MRC1 also demonstrated OS benefit when receiving Vigil compared to placebo (median not reached vs 48.7 months, p = 0.019; not reached vs 41.4 months, p = 0.027; not reached vs 41.4, p = 0.005 respectively). Similar results demonstrating RFS benefit were observed (not achieved vs 8.4 months p = 0.006; 19.4 vs 8.1 months, p = 0.010; not achieved vs 8.1 months, p = 0.001 respectively). The two-sided p values of the interaction term in the Cox model and one-sided p values of log rank test comparing the OS and RFS KM curves are shown in Tables 2 and 3.
Fig. 3. Overall survival Kaplan Meier curves of all four genes analyzed.
Overall survival Kaplan Meier curves of ENTPD1/CD39 A, CCL13 B, CD79B C, and MRC1 D expression <median in Vigil (n = 24, 23, 21, 26 respectively) and placebo (n = 21, 22, 24, 19 respectively) and ENTPD1/CD39, CCL13, CD79B and MRC1 expression ≥median in Vigil (n = 23, 24, 26, 21, respectively) and placebo (n = 23, 22, 20, 25, respectively) treated patients.
Fig. 4. Relapse free survival Kaplan Meier curves of all four genes analyzed.
Relapse free survival Kaplan Meier curves of ENTPD1/CD39 A, CCL13 B, CD79B C, and MRC1 D expression <median in Vigil (n = 24, 23, 21, 26 respectively) and placebo (n = 21, 22, 24, 19 respectively) and ENTPD1/CD39, CCL13, CD79B, and MRC1 expression ≥median in Vigil (n = 23, 24, 26, 21, respectively) and placebo (n = 23, 22, 20, 25, respectively) treated patients. P values are one sided.
Table 2.
Two-sided p values of the interaction term in the Cox model.
| Interaction term (continuous)* OS | Interaction term (continuous)* RFS | Interaction term (binary)** OS | Interaction term (binary)** RFS | |
|---|---|---|---|---|
| ENTPD1/CD39 | 0.00751 | 0.00375 | 0.0158 | 0.00014 |
| CCL13 | 0.0190 | 0.00271 | 0.044 | 0.00998 |
| CD79B | 0.00426 | 0.00280 | 0.0303 | 0.0152 |
| MRC1 | 0.01040 | 0.0169 | 0.0173 | 0.000822 |
*Analyzed with genes as raw continuous data.
**Analyzed with genes dichotomized.
Table 3.
One-sided p values of log-rank test comparing two KMs and hazard ratios and 90% CI from the univariate Cox proportional hazards model based on four predicted genes from multivariate analysis.
| Vigil ≥ median vs. Vigil < median OS | Vigil ≥ median vs. Vigil < median RFS | Vigil ≥ median vs. placebo ≥ median OS | Vigil ≥ median vs. placebo ≥ median RFS | |||||
|---|---|---|---|---|---|---|---|---|
| P value | HR | P value | HR | P value | HR | P value | HR | |
| ENTPD1/CD39 | 0.002 | 0.177 [0.059, 0.524] | 0.0003 | 0.238 [0.114, 0.498] | 0.013 | 0.257 [0.087, 0.761] | 0.00007 | 0.200 [0.094, 0.427] |
| CCL13 | 0.0005 | 0.119 [0.034, 0.423] | 0.0003 | 0.236 [0.113, 0.493] | 0.019 | 0.228 [0.063, 0.824] | 0.006 | 0.338 [0.161, 0.709] |
| CD79B | 0.006 | 0.248 [0.092, 0.670] | 0.014 | 0.423 [0.219, 0.817] | 0.027 | 0.324 [0.118, 0.892] | 0.010 | 0.421 [0.224, 0.793] |
| MRC1 | 0.0001 | 0.058 [0.010, 0.325] | 0.0004 | 0.229 [0.105, 0.502] | 0.005 | 0.109 [0.019, 0.613] | 0.001 | 0.245 [0.112, 0.535] |
Multivariate Analysis Vigil Patients Only
To further select significant gene associations with OS or RFS in Vigil treated patients, the my.stepwise.coxph function in R was used as the stepwise variable selection procedure (with iterations between the ‘forward’ and ‘backward’ steps) including the 4 genes showing RFS and OS advantage to Vigil treatment over placebo. Two common strategies for adding or removing variables in a multiple regression model are backward elimination and forward selection. Backward elimination begins with all genes included in the model and eliminates variables one-by-one until the model cannot be improved per the model fitting criterion. Forward selection starts with no variables included in the model, then adds variables according to importance (e.g., based on p values) until no other significant variables are found. The significance level for variable entry in the model was set at 0.01 and for variable stay was set at 0.01 to account for potential multiplicity in the model selection process. ENTPD1/CD39 was the only gene identified through this stepwise model selection process for both OS and RFS (p value < 0.001).
Subgroup Vigil/Placebo: HRP, ENTPD1/CD39
Twenty of the 91 patients (22%) enrolled into the VITAL trial (11 Vigil, 9 placebo) had HRP molecular profile5–7 and ENTPD1/CD39 “high” expression. Note HRP status and TP53 mutations have been identified in previous analyses as predictive of Vigil response6,7,20. OS advantage was demonstrated (Fig. 5A) in the Vigil treated HRP/high ENTPD1/CD39 patients relative to placebo (not achieved vs 27 months, HR = 0.23, p = 0.025). In the same subset, the median RFS for Vigil was 21.1 months and 5.6 months for placebo (HR = 0.18, p = 0.004) (Fig. 5B). Despite small sample size, these subgroup results support additional survival benefit in patients whose tumors demonstrate ENTPD1/CD39 high expression in the HRP subgroup. In order to assess the impact of HRP and ENTPD1/CD39 on outcomes in patients treated with Vigil, multivariate analyses including HRP status and ENTPD1/CD39 as factors were conducted on OS and RFS for all Vigil patients. For OS, based on the multivariate Cox model for Vigil patients including both HRP status and ENTPD1/CD39 as factors, the p values for HRP status and ENTPD1/CD39 status are 0.30 and 0.007 respectively. For RFS, based on the multivariate Cox model for Vigil patients including both HRP status and ENTPD1/CD39 as factors, the p values for HRP status and ENTPD1/CD39 status are 0.15 and 0.0005 respectively. This demonstrates that within the Vigil patients, after adjusting for HRP status, ENTPD1/CD39 is still a statistically significant factor. ENTPD1/CD39 high Vigil patients demonstrated improved OS and RFS outcomes compared with ENTPD1/CD39 low Vigil patients.
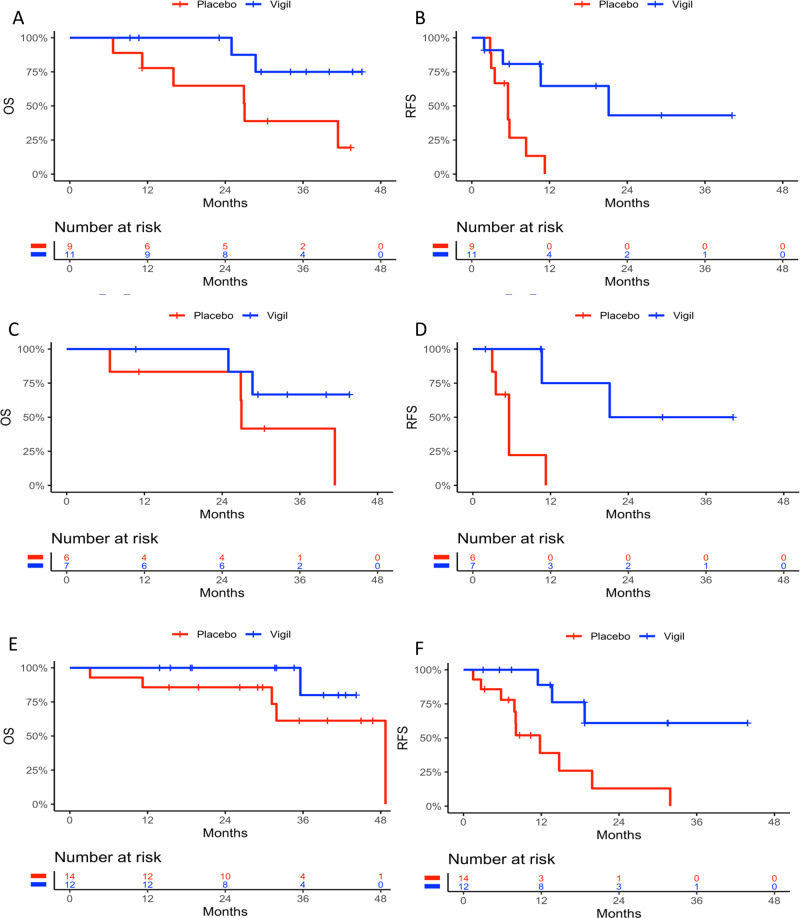
Fig. 5. Stratification of patient population by homologous recombination and p53 mutation status.
Kaplan Meier (KM) curves of ENTPD1/CD39 ≥ median expression in Vigil (n = 11) versus placebo (n = 9) patients for overall survival A and Relapse free survival B in the homologous recombination proficient (HRP) population. HRP, p53 mutant with high ENTPD1/CD39 expression KM curves in Vigil (n = 7) versus placebo (n = 6) are presented for overall survival C and relapse free survival D. KM curves of homologous recombination deficient (HRD) patients with high ENTPD1/CD39 expression in Vigil (n = 12) versus placebo (n = 14) for overall survival E and relapse free survival F. P values are one sided.
Subgroup Vigil/Placebo: HRP, p53, ENTPD1/CD39
Evidence of survival advantage was further suggested in patients with tumors demonstrating high ENTPD1/CD39 expression and of HRP/p53-mu profile. Despite the small sample size (n = 13), a trend toward OS benefit with Vigil therapy (median not reached vs 27 months, HR = 0.34, p = 0.099) and robust RFS benefit (21.1 vs 5.6 months, HR = 0.09, p = 0.004) was suggested (Fig. 5C, D).
Subgroup Vigil/Placebo: HRD, ENTPD1/CD39
Twenty-six of the 91 patients (29%) had tumors with elevated ENTPD1/CD39 expression that were also HRD (including BRCA1/2-mutation and BRCA1/2-wt/HRD). There appeared to be a trend towards improved OS with Vigil therapy (median not reached vs 48.7 months, HR = 0.24, p = 0.08) (Fig. 5E) and RFS difference between Vigil and placebo was highly significant in this population (median not reached vs 11.8 months, HR = 0.21, p = 0.005) (Fig. 5F).
Discussion
Using NanoString PanCancer Immuno-Oncology 360TM molecular profiles derived from patient tumor samples in conjunction with NSA, we identified that high expression of ENTPD1/CD39 was associated with a significant and independent improvement in OS and RFS with Vigil maintenance therapy in the VITAL study. ENTPD1/CD39 is highly expressed in OC cell-lines22, and functions as a master regulator to maintain the balance between proinflammatory and immunosuppressive regulatory function23. The latter largely due the role of ENTPD1/CD39 as the rate limiting step in the conversion of ATP to ADP in the adenosine pathway. Adenosine inhibits both T-cell and NK-cell anti-tumor function. Although adenosine can be exported from the tumor into the extracellular space by nucleoside transport proteins, it is primarily formed via the action of membrane ectoenzymes by phosphohydrolysis from dead cells24. In addition, ENTPD1/CD39 is present on cancer extracellular vesicles (ECVs)24. ENTPD1/CD39 is ubiquitously expressed in the vasculature, B cells, NK cells, dendritic cells, monocytes, macrophages, regulatory T cells and monocyte derived suppressor cells in the TME25,26. CD8 + T cells demonstrate T cell exhaustion signatures with malignant upregulation of CD39 in the tumor microenvironment27–29. Moreover, T regulatory (Treg) cell upregulation of ENTPD1/CD39 within the tumor microenvironment generates immunosuppressive activity thereby facilitating malignant growth and survival30,31. Inhibition of ENTPD/1CD39 in murine cancer models induces anticancer activity and ENTPD1/CD39 deficient mice demonstrated a reduction in tumor growth32–35. Furthermore, anti-ENTPD1/CD39 increased cytotoxicity of alloreactive primed T-cell towards fresh OvCA cells22.
In the current study, Vigil treated patients with baseline elevated tumor expression of ENTPD1/CD39 was associated with a significantly improved response compared to those patients with tumors with low expression and to those with high tumor expression treated with placebo. The primary VITAL study results suggest that Vigil induction of GMCSF, knock down of TGFβ1 and TGFβ2 and induced CD8 + T cell activity targeted to tumor-specific cancer neoantigens provide anticancer activity beneficially impacts OS and RFS in newly diagnosed Stage III/IV OC patients receiving Vigil as maintenance therapy. This activity appears to be correlated to high ENTPD1/CD39 expression—a presumptive predictive marker. Interestingly, in a murine model, high levels of TGFβ were associated with immunosuppressive CD39 + myeloid derived suppressor cells (MDSC)36. Notably, placebo treated patients from the VITAL study with high ENTPD1/CD39 expression tended to show poorer survival compared to patients with lower expression, presumably reflecting the immunosuppressive role of ENTPD1/CD39 in these patients. It is also of interest that ENTPD1/CD39 promotes tumor cell survival in hypoxic regions characterized by increased levels of ATP and high concentrations of vascular endothelial growth factor (VEGF), thereby supporting the consideration of a combination of Vigil and a VEGF inhibitor in therapeutic trial22.
Previously, Vigil has shown the ability to activate a systemic immune response. In Phase IIA clinical testing, all Vigil treated patients (n = 31) demonstrated immune activation through γIFN-ELISPOT assay which correlated with durable overall survival benefit1,3. Vigil also demonstrated in a small number of patients increase in the number of circulating CD3 + /CD8 + T cells following treatment37. In the VITAL trial we demonstrated RFS and OS benefit in patients with HRP molecular profile6,7. We also suggested that the presence of mutant p53 may further improve delineation of Vigil responsive patients20. Results of mRNA expression via NanoString signature also indicate enhanced OS and/or RFS endpoint benefits of Vigil maintenance in both these groups. These results support the need for further verification of ENTPD1/CD39 as a biomarker of sensitivity to Vigil treatment in OC and possibly other solid tumors with high ENTPD1/CD39 expression.
The presence of ENTPD1/CD39 in multiple cell types other than certain cancers (e.g., CD4 + / Treg, CD8 + and MDSC) supports the consideration of therapeutic assessment of combined ENTPD1/CD39 inhibition and Vigil in patients with ENTPD1/CD39high tumor expression. ENTPD1/CD39 monoclonal antibodies have demonstrated anticancer activity in murine models as single agents and in combination with checkpoint inhibitors and autologous EBV-specific human T cells38. Currently, there are a number of different CD39 targeting agents in early Phase I clinical trials under evaluation39–41.
It is also possible that in a larger patient population receiving Vigil, supportive evidence demonstrated with the other immune modulatory signals identified by NanoString analysis (i.e., CXCL1342,43, CD79B44, MRC145) will also be found to have further impact on OS and RFS. All three of these genes also perform important immunologic functions. MRC1 is expressed on tumor associated macrophages (TAMs) with M2 phenotype. Once activated MRC1 directs TAM’s to M1 phenotype thereby activating the innate response46. Recent work has shown that high CXCL13 expression in high-grade serous ovarian cancer correlates with increased survival by maintaining CXCR5 + /CD8 + T cells with in tertiary lymphoid structures47. CD79b expression is limited to B cells. B cells play an important role in anti-tumor immunity through secretion of cytokines and antigen presentation48. Such results may further direct research towards a multiplex of biomarker sensitivity and may even direct novel combination therapeutic approaches with Vigil, including combination treatment regimens based on various molecular signal expression patterns and immune related signal pathways that are relevant to Vigil related benefit.
Molecular biomarker assessment to optimize the proportion of responsive patient populations to Vigil therapy will involve more comprehensive analyses including p53mu, BRCA1/2-wt, ENTPD1/CD39 and HRP molecular profiles. Evidence provided in this report suggests these gene expression signals can act independently in defining sensitive subpopulations and also appears to support the possibility that the combined use of predictive biomarkers can suggest if not identify additive and possibly, synergistic therapeutic activity combinations. Clearly, statistical analyses such as those applied to the VITAL study here will likely continue to help identify optimal subpopulations with potential to benefit via treatment with Vigil as well as suggest the direction for continued Vigil combination studies. Results also justify trial consideration of Vigil in other solid tumor patients with HRP profile, p53mu and those with ENTPD1/CD39 high expression by NanoString PanCancer Immuno-Oncology 360TM CodeSet analysis.
In conclusion, we identified gene signatures indicative of response to Vigil maintenance therapy as part of frontline treatment in newly diagnosed OC patients. Interestingly, NSA identified ENTPD1/CD39, a gene signal associated with an immunosuppressive tumor microenvironment, was most highly predictive of Vigil responsiveness. Previous work has indicated TGFβ may upregulate ENTPD1/CD39 in immunosuppressive myeloid cells, such that Vigil’s effect in downregulating TGFβ may counter this effect and account for its activity in patients with high tumor expression of ENTPD1/CD39. Combining previously identified biomarkers of Vigil response, such as HRP and mutant p53, with ENTPD1/CD39 expression may allow for refined identification of Vigil responsive populations—ultimately allowing a transition from predictive analysis to prescriptive analytics. Such an approach can be more broadly applied to assess for correlations between gene expression signals and survival benefits as well as widen the therapeutic index by optimizing patient selection and treatment allocation with other targeted therapies.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to acknowledge Brenda Marr for her competent and knowledgeable assistance in the preparation of the manuscript.
Author contributions
R.P.R. was responsible for conceptualization, methodology, investigation, resources, writing – review and editing. L.S. and M.T. were responsible for conceptualization, methodology, validation, formal analysis, investigation, resources, writing – original draft, review and editing. L.M.d.S. was responsible for methodology, data generation, investigation, writing – editing. A.W. was responsible for methodology, supervision, and writing – review and editing. B.J.M., T.J.H., R.L.C., N.S. and S.B. were responsible for supervision and writing – review and editing. L.M., G.W., S.H. and E.B. contributed to investigation, resources and writing – review and editing. J.N. was responsible for conceptualization, investigation, formal analysis and writing – original draft, review and editing.
Peer review
Peer review information
Communications Medicine thanks Joshy George and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Supplementary Data 1 contains source data for the main figures (Figs. 3–5) in this manuscript. Additional data that support the findings of this study, and that do not involve proprietary or similar commercial information, will be made available to appropriate parties following an approved data sharing request sent to Laura Nejedlik (lnejedlik@gradalisinc.com) at Gradalis, Inc. Requests that may involve a conflict of interest or a competitive risk maybe declined by Gradalis, Inc. in its sole discretion.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/5/2023
A Correction to this paper has been published: 10.1038/s43856-023-00290-0
Supplementary information
The online version contains supplementary material available at 10.1038/s43856-022-00163-y.
References
- 1.Senzer N, et al. Long term follow up: phase I trial of “bi-shRNA furin/GMCSF DNA/Autologous Tumor Cell” Immunotherapy (FANG™) in Advanced Cancer. Journal of Vaccines & Vaccination. 2013;4:209. [Google Scholar]
- 2.Nemunaitis J. Multifunctional vaccines in cancer: the ʻtriadʼ approach. Expert Rev. Vaccines. 2011;10:713–715. doi: 10.1586/erv.11.78. [DOI] [PubMed] [Google Scholar]
- 3.Senzer N, et al. Phase I trial of “bi-shRNAi(furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol. Ther. 2012;20:679–686. doi: 10.1038/mt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maples P, et al. FANG vaccine: autologous tumor cell vaccine genetically modified to express GM-CSF and block production of furin. BioProcessing J. 2010;8:4–14. doi: 10.12665/J84.Maples. [DOI] [Google Scholar]
- 5.Rocconi RP, et al. Gemogenovatucel-T (Vigil) immunotherapy as maintenance in frontline stage III/IV ovarian cancer (VITAL): a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Oncol. 2020;21:1661–1672. doi: 10.1016/S1470-2045(20)30533-7. [DOI] [PubMed] [Google Scholar]
- 6.Rocconi RP, et al. Gemogenovatucel-T (Vigil) immunotherapy demonstrates clinical benefit in homologous recombination proficient (HRP) ovarian cancer. Gynecol. Oncol. 2021;161:676–680. doi: 10.1016/j.ygyno.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Walter A, et al. Gemogenovatucel-T (Vigil) maintenance immunotherapy: 3-year survival benefit in homologous recombination proficient (HRP) ovarian cancer. Gynecol. Oncol. 2021;163:459–464. doi: 10.1016/j.ygyno.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Oh J, et al. Long-term follow-up of Phase 2A trial results involving advanced ovarian cancer patients treated with Vigil(R) in frontline maintenance. Gynecol. Oncol. Rep. 2020;34:100648. doi: 10.1016/j.gore.2020.100648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morand, S., Devanaboyina, M., Staats, H., Stanbery, L. & Nemunaitis, J. Ovarian cancer immunotherapy and personalized medicine. Int. J. Mol. Sci.22, 10.3390/ijms22126532 (2021). [DOI] [PMC free article] [PubMed]
- 10.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat. Med. 2019;25:389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledermann JA, et al. LBA 25 - Scientific Plenary: Avelumab in combination with and/or following chemotherapy vs chemotherapy alone in patients with previously untreated epithelial ovarian cancer: Results from the phase 3 javelin ovarian 100 trial. Gynecol. Oncol. 2020;159:13–14. doi: 10.1016/j.ygyno.2020.06.025. [DOI] [Google Scholar]
- 12.Moore, K. N. et al. Atezolizumab, bevacizumab, and chemotherapy for newly diagnosed stage III or IV Ovarian Cancer: Placebo-Controlled Randomized Phase III Trial (IMagyn050/GOG 3015/ENGOT-OV39. J. Clin. Oncol. 39, 1842–1855 (2021). [DOI] [PMC free article] [PubMed]
- 13.Sabbatini P, et al. Abagovomab as maintenance therapy in patients with epithelial ovarian cancer: a phase III trial of the AGO OVAR, COGI, GINECO, and GEICO–the MIMOSA study. J. Clin. Oncol. 2013;31:1554–1561. doi: 10.1200/JCO.2012.46.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergote I, et al. A randomized, double-blind, placebo-controlled, phase III study to assess efficacy and safety of weekly farletuzumab in combination with carboplatin and taxane in patients with ovarian cancer in first platinum-sensitive relapse. J. Clin. Oncol. 2016;34:2271–2278. doi: 10.1200/JCO.2015.63.2596. [DOI] [PubMed] [Google Scholar]
- 15.Monk BJ, et al. A phase 2, randomized, double-blind, placebo- controlled study of chemo-immunotherapy combination using motolimod with pegylated liposomal doxorubicin in recurrent or persistent ovarian cancer: a Gynecologic Oncology Group partners study. Ann. Oncol. 2017;28:996–1004. doi: 10.1093/annonc/mdx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choucair K, et al. TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther. 2020;27:841–853. doi: 10.1038/s41417-020-0174-y. [DOI] [PubMed] [Google Scholar]
- 17.Kandalaft LE, Powell DJ, Jr., Singh N, Coukos G. Immunotherapy for ovarian cancer: what’s next? J. Clin. Oncol. 2011;29:925–933. doi: 10.1200/JCO.2009.27.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matulonis UA, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019;30:1080–1087. doi: 10.1093/annonc/mdz135. [DOI] [PubMed] [Google Scholar]
- 19.Rocconi RP, et al. Long-term follow-up of gemogenovatucel-T (Vigil) survival and molecular signals of immune response in recurrent ovarian cancer. Vaccines (Basel) 2021;9:894. doi: 10.3390/vaccines9080894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sliheet, E. et al. Network based analysis identifies TP53m-BRCA1/2wt-homologous recombination proficient (HRP) population with enhanced susceptibility to Vigil immunotherapy. Cancer Gene Therapy, 10.1038/s41417-021-00400-x (2021). [DOI] [PMC free article] [PubMed]
- 21.International Harvard Statistical Consulting Company. My.stepwise.package (2017).
- 22.Hausler SF, et al. Anti-CD39 and anti-CD73 antibodies A1 and 7G2 improve targeted therapy in ovarian cancer by blocking adenosine-dependent immune evasion. Am. J. Transl. Res. 2014;6:129–139. [PMC free article] [PubMed] [Google Scholar]
- 23.Takenaka MC, Robson S, Quintana FJ. Regulation of the T cell response by CD39. Trends Immunol. 2016;37:427–439. doi: 10.1016/j.it.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J. Immunol. 2011;187:676–683. doi: 10.4049/jimmunol.1003884. [DOI] [PubMed] [Google Scholar]
- 25.Koziak K, Sevigny J, Robson SC, Siegel JB, Kaczmarek E. Analysis of CD39/ATP diphosphohydrolase (ATPDase) expression in endothelial cells, platelets and leukocytes. Thromb Haemost. 1999;82:1538–1544. doi: 10.1055/s-0037-1614868. [DOI] [PubMed] [Google Scholar]
- 26.Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013;19:355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thelen M, Lechner A, Wennhold K, von Bergwelt-Baildon M, Schlosser HA. CD39 expression defines cell exhaustion in tumor-infiltrating CD8(+) T cells-letter. Cancer Res. 2018;78:5173–5174. doi: 10.1158/0008-5472.CAN-18-0873. [DOI] [PubMed] [Google Scholar]
- 28.Canale FP, et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8+ T cells. Cancer Res. 2018;78:115–128. doi: 10.1158/0008-5472.CAN-16-2684. [DOI] [PubMed] [Google Scholar]
- 29.Simoni Y, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 30.Borsellino G, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110:1225–1232. doi: 10.1182/blood-2006-12-064527. [DOI] [PubMed] [Google Scholar]
- 31.Gu J, et al. Human CD39 hi regulatory T cells present stronger stability and function under inflammatory conditions. Cell. Mol. Immunol. 2017;14:521–528. doi: 10.1038/cmi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X, et al. CD39/ENTPD1 expression by CD4+Foxp3+ regulatory T cells promotes hepatic metastatic tumor growth in mice. Gastroenterology. 2010;139:1030–1040. doi: 10.1053/j.gastro.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng L, et al. Vascular CD39/ENTPD1 directly promotes tumor cell growth by scavenging extracellular adenosine triphosphate. Neoplasia. 2011;13:206–216. doi: 10.1593/neo.101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, et al. Disordered purinergic signaling and abnormal cellular metabolism are associated with development of liver cancer in Cd39/ENTPD1 null mice. Hepatology. 2013;57:205–216. doi: 10.1002/hep.25989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. The role of NK cells and CD39 in the immunological control of tumor metastases. Oncoimmunology. 2019;8:e1593809. doi: 10.1080/2162402X.2019.1593809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryzhov SV, et al. Role of TGF-beta signaling in generation of CD39+CD73+ myeloid cells in tumors. J. Immunol. 2014;193:3155–3164. doi: 10.4049/jimmunol.1400578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herron J, et al. Vigil: personalized immunotherapy generating systemic cytotoxic T cell response. Cancer Sci. Res. 2020;1:210–221. [Google Scholar]
- 38.Li XY, et al. Targeting CD39 in cancer reveals an extracellular ATP- and inflammasome-driven tumor immunity. Cancer Discov. 2019;9:1754–1773. doi: 10.1158/2159-8290.CD-19-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerner AG, et al. Abstract 5012: targeting CD39 with a first-in-class inhibitory antibody prevents ATP processing and increases T-cell activation. Cancer Res. 2019;79:5012. doi: 10.1158/1538-7445.AM2019-5012. [DOI] [Google Scholar]
- 40.Perrot I, et al. Blocking antibodies targeting the CD39/CD73 immunosuppressive pathway unleash immune responses in combination cancer therapies. Cell Rep. 2019;27:2411–2425.e2419. doi: 10.1016/j.celrep.2019.04.091. [DOI] [PubMed] [Google Scholar]
- 41.Das SG, et al. Abstract 6639: SRF617, a potent enzymatic inhibitor of CD39, demonstrates single-agent activity and cooperates with various cancer therapies in both solid tumor and hematologic malignancies. Cancer Res. 2020;80:6639–6639. doi: 10.1158/1538-7445.AM2020-6639. [DOI] [Google Scholar]
- 42.Kazanietz MG, Durando M, Cooke M. CXCL13 and its receptor CXCR5 in cancer: inflammation, immune response, and beyond. Front. Endocrinol. (Lausanne) 2019;10:471. doi: 10.3389/fendo.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu D, Ye W, Jiang J. Clinical significance of CXCL13/CXCR5 axis in human cancers. Transl. Cancer Res. 2018;7:1737–1742. doi: 10.21037/tcr.2018.11.26. [DOI] [Google Scholar]
- 44.Ferreri AJM. Targeted therapies make room, anti-CD79b agents are coming. Lancet Oncol. 2019;20:898–900. doi: 10.1016/S1470-2045(19)30182-2. [DOI] [PubMed] [Google Scholar]
- 45.von Ehr A, et al. Inhibition of microglial TGFbeta signaling increases expression of Mrc1. Front. Cell Neurosci. 2020;14:66. doi: 10.3389/fncel.2020.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaynes, J. M. et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci. Transl. Med.12, 10.1126/scitranslmed.aax6337 (2020). [DOI] [PMC free article] [PubMed]
- 47.Yang M, et al. CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J. ImmunoTherapy Cancer. 2021;9:e001136. doi: 10.1136/jitc-2020-001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinker, G. S. et al. B cell orchestration of anti-tumor immune responses: a matter of cell localization and communication. Front. Cell Dev. Biol.9, 10.3389/fcell.2021.678127 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Supplementary Data 1 contains source data for the main figures (Figs. 3–5) in this manuscript. Additional data that support the findings of this study, and that do not involve proprietary or similar commercial information, will be made available to appropriate parties following an approved data sharing request sent to Laura Nejedlik (lnejedlik@gradalisinc.com) at Gradalis, Inc. Requests that may involve a conflict of interest or a competitive risk maybe declined by Gradalis, Inc. in its sole discretion.