Abstract
The endogenous plasmid pRA2 from Pseudomonas alcaligenes NCIB 9867 was determined to have 32,743 bp with a G+C content of 59.8%. Sequence analysis predicted a total of 29 open reading frames, with approximately half of them contributing towards the functions of plasmid replication, mobilization, and stability. The Pac25I restriction-modification system and two mobile elements, Tn5563 and IS1633, were physically localized. An additional eight open reading frames with unknown functions were also detected. pRA2 was genetically tagged with the ΩStrr/Spcr gene cassette by homologous recombination. Intrastrain transfer of pRA2-encoded genetic markers between isogenic mutants of P. alcaligenes NCIB 9867 were observed at high frequencies (2.4 × 10−4 per donor). This transfer was determined to be mediated by a natural transformation process that required cell-cell contact and was completely sensitive to DNase I (1 mg/ml). Efficient transformation was also observed when pRA2 DNA was applied directly onto the cells, while transformation with foreign plasmid DNAs was not observed. pRA2 could be conjugally transferred into Pseudomonas putida RA713 and KT2440 recipients only when plasmid RK2/RP4 transfer functions were provided in trans. Plasmid stability analysis demonstrated that pRA2 could be stably maintained in its original host, P. alcaligenes NCIB 9867, as well as in P. putida RA713 after 100 generations of nonselective growth. Disruption of the pRA2 pac25I restriction endonuclease gene did not alter plasmid stability, while the pRA2 minireplicon exhibited only partial stability. This indicates that other pRA2-encoded determinants could have significant roles in influencing plasmid stability.
The adaptive abilities of Pseudomonas species that allow them to grow in the polluted environment have made them ideal biocatalysts in bioremediation studies. Genetic diversity greatly enhances the adaptive abilities of bacteria, and this diversity can be accelerated by the horizontal exchange of genetic information, processes in which plasmids play a prominent role. Horizontal transfer of plasmids can occur by conjugation, transformation, and transduction. Both conjugation and transformation are active bacterial processes that require genetic information that is provided by either plasmids or their bacterial hosts.
Investigations into the processes of conjugal transfer among gram-negative bacteria have shown that conjugative plasmids require a cis-acting component known as the origin of transfer (oriT) and numerous genes involved in DNA processing and mating pair formation (52). In IncP and related plasmids, the process of conjugal transfer is initiated by the binding of TraJ to oriT (53), followed by strand-specific nicking by TraI (27). A third DNA-binding protein, TraK, was also found to be important to the formation of this protein-DNA complex (55), and when it is assembled, it is often referred to as the relaxosome. These plasmid-encoded proteins seem to have developed a specific activity for their own oriT sequences and are not effective in mobilizing plasmids with divergent oriT sequences. Mobile plasmids lack some of the tra genes that are essential for conjugal transfer but can be transferred between bacteria if a coexisting plasmid provides the necessary proteins. A mobile plasmid usually requires its own oriT in cis and the products of its own specific relaxosome genes before it can be effectively transferred by a conjugative plasmid.
Transformation is another mechanism of horizontal genetic transfer that occurs in bacteria, a process in which extracellular DNA is actively imported from the environment. Double-stranded DNA is hydrolyzed into a single-stranded form and transported across the membrane into the cytoplasm, where it may recombine with the host genome (24). Natural transformation in Pseudomonas has previously been reported for Pseudomonas stutzeri ATCC 17587, Pseudomonas mendocina ATCC 25411-13, Pseudomonas alcaligenes ATCC 12815, and Pseudomonas pseudoalcaligenes ATCC 17443 (7). In P. stutzeri, it was found that only chromosomal DNA was effective in natural transformation, while foreign plasmid DNA was not (7, 8). Transformation was found to be more efficient when the DNA was provided as a component of intact cells rather than in a solution (38).
P. alcaligenes NCIB 9867, which harbors the cryptic plasmid pRA2, is capable of utilizing 2,5-xylenol, 3,5-xylenol, and m-cresol as a sole carbon and energy source (16). Previous reports on pRA2 described a replication region that was novel in sequence (23), a functional transposon, Tn5563 (48), and the Pac25I restriction-modification (R-M) system (47). Here we report the complete nucleotide sequence of the plasmid and discuss the genetic organization and the predicted gene products that are encoded by pRA2. We found that while pRA2 is not self-transmissible, plasmid-carried genetic markers can be transferred efficiently between isogenic mutants of P. alcaligenes NCIB 9867 via a natural transformation process.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The relevant properties and sources of bacterial strains and plasmids are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria broth (LB), and Pseudomonas species were grown at 32°C in either LB or minimal salts media (MMB) (15) supplemented with sodium lactate (20 mM). All liquid cultures were grown in flasks and placed in an orbital-shaking incubator set at 200 rpm. P. alcaligenes NCIB 9867 was maintained on MMB with 2,5-xylenol (2.5 mM) as the sole carbon source. Oxoid purified agar (15 g/liter) was added to the media when required. When necessary, media were supplemented with antibiotics at the following concentrations: ampicillin (AMP), 100 μg/ml; kanamycin (KAN), 25 μg/ml; spectinomycin (SPC), 100 μg/ml; streptomycin (STR), 100 μg/ml; tetracycline (TET), 15 μg/ml; rifampin (RIF), 100 μg/ml; nalidixic acid (NAL), 50 μg/ml; and chloramphenicol (CAM), 15 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Pseudomonas | ||
| P. alcaligenes NCIB 9867 | Wild type, 2,5-xylenol+ 3,5-xylenol+m-cresol+ | 16 |
| P. alcaligenes P25X14 | Strr/Spcr, pRA14 | This study |
| P. alcaligenesc P25X19 | Strr/Spcr, pRA19 | This study |
| P. alcaligenes P25X20 | Kanr, Tn5 insertion into NCIB 9867 | This study |
| P. putida RA713 | Plasmid-free derivative of NCIB 9869 | 17 |
| P. putida RA713RifNal | Rifr Nalr, spontaneous mutant of RA713 | This study |
| P. putida KT2440 | Restriction-deficient derivative of P. putida mt-2 | 2 |
| P. putida KT2440RifNal | Rifr Nalr, spontaneous mutant of KT2440 | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | 32 |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 Δ(lac)X74 deoR recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG | 25 |
| DF1 | F mcrB mrr hsdS20(rB− mB−) supE44 ara-14 galK2 lacY1 proA2 rpsL20 (Strr) xyl-5 λ− leuB6 mtl-1 Δ(srl-recA)306 γδ−tnpA+ Camr | 43 |
| S17-1 | thi pro hsdR hsdM+ recA RP4 tra | 36 |
| Plasmids | ||
| pRA2 | Cryptic endogenous plasmid from P. alcaligenes NCIB 9867 | 23 |
| pVK182 | pRA2 minireplicon: pRA2 replication region cloned into BamHI and gene cassette from Tn5 conferring KAN resistance cloned into the HindIII sites of pUC18 | 23 |
| pHP45Ω | Source of ΩStrr/Spcr | 30 |
| pSUP202 | pBR325 derivative, Ampr Camr Tetrmob+ | 36 |
| pRA14 | pRA2 pac25I.R::ΩStrr/Spcr (replaced nt 20373 to 20396) | This study |
| pRA19 | pRA2 Tn5563::ΩStrr/Spcr (inserted at nt 13355) | This study |
| pRK415 | IncP TetrlacZ mob, broad-host-range cloning vector | 21 |
| pPSK1 | 19,386-bp PstI fragment from pRA2 cloned into pRK415 | This study |
| pPSK2 | 6,311-bp PstI fragment from pRA2 cloned into pRK415 | This study |
| pPSK3 | 3,405-bp PstI fragment from pRA2 cloned into pRK415 | This study |
| pPSK4 | 2,794-bp PstI fragment from pRA2 cloned into pRK415 | This study |
| pPSK5 | 847-bp PstI fragment from pRA2 cloned into pRK415 | This study |
| pDelta2 | pBR322 replicon, Ampr Kanr Sucs StrslacZ | 43 |
| pVLT33 | IncQ KanrPtac lacIqmob, broad-host-range cloning vector | 10 |
| pMMB67EH | IncQ AmprPtac lacIqmob, broad-host-range cloning vector | 12 |
| RK2/RP4 | IncPα Ampr Kanr Tetrtra, endogenous plasmid | 9 |
Ampr, Kanr, Strr, Spcr, Tetr, Camr, Rifr, and Nalr indicate resistance to the respective antibiotics. Strs and Sucs indicate sensitivity to STR and sucrose, respectively. mob, mobilized by RK2/RP4 transfer functions. tra, RK2/RP4 transfer functions. nt, nucleotide.
DNA manipulation.
Plasmid pRA2 was isolated by alkaline lysis and purified through a cesium chloride-ethidium bromide density gradient as described previously (32). Restriction endonucleases, polymerases, T4 ligase, and calf intestinal phosphatase were purchased from New England Biolabs, Inc. (Beverly, Mass.) and used according to the manufacturer's instructions. DNA cloning procedures were performed by standard techniques (32). PCR primers were synthesized on an ABI 392 DNA synthesizer (Perkin-Elmer, Norwalk, Conn.) and used in PCR with DyNAzyme II DNA polymerase (Finnzymes, Riihitontuntie, Finland) on a DNA Thermal Cycler 480 (Perkin-Elmer). PCR products were purified with GFX spin columns (Amersham Pharmacia Biotech AB, Uppsala, Sweden) and cloned with the pGEM-T Easy Vector System I (Promega, Madison, Wis.).
DNA sequence determination.
Plasmid libraries were obtained by cloning pRA2 PstI and AgeI restriction fragments into pUC18. Cloned DNA was purified with the Wizard Plus Minipreps Kit (Promega). Large DNA fragments (>10 kb) were recloned into pDELTA2, and nested deletions were obtained with the Deletion Factory System (Life Technologies, Inc., Rockville, Md.). Deletion clones were purified by the modified alkaline lysis method (5). DNA templates were labeled for sequencing with the ABI Taq Dye Deoxy Cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.) and sequencing was performed with an ABI 373 DNA sequencer (PE Applied Biosystems). The terminal regions of pUC18-cloned DNA were sequenced with the M13FWD (5′-GTA AAA CGA CGG CCA GT-3′) and M13REV (GAT AAC AAT TTC ACA CAG GA) primers. Deletion clones in pDELTA2 were sequenced with primers designed from the SP6 (ATT TAG GTG ACA CTA TAG) and T7 (TAA TAC GAC TCA CTA TAG GG) promoters. Additional sequencing primers were manufactured on an ABI 392 DNA synthesizer (PE Applied Biosystems) and were used to complete the plasmid sequence in both strands.
DNA sequence analysis.
DNA sequences were edited and connected, and restriction sites and potential open reading frames (ORFs) were located by the DNASIS program (Hitachi Software, San Francisco, Calif.). Potential Shine-Dalgarno sequences were located manually, and ORFs that were considered likely to be transcribed were analyzed with BLAST (1) (http://www.ncbi.nlm.nih.gov/BLAST/). Comparisons of predicted pRA2 protein sequences in relation to other protein sequences were expressed in percentage identity. Calculations were done by aligning sequences with the DDBJ malign and clustal alignment programs (http://www.ddbj.nig.ac.jp/searches-e.html), with a gap penalty of 8 and a gap length of 3 and by dividing the total number of matches by the total number of residues in the shorter sequence.
Insertion of ΩStrr/Spcr into plasmid pRA2.
Plasmid pRA2 was genetically labeled with the ΩStrr/Spcr gene cassette (30) by first cloning the 3.4-kb PstI fragment from pRA2 into pUC18, giving rise to pSK183. The ScaI restriction site located in the central region of the insert was a suitable site for the insertion of ΩStrr/Spcr (2.0 kb) that had been cleaved with SmaI. The selection of transformants with media containing AMP, STR, and SPC allowed for the isolation of the plasmid, pSK183Ω, which was then digested with PstI. The cleaved 5.4-kb PstI restriction fragment was purified and cloned into pSUP202, forming the suicide construct pSUP3Ω. pSUP3Ω was transformed into S17-1, which was then diparentally conjugated with P. alcaligenes NCIB 9867. Strr/Spcr transconjugants appeared at an approximate frequency of 10−5 per donor cell. To distinguish between single crossover and double crossover events, transconjugants were replica plated onto LB plates with and without tetracycline. Plasmids were examined by restriction analysis from transconjugants that were tetracycline sensitive.
Conjugation experiments.
Diparental matings were achieved by growing donor and recipient cells separately in 10 ml of LB medium containing the relevant antibiotics until stationary phase. Cells were harvested, washed twice with sterile LB, and resuspended in 1 ml of fresh LB. One hundred microliters of the recipient suspension was spotted onto a filter (Whatman cellulose nitrate membrane, 0.45-μm pore size, 25-mm diameter) placed on a prewarmed LB agar plate. When excess liquid had been absorbed through the filter (5 to 10 min), an equal volume of the bacterial donor was placed onto the recipient cells. The filter was incubated at 32°C for 24 h, removed from the LB plate, placed in 5 ml of sterile MMB (without carbon source), and vortexed vigorously to resuspend the cells. The suspension was then centrifuged, and the supernatant was discarded. Cells were resuspended in 1 ml of MMB, and aliquots of 100 μl were plated onto the respective selective media. Transfer frequencies were expressed as the number of transconjugants per donor CFU after the mating period.
Transformation of plasmid-encoded genetic markers.
DNA exchange experiments between P25X19 (Strr/Spcr) and P25X20 (Kanr) isogenic mutants were performed by mixing the two strains on a solid surface and in liquid medium and also under conditions that prevented cell-cell contact. In all of these experiments, bacterial strains were grown in LB until they reached stationary phase, harvested, and resuspended in 1 ml of fresh LB. For mating experiments on a solid surface, the procedures were the same as in the conjugation experiments. In liquid experiments, donor and recipient cell suspensions in LB were mixed in an Eppendorf tube. For DNA exchange under conditions that prevented cell-cell contact in liquid media, a 24-well tissue culture plate (Transwell 3423; Costar, Cambridge, Mass.) that provided adjacent chambers separated by a polycarbonate membrane of 0.1-μm pore size was used. The membrane should allow for the passage of DNA but not bacterial cells. Bacterial donors (100-μl aliquots) were placed in the upper chamber while recipients (100-μl aliquots) were placed in the lower chamber with an additional 500 μl of fresh LB. When required, DNase I in 20 mM Tris-HCl (pH 7.5) containing 1 mM MgCl2 was added to mating experiments. Cultures from all of these experiments were incubated at 32°C for 24 h, without shaking. Cells used as recipients in all DNA transfer experiments were washed twice with sterile MMB. Serial dilutions were then plated onto both selective and nonselective media. Experiments involving direct application of purified plasmid DNA were performed under the same conditions as described above except that the appropriate amount of plasmid DNA was suspended in sterile LB before addition to the recipients.
Plasmid stability analysis.
Pseudomonas strains containing the plasmid to be assayed were grown overnight in LB medium with antibiotic selection for the plasmid. The culture was then diluted 103-fold in fresh media without antibiotics and grown to stationary phase. Since the doubling of cell mass is equivalent to one generation, a 103-fold increase in cell mass is approximately 10 generations. At intervals of 20 generations, cultures were serially diluted, plated onto nonselective media, and incubated overnight at 32°C. The dilution step was repeated until the cultures had grown for 100 generations. Colonies were randomly transferred to selective media, and after incubation, the colonies were quantitated for plasmid retention.
Nucleotide sequence accession number.
The complete DNA sequence of plasmid pRA2 can be found in GenBank under the accession no. U88088.
RESULTS
DNA sequence analysis. (i) Nucleotide sequence and genetic organization of plasmid pRA2.
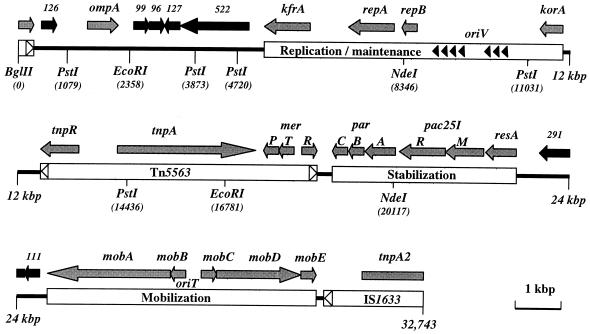
Plasmid pRA2 is a circular DNA molecule consisting of 32,743 bp and has a total G+C content of 59.8%. The deduced physical and genetic maps of pRA2 are illustrated in Fig. 1. pRA2 is predicted to contain a total of 29 ORFs that are likely to give rise to functional proteins. The predicted characteristics of pRA2-encoded gene products are listed in Table 2. Consensus promoter sequences, such as the E. coli −10 and −35 transcriptional signals, could not be identified. However, Shine-Dalgarno sequences were found between 5 and 14 nucleotides upstream of the majority of potential start codons (Met, 27 out of 29; Val, 2 out of 29). Generally, the pRA2 coding regions seem to be transcribed divergently away from the putative oriT sequence, with the exceptions being tnpA and merR, which are both within Tn5563 (Fig. 1). Genes appear to converge onto a 77-nucleotide intergenic region that contains a stem-loop structure flanked by two poly(A) sequences, and this may represent a transcriptional terminator for both orf96 and orf127. Other potential terminators lie after repB, pac25I.R, parC, and ompA and are characterized by a G+C-rich stem of six or more pairings with a loop of 3 to 6 nucleotides that is followed by a string of consecutive thymine nucleotides. Additional palindromic sequences that might be associated with transcriptional regulation are found in the promoter regions of korA, kfrA, orf522, and orf111. Another stem-loop structure is found 158 nucleotides upstream of the Tn5563 tnpR gene and may function as a res site for the transposon. The distal arrangement of genes is compact in the regions associated with mobilization and plasmid stability, while in the replication region, genes are spaced further apart. According to our ORF analysis, approximately 70% of the pRA2 DNA sequence encodes proteins.
FIG. 1.
Genetic and physical maps of plasmid pRA2 as deduced from the DNA sequence. The complete genome is represented by three horizontal lines that are marked in kilobase pairs (kbp) and drawn to scale. Predicted gene products are illustrated above the genome by shaded arrows (function predicted) and solid arrows (unknown function). The plasmid has been separated into regions that are involved in replication, stability, and mobilization. The mobile elements Tn5563 and IS1633 are also shown, and their terminal inverted repeats are marked by unshaded arrowheads. Solid arrowheads represent the seven iterons of the replication region. All of the PstI, EcoRI, NdeI, and BglII restriction sites are shown (positions in base pairs [bp]). The BglII site was arbitrarily taken as coordinate 1 and appears to cleave IS1633 and its gene, tnpA2, but both of these elements are in fact continuous.
TABLE 2.
Predicted gene products of pRA2a
| Predicted product | Coding position (start–stop) | Predicted molecular mass (kDa) | Predicted pI | Predicted function and product description (GenBank accession no. of closest relative) |
|---|---|---|---|---|
| Replication and maintenance | ||||
| RepA | 8075–7245 | 30.3 | 10.77 | Plasmid replication protein, no protein relatives |
| RepB | 8481–8239 | 8.8 | 10.16 | Hypothetical plasmid replication protein |
| KorA | 11844–11419 | 15.4 | 10.56 | Plasmid maintenance, KorA homolog (P03052) |
| KfrA | 6327–5302 | 37.1 | 4.92 | Plasmid maintenance, KfrA homolog (JQ0559) |
| Stability | ||||
| ResA | 22800–22186 | 22.9 | 9.89 | Plasmid multimer resolution, resolvase (AF015307) |
| Pac25I.M | 22171–21269 | 33.6 | 7.22 | Pac25I methyltransferase (AF051091) |
| Pac25I.R | 21272–20271 | 36.8 | 5.81 | Pac25I restriction endonuclease (AF051091) |
| ParA | 20117–19479 | 22.6 | 4.98 | Plasmid partition, ParA homolog (AAD19678) |
| ParB | 19457–19236 | 7.9 | 10.17 | Hypothetical plasmid stability protein |
| ParC | 19202–18816 | 14.4 | 9.81 | Hypothetical plasmid stability protein |
| Mobilization | ||||
| MobA | 27275–24711 | 98.5 | 9.60 | Plasmid relaxase/nikase, TraI homolog (S23001) |
| MobB | 27592–27278 | 11.5 | 8.83 | Plasmid relaxase/nikase, TraJ homolog (P17909) |
| MobC | 27987–28337 | 12.6 | 10.13 | Plasmid relaxase/nikase, TraK homolog (P17910) |
| MobD | 28343–30130 | 66.2 | 5.63 | Hypothetical plasmid mobilization protein |
| MobE | 30143–30421 | 10.4 | 9.30 | Hypothetical plasmid mobilization protein |
| Mobile elements | ||||
| TnpA | 14203–17196 | 111.4 | 7.93 | Tn5563 transposase (CAA97955) |
| TnpR | 13348–12419 | 34.5 | 9.35 | Tn5563 resolvase (CAA97956) |
| MerR | 18142–18540 | 14.5 | 7.80 | Tn5563 MerR homolog, regulatory (S51720) |
| MerT | 18073–17717 | 12.5 | 9.52 | Tn5563 heavy-metal transport protein (CAA70228) |
| MerP | 17704–17429 | 9.7 | 8.18 | Tn5563 periplasmic transport protein (P04129) |
| TnpA2 | 31426–290 | 62.3 | 10.04 | IS1633 transposase (U25434) |
| Miscellaneous gene products | ||||
| OmpA | 1509–2171 | 22.9 | 9.07 | Outer membrane protein, OmpA homolog (P37665) |
| Orf 126 | 432–812 | 13.9 | 4.96 | Hypothetical protein, no relatives |
| Orf 96 | 2725–3015 | 10.4 | 4.84 | Conserved hypothetical protein (P44191) |
| Orf 99 | 2429–2728 | 11.2 | 10.28 | Conserved hypothetical protein (P44190) |
| Orf 127 | 3476–3093 | 13.9 | 6.82 | Hypothetical protein, no relatives |
| Orf 522 | 5041–3473 | 57.4 | 6.61 | Conserved hypothetical protein (S45085) |
| Orf 291 | 24149–23274 | 32.7 | 5.64 | Hypothetical protein, no relatives |
| Orf 111 | 24481–24146 | 13.0 | 4.64 | Hypothetical protein, no relatives |
Predicted gene products of pRA2 are listed in groups based on their likely role. Proteins have been named according to their closest relative, probable function, or length in amino acids. Coding region positions are numbered with respect to the unique BglII restriction site (taken as coordinate 1) and from the first base of the start codon to the last base of the stop codon. Ascending coding positions indicate that the gene is located on the positive strand while descending coding positions indicate the gene is found on the negative strand. The DNASIS Program (Hitachi Software) was used to make predictions of coding regions, molecular weights, and isoelectric points. The GenBank BLAST algorithm (1) was used to conduct homology searches.
(ii) The novel replication region of plasmid pRA2 is flanked by korA and kfrA homologs.
A minireplicon of pRA2 was previously characterized, and it had been suggested that pRA2 was an iteron-regulated plasmid because of the presence of seven 72-bp direct repeats and an ORF that gives rise to RepA (formerly designated ORF1), which is essential for replication (23). The deduced product of pRA2 RepB contains a predicted helix-turn-helix domain, suggesting that it might be a DNA-binding protein. The location of repB immediately upstream from repA (Fig. 1) raises the possibility that repB may be involved in the regulation of plasmid replication. Neither RepA nor RepB showed any sequence homology to protein sequences in the databases.
Two genes, korA and kfrA, flank the pRA2 replication region. Neither of these genes were found to be essential for the replication of a pRA2 minireplicon (23). Instead, their gene products showed considerable amino acid sequence similarities to plasmid RK2/RP4 KorA (3) and KfrA (42) with 37 and 41% identity, respectively. In RK2/RP4, korA is a global regulator coordinating its own expression, kfrA expression, and that of six other genes involved in replication and stable inheritance functions (34, 37, 40–42, 50, 51). The operator sequence that KorA recognizes contains the palindrome GTT TAG CTA AAC, and in the trfA promoter, this operator overlaps the −10 box of the promoter (20, 37). In pRA2, palindromic sequence GCA AAG GGC GCG ACT TAT CGC GCC CCT TTG C is found 37 nucleotides upstream from korA and may serve as the point of autoregulation. RK2/RP4 KfrA has a predicted role in plasmid partitioning during cell division and also represses its own transcription (19). Located 26 nucleotides upstream from pRA2 kfrA is a palindromic sequence, AAT AAT AAT AAT ATG ATA TTA TTA TTA TT. This sequence is very different from the pRA2 korA palindromic sequence but may serve as the pRA2 kfrA point of autorepression. No homologs of the other genes that are regulated by RK2/RP4 KorA, namely, klaA, kleC, kleA, klcA, trfA, and trbA (34, 37, 40, 42, 50, 51), were detected in the pRA2 DNA sequence. Furthermore, we could not identify another pRA2 consensus sequence that was common to the promoter regions of korA and kfrA or, in fact, to any of the other potential pRA2 coding regions. This suggests that, unlike its homolog in RK2/RP4, pRA2 KorA probably does not play a global regulatory role in pRA2.
(iii) Conjugation-like ORFs of pRA2.
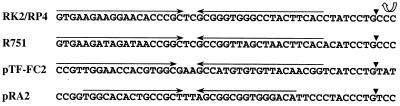
The mobilization region of plasmid pRA2 most closely resembles the cis regions and several of the trans-acting regions that are responsible for plasmid nicking and relaxation prior to conjugal transfer in the IncP plasmids RK2/RP4 and R751. Three of the predicted gene products from pRA2, i.e., MobA, MobB, and MobC, show significant similarities to the TraI (25% identity), TraJ (32% identity), and TraK (17% identity) proteins from RK2/RP4, respectively, with comparable arrangements of their genes (54). In RK2/RP4, traJ and traK are divergently transcribed away from oriT, and there is a similar arrangement in pRA2 for the mobB and mobC genes. The 394-bp region between mobB and mobC is the DNA sequence most likely to constitute the pRA2 oriT. This region is rich in direct and inverted repetitive DNA sequences. For example, the direct repeat sequence G(G/C)C GGT GG(G/C) ACA is found in the promoter region of mobB at position −17 and is also found at two other positions in the putative oriT region, both of which overlap the inverted sequence TGG CAC ACT GCC GCT TTA GCG GCG GTG GGA CA (Fig. 2). If the direct repeat sequence was a binding site for MobB, then it could conceivably regulate its own expression and the events leading to plasmid relaxation. When the DNA sequences of the oriT regions of RK2/RP4, R751, and pTF-FC2 were compared with that of pRA2, it was found that in each case the nic site is preceded by an inverted repeat sequence that is capable of forming a stem-loop structure (Fig. 2). The inverted repeat sequence in RK2/RP4 is the site at which the relaxosome gene products bind to initiate plasmid nicking (53). The RK2/RP4 traH gene is found within the traI coding region in a second reading frame. No traH homolog was detected in the pRA2 sequence. The remaining genes found in the Tra1-encoding region in IncP conjugative plasmids that are associated with further DNA processing (52) are not present on pRA2, nor are any of the other loci that are required for mating pair formation. The pRA2 Mob proteins also share sequence similarities with the MobA, MobB, and MobC proteins from plasmid pTF-FC2, with 27, 32, and 25% identity, respectively. pTF-FC2 has been shown to be mobilized by RP4 (31); however, the pTF-FC2-encoded MobA is 409 amino acids (aa), compared with 854 aa encoded by pRA2 and 732 aa encoded by RK2/RP4. Downstream of pRA2 mobC are two ORFs that have been designated mobD and mobE. The predicted products from these genes did not show any homology to protein sequences in GenBank but appeared to be transcriptionally coupled to mobC with intergenic regions of 5 and 12 nucleotides, respectively.
FIG. 2.
DNA sequence comparison of the oriT regions of RP4/RK2, R751, pTF-FC2, and pRA2. Inverted repeat sequences are marked with arrows, and the determined or predicted nic sites are indicated by wedges. The direction of transfer of the leading strand of RK2/RP4 is represented by the curved arrow.
(iv) The pRA2 stability region.
We have identified six pRA2 genes, resA, pac25I.M, pac25I.R, parA, parB, and parC, that are located in a contiguous DNA region found between the pRA2 transfer genes and Tn5563 (Fig. 1). These genes may contribute to pRA2 stability, and their compact arrangement suggests that at least several of these genes are transcriptionally linked to one another. The pRA2 resA gene was predicted to encode a product having similarities with a superfamily of resolvases, which includes plasmid resolvases. Enzymes from resolvase superfamilies have been shown, in some cases, to promote segregational stability of plasmids, probably by facilitating efficient partitioning through a conversion of plasmid multimers into monomers (11, 39). Located 17 nucleotides downstream from resA is the Pac25I R-M system, which encodes both a site-specific restriction endonuclease and the corresponding methyltransferase, which protects the DNA from cleavage (47). It is thought that R-M systems in bacteria have evolved to protect cells from the invasion of foreign DNA, but more recently they have also been implicated in plasmid stability (22, 26).
The parABC operon lies 147 nucleotides downstream from the Pac25I R-M system (Fig. 1). The predicted product of pRA2 parA belongs to a large family of proteins that includes plasmid and chromosomal partitioning proteins, which are thought to physically separate DNA molecules and provide equal distribution of genetic material upon cell division. The IncC protein encoded by RK2/RP4 also belongs to this family of partition proteins. Two ORFs designated parB and parC are found downstream from parA and have intergenic regions of 15 and 33 nucleotides, respectively. ParB and ParC did not show sequence homology to protein sequences in GenBank but could be involved in pRA2 stability because their coding regions seem to be transcriptionally linked to parA.
(v) pRA2 mobile elements.
Tn5563 and IS1633 are two mobile genetic elements in the plasmid sequence that have been identified (Fig. 1). Tn5563 encodes a transposase (TnpA) and a resolvase (TnpR), and has three additional genes that are predicted to give rise to proteins with similarity to the mercuric ion transport proteins, MerP and MerT, and the regulatory protein MerR, which are constituents of many bacterial mercury resistance operons. Even though these genes are present, Tn5563 does not confer resistance to mercuric ions (48).
IS1633 is a putative 2,598-bp insertion element with 47-bp imperfect terminal repeats. The transposase of IS1633 (tnpA2) showed up to 20% identity to the transposases of IS1181, IS1251, IS1165, and IS1001, all belonging to the ISL3 family of insertion sequences. In pRA2, IS1633 was flanked by what appears to be a 5-bp (TTTAT) target duplication that was probably generated on insertion. We have amplified IS1633 and used the PCR product to probe P. alcaligenes NCIB 9867 genomic DNA. IS1633 is not distributed throughout the genome like other IS elements found in this organism (45, 49) and appears to be located only in the plasmid (data not shown).
(vi) pRA2 ORFs with unknown functions.
Seven of the pRA2 genes have no detectable homologs in GenBank, and their products do not appear to be involved in plasmid replication, stability, or mobilization. Five of these genes, orf126, orf99, orf96, orf127, and orf522, are clustered around an ompA-like gene (Fig. 1), which encodes a member of the porin-forming, outer membrane protein family of gram-negative bacteria. The oprF gene product of Pseudomonas aeruginosa forms a transmembrane channel with a permeability barrier that allows the uptake of molecules as large as a tetrasaccharide (4). orf99 and orf96 are similar to HI1419 (55% identity) and HI1420 (35% identity), respectively, which are two putative genes from the Haemophilus influenzae genome with unknown function. These gene pairs occur in tandem in both pRA2 and H. influenzae. Two other ORFs, orf111 and orf291, are found in an area between the stability and mobilization regions of pRA2 and overlap each other in a head-to-tail formation by one nucleotide. It is possible that their products are involved in either plasmid stabilization or mobilization.
Mechanism of plasmid pRA2 transfer. (i) Genetic tagging of plasmid pRA2.
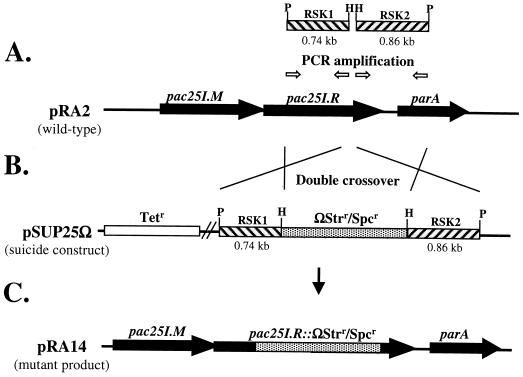
Plasmid pRA2 was cryptic and did not have any selective markers that would enable plasmid stability and transfer processes to be analyzed. We incorporated the ΩStrr/Spcr gene cassette (30) into pRA2 by homologous recombination as described in Materials and Methods. The precise location of the insertion target site was evaluated with the DNA sequence data. Two tagged pRA2 plasmids, designated pRA14 and pRA19, were generated and used in this study (Table 1). In pRA19, the ΩStrr/Spcr gene cassette was inserted at nucleotide position 13345, which was within Tn5563 and therefore unlikely to affect general plasmid functions. While in pRA14, the ΩStrr/Spcr gene cassette was inserted at position 20372, a mutation that disrupted the ORF of the Pac25I restriction endonuclease. Sequencing of the regions covering the targeted insertion sites confirmed both mutation events to be precise. Both pRA19 and pRA14 could be transformed into and stably maintained in artificially competent Pseudomonas putida strains RA713 and KT2440 but could not be transformed into artificially competent E. coli DH5α, suggesting that pRA2 has a narrow host range.
(ii) Stability analysis of pRA14 and pRA19 in homologous and heterologous hosts.
The two genetically tagged plasmids were also examined for segregational stability. When analyzed, both pRA14 and pRA19 were stably maintained in their native host, P. alcaligenes NCIB 9867, with 100% of the cells retaining the plasmid after 100 generations of nonselective growth. In the heterologous host, P. putida RA713, retention of both pRA14 and pRA19 also remained at 100% after 100 generations of nonselective growth. The segregational stability of the pRA2 minireplicon, pVK182, was also assayed in P. putida. In contrast, pVK182 had a reduced stability, being present in approximately 30% of the cells after 100 generations of nonselective growth.
(iii) pRA2 was not self-transmissible but could be mobilized by RP4.
The ability of pRA14 and pRA19 to conjugally transfer from P. alcaligenes NCIB 9867 into two other P. putida strains was examined. RA713RifNal and KT2440RifNal were used as recipients in diparental filter matings. Neither pRA14 nor pRA19 could be transferred into either P. putida strains by conjugation experiments (Table 3). The possibility that pRA2 could be mobilized by a conjugative plasmid by providing RP4 conjugal transfer functions in trans was examined. This was done by introducing RK2/RP4 into P. alcaligenes strains P25X14 and P25X19 (which contained pRA14 and pRA19, respectively) by conjugation and selecting transconjugants on MMB supplemented with 2,5-xylenol, AMP, KAN, and TET. Transconjugants were designated P25X14T and P25X19T, respectively, and these strains were used as donors in diparental filter matings. The Strr/Spcr phenotype of pRA19 and pRA14 was observed to transfer into RA713RifNal and KT2440RifNal at frequencies of 10−6 and 10−7, respectively (Table 3). Replica plating analysis of the transconjugants showed that the phenotypes of the genetically tagged pRA2 plasmids (Strr/Spcr) and RK2/RP4 (Kanr) were not genetically linked and that both plasmids existed as individual replicons.
TABLE 3.
Conjugation and mobilization frequencies of pRA2 during filter matings
| Donor strain | Donor plasmid | Frequency of transfer intoa:
|
|
|---|---|---|---|
| RA713RifNal | KT2440RifNal | ||
| Conjugation | |||
| P25X19 | pRA19 | <1.0 × 10−9 | <1.0 × 10−9 |
| P25X14 | pRA14 | <1.0 × 10−9 | <1.0 × 10−9 |
| Mobilizationb | |||
| P25X19T | pRA19 | 2.6 × 10−6 | 1.0 × 10−7 |
| P25X14T | pRA14 | 4.6 × 10−6 | 6.0 × 10−8 |
Frequencies of transfer are given as the number of transconjugants per donor CFU.
Plasmid RK2/RP4 was introduced into strains prior to mating to provide Tra functions in trans.
(iv) Interstrain transfer of pRA2-carried genetic markers was sensitive to DNase I.
When interstrain filter matings between P25X19 (Strr/Spcr) and P25X20 (Kanr) were carried out, high frequencies of doubly resistant progeny (10−4) were observed (Table 4). These bacterial matings were repeated in the presence of 200 μg of DNase I per ml. It was observed that the frequency of doubly resistant progeny was reduced approximately 500-fold. By varying the amount of DNase I that was present in the matings, a relationship in which the amount of DNase I was inversely proportional to the gene transfer frequency was established. When the level of DNase I was increased to 1 mg/ml, the occurrence of doubly resistant progeny was abolished (Table 4) without affecting the viability of the bacterial cells. The number of bacterial donors that was used in each filter mating was determined to be (3.95 ± 0.88) × 109 while addition of DNase I to a final concentration of 1 mg/ml resulted in (3.60 ± 0.63) × 109 viable cells. Spontaneous Strr/Spcr or Kanr mutants of P. alcaligenes strains were not detected under our experimental conditions (<10−9).
TABLE 4.
Effect of DNase I on the frequency of transformation between P. alcaligenes NCIB 9867 isogenic strains in solid-surface bacterial matings
| Strain (phenotype) and mating | DNase I (μg/ml)a | No. of transformantsb | Transformation frequencyc |
|---|---|---|---|
| P25X19 (Strr/Spcr) | 0 | 0 | <1.0 × 10−9 |
| P25X20 (Kanr) | 0 | 0 | <1.0 × 10−9 |
| P25X19 × P25X20 | 0 | 7.8 × 105 | 2.4 × 10−4 |
| 100 | 4.6 × 103 | 1.5 × 10−6 | |
| 200 | 1.7 × 103 | 6.3 × 10−7 | |
| 500 | 52 | 2.3 × 10−8 | |
| 1000 | 0 | <1.0 × 10−9 |
DNase I was added to cell suspensions at the concentrations indicated before suspensions were added to filters.
Transformants were selected on STR-, SPC-, and KAN-containing media. Average numbers of transformants are based on the results of three independent experiments.
Transformation frequencies were calculated by dividing the number of transformants by the number of recipients.
(v) Plasmid DNA uptake by P. alcaligenes strains.
When purified pRA19 (Strr/Spcr) DNA was applied directly to P25X20 (Kanr) recipient cells, uptake of the Strr/Spcr marker was observed. DNA uptake was shown to be linearly proportional to the amount of plasmid DNA applied (Table 5). For plasmid pRA19, frequencies were calculated to be approximately 106 transformants per microgram of DNA when applied to cells on a filter. Similar results were observed in experiments in which plasmid pRA14 DNA (Strr/Spcr) was directly applied to P25X20 (Kanr) and also those in which both plasmids were applied to the wild-type strain, NCIB 9867. Attempts to introduce broad-host-range plasmids, such as pRK415, pMMB67EH, and pVLT33, into P. alcaligenes NCIB 9867 were unsuccessful despite the fact that these plasmids could be conjugally transferred into this strain from S17-1. Each of the five fragments that was generated when pRA2 was digested with PstI was cloned into pRK415. The clones were designated pPSK1 for the largest cloned fragment to pPSK5 for the smallest fragment (Table 1). The purified plasmid DNA from each of these clones was then applied separately to P. alcaligenes NCIB 9867 cells on filters. The pRK415 Tetr phenotype was taken up at observed frequencies of approximately 10−6 for pPSK1, 10−9 for pPSK2, and at undetectable levels (<10−9) for pPSK3, pPSK4, and pPSK5.
TABLE 5.
Comparison of the transformation frequencies of P. alcaligenes NCIB 9867 strains under different mating conditions
| Strain(s) | Transformation frequency ona:
|
||
|---|---|---|---|
| Solid surfaceb | Liquid mediac | Membraned | |
| Intrastrain or interstrain transfer | |||
| P25X19 (Strr/Spcr) | <3.0 × 10−9 | <3.0 × 10−9 | <3.0 × 10−9 |
| P25X20 (Kanr) | <3.0 × 10−9 | <3.0 × 10−9 | <3.0 × 10−9 |
| P25X19 (Strr/Spcr) × P25X20 (Kanr) | 2.4 × 10−4 | 1.0 × 10−9 | <3.0 × 10−9 |
| Exogenous DNA uptake | |||
| P25X20 + pRA19 DNA (10 ng) | 5.0 × 10−6 | 6.0 × 10−7 | 5.0 × 10−7 |
| P25X20 + pRA19 DNA (100 ng) | 1.2 × 10−4 | 7.6 × 10−5 | 2.0 × 10−6 |
| P25X20 + pRA19 DNA (500 ng) | 6.6 × 10−4 | 1.5 × 10−5 | 2.4 × 10−6 |
Transformation frequency is the number of transformants per bacterial recipient and is the average of three independent experiments.
Solid surfaces were filters on LB plates.
Liquid experiments were conducted in LB.
A membrane with 0.1-μm pores was used to separate cells in LB.
(vi) Plasmid DNA uptake under different mating conditions.
The transfer of genetic markers between P25X19 (Strr/Spcr) and P25X20 (Kanr) and the uptake from an exogenous source of plasmid DNA by P25X20 (Kanr) were also examined in liquid media. In liquid experiments in which donor and recipient strains were used, doubly resistant progeny occurred at a 105-fold lower frequency than that observed in filter matings. However, when pRA19 plasmid DNA was added to recipient cells in liquid media, transformation was reduced only 10-fold compared to the analogous experiments on filters. A membrane was used to separate donor and recipient cells during isogenic matings, and when in place, DNA transfer was no longer observed (<10−9). In control experiments, in which DNA was placed on one side of the membrane and recipient cells were placed on the other side, the passage of pRA19 plasmid DNA through the membrane was found to transform at reduced frequencies (Table 5). When 500 ng of pRA19 DNA was separated from P25X20 (Kanr) recipient cells by the membrane, frequencies dropped about 100-fold compared to those of liquid DNA exchange without a membrane; however, when 10 ng of pRA19 DNA was used, the frequency appeared to remain at a comparable level (Table 5). When supernatants from the donor culture P25X19 (Strr/Spcr) were used to transform P. alcaligenes NCIB 9867 or P25X20 (Kanr) cells that had been placed on filters, no transformants were observed.
DISCUSSION
The complete nucleotide sequence has allowed us to construct both genetic and physical maps of pRA2 which provide an overview of the genetic organization of the plasmid. We have identified three separate regions that are likely to be associated with functions of plasmid replication, stability, and mobilization. The genes of the predicted products associated with replication, RepA and RepB, and the seven 72-bp iterons still do not have any homology to sequences in GenBank. Due to the presence of direct repeats in oriV, pRA2 may be classified as an iteron-regulated plasmid. The kfrA and korA ORFs flank the replication region and are likely to give rise to proteins that are similar in amino acid sequence to those encoded by the respective genes in plasmid RK2/RP4 (3, 42). The lack of conserved binding sites in pRA2 suggests that pRA2 KorA does not have a global regulatory role such as it does in RK2/RP4, but the presence of a palindromic sequence in the korA promoter may allow autorepression. Similarly, pRA2 kfrA expression may be self-regulated. The functions of both of these pRA2 ORFs remain unknown, but it is possible that they may be involved in plasmid maintenance.
The genes resA, parA, parB, and parC are located in a separate region and are thought to be involved in pRA2 stability. Incorporation of the ΩStrr/Spcr gene cassette into the pRA2 transposon Tn5563, resulting in pRA19, allowed us to monitor the presence of pRA2 without affecting plasmid functions. The segregational stability results established that pRA19 is extremely stable in P. alcaligenes NCIB 9867 and P. putida RA713 when grown under nonselective conditions. Stability assays of the minireplicon, pVK182, in P. putida RA713 demonstrated that the pRA2 replication region provided only partial stability (30% of cells retained the plasmid after 100 generations) when grown under the same conditions. This data shows that other genes, in addition to those carried by the minireplicon, contribute to pRA2 stability. ResA and ParA are likely to increase plasmid stability by resolving plasmid multimers and by providing active partitioning functions, respectively. However, the lack of homologous proteins in GenBank make the roles of ParB and ParC less obvious. An example of the high stability of pRA2 was demonstrated by introducing a plasmid encoding the pRA2 replicative functions into P. alcaligenes NCIB 9867. The introduced plasmid and pRA2 were expected to be incompatible; however, selection for the incoming plasmid did not result in the loss of pRA2 but rather resulted in its integration into the chromosome (46). This phenomenon suggested that pRA2 may encode a protein that was essential for cell viability or may encode a plasmid poison-antidote stability system. Poison-antidote systems, such as the ccd of F, pem/parD of R100/R1, phd/doc of prophage P1, and parDE of RK2/RP4, are comprised of two genes that encode the poison and antidote enzymes. The size of the antidote component ranges from 70 to 93 aa in length while the poison component ranges from 90 to 127 aa in length. The poison was always located downstream of the antidote (18). We are currently investigating the possibility that pRA2 parB (73 aa) and parC (128 aa) may constitute a poison-antidote stability system.
Three type II R-M systems (PaeR71, Bsp6I, and EcoRI) have been shown to have a stabilizing effect when they are located on a plasmid (22, 26). R-M systems presumably function in a similar way to that of the poison-antidote stability systems. The methylase may act as an antidote that prevents cleavage of the cellular DNA by the restriction endonuclease or poison. A pRA2 mutant (pRA14) which carried ΩStrr/Spcr inserted into the pac25I restriction endonuclease gene was generated. When assayed for segregational stability, pRA14 was found to be equally as stable as pRA19. Therefore, the Pac25I R-M system did not appear to be critical for pRA2 stability.
There are clear similarities, in both sequence and the arrangement of their genes, between MobA, MobB, and MobC and the TraI, TraJ, and TraK proteins in RK2/RP4, respectively, which bind to oriT and initiate events that are essential for conjugal transfer (27, 53, 55). We propose a similar role for the pRA2 Mob proteins; they are likely to act in a plasmid-specific manner, modifying the plasmid structure such that it can be transferred by a conjugative plasmid. Despite the high degree of homology between IncP plasmids R751 and RK2/RP4, the oriT of the latter plasmid cannot be transferred by R751. Transfer of the RK2/RP4 oriT only occurred when RK2/RP4 traI, traJ, and traK were also present (13, 44). Therefore, the Mob proteins that are encoded by pRA2 are likely to be essential for pRA2 mobilization by RK2/RP4 Tra functions. Alignment of the pRA2 oriT sequence with the plasmid nicking regions of IncP plasmids suggests that the potential nic site for pRA2 occurs between positions 27767 and 27768. The traK homolog, mobC, is likely to be the first gene on the leading strand to enter the recipient cell during pRA2 transfer. MobD and MobE may also contribute to pRA2 mobilization, but both are novel in amino acid sequence and do not have any homologs in GenBank.
The transfer of pRA2-carried genetic markers was found to occur between isogenic P. alcaligenes strains. Transfer was sensitive to DNase I, and concentrations of 1 mg/ml of DNase I was sufficient to abolish plasmid transfer. Sensitivity to DNase I has not been observed for processes such as conjugal transfer or transduction, in which plasmids would be protected from extracellular nucleases by pilus structures extending from the donor cell to the recipient cell or by viral particles. Intrastrain pRA2 transfer was characteristic of natural transformation processes, and sensitivity to DNase I indicates that at some stage during transfer, pRA2 is exposed to the external environmental conditions. When the P. alcaligenes strains P25X19 (Strr/Spcr) and P25X20 (Kanr) were mated in liquid media, DNA exchange was observed to be 105-fold lower than the frequency achieved from matings on a solid surface. When donor and recipient cells were separated by a membrane with 0.1-μm pores, DNA exchange was undetectable (<10−9). These observations suggest that cell-cell contact was required for pRA2 transfer. Transfer processes that are both DNase I sensitive and require cell-cell contact have been reported for plasmid transfer between Streptococcus pyogenes and Streptococcus sanguis (6) and also between E. coli and marine Vibrio species (28). In these bacteria, plasmid DNA seems to be externalized, but it is unclear whether the plasmid DNA is permanently associated with the outer membrane or whether externalization is triggered by cell-cell interactions. The requirement for cell-cell contact and the fact that supernatants from donor cultures did not have any transforming activity argue against DNA release due to cell lysis being a major contributing factor towards plasmid transformation in P. alcaligenes NCIB 9867.
pRA2-derived plasmids that had been extracted and purified were also found to have transforming activity when added to P. alcaligenes NCIB 9867 cells. Frequencies of 106 transformants per microgram of plasmid DNA were observed on solid surfaces. The same plasmids could transform cells in liquid media; however, a 10-fold reduction in transformation frequency was observed. In P. stutzeri, addition of extracted chromosomal DNA was found to be 103-fold less efficient in transformation when compared to the same amount of DNA supplied as a component of intact donor cells (38). A similar observation was made for liquid transformation in Vibrio strains in which plasmid transfer from donors was found to be more efficient than addition of purified plasmid DNA into the media (28). In contrast, for P. alcaligenes NCIB 9867, there was no reduction in transformation activity when plasmid DNA that was supplied exogenously was compared to plasmid DNA as a component of intact donors.
In order to explain our observations of pRA2 transfer under different mating conditions, we propose that cell-cell contact must be achieved at two specific stages during transformation. First, cell-cell contact between the donor and the recipient may be required to trigger plasmid export from the cytoplasm of the donor to a position on the outer membrane. Second, cell-cell contact, or at least cell intimacy, is required for the recipient to capture the plasmid DNA from the cell surface of the donor. In P. alcaligenes NCIB 9867, cell-cell contact may be sustained long enough for both of these processes to occur when matings are carried out on filters. However, in liquid media, firm cell-cell contact between the donor and recipient may occur at much lower frequencies or may not be sustained for the required time period. This would account for the dramatic decrease in transformation frequencies observed in liquid media. Under these circumstances, inhibition of cell-cell contact by a membrane with 0.1-μm pores should completely abolish transfer, which was the observation made in our experiments. On the other hand, addition of purified plasmid DNA would eliminate the requirement for cell-cell contact, and transformation would then become independent of donor externalization and then may only be dependent upon DNA-cell interactions. In other words, transformation can occur when a free plasmid molecule comes into contact with the recipient cell.
Several broad-host-range plasmids, including pRK415, did not have any transforming activity when applied directly to cells. However, when the 19.3-kb PstI pRA2 fragment was cloned into pRK415, the resulting plasmid (pPSK1) demonstrated approximately 1% of the transforming activity of pRA19 plasmid DNA. pRK415 carrying smaller pRA2 fragments had much less transforming activity. It is possible that the 19.3-kb pRA2 fragment could contain a specific DNA uptake sequence, such as has been observed in H. influenzae (35) and Neisseria gonnorhoeae (14). However, in this case, one would expect the transforming activity of pPSK1 to be similar to the transforming activity of pRA19 plasmid. A specific DNA uptake sequence could not account for the partial transforming activity that was observed for pPSK1. It may be more plausible to expect that transforming activity is somehow related to the size of the plasmid. A model for plasmid transformation in Streptococcus pneumoniae was proposed by Saunders and Guild (33). They suggested that two single-stranded DNA molecules that have entered the cell separately associate to form a duplex and that any single-stranded gaps can be regenerated by the cell's DNA repair systems. In P. stutzeri, plasmid DNA could not undergo transformation and only hybrid plasmids carrying chromosomal inserts were able to transform. Transformation frequencies were also found to increase with an increase in insert size (8).
Previously, a genetic system in P. alcaligenes NCIB 9867 was identified with both auxotrophic and catabolic chromosomal markers (29). Transfer of chromosomal DNA was established to occur in a bidirectional manner at frequencies ranging from 10−5 for auxotrophic markers to 10−7 for catabolic markers. In comparison, our study found that plasmid-carried genetic determinants were transformed at a frequency of 10−4 in P. alcaligenes NCIB 9867, and it remains unclear whether the transfer of chromosomal and plasmid-carried markers occurs by the same transformation mechanism.
FIG. 3.
Directed mutagenesis of the pac25I.R gene by homologous recombination. (A) PCR products RSK1 (0.74 kb) and RSK2 (0.86 kb) were amplified from sequences flanking the insertion site with primer pairs RSK1F (5′-CTG CAG CTT GCC GAG TAC G-3′) and RSK1R (5′-AAG CTT CGC TGC GGG CCT TCA-3′) and RSK2F (5′-AAG CTT CGC TGG CCG CCC T-3′) and RSK2R (5′-CTG CAG CGC CCG GAT TTC CT-3′). These primers were designed to incorporate HindIII restriction sites into the ends of the PCR products adjacent to the target site and PstI restriction sites at the opposing ends. Both RSK1 and RSK2 were cloned so that the terminal restriction sites would be cleaved efficiently before ligation. (B) A ligation reaction was set up with pUC18 cleaved with PstI, ΩStrr/Spcr cleaved with HindIII, and the two cloned PCR products, RSK1 and RSK2, digested with both enzymes. The orientation of the inserts in the desired construct was monitored by restriction analysis. The PstI fragment from this construct, which consisted of the ΩStrr/Spcr gene cassette flanked by pRA2 DNA sequences, was recloned into pSUP202, resulting in pSUP25Ω. (C) pSUP25Ω was introduced into P. alcaligenes NCIB 9867 by conjugation via S17-1. Transconjugants were selected on STR- and SPC-containing media and subsequently screened for tetracycline sensitivity. In the tetracycline-sensitive transconjugant P25X14, sequencing confirmed that the ΩStrr/Spcr gene cassette had replaced pRA2 DNA from nucleotide 20373 to 20396 within pac25I.R, and this plasmid was designated pRA14.
ACKNOWLEDGMENTS
This work was supported by the National University of Singapore Academic Research Fund no. RP3-95-0383 to C. L. Poh. S. M. Kwong was supported by a National University of Singapore Postgraduate Scholarship.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific purpose cloning vectors. II. Broad-host-range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Bechhofer D H, Figurski D H. Map location and nucleotide sequence of korA, a key regulatory gene of promiscuous plasmid RK2. Nucleic Acids Res. 1983;21:7453–7469. doi: 10.1093/nar/11.21.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellido F, Martin N L, Siehnel R J, Hancock R E W. Reevaluation, using intact cells, of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J Bacteriol. 1992;174:5196–5203. doi: 10.1128/jb.174.16.5196-5203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buu-Hoï A, de Cespédès G, Horaud T. Deoxyribonuclease-sensitive transfer of an R plasmid in Streptococcus pyogenes (group A) FEMS Microbiol Lett. 1985;30:407–410. [Google Scholar]
- 7.Carlson C A, Pierson L S, Rosen J J, Ingraham J L. Pseudomonas stutzeri and related species undergo natural transformation. J Bacteriol. 1983;153:93–99. doi: 10.1128/jb.153.1.93-99.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson C A, Steenbergen S M, Ingraham J L. Natural transformation of Pseudomonas stutzeri by plasmids that contain cloned fragments of chromosomal deoxyribonucleic acid. Arch Microbiol. 1984;140:134–138. [Google Scholar]
- 9.Datta N, Hedges R W, Shaw E J, Sykes R B, Richmond M H. Properties of an R-factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo Z, Herrero M, Nielands J B. IS1-mediated mobility of the aerobactin system of pColV-K30 in E. coli. Mol Gen Genet. 1983;213:487–490. doi: 10.1007/BF00339620. [DOI] [PubMed] [Google Scholar]
- 11.Easter C L, Schwab H, Helinski D R. Role of the parCBA operon of the broad-host-range plasmid RK2 in stable plasmid maintenance. J Bacteriol. 1998;180:6023–6030. doi: 10.1128/jb.180.22.6023-6030.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fürste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 priming region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 13.Fürste J P, Pansegrau W, Ziegelin G, Kröger M, Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci USA. 1989;86:1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegeman G D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966;91:1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopper D J, Chapman P J. The enzyme degradation of alkyl-substituted gentisates, maleates and malates. Biochem J. 1971;122:29–40. doi: 10.1042/bj1220029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain R K, Bayly R C, Skurray R A. Characterization and physical analysis of a 3,5-xylenol degradative plasmid in Pseudomonas putida. J Gen Microbiol. 1984;130:3019–3028. [Google Scholar]
- 18.Jensen R B, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 19.Jugara-Burdzy G, Thomas C M. kfrA gene of broad host range plasmid RK2 encodes a novel DNA-binding protein. J Mol Biol. 1992;225:651–660. doi: 10.1016/0022-2836(92)90392-w. [DOI] [PubMed] [Google Scholar]
- 20.Jugara-Burdzy G, Thomas C M. Purification of KorA protein from broad host range plasmid RK2: definition of a hierarchy of KorA operators. J Mol Biol. 1995;265:3507–3518. doi: 10.1006/jmbi.1995.0534. [DOI] [PubMed] [Google Scholar]
- 21.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 22.Kulakauskas S, Lubys A, Ehrlich S D. DNA restriction-modification systems mediate plasmid maintenance. J Bacteriol. 1995;177:3451–3454. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong S M, Yeo C C, Chuah D, Poh C L. Sequence analysis of plasmid pRA2 reveals a novel replication region. FEMS Microbiol Lett. 1998;158:159–165. doi: 10.1111/j.1574-6968.1998.tb12815.x. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorow D, Jessee J. Max efficiency DH10BTM: a host for cloning methylated DNA. FOCUS (Life Technologies) 1990;19:28–29. [Google Scholar]
- 26.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 27.Pansegrau W, Balzer D, Kruft V, Lurz R, Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci USA. 1990;87:11538–11542. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul J H, Thurmond J M, Frischer M E, Cannon J P. Intergeneric natural plasmid transformation between E. coli and a marine Vibrio species. Mol Ecol. 1992;1:37–46. doi: 10.1111/j.1365-294x.1992.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 29.Poh C L, Tham J M. Genetic system in Pseudomonas alcaligenes NCIB 9867. FEMS Microbiol Lett. 1993;106:253–258. [Google Scholar]
- 30.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 31.Rohrer J, Rawlings D E. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J Bacteriol. 1992;174:6230–6237. doi: 10.1128/jb.174.19.6230-6237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Saunders C W, Guild W R. Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol Gen Genet. 1981;181:57–62. doi: 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- 34.Shingler V, Thomas C M. Transcription in the trfA region of broad-host-range RK2 is regulated by trfB and korB. Mol Gen Genet. 1984;195:523–529. doi: 10.1007/BF00341457. [DOI] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 36.Sisco K L, Smith H O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci USA. 1979;76:972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C A, Shingler V, Thomas C M. The trfA and trfB promoter regions of broad-host-range plasmid RK2 share common potential regulatory sequences. Nucleic Acids Res. 1984;12:3619–3630. doi: 10.1093/nar/12.8.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart G J, Carlson C A, Ingraham J L. Evidence for an active role of donor cells in natural transformation of Pseudomonas stutzeri. J Bacteriol. 1983;156:30–35. doi: 10.1128/jb.156.1.30-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinfield T J, Janniere L, Ehrlich S D, Minton N P. Characterization of a region of the Enterococcus faecalis plasmid pAMβ1 which enhances the segregational stability of pAMβ1-derived cloning vectors in Bacillus subtilus. Plasmid. 1991;26:209–221. doi: 10.1016/0147-619x(91)90044-w. [DOI] [PubMed] [Google Scholar]
- 40.Theophilus B D M, Cross M A, Smith C A, Thomas C M. Regulation of the trfA and trfB promoters of broad-host-range plasmid RK2: identification of sequences essential for regulation by trfA/korA/korD. Nucleic Acids Res. 1985;13:8129–8142. doi: 10.1093/nar/13.22.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas C M, Ibbotson J P, Wang N, Smith C A, Tipping R, Loader N M. Gene regulation on broad host range plasmid RK2: identification of three novel operons whose transcription is repressed by both KorA and KorC. Nucleic Acids Res. 1988;16:5345–5359. doi: 10.1093/nar/16.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas C M, Theophilus B D M, Johnston L, Jugara-Burdzy G, Schilf W, Lurz R, Lanka E. Identification of a seventh operon on plasmid RK2 regulated by the korA gene product. Gene. 1990;89:29–35. doi: 10.1016/0378-1119(90)90202-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Berg C M, Chen J, Young A C, Blakesley R W, Lee L Y, Berg D E. Creating nested deletions for sequencing cosmid DNAs. FOCUS (Life Technologies) 1993;15:47. [Google Scholar]
- 44.Yakobson E, Guiney D. Homology in the transfer origins of broad host range IncP plasmids: definition of two subgroups of P plasmids. Mol Gen Genet. 1983;192:436–438. doi: 10.1007/BF00392187. [DOI] [PubMed] [Google Scholar]
- 45.Yeo C C, Poh C L. IS1394 from Pseudomonas alcaligenes NCIB 9867: identification and characterization of a member of the IS30 family of insertion elements. Gene. 1996;175:109–113. doi: 10.1016/0378-1119(96)00133-3. [DOI] [PubMed] [Google Scholar]
- 46.Yeo C C. Mobile genetic elements of Pseudomonas alcaligenes NCIB 9867. Ph.D. thesis. Singapore: National University of Singapore; 1997. [Google Scholar]
- 47.Yeo C C, Tham J M, Kwong S M, Poh C L. Characterization of the pac25I restriction-modification genes isolated from the endogenous pRA2 plasmid of Pseudomonas alcaligenes NCIB 9867. Plasmid. 1998;40:203–213. doi: 10.1006/plas.1998.1365. [DOI] [PubMed] [Google Scholar]
- 48.Yeo C C, Tham J M, Kwong S M, Yiin S, Poh C L. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol Lett. 1998;165:253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x. [DOI] [PubMed] [Google Scholar]
- 49.Yeo C C, Wong D T S, Poh C L. IS1491 from Pseudomonas alcaligenes NCIB 9867: characterization and distribution among Pseudomonas species. Plasmid. 1998;39:187–195. doi: 10.1006/plas.1997.1331. [DOI] [PubMed] [Google Scholar]
- 50.Young C, Prince A S, Figurski D H. KorA function of promiscuous plasmid RK2: an autorepressor that inhibits expression of host-lethal gene kilA and replication gene trfA. Proc Natl Acad Sci USA. 1985;82:7374–7378. doi: 10.1073/pnas.82.21.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young C, Burlage R S, Figurski D H. Control of the kilA gene of the broad-host-range plasmid RK2: involvement of korA, korB, and a new gene, korE. J Bacteriol. 1987;169:1315–1320. doi: 10.1128/jb.169.3.1315-1320.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zatyka M, Thomas C M. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol Rev. 1998;21:291–319. doi: 10.1111/j.1574-6976.1998.tb00355.x. [DOI] [PubMed] [Google Scholar]
- 53.Ziegelin G, Fürste J P, Lanka E. TraJ protein of plasmid RP4 binds to a 19 base pair inverted sequence repetition within transfer origin. J Biol Chem. 1989;264:11989–11994. [PubMed] [Google Scholar]
- 54.Ziegelin G, Pansegrau W, Strack B, Balzer D, Kröger M, Kruft V, Lanka E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Sequence. 1991;1:303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]
- 55.Ziegelin G, Pansegrau W, Lurz R, Lanka E. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J Biol Chem. 1992;267:17279–17286. [PubMed] [Google Scholar]