Abstract
Background
Given the plethora of pathophysiologic mechanisms described in idiopathic pulmonary fibrosis (IPF), we hypothesize that the mechanisms driving fibrosis in IPF may be different from one patient to another.
Research Question
Do IPF endotypes exist and are they associated with outcome?
Study Design and Methods
Using a publicly available gene expression dataset retrieved from BAL samples of patients with IPF and control participants (GSE70867), we clustered IPF samples based on a dimension reduction algorithm specifically designed for -omics data, called DDR Tree. After clustering, gene set enrichment analysis was performed for functional annotation, associations with clinical variables and prognosis were investigated, and differences in transcriptional regulation were determined using motif enrichment analysis. The findings were validated in three independent publicly available gene expression datasets retrieved from IPF blood samples.
Results
One hundred seventy-six IPF samples from three centers were clustered in six IPF clusters, with distinct functional enrichment. Although clinical characteristics did not differ between the clusters, one cluster conferred worse sex-age-physiology score-corrected survival, whereas another showed a numeric trend toward worse survival (P = .08). The first was enriched for increased epithelial and innate and adaptive immunity signatures, whereas the other showed important telomere and mitochondrial dysfunction, loss of proteostasis, and increased myofibroblast signatures. The existence of these two endotypes, including the impact on survival of the immune endotype, was validated in three independent validation cohorts. Finally, we identified transcription factors regulating the expression of endotype-specific survival-associated genes.
Interpretation
Gene expression-based endotyping in IPF is feasible and can inform clinical evolution. As endotype-specific pathways and survival-associated transcription factors are identified, endotyping may open up the possibility of endotype-tailored therapy.
Key Words: endotyping, gene expression, IPF
Abbreviations: HR, hazard ratio; IPF, idiopathic pulmonary fibrosis
FOR EDITORIAL COMMENT, SEE PAGE 1440
Take-home Points.
Study Question: Given the plethora of pathophysiologic mechanisms described in idiopathic pulmonary fibrosis (IPF), do IPF endotypes exist and are they associated with outcome?
Results: Using a publicly available gene expression dataset, we clustered IPF BAL samples into six endotypes with distinct functional enrichment, one endotype of which, enriched for epithelial and immune signatures, conferred worse gender-age-physiology-corrected outcomes.
Interpretation: IPF endotyping based on BAL gene expression predicts prognosis and may open up the possibility of novel endotype-specific therapies.
Idiopathic pulmonary fibrosis (IPF) is a relentless fibrotic disease with a median survival of 3 to 5 years if left untreated. Its pathophysiologic characteristics remain incompletely understood. Alveolar type 2 cells (type 2 pneumocytes) injured by alveolar stressors develop an apoptosis-resistant and senescent phenotype,1,2 including progressive shortening of its telomeres,3,4 endoplasmic reticulum stress,5,6 and mitochondrial dysfunction.7,8 These type 2 pneumocytes as well as alveolar macrophages9 have been shown to secrete profibrotic mediators, activating fibroblasts and inducing myofibroblast subtypes, with the transforming growth factor β pathway playing a quintessential role.10 Recently, aberrant basaloid cells were found lining myofibroblast foci and were characterized by an intense profibrotic gene signature.11,12 Epigenetic changes, including histone acetylation and methylation,13,14 reactivation of developmental pathways,15, 16, 17 and metabolic changes,18 also were found to be important.
Development of effective treatment options for this dismal disease has proven to be a years-long struggle. Many proposed molecules with promising preclinical results failed in large phase 3 trials.19, 20, 21, 22, 23, 24, 25, 26, 27 In 2014, two drugs were found to be effective28,29 (ie, pirfenidone and nintedanib), both with broad and incompletely understood working mechanisms.30,31 Their inhibition of the transforming growth factor β pathway is thought to be essential for efficacy,32 although effects on angiogenesis, profibrotic growth factors, immune pathways, and apoptosis also are recognized.30,31
Although the quest for effective treatment options in non-small cell lung cancer has been equally frustrating, since non-small cell lung cancer cases have been stratified according to their driving mechanisms in multiple endotypes, new endotype-specific drugs were developed with impressive treatment effects that seemed impossible 20 years ago.33, 34, 35
The aim of this study was to assess diversity in gene expression profiles of patients with IPF and to investigate the existence of endotypes impacting patient prognosis characterized by differential expression patterns that might suggest distinct endotype-specific mechanisms driving fibrosis. Therefore, we analyzed gene expression data from an already published multicenter cohort including BAL samples of 176 patients with IPF.12 We assessed variability of gene expression signatures, clustered cases based on their transcriptomic profile, and assessed their association on clinical outcome. We also functionally enriched these endotypes, determined endotype-specific transcription factors (which may serve as a rationale for endotype-tailored treatment in the future), and finally validated the existence of these endotypes in three independent cohorts.
Study Design and Methods
The methods are described in more detail in e-Appendix 1.
Patient Cohort
We used BAL gene expression and clinical data included in the GSE70867 dataset, consisting of 20 control participants and 176 patients with IPF whose disease was diagnosed in a multidisciplinary discussion.36 BAL sampling was performed in three centers (Freiburg, Siena, and Leuven), all before initiation of antifibrotic or immunosuppressive treatment (for cases diagnosed before the antifibrotics era). Ethical approval was granted at the moment of the initial study.12
Gene Expression Data and Clustering
After downloading this publicly available gene expression dataset from the GEO database (GSE70867), batch effects associated with the cohort were diagnosed and removed using the Combat functionality37 (e-Fig 1). DDR Tree is a dimension reduction technique specifically designed for gene expression data sets to maximally preserve the intrinsic structure of the data.38 First, a reverse graph embedding is applied to determine the intrinsic graph structure of the original high-dimension dataset. Second, dimension reduction is performed, preserving distances of the data points in the resulting low-dimension space. Because the goals of the study were to explore the variability of the data and to assess whether clustering could be meaningful, the number of clusters chosen as the maximum number of clusters possible, with a minimum of 10 samples per cluster. For every cluster, differential gene expression vs control samples was calculated using linear mixed models, correcting for age, sex, and the center where the sample was obtained.
Functional Enrichment
To evaluate the functional annotations of the clusters, gene ontology enrichment was performed using generally applicable gene set enrichment analysis.39 To obtain a broad overview of the functional enrichments of each cluster, we analyzed differential expression of gene ontologies for every cluster between the cluster and all other clusters.
In a second phase, to assess direction of differential expression of these pathways, generally applicable gene set enrichment analysis was used to determine differential expression of gene ontologies for each cluster compared with control samples. Expression of gene ontologies associated with the most important pathways were visualized using density plots.
Moreover, we performed cellular subtype deconvolution using xCell,40 which is based on several single-cell dataset-based gene signatures, using a spillover compensation technique to reduce associations of closely related cell types, which was validated using cytometry, immunophenotyping and in-silico simulations.
Analysis of Transcription Factors Regulating the Expression of Survival-Associated Genes
To evaluate whether distinct gene expression signatures would impact survival directly, we assessed the expression of 1,381 genes shown to be associated with survival in the initial study of Prasse et al.12 We evaluated the expression of these genes in each cluster vs all other IPF samples. We evaluated whether these endotype-specific survival-associated gene sets were modulated by distinct motifs, transcription factors, or both using iRegulon and the Rcistarget package.41
Endotype Validation in Three Independent Validation Cohorts
To validate the existence of these endotypes, gene expression of the 250 most differentially expressed genes was determined in three independent gene expression datasets retrieved from IPF blood samples: GSE2822142 (n = 75), GSE9360643 (n = 57), and GSE13260744 (n = 74). Gene expression was visualized using heatmaps, which allowed the identification of these endotypes by hierarchical clustering. Survival differences between cases with and without an endotype-specific transcriptome in these cohorts were analyzed using multivariate Cox proportional hazards models, correcting for age and sex.
Results
Baseline Characteristics
This study included 176 patients with IPF and 20 healthy volunteers (serving as control subjects) involved in the GSE70867 dataset, originally included in a BAL gene expression study by Prasse et al.12 All patients and control participants underwent BAL sampling. Patients with IPF were significantly older compared with control participants (patients with IPF, 68.1 ± 9.5 years; control participants, 61.3 ± 8.3 years; P = .002). Both patients with IPF and control participants were predominantly men (ratio of men to women: patients with IPF, 144:32; control participants, 16:4; P = .76). One hundred patients with IPF (56.8% of patients with IPF) died during a mean follow-up of 699 ± 520 days. Mean gender-age-physiology score was 4.3 ± 1.7.
Clustering and Association With Clinical Outcome
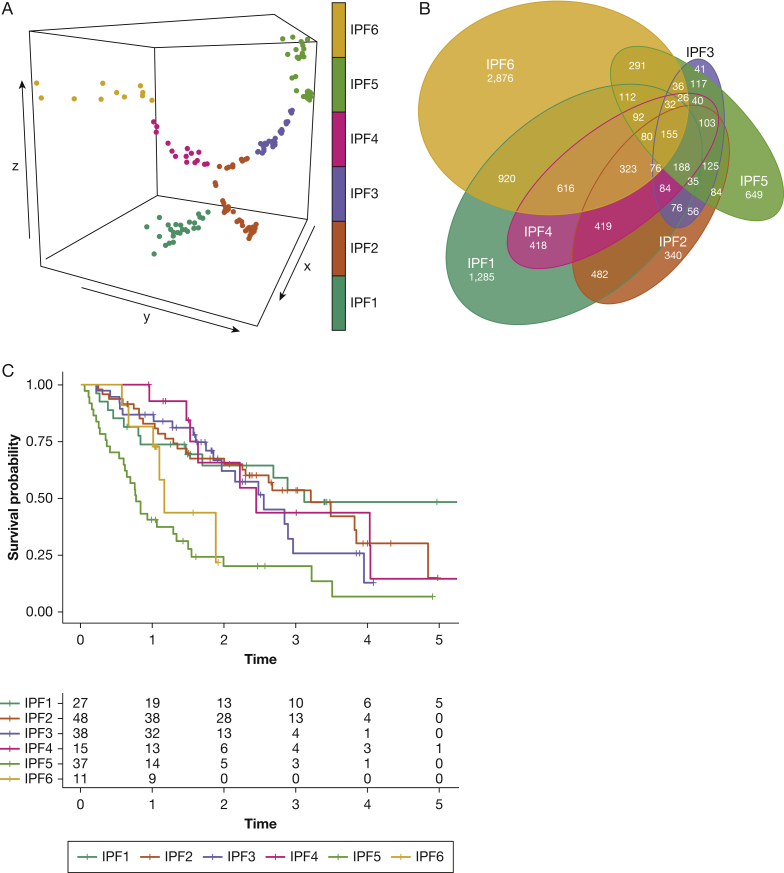
DDR Tree dimension reduction retrieved six clusters (Fig 1A), labelled IPF1 to IPF6. Baseline characteristics of the six clusters are shown inTable 1. Broadly, no significant differences were observed for baseline characteristics, pulmonary function test results, smoking status, high-resolution CT scan findings, BAL lymphocytosis, and lag time between symptoms onset and BAL procedure and between IPF diagnosis and BAL procedure. However, BAL neutrophilia was higher in the IPF5 cluster compared with the others (P = .001). All clusters included patients from the three centers, although imbalances occurred (P = .003).
Figure 1.
Clustering of IPF samples based on gene expression data and association with survival. IPF samples were clustered based on gene expression data using the DDR Tree algorithm. A, Three-dimensional scatterplot of the three DDR Tree dimensions and clustering based on Euclidean distance. B, Euler diagram showing differentially expressed genes compared with those of control participants. C, Kaplan-Meier curves showing IPF endotypes. IPF = idiopathic pulmonary fibrosis.
Table 1.
Baseline Characteristics of the Six IPF Clusters
| Characteristic | IPF1 | IPF2 | IPF3 | IPF4 | IPF5 | IPF6 | P Value |
|---|---|---|---|---|---|---|---|
| No. (%) | 27 (15.3) | 48 (27.3) | 38 (21.6) | 15 (8.5) | 37 (21.0) | 11 (6.3) | … |
| Age, y | 68.6 ± 8.8 | 69.3 ± 8.7 | 67.4 ± 10.7 | 63.5 ± 10.2 | 68.5 ± 10.0 | 68.8 ± 7.6 | .46 |
| Male sex | 20 (74) | 37 (77) | 32 (84) | 15 (100) | 30 (81) | 10 (91) | .30 |
| FVC, percent predicted | 75.6 ±17.4 | 75.3 ± (21.6) | 67.0 ± 19.7 | 69.1 ± 18.2 | 64.3 ± 22.6 | 75.5 ± 22.4 | .11 |
| FEV1, percent predicted | 72.3 ± 15.4 | 75.6 ± 20.7 | 67.4 ± 19.7 | 72.3 ± 17.3 | 64.9 ± 20.2 | 77.7 ± 19.5 | .31 |
| Dlco, percent predicted | 47.2 ± 11.8 | 44.6 ± 15.3 | 48.2 ± 14.8 | 44.1 ± 14.6 | 39.7 ± 13.1 | 43.6 ± 15.1 | .11 |
| GAP | 3.9 ± 1.7 | 4.3 ± 1.8 | 4.1 ± 1.7 | 3.8 ± 1.4 | 4.9 ± 1.6 | 4.3 ± 1.2 | .11 |
| Smoking status | .87 | ||||||
| Active | 1 | 2 | 1 | 0 | 1 | 1 | |
| Former | 17 | 32 | 21 | 12 | 23 | 7 | |
| Never | 9 | 14 | 16 | 3 | 13 | 3 | |
| BAL lymphocytosis | 6.9 ± 7.4 | 9.3 ± 8.5 | 10.5 ± 8.5 | 8.1 ± 6.4 | 12.7 ± 13.1 | 10.5 ± 9.5 | .23 |
| BAL neutrophilia | 8.7 ± 12.5 | 8.4 ± 7.5 | 10.1 ± 13.6 | 10.2 ± 9.1 | 20.1 ± 20.3 | 7.3 ± 6.1 | .001 |
| HRCT scan pattern | .25 | ||||||
| Definite | 17 | 36 | 29 | 12 | 29 | 10 | |
| Probable | 7 | 10 | 3 | 3 | 7 | 0 | |
| Possible | 3 | 2 | 5 | 0 | 1 | 0 | |
| Emphysema | 6 (22) | 8 (17) | 8 (22) | 4 (27) | 5 (14) | 3 (30) | .72 |
| Time between symptoms onset and BAL, moa | 20.5 (30.6) | 16.2 (9.2) | 25.4 (21.4) | 13.5 (17.6) | 13.6 (21.1) | 5.0 (2.0) | .20 |
| Time between initial diagnosis and BAL, moa | 3.7 (8.4) | 2.7 (6.8) | 12.9 (25.6) | 9.0 (17.0) | 2.5 (10.3) | 3.5 (1.5) | .29 |
| Cohort | .003 | ||||||
| Freiburg | 15 | 10 | 16 | 5 | 15 | 1 | |
| Leuven | 8 | 14 | 17 | 6 | 15 | 4 | |
| Siena | 4 | 24 | 5 | 4 | 7 | 6 |
Data are presented as No. (%), No., mean ± SD, or median (interquartile range), unless otherwise indicated. Differences between patients with IPF and control participants were calculated using Student t tests and Fisher exact tests for continuous and categorical variables, respectively. Differences between clusters regarding clinical variables were calculated using analyses of variance for continuous variables and Fisher exact tests for categorical variables. Dlco = diffusing capacity of the lung for carbon monoxide; GAP = gender-age-physiology; HRCT = high-resolution CT scan.
Timing data available only for the Leuven cohort. Because of small numbers, data are presented as median (interquartile range). P values are determined using the Kruskal-Wallis test for these variables because of low numbers.
From the 18,604 genes analyzed, 12,467 genes (67.0%) were expressed differentially compared with genes from control participants in at least 1 cluster, whereas only 155 genes were differentially expressed in all clusters. Overlapping numbers of differentially expressed genes are visualized in Figure 1B. The proportion of genes that were differentially expressed specifically in one specific cluster was 20.5%, 9.4%, 2.8%, 8.7%, 22.1%, and 41.2% for IPF1, IPF2, IPF3, IPF4, IPF5, and IPF6, respectively. Clustering predicted survival, both in univariate analysis (P < .001) and after correction for gender-age-physiology score (P = .002). IPF5 conferred worse survival vs all other patients with IPF (hazard ratio [HR], 3.35; 95% CI, 2.15-5.21; P < .001) (Fig 1C). Although a numeric trend was observed, IPF6 did not confer worse survival vs all other patients with IPF (HR, 2.13; 95% CI, 0.91-5.0; P = .08), but did vs the four endotypes with the best survival (IPF6 vs IPF1-4: HR, 2.4; 95% CI, 1.02-5.79; P = .045). Because IPF5 conferred worse survival vs all other endotypes, further analysis was focused on this cluster. Similar analyses regarding IPF6 can be found in e-Appendix 1. Top differentially expressed genes for IPF5 and IPF6 are provided in e-Tables 1A and 1B, respectively. Top differentially expressed gene ontologies are provided in e-Table 2.
Functional Enrichment
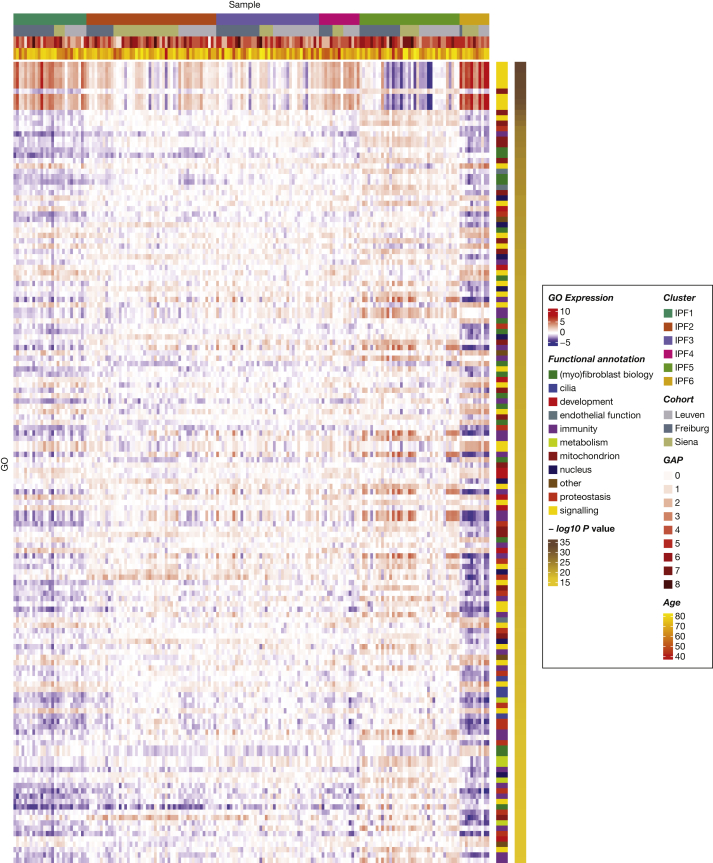
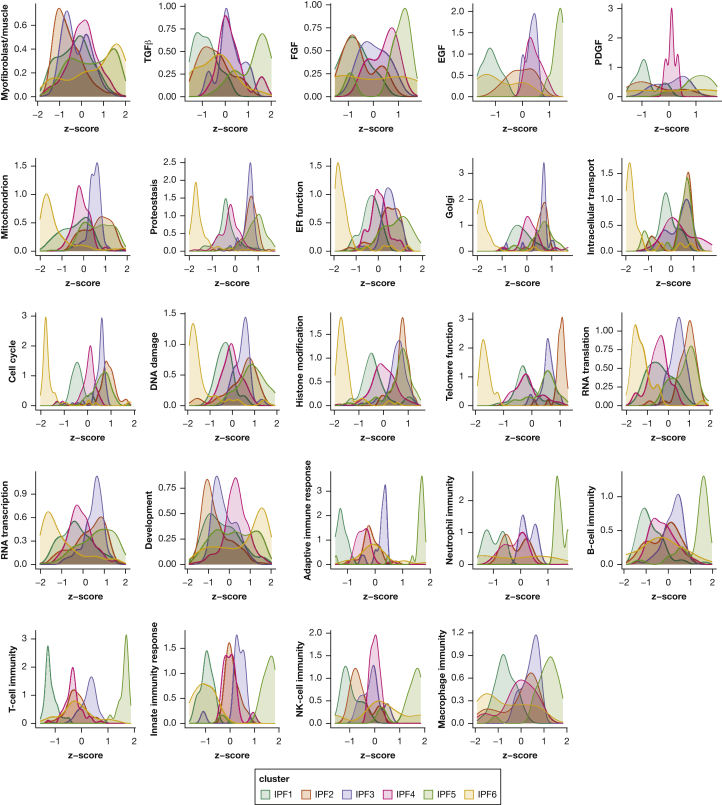
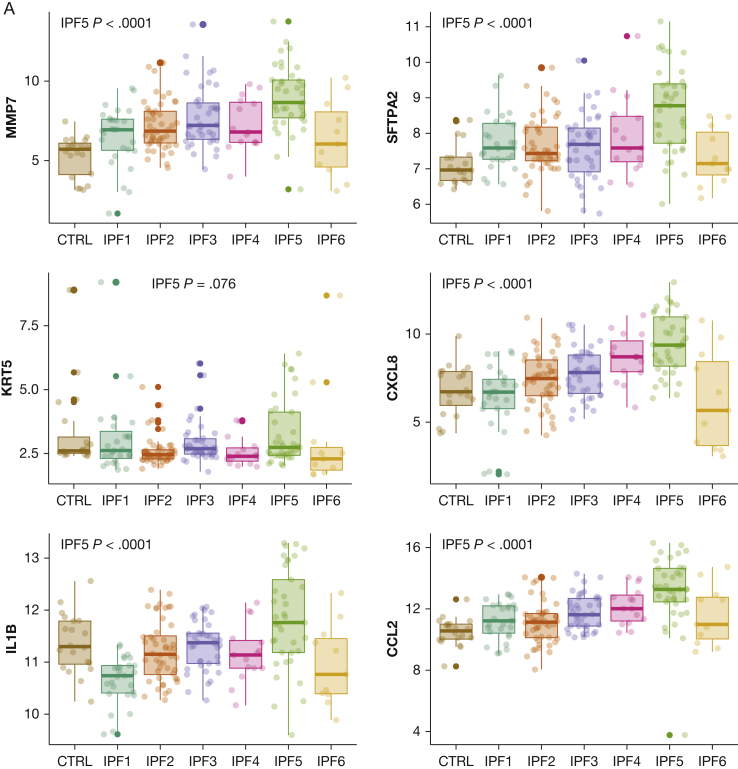
Gene enrichment analysis revealed 6,493 gene ontologies to be differentially expressed between the different endotypes. The 150 gene ontologies most differentially expressed between endotypes largely were involved in myofibroblast, mitochondrial, developmental, immune, and signalling functions (Figs 2, 3). Whereas IPF1 was characterized by decreased immune signatures, IPF2 showed increased expression in cell cycle and cell homeostasis signatures. IPF3 and IPF4 were intermediary endotypes without functional enrichment vs the other endotypes. IPF5 showed increased expression of immune pathways and genes involved in Golgi apparatus and endoplasmic reticulum functions, as well as signatures associated with epithelial and ciliated cells (Fig 4). Hence, the epithelial markers MMP7, SFTPA2, and KRT5 as well as immune markers CXCL8, IL1β, and CCL2 were upregulated vs all other IPF samples (Fig 4). IPF6 showed important telomere dysfunction (e-Fig 2) and associated mitochondrial dysfunction, loss of proteostasis, cell cycle dysfunction, and increases of developmental, signalling, and myofibroblast pathways. These enrichments associated with upregulation of the myofibroblast markers45,46 CTHRC1, ASPN, and COL1A1 and downregulation of GOLGA2 (a Golgi function marker) and ATF6 (member of the unfolded protein response47), and upregulation of CXCL14, a marker of the Hedgehog developmental pathway48 (e-Fig 3).
Figure 2.
Top 100 gene ontologies most differentially expressed between endotypes. Each row represents the expression of a specific gene ontology, each column represents one sample. Functional annotations are manually adjudicated. P values are determined based on differential expression between endotypes. GAP = gender-age-physiology; Go = gene ontology.
Figure 3.
Density plots showing z scores for expression of gene ontologies involved in specific functions. For each specific function, density plots stratified per cluster are shown. The density plots include the z scores of gene ontology expression of gene ontologies included in that specific function. EGF = epidermal growth factor; ER = endoplasmic reticulum; FGF = fibroblast growth factor; IPF = idiopathic pulmonary fibrosis; NK: natural killer cells;PDGF = platelet-derived growth factor; TGFβ = transforming growth factor β.
Figure 4.
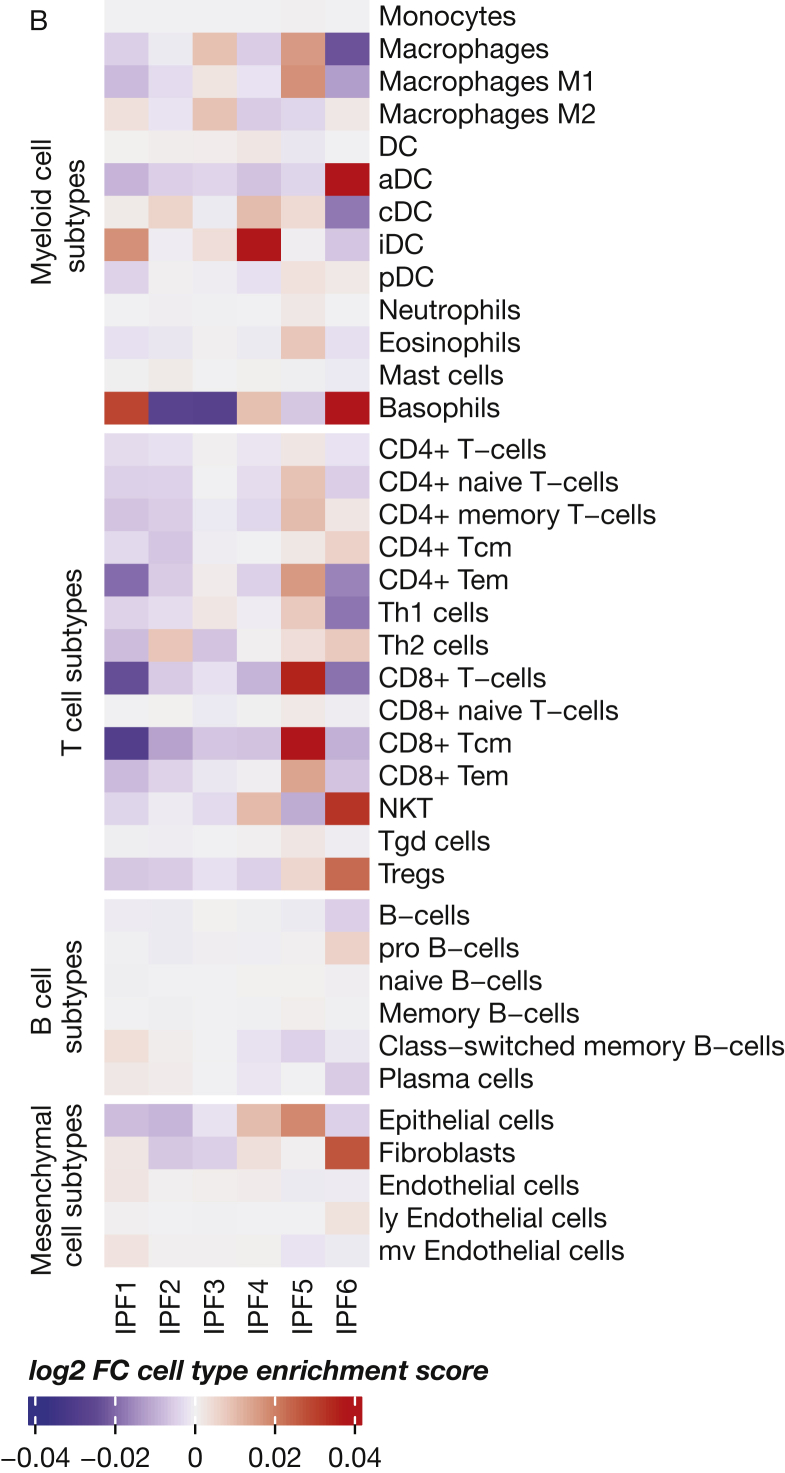
Gene expression of marker genes associated with enriched functions of IPF5 and cell type deconvolution. A, Gene expression of several epithelial and immune archetypical genes. P values were determined vs all other idiopathic pulmonary fibrosis samples. B, Cell subtype deconvolution using xCell based on bulk transcriptome datasets. aDC = activated DC; cDC = conventional DC; DC = dendritic cells; iDC = immature DC; IPF = idiopathic pulmonary fibrosis; ly = lymphatic; mv = microvascular; NKT = natural killer T-cells; pDC = plasmacytoid DC; Tcm = central memory T-cells; Tem = effector memory T-cells; Tgd = gamma-delta T-cells; Th = T-helper cells; Tregs = regulatory T-cells.
To assess further the functional enrichment of these endotypes, we performed xCell, a cell deconvolution technique using enrichments scores (Fig 4B). IPF1 showed remarkable decreased enrichment scores for several T-cell subsets and innate immune subsets, whereas IPF5 was enriched for epithelial cells as well as CD4-positive and CD8-positive T-cell subpopulations. Finally, IPF6 showed increased fibroblast enrichment scores and decreased innate and adaptive immune cell enrichment scores.
Analysis of Transcription Factors Regulating the Expression of Survival-Associated Genes
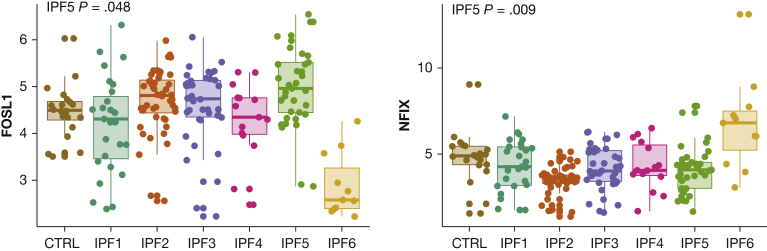
The original Prasse et al12 study derived 1,582 genes associated with survival (988 associated with worse survival, 594 associated with better survival). From the original genes associated with worse survival, 701 were upregulated in IPF5. From the original genes associated with better survival, 174 genes were downregulated in IPF5. We evaluated the transcriptional regulation of these sets of survival-associated genes. Upregulated genes associated with worse survival were regulated by the SOX family as well as the AP-1 transcription factor subunits Jun and FOS. One member of the AP-1 complex, FOSL1, showed increased expression in IPF5, but not in IPF1, IPF2, IPF3, or IPF4, and showed decreased expression in IPF6. Downregulated genes associated with better survival were regulated by nuclear factor IA and nuclear factor IX (NFIX), the latter of which was downregulated in IPF5 and IPF2, but not in the other endotypes (Fig 5). Interestingly, a completely different transcriptional regulation profile was found for IPF6 (e-Appendix 1, e-Fig 4).
Figure 5.
Gene expression of transcription factors regulating endotype-specific survival genes. P values were determined vs control participants. The y-axis represents normalized expression. IPF = idiopathic pulmonary fibrosis.
Validation of Endotype Existence and Its Association With Survival
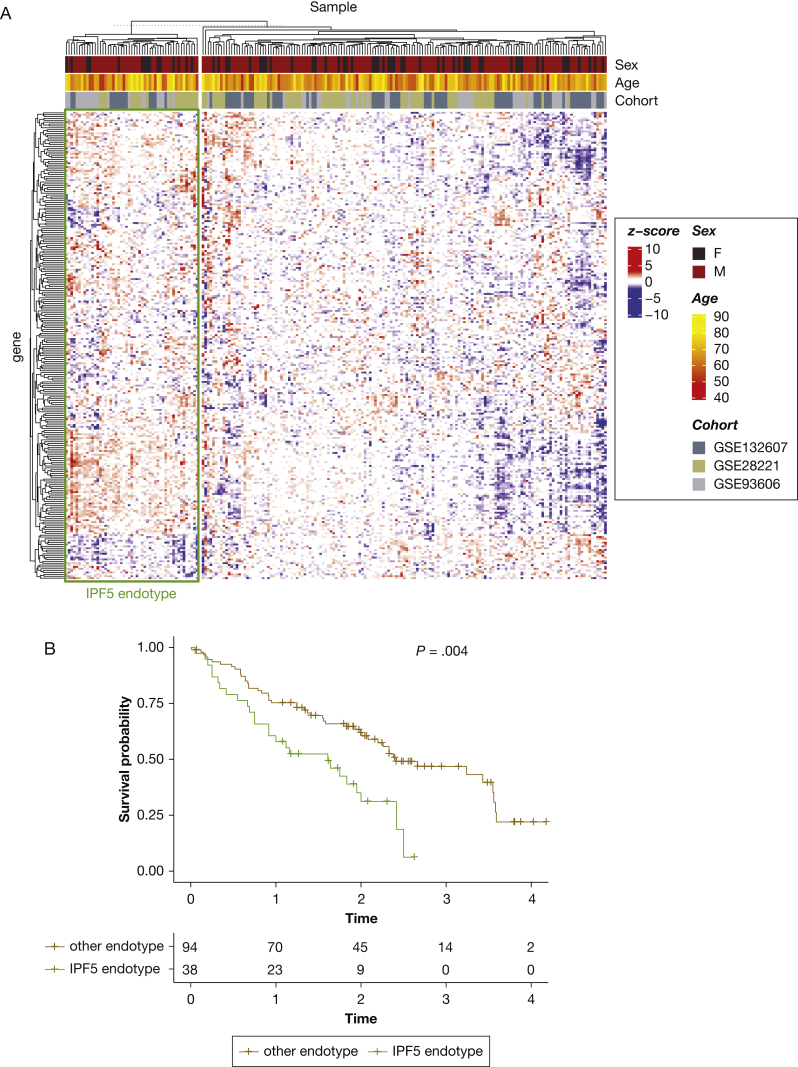
The existence of the IPF5 endotype was evaluated in three independent datasets: GSE2822142 (n = 75), GSE9360643 (n = 57), and GSE13260744 (n = 74), consisting of 206 IPF blood samples at diagnosis. Based on the gene expression of the 250 top differentially expressed genes for IPF5 (compared with all other patients with IPF), 51 patients (24.8%) in the validation cohort showed an IPF5 transcriptome signature (Fig 6A). IPF5 endotype conferred worse survival (HR, 2.1; P = .003) in the two cohorts with survival data (ie, GSE28221 and GSE93606) (Fig 6B). The presence of the IPF6 endotype was validated as well, but did not confer worse survival in the validation cohorts (e-Appendix 1, e-Fig 5).
Figure 6.
IPF endotype validation in separate blood gene expression datasets. A, Heatmap showing gene expression of most differentially expressed genes for IPF5 in three independent gene expression datasets based on blood RNA from patients with IPF. B, Patients with a IPF5-like gene expression in the validation cohorts showing worse survival compared with other patients. IPF = idiopathic pulmonary fibrosis.
Discussion
In this study, we explored the presence of IPF endotypes characterized by distinct gene expression profiles. Using a publicly available gene expression dataset retrieved from BAL samples of 176 patients with IPF and 20 control participants, we determined six endotypes (ie, IPF1-IPF6), of which one showed worse survival (IPF5). Median survival was only 0.77 years for IPF5. Endotypes showed distinct functional enrichment, with IPF1 showing dramatically decreased immune signatures and IPF2 revealing increased cell cycle signatures. IPF5 showed important increases in immune-associated profiles and epithelial signatures. IPF6 revealed important telomere dysfunction and associated mitochondrial dysfunction, loss of proteostasis, and cell cycle dysfunction and increases of developmental, signalling, and myofibroblast pathways. Based on analyses of the respective motifs and transcription factors regulating these survival-associated gene sets, we explored targets for endotype-tailored therapy. Finally, the existence of endotypes and their impact on survival were validated in three independent cohorts consisting of blood samples from patients with IPF.
Some observations deserve further attention. First, the notion of IPF endotypes was suggested some years ago.49 Hoffmann-Vold et al50 suggested the presence of endotypes based on the relative increase of PDGF, VEGF, and EGFR expression. Yang et al51 were the first to cluster IPF based on gene expression patterns, clustering IPF biopsies in two groups: one group with more classical fibroblast and extracellular matrix signatures, and another group showing increased expression of cilia-associated genes.
The upregulation of immune-associated gene ontology signatures in IPF5 may be surprising because IPF has been regarded as the consequence of an aberrant wound-healing mechanism, rather than immune activation. However, the relevance of immunologic involvement has never been abandoned totally: multiple studies have shown associations between IPF outcome and B cells,52 T cells,53 macrophage- and neutrophil-derived proteins,46,54 monocytes,55 and neutrophils.56 Finally, CXCL8 and IL1β levels were associated with microbial dysbiosis.57 Moreover, the association between BAL neutrophilia and poor prognosis has been known for some time.56,58
Finally, our endotype approach may provide a rationale for the high failure rate of large phase 3 trials in IPF: although these drugs may have a beneficial effect in a specific IPF endotype, this effect may be absent in other endotypes, rendering futility of efficacy when tested in a broad IPF cohort. Moreover, we appreciate our results as a warning sign for further clinical studies evaluating the efficacy of molecules with preclinical potential. If the targeted pathways do not prevail in all patients with IPF, the risks of failure may increase, and a promising drug may be lost. However, these data open the horizon for endotype-specific therapies. SOX11, SOX4, and the AP-1 complex (including FOS and Jun), all important transcription factors of survival-associated genes upregulated in IPF5, bind to motifs in the same genomic region,59 thus providing an interesting treatment target for this specific endotype. Moreover, because mitochondrial dysfunction seems to be an important (and rather specific) finding of IPF6, Sobetirome may be an interesting molecule for this endotype.8 Because the expression of genes involved in histone modifications is severely reduced in IPF6 with transcriptomic evidence of reduced RNA transcription and translation, as well as intracellular metabolism and endoplasmic reticulum function, Histone deacetylase (HDAC) inhibitors13 may be of specific interest for this endotype as well.
We acknowledge the limitations of this study. First, the paucity of clinical data provided with the gene expression data limits the exploration of clinical variables potentially associated with these endotypes. Because no data regarding the therapy administered to these patients with IPF are available, we cannot exclude an important effect of imbalances regarding the use of antifibrotics between IPF endotypes, which could explain survival differences as well.60, 61, 62, 63 Second, the cases originated from three centers. Although clustering was blinded to clinical information, some clusters were enriched for cases from a specific center (eg, IPF2 cluster was highly enriched for patients from Siena). Therefore, gene expression analysis was corrected for the center as a random effect. Third, although all diagnoses were reached during a multidisciplinary meeting, only 30% of patients showed histopathologic proof of usual interstitial pneumonia (UIP). Misclassification of diagnoses based on the absence of pathologic data theoretically is not excluded (which also may serve as a source of clinical variability). However, the three centers who provided the BAL samples are expert in ILD diagnosis, and multidisciplinary discussion is the gold standard in IPF diagnosis.36 Fourth, because the original data consisted of bulk transcriptome data, the cellular origins of the retrieved signals are very difficult to verify and the xCell deconvolution technique should be regarded as exploratory only. Fifth, although clustering was performed on an impressive dataset of 176 patients with IPF, the smallest clusters contained only 11 (IPF6) and 15 (IPF2) samples, which limits statistical power in cluster-specific analyses. Sixth, the clustering process importantly is dependent on the used dimension reduction technique, as well as distance metrics. Whether the retrieved clusters represent clinically distinct IPF endotypes should be assessed in prospective work, ascertaining whether the distinct gene expression signatures represent endotype-specific driving mechanisms. Finally, the endotype validation was performed on blood transcriptome datasets (because of the absence of other BAL transcriptome datasets). We acknowledge that these two compartments have a very different cell composition that may affect the results. Hence, validation in BAL gene expression datasets is needed, and further assessment of blood transcriptome profiles in patients with concomitant BAL gene expression assessment would be very useful.
Moreover, the added value of endotyping patients with IPF largely will be dependent on its potential for therapeutic consequences. Whether therapeutically targeting endotype-specific mechanisms is beneficial for IPF subsets remains to be investigated. This study aimed merely to provide evidence for feasibility, rather than determining fixed IPF endotypes.
Interpretation
In conclusion, we explored the potential existence of IPF endotypes by clustering IPF samples based on their gene expression patterns. Two important endotypes were identified with unique functional enrichment, both conferring worse survival compared with the other IPF samples, and were validated in three independent cohorts. We identified endotype-specific survival-associated genes regulated by endotype-specific transcription factors that could serve as targets for endotype-tailored therapies.
Acknowledgments
Author contributions: L. J. D. S. is the guarantor of the content of the study, including the data and the analysis. L. D. J. S. and W. A. W. conceptualized the study. L. J. D. S. and A. P. contributed to data accumulation. L. J. D. S. contributed to formal analysis. A. P. and W. A. W. contributed to funding acquisition. L. J. D. S., J. C. S., J. E. M., S. E. V., A. P., and W. A. W. contributed to investigation. L. J. D. S., S. E. V., J. C. S., and J. E. M. contributed to methodology. W. A. W. contributed to project administration. W. A. W., S. E. V., and A. P. contributed resources. L. J. D. S. contributed software. W. A. W. and A. P. supervised the study. L. J. D. S. contributed to validation. L. J. D. S. and S. E. V. contributed to visualization. L. J. D. S. wrote the original draft. All authors contributed to reviewing and editing the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: L. J. D. S. reports nonfinancial support from Roche and Boehringer-Ingelheim, outside the submitted work. S. E. V. was a consultant for Boehringer-Ingelheim outside the submitted work. E. B. reports grants from Boehringer-Ingelheim and Roche. A. P. reports personal fees from Boehringer Ingelheim, Roche, Novartis, Astra Zeneca, Chiesi, PHWC, Veracyte, and Pliant. N. K. served as a consultant to Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Theravance, LifeMax, Three Lake Partners, Optikira, Astra Zeneca, and Augmanity over the previous 3 years; reports equity in Pliant and a grant from Veracyte and Boehringer Ingelheim; and reports nonfinancial support from MiRagen and AstraZeneca. N. K. served as primary investigator on novel biomarkers and therapeutics in IPF licensed to Biotech. W. A. W. reports grants from Roche, grants from Boehringer-Ingelheim, all paid to his institution (University Hospitals Leuven), outside the submitted work. None declared (T. G., J. C. S., J. E. M., J. Y.).
Role of sponsors: The sponsors were not involved in study design, data collection, data analysis, data interpretation, manuscript writing, or the decision to submit the article for publication.
Other contributions: The authors thank all patients who participated in the study.
Additional information: The e-Appendix, e-Figures, and e-Tables are available online under “Supplementary Data.”
Footnotes
FUNDING/SUPPORT: J. C. S. is supported by the US Department of Defense [Grant W81XWH-19-1-0131]. T. G. was supported by Fonds wetenschappelijk onderzoek [Grant 12X9620N]. A. P. is funded by ERA-Net on rare diseases project, joint research centre 2011 acute exacerbation of idiopathic pulmonary fibrosis [Grant DLR 01GM1210A], and DZL-BREATH (German Center for Lung Research). N. K. is supported by the National Institutes of Health [National Heart, Lung, and Blood Institute Grants R01HL127349, R01HL141852, U01HL145567, and UH2HL123886] and a gift from Three Lakes Partners. W. A. W. is supported by FWO [Grant 1832512N] and the National Institutes of Health.
Supplementary Data
References
- 1.Schafer M.J., White T.A., Iijima K., et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest. 2013;123(3):996–1002. doi: 10.1172/JCI66370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snetselaar R., Batenburg AA van, Oosterhout MFM Van, et al. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS One. 2017;12(12):1–15. doi: 10.1371/journal.pone.0189467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armanios M., Alder J.K., Chen J.J.-L., et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 2008;105(35):13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burman A., Tanjore H., Blackwell T.S. Endoplasmic reticulum stress in pulmonary fibrosis. Matrix Biol. 2018;68-69:355–365. doi: 10.1016/j.matbio.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kropski J.A., Blackwell T.S. Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. J Clin Invest. 2018;128(1):64–73. doi: 10.1172/JCI93560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mora A.L., Bueno M., Rojas M. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J Clin Invest. 2017;127(2):405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu G., Tzouvelekis A., Wang R., et al. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2018;24(1):39–49. doi: 10.1038/nm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Wang Y., Wu G., Xiong W., Gu W., Wang C.Y. Macrophages: friend or foe in idiopathic pulmonary fibrosis? Respir Res. 2018;19(1):1–10. doi: 10.1186/s12931-018-0864-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lederer D.J., Martinez F.J. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378(19):1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 11.Adams T.S., Schupp J.C., Poli S., et al. Single cell RNA-seq reveals ectopic and aberrant lung resident cell populations in idiopathic pulmonary fibrosis. bioRxiv. 2019:759902. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasse A., Binder H., Schupp J.C., et al. BAL cell gene expression is indicative of outcome and airway basal cell involvement in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(5):622–630. doi: 10.1164/rccm.201712-2551OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubio K., Singh I., Dobersch S., et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nat Commun. 2019;10(1):2229. doi: 10.1038/s41467-019-10066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu X., Hu M., Peng J., Zhang X., Sanders Y.Y. HDAC inhibitors as antifibrotic drugs in cardiac and pulmonary fibrosis. Ther Adv Chronic Dis. 2019;10(6) doi: 10.1177/2040622319862697. 204062231986269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann M., Hu Q., Hu Y., et al. Chronic WNT/β-catenin signaling induces cellular senescence in lung epithelial cells. Cell Signal. 2020;70(February):109588. doi: 10.1016/j.cellsig.2020.109588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson W.R., Chi E.Y., Ye X., et al. Inhibition of Wnt/β-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci U S A. 2010;107(32):14309–14314. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baarsma H.A., Königshoff M. ‘WNT-er is coming’ : WNT signalling in chronic lung diseases. Thorax. 2017;72(8):746–759. doi: 10.1136/thoraxjnl-2016-209753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y.P., Lee S.B., Lee J.M., et al. Metabolic profiling regarding pathogenesis of idiopathic pulmonary fibrosis. J Proteome Res. 2016;15(5):1717–1724. doi: 10.1021/acs.jproteome.6b00156. [DOI] [PubMed] [Google Scholar]
- 19.Raghu G., Brown K.K., Bradford W.Z., et al. A placebo-controlled trial of interferon gamma-1b in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2004;350(2):125–133. doi: 10.1056/NEJMoa030511. [DOI] [PubMed] [Google Scholar]
- 20.King T.E., Albera C., Bradford W.Z., et al. Effect of interferon gamma-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet. 2009;374(9685):222–228. doi: 10.1016/S0140-6736(09)60551-1. [DOI] [PubMed] [Google Scholar]
- 21.Daniels C.E., Lasky J.A., Limper A.H., et al. Imatinib treatment for idiopathic pulmonary fibrosis: randomized placebo-controlled trial results. Am J Respir Crit Care Med. 2010;181(6):604–610. doi: 10.1164/rccm.200906-0964OC. [DOI] [PubMed] [Google Scholar]
- 22.King T.E., Brown K.K., Raghu G., et al. BUILD-3: a randomized, controlled trial of bosentan in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;184(1):92–99. doi: 10.1164/rccm.201011-1874OC. [DOI] [PubMed] [Google Scholar]
- 23.Noth I., Anstrom K.J., Calvert S.B., et al. A placebo-controlled randomized trial of warfarin in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186(1):88–95. doi: 10.1164/rccm.201202-0314OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghu G., Million-Rousseau R., Morganti A., et al. Macitentan for the treatment of idiopathic pulmonary fibrosis: the randomised controlled MUSIC trial. Eur Respir J. 2013;42(6):1622–1632. doi: 10.1183/09031936.00104612. [DOI] [PubMed] [Google Scholar]
- 25.Raghu G., Behr J., Brown K.K., et al. Treatment of idiopathic pulmonary fibrosis with ambrisentan: a parallel, randomized trial. Ann Intern Med. 2013;158(9):641–649. doi: 10.7326/0003-4819-158-9-201305070-00003. [DOI] [PubMed] [Google Scholar]
- 26.Behr J., Bendstrup E., Crestani B., et al. Safety and tolerability of acetylcysteine and pirfenidone combination therapy in idiopathic pulmonary fibrosis: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2016;4(6):445–453. doi: 10.1016/S2213-2600(16)30044-3. [DOI] [PubMed] [Google Scholar]
- 27.Raghu G., Brown K.K., Collard H.R., et al. Efficacy of simtuzumab versus placebo in patients with idiopathic pulmonary fibrosis: a randomised, double-blind, controlled, phase 2 trial. Lancet Respir Med. 2017;5(1):22–32. doi: 10.1016/S2213-2600(16)30421-0. [DOI] [PubMed] [Google Scholar]
- 28.Richeldi L., Bois RM du, Raghu G., et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 29.Noble P.W., Albera C., Bradford W.Z., et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377(9779):1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 30.Wollin L., Wex E., Pautsch A., et al. Mode of action of nintedanib in the treatment of idiopathic pulmonary fibrosis. Eur Respir J. 2015;45(5):1434–1445. doi: 10.1183/09031936.00174914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwapiszewska G., Gungl A., Wilhelm J., et al. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF. Eur Respir J. 2018;52(5):1800564. doi: 10.1183/13993003.00564-2018. 1800564. [DOI] [PubMed] [Google Scholar]
- 32.Lehtonen S.T., Veijola A., Karvonen H., et al. Pirfenidone and nintedanib modulate properties of fibroblasts and myofibroblasts in idiopathic pulmonary fibrosis. Respir Res. 2016;17:14. doi: 10.1186/s12931-016-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch T.J., Bell D.W., Sordella R., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 34.Shaw A.T., Kim D.W., Nakagawa K., et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 35.Planchard D., Besse B., Groen H.J.M., et al. Dabrafenib plus trametinib in patients with previously treated BRAFV600E-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17(7):984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raghu G., Remy-Jardin M., Myers J.L., et al. Diagnosis of idiopathic pulmonary fibrosis an official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Med Crit Care. 2018;198(5):e44–e68. doi: 10.1164/rccm.201807-1255ST. [DOI] [PubMed] [Google Scholar]
- 37.Leek J.T., Evan Johnson W., Parker H.S, Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu X, Trapnell C, Mao Q, Wang L. DDRTree: learning principal graphs with DDRTree. 2017.
- 39.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009;10:1–17. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biology. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aibar S., González-Blas C.B., Moerman T., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herazo-Maya J.D., Noth I., Duncan S.R., et al. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med. 2013;5(205) doi: 10.1126/scitranslmed.3005964. 205-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molyneaux P.L., Willis-Owen S.A.G., Cox M.J., et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;195(12):1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y., Ma S.-F., Espindola M.S., et al. Microbes associate with host innate immune response in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196(2):208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukui T., Sun K.H., Wetter J.B., et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun. 2020;11(1):1920. doi: 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayr C.H., Simon L.M., Leuschner G., et al. Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers. EMBO Mol Med. 2021;13(4):e12871. doi: 10.15252/emmm.202012871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marciniak S.J. Endoplasmic reticulum stress in lung disease. Eur Respir Rev. 2017;26(144):170018. doi: 10.1183/16000617.0018-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia G., Chandriani S., Abbas A.R., et al. CXCL14 is a candidate biomarker for hedgehog signalling in idiopathic pulmonary fibrosis. Thorax. 2017;72(9):780–787. doi: 10.1136/thoraxjnl-2015-207682. thoraxjnl-2015-207682. [DOI] [PubMed] [Google Scholar]
- 49.Goodwin A.T., Jenkins G. Molecular endotyping of pulmonary fibrosis. Chest. 2016;149(1):228–237. doi: 10.1378/chest.15-1511. [DOI] [PubMed] [Google Scholar]
- 50.Hoffmann-Vold A.M., Weigt S.S., Saggar R., et al. Endotype-phenotyping may predict a treatment response in progressive fibrosing interstitial lung disease. EBioMedicine. 2019;50:379–386. doi: 10.1016/j.ebiom.2019.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang I.V., Coldren C.D., Leach S.M., et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax. 2013;68(12):1114–1121. doi: 10.1136/thoraxjnl-2012-202943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DePianto D.J., Chandriani S., Abbas A.R., et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax. 2015;70(1):48–56. doi: 10.1136/thoraxjnl-2013-204596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y., Ma S.-F., Vij R., et al. A functional genomic model for predicting prognosis in idiopathic pulmonary fibrosis. BMC Pulm Med. 2015;15:147. doi: 10.1186/s12890-015-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foster M.W., Morrison L.D., Todd J.L., et al. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res. 2015;14(2):1238–1249. doi: 10.1021/pr501149m. [DOI] [PubMed] [Google Scholar]
- 55.Scott M.K.D., Quinn K., Li Q., et al. Increased monocyte count as a cellular biomarker for poor outcomes in fibrotic diseases: a retrospective , multicentre cohort study. Lancet Respir Med. 2019;7(6):497–508. doi: 10.1016/S2213-2600(18)30508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinder B.W., Brown K.K., Schwarz M.I., Ix J.H., Kervitsky A., King T.E. Baseline BAL neutrophilia predicts early mortality in idiopathic pulmonary fibrosis. Chest. 2008;133(1):226–232. doi: 10.1378/chest.07-1948. [DOI] [PubMed] [Google Scholar]
- 57.O’Dwyer D.N., Ashley S.L., Gurczynski S.J., et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(9):1127–1138. doi: 10.1164/rccm.201809-1650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziegenhagen M.W., Zabel P., Zissel G., Schlaak M., Müller-Quernheim J. Serum level of interleukin 8 is elevated in idiopathic pulmonary fibrosis and indicates disease activity. Am J Respir Crit Care Med. 1998;157(3 part I):762–768. doi: 10.1164/ajrccm.157.3.9705014. [DOI] [PubMed] [Google Scholar]
- 59.Miao Q., Hill M.C., Chen F., et al. SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat Commun. 2019;10(1):4042. doi: 10.1038/s41467-019-11880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher M., Nathan S.D., Hill C., et al. Predicting life expectancy for pirfenidone in idiopathic pulmonary fibrosis. J Manag Care Spec Pharm. 2017;23(3b):S17–S24. doi: 10.18553/jmcp.2017.23.3-b.s17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nathan S.D., Albera C., Bradford W.Z., et al. Effect of pirfenidone on mortality: pooled analyses and meta-analyses of clinical trials in idiopathic pulmonary fibrosis. Lancet Respir Med. 2017;5(1):33–41. doi: 10.1016/S2213-2600(16)30326-5. [DOI] [PubMed] [Google Scholar]
- 62.Lancaster L., Crestani B., Hernandez P., et al. Safety and survival data in patients with idiopathic pulmonary fibrosis treated with nintedanib: pooled data from six clinical trials. BMJ Open Respir Res. 2019;6(1):e000397. doi: 10.1136/bmjresp-2018-000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Behr J., Prasse A., Wirtz H., et al. Survival and course of lung function in the presence or absence of antifibrotic treatment in patients with idiopathic pulmonary fibrosis: long-term results of the INSIGHTS-IPF registry. Eur Respir J. 2020;56(2):1902279. doi: 10.1183/13993003.02279-2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.