Abstract
Few studies have evaluated the role of autophagy in the development of oxaliplatin (OXT) resistance in colon cancer cells. In this study, we compared the role of autophagy between SNU-C5 colon cancer cells and OXT-resistant SNU-C5 (SNU-C5/OXTR) cells. At the same concentration of OXT, the cytotoxicity of OXT or apoptosis was significantly reduced in SNU-C5/OXTR cells compared with that in SNU-C5 cells. Compared with SNU-C5 cells, SNU-C5/OXTR cells exhibited low levels of autophagy. The expression level of important autophagy proteins, such as autophagy-related protein 5 (Atg5), beclin-1, Atg7, microtubule-associated proteins 1A/1B light chain 3B I (LC3-I), and LC3-II, was significantly lower in SNU-C5/OXTR cells than that in SNU-C5 cells. The expression level of the autophagy-essential protein p62 was also lower in SNU-C5/OXTR cells than in SNU-C5 cells. In SNU-C5/OXTR cells, the production of intracellular reactive oxygen species (ROS) was significantly higher than that in SNU-C5 cells, and treatment with the ROS scavenger N-acetylcysteine restored the reduced autophagy levels. Furthermore, the expression of antioxidant-related nuclear factor erythroid 2-related factor 2 transcription factor, heme oxygenase-1, and Cu/Zn superoxide dismutase were also significantly increased in SNU-C5/OXTR cells. These findings suggest that autophagy is significantly reduced in SNU-C5/OXTR cells compared with SNU-C5 cells, which may be related to the production of ROS in OXT-resistant cells.
Keywords: Autophagy, Oxaliplatin, Oxaliplatin-resistant SNU-C5 cells, Reactive oxygen species, Colon cancer
INTRODUCTION
Oxaliplatin (OXT) is a third-generation platinum-based anticancer agent that contains 1,2-diaminocyclohexane in its structure. It binds to DNA as a target site and the platinum atom in its structure forms a cross-link with DNA molecules to inhibit replication and transcription, leading to apoptotic cell death (Alcindor and Beauger, 2011; Huang et al., 2019; Ray et al., 2019). OXT is widely used clinically in combination chemotherapy, such as with 5-fluorouracil and leucovorin, mainly for the treatment of malignant tumors of the digestive system, such as colorectal, gastric, and pancreatic cancers (Kornmann et al., 2000; Shiragami et al., 2013; Lin et al., 2019; Wang et al., 2020). OXT-based adjuvant chemotherapy represents the standard treatment for stage III colon cancer (Auclin et al., 2017). However, most patients develop resistance to OXT after prolonged use, which limits its therapeutic efficacy (Huang et al., 2019). OXT resistance can be caused by reduced intracellular uptake or inactivation via structural changes and is associated with DNA repair, such as mismatch repair or nucleotide excision repair mechanisms (Alcindor and Beauger, 2011). Cancer cell resistance represents the major cause of failure of OXT therapy (Holohan et al., 2013; Huang et al., 2019). Therefore, revealing the mechanisms that cause OXT resistance is necessary to develop more effective treatment strategies to overcome resistance and enhance the efficacy of OXT.
Autophagy is a cellular degradation pathway that mediates the clearance and recycling of damaged or long-lived proteins and organelles. It plays a critical role in homeostasis by maintaining the quality of proteins and organelles, and also eliminates pathogens (Yun and Lee, 2018; Chen et al., 2019; Li et al., 2020). Autophagy is generally induced in response to cellular stress, starvation, hypoxia, or DNA damage (Martinez-Balibrea et al., 2015). Autophagy frequently occurs during tumorigenesis and chemotherapy. Typically, autophagy protects cancer cells from apoptosis during chemotherapy, leading to drug-resistant and refractory cancers. However, autophagy can also lead to cell death and inhibit cell growth, depending on the tissue type, stage of tumor development, and the degree of autophagic activity (Xie et al., 2020). Reactive oxygen species (ROS) play an important role in the process of autophagy. ROS can promote the process of autophagy, and on the other hand, autophagy reduces ROS by removing damaged mitochondria and endoplasmic reticulum stress (ER), and other substances that contribute to ROS production (Xie et al., 2020).
SNU-C5 colon cancer cells contain two missense mutations in the p53 gene, resulting in the complete loss of normal p53 function (Rand et al., 1996), and have been used in many studies, including multidrug resistance and anti-cancer drug screening studies (Ku and Park, 2005). An OXT-resistant colon cancer cell line (SNU-C5/OXTR) has been derived from a parental wild-type colon cancer cell line (SNU-C5) via chronic exposure to OXT (Jung et al., 2007). A few studies have evaluated OXT resistance using SNU-C5/OXTR cells. The combined application of betulinic acid, a chemosensitizer, and OXT induces apoptosis of SNU-C5/OXTR via the mitochondrial pathway (Jung et al., 2007), and co-treatment with OXT and melatonin increases ER stress and apoptosis in SNU-C5/OXTR cells (Lee et al., 2018).
However, few studies have determined the role of autophagy in the development of OXT resistance in colon cancer cells. In this study, we aimed to evaluate the effect of autophagy on SNU-C5/OXTR cells compared with that on SNU-C5 cells to reveal a potential approach to overcome chemotherapy resistance in colon cancer.
MATERIALS AND METHODS
Materials
OXT, N-acetylcysteine (NAC), Hoechst 33342, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), and actin antibodies were obtained from Sigma-Aldrich (St. Louis, MO, USA). Thiazolyl blue tetrazolium bromide (MTT) was purchased from Amresco (Solon, OH, USA). Acridine orange was purchased from Invitrogen (Madison, WI, USA). Antibody against Atg5 (C-term) was purchased from Abgent (San Diego, CA, USA). Antibodies against beclin-1, Atg7, LC3, p62, and phosphorylated Nrf2 (phospho-Nrf2) were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies against Nrf2 and heme oxygenase-1 (HO-1) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibody against TATA-binding protein (TBP) was purchased from Abcam (Cambridge, UK). Antibody against Cu/Zn superoxide dismutase (SOD) was purchased from Biodesign International (Saco, ME, USA). All other chemicals and reagents used were of analytical grade.
Cell culture
SNU-C5 cells were obtained from the Korean Cell Line Bank (Seoul, Korea). The cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C, 5% CO2, and 95% humidity. The established SNU-C5/OXTR cells were obtained from the Research Center for Resistant Cells, Chosun University (Gwangju, Korea). SNU-C5/OXTR cells were stabilized by subculturing them weekly for at least 6 months in a medium containing 7.14 μM OXT (Kang et al., 2016).
MTT assay
The susceptibility of SNU-C5 and SNU-C5/OXTR cells to OXT-induced cytotoxicity was determined by plating 1.0×105 cells/well in a 24-well culture plate containing media supplemented with various concentrations of OXT (10, 25, 50, 100, and 200 μM), following incubation at 37°C for 48 h. After adding 125 μL of the MTT stock solution to each well and incubating for 4 h, the supernatant was aspirated. The formazan crystals in each well were dissolved in dimethyl sulfoxide (350 μL), and the absorbance was measured at 540 nm using a multi-mode microplate reader.
Hoechst 33342 staining
Cells were seeded at 1.0×105 cells/well in a 24-well culture plate and incubated for 48 h. Cells were treated with Hoechst 33342 cell-permeable nuclear counterstain dye for 10 min, and images were acquired using a fluorescence microscope equipped with a CoolSnap-Pro color digital camera (Media Cybernetics, Rockville, MD, USA). The apoptotic index was calculated using a formula as described previously (Piao et al., 2019).
Cell morphology analyses
Cells were seeded into a 60 mm culture dish at a density of 2.5×105 cells/well and cultured for 1, 2, and 3 days. Changes in cell morphology were evaluated at 1, 2, and 3 days using a phase-contrast inverted microscope (DP71 digital microscope camera; Olympus, Tokyo, Japan). All images were acquired at a magnification of 200×.
Acridine orange staining
Cells were seeded in a 35 mm culture plate at a density of 1.5×105 cells/well. After 48 h, the cells were stained with 10 μg/ml acridine orange for 15 min at 37°C. Subsequently, cells were washed with phosphate-buffered saline (PBS) and observed under a fluorescence microscope. Depending on the acidity, autophagic lysosomes appeared as orange/red fluorescent cytoplasmic vesicles, whereas the nuclei were stained green. Three different images, with 20 cells per image, were used to calculate the percentage of acidic vesicular organelles (AVO; dots with clear orange-red fluorescence).
Green fluorescent protein (GFP)-microtubule-associated proteins 1A/1B light chain 3B (LC3) transfection and detection of GFP-LC3 dots
Autophagy was determined based on the formation of puncta LC3-positive structures, which are essential for the dynamic process of autophagosome formation (Steinmetz et al., 2021). GFP-tagged LC3 plasmids were used to transfect cells using Lipofectamine reagent (Invitrogen). GFP-LC3 fluorescence was imaged using a confocal microscope equipped with a laser scanning microscope 5 PASCAL program (Carl Zeiss, Oberkochen, Germany). The numbers of GFP-LC3 dots in each sample were counted.
Determination of protein expression
Protein expression was detected via western blotting. The total and nuclear proteins in harvested cells were extracted using a total protein extraction solution (iNtRON Biotechnology, Seoul, Korea) and a nuclear extraction kit (Cayman Chemical Company, Ann Arbor, MI, USA), respectively, following protein level quantification. Cell lysates were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the separated proteins were transferred onto nitrocellulose membranes. The protein-coated membranes were blocked with 3% bovine serum albumin and incubated with primary antibodies against Atg5, beclin-1, Atg7, LC3, p62, HO-1, Cu/Zn SOD, actin, phospho-Nrf2, Nrf2, and TBP for 2 h. Membranes were washed thrice with 1×Tris-buffered saline and Tween 20 (TTBS), incubated with secondary antibodies for 1 h, and then washed with 1× TTBS. Protein bands were detected using the enhanced chemiluminescence plus western blotting detection system (GE Healthcare Life Sciences, Buckinghamshire, UK).
Detection of intracellular ROS levels
Cells were seeded in a 6-well culture plate at a density of 2.5×105 cells/well and incubated for 16 h. The cells were then treated with 2 mM NAC and incubated for an additional 24 h, and then treated with 20 μM H2DCFDA fluorescent reagent for 30 min. The culture medium was aspirated, washed with PBS, and trypsinized. Subsequently, the fluorescence intensity of the harvested cells was measured using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). For confocal microscopy, cells were seeded in 4-well chamber slides at a density of 1.5×105 cells/well, incubated for 16 h, and then treated with 2 mM NAC for an additional 24 h. The cells were treated with 20 μM H2DCFDA for 30 min, washed with PBS, mounted with 4′,6-diamidino-2-phenylindole-containing mounting medium (DAKO, Carpinteria, CA, USA), and photographed using a confocal microscope (Carl Zeiss).
Statistical analysis
All tests were performed in triplicate, and all values are expressed as the mean ± standard error of the mean. The results were subjected to an analysis of variance followed by Tukey’s post hoc test to analyze the differences between conditions; significance was set at p<0.05.
RESULTS
SNU-C5/OXTR cells exhibited decreased autophagy
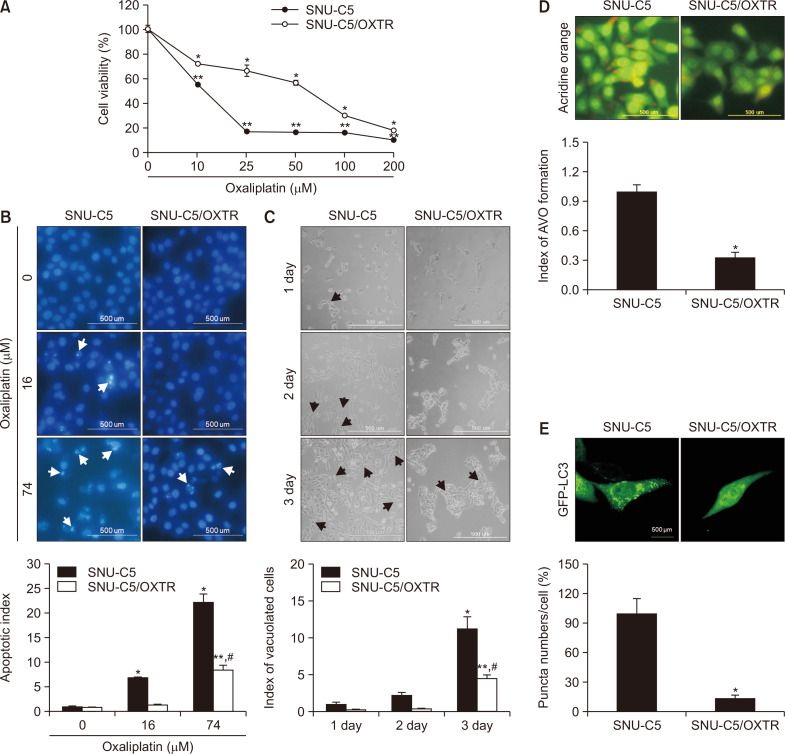
MTT assay was performed to compare the cytotoxicity of OXT in SNU-C5 cells and their resistant variant, SNU-C5/OXTR cells. The concentration of OXT that yielded 50% growth inhibition (IC50) was 16 μM in SNU-C5 cells and 74 μM in SNU-C5/OXTR cells, in which resistance was induced via continuous culture with 7.14 μM OXT (Fig. 1A). The IC50 treatment of OXT in SNU-C5 and SNU-C5/OXTR cells induced apoptotic cell death, as determined via Hoechst 33342 staining. The treatment of 16 μM OXT (IC50 in SNU-C5) yielded an apoptotic index of 6.8 for SNU-C5 cells compared to 1.4 for SNU-C5/OXTR (Fig. 1B). The apoptotic cell population after treatment with 74 μM OXT (IC50 in SNU-C5/OXTR) had an apoptotic index of 22.2 for SNU-C5 cells compared to 8.5 for SNU-C5/OXTR cells (Fig. 1B). Accumulation of vacuolated cells, a marker of autophagy, was diminished in SNU-C5/OXTR cells relative to that in SNU-C5 cells, as determined via phase-contrast microscopy (Fig. 1C). AVO with autophagic vacuoles was stained red with acridine orange. The number of acridine orange-stained vacuolated cells was lower among SNU-C5/OXTR cells than that among SNU-C5 cells (Fig. 1D). Furthermore, GFP-LC3 expression was decreased in SNU-C5/OXTR cells compared to that in SNU-C5 cells (Fig. 1E).
Fig. 1.
SNU-C5/OXTR cells exhibit reduced autophagy. (A) Cell viability following treatment with OXT in SNU-C5 and SNU-C5/OXTR cells for 48 h was assessed via MTT assay. *,**Significantly different from OXT-untreated SNU-C5 or OXT-untreated SNU-C5/OXTR cells (p<0.05). (B) Apoptotic body formation (white arrows) in SNU-C5 or SNU-C5/OXTR cells was determined via fluorescence microscopy after Hoechst 33342 staining. *,**Significantly different from OXT-untreated SNU-C5 or OXT-untreated SNU-C5/OXTR cells (p<0.05), #significantly different from 74 μM OXT-treated SNU-C5 cells (p<0.05). (C) After 16 h of culture, the cells were imaged using a phase-contrast microscope every day for 3 days, and the vacuolated cells/20 cells were quantified. The arrows indicate vacuolated cells. *,**Significantly different from SNU-C5 or SNU-C5/OXTR cells at 1 day (p<0.05), #significantly different from SNU-C5 cells at 3 day (p<0.05). (D) Cells stained by acridine orange were imaged via fluorescence microscopy and quantified as acridine orange-stained AVO. Acridine orange-red fluorescence indicated the autophagy-induced formation of AVO, and acridine orange-green fluorescence indicated nucleus and cytoplasm in cells. (E) After 24 h of transfection with GFP-LC3 reagent, cells were imaged and quantified via fluorescence microscopy. (D, E) *Significantly different from SNU-C5 cells (p<0.05).
Oxaliplatin reduced the expression of autophagy-related proteins
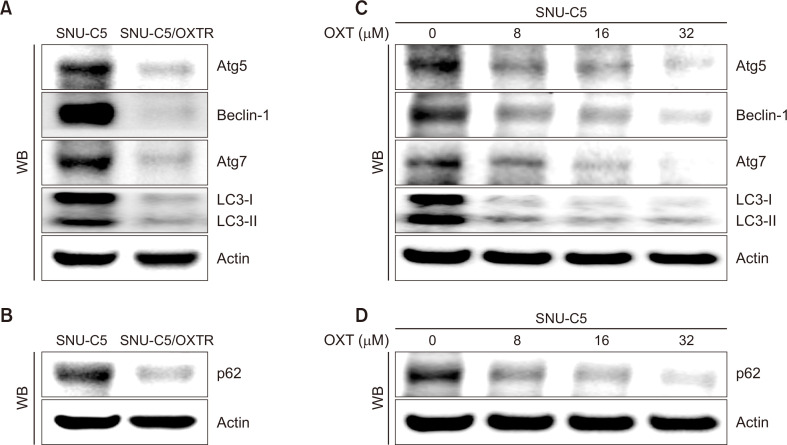
Autophagy is characterized by the formation of autophagosomes, which depend on the recruitment of Atg proteins. The expression levels of Atg proteins provide key information regarding the autophagy state of a cell. The expression levels of Atg5, beclin-1, Atg7, LC3-I, and LC3-II, which are well-characterized hallmarks of autophagy, were lower in SNU-C5/OXTR cells than those in SNU-C5 cells (Fig. 2A). As the most extensive substrate for autophagic degradation, p62 is an essential protein for autophagy that targets the packaging and delivery of proteins for autophagic digestion. It has been identified as a crossroads between apoptosis, autophagy, and cancer (Koustas et al., 2019). p62 expression level in SNU-C5/OXTR cells was lower than that in SNU-C5 cells (Fig. 2B). In addition, to evaluate whether OXT directly inhibited autophagy, we examined the levels of autophagy-related proteins at various concentrations of OXT in SNU-C5 cells. The expression levels of Atg5, beclin-1, Atg7, LC3-I, LC3-II, and p62 decreased in a dose-dependent manner (Fig. 2C, 2D). Therefore, OXT treatment reduced autophagy in SNU-C5 cells.
Fig. 2.
OXT reduces the expression level of autophagy-related proteins. (A, C) Cells were harvested, and protein expression levels of Atg5, beclin-1, Atg7, and LC3 were assessed via western blotting. (B, D) Cells were lysed and the level of p62 protein was analyzed via western blotting. Actin was used as a total protein loading control.
Intracellular ROS levels and the antioxidant system were higher in SNU-C5/OXTR cells than in SNU-C5 cells
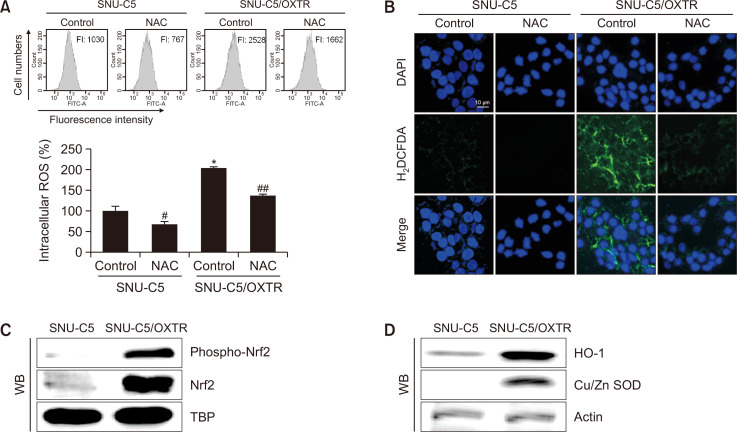
To evaluate the relationship between ROS levels and OXT-resistant cells, ROS levels were detected using flow cytometry and confocal microscopy after staining with H2DCFDA. ROS levels were higher in SNU-C5/OXTR cells than in SNU-C5 cells; however, treatment with the ROS scavenger NAC attenuated ROS levels (Fig. 3A, 3B). The expression levels of the main transcription factor of antioxidant enzymes, nuclear Nrf2, and its active form phospho-Nrf2, were higher in SNU-C5/OXTR cells than in SNU-C5 cells (Fig. 3C), leading to higher expression of target proteins HO-1 and Cu/Zn SOD in SNU-C5/OXTR cells (Fig. 3D).
Fig. 3.
Intracellular ROS levels and antioxidant systems are higher in SNU-C5/OXTR cells than in SNU-C5 cells. Cells were treated with 2 mM NAC for 24 h. ROS levels were assessed via (A) flow cytometry and (B) confocal microscopy after H2DCFDA staining. DAPI staining was performed to determine the number of nuclei and to assess gross cell morphology. *Significantly different from SNU-C5 control cells (p<0.05), #,##significantly different from SNU-C5 or SNU-C5/OXTR control cells (p<0.05). (C) Cells were harvested, and levels of phospho-Nrf2, Nrf2, and TBP were assessed via western blotting. TBP was used as a nuclear loading control. (D) HO-1, Cu/Zn SOD, and actin levels were assessed via western blotting. Actin was used as a total protein loading control.
ROS regulated autophagy in SNU-C5/OXTR cells
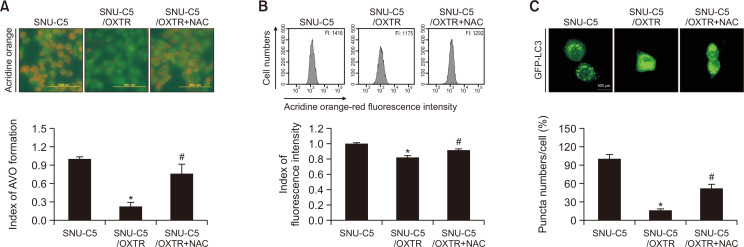
ROS are considered major regulators of autophagy (Guan et al., 2019; Xue et al., 2020). After the addition of NAC, the red fluorescence intensity based on acridine orange staining in SNU-C5/OXTR cells recovered to that of untreated SNU-C5 cells (Fig. 4A, 4B). In addition, GFP-LC3 expression was increased in SNU-C5/OXTR cells treated with NAC compared to that in untreated SNU-C5/OXTR cells (Fig. 4C). These results suggest that ROS are associated with autophagy in SNU-C5/OXTR cells.
Fig. 4.
ROS regulates autophagy in SNU-C5/OXTR cells. (A) Cells stained with acridine orange were imaged via fluorescence microscopy and quantified as acridine orange-stained AVO. Acridine orange-red fluorescence indicated the autophagy-induced formation of AVO, and acridine orange-green fluorescence indicated nucleus and cytoplasm in cells. (B) The acridine orange-stained cells were assessed via flow cytometry. FI: fluorescence intensity. (C) After 24 h of transfection using a GFP-LC3 reagent, the cells were imaged and quantified via fluorescence microscopy. (A-C) *Significantly different from SNU-C5 cells (p<0.05), #significantly different from SNU-C5/OXTR cells (p<0.05).
DISCUSSION
OXT is an important platinum-based drug used in the treatment of colorectal cancer. A major cause of treatment failure involves intrinsic or acquired resistance to OXT (Martinez-Balibrea et al., 2015). Few studies to date have investigated the role of autophagy in the development of OXT resistance in colon cancer cells after prolonged OXT treatment. In the present study, to elucidate the cause of OXT resistance and find a way to overcome it, we focused on the role of autophagy in OXT resistance. We demonstrated that the same concentration of OXT showed a significant difference in the survival rate of SNU-C5 and SNU-C5/OXTR cells, and the sensitivity of SNU-C5/OXTR cells was much lower than that of SNU-C5 cells (SNU-C5 IC50: 16 μM, SNU-C5/OXTR IC50: 74 μM). In cells cultured on day 3, autophagosomes were readily observed in SNU-C5 cells, whereas only a small amount of autophagosomes were found in SNU-C5/OXTR cells.
Double-membrane vesicles called autophagosomes are formed in the cytoplasm during autophagy. These vesicles fuse with lysosomes to form autolysosomes (Yim and Mizushima, 2020). Acridine orange is a fluorescent dye that stains acidic vacuoles, such as autophagosomes and autolysosomes, in cells (Fan et al., 2006). We detected fewer vacuoles in SNU-C5/OXTR cells than those in SNU-C5 cells, indicating a relatively lower level of autophagy in SNU-C5/OXTR cells.
Autophagosome formation involves initiation, nucleation, elongation, and recycling processes, each of which depends on specific proteins. LC3 is conjugated to phosphatidylethanolamine via an enzymatic cascade containing Atg3, Atg7, and Atg5-Atg12 complex; the resulting lipid-conjugated form is targeted to the autophagosome membrane (Mizushima, 2020). Therefore, Atg5 and LC3 levels can evaluate used as autophagy markers. p62 is also an essential protein that is involved in autophagy. Our results showed that the levels of Atg5, beclin-1, Atg7, LC3-I, LC3-II, and p62 were lower in SNU-C5/OXTR cells than in SNU-C5 cells. Based on these findings, we presume that chemoresistance in SNU-C5/OXTR cells is associated with the attenuation of autophagy.
Under normal physiological conditions, autophagy occurs at a basal rate to maintain cell viability and homeostasis. OXT treatment in colorectal cancer cells induces autophagy, leading to cell death (Jeong et al., 2019). However, our data showed that the occurrence of autophagy was decreased in SNU-C5/OXTR cells compared to that in SNU-C5 cells, leading to resistance to OXT.
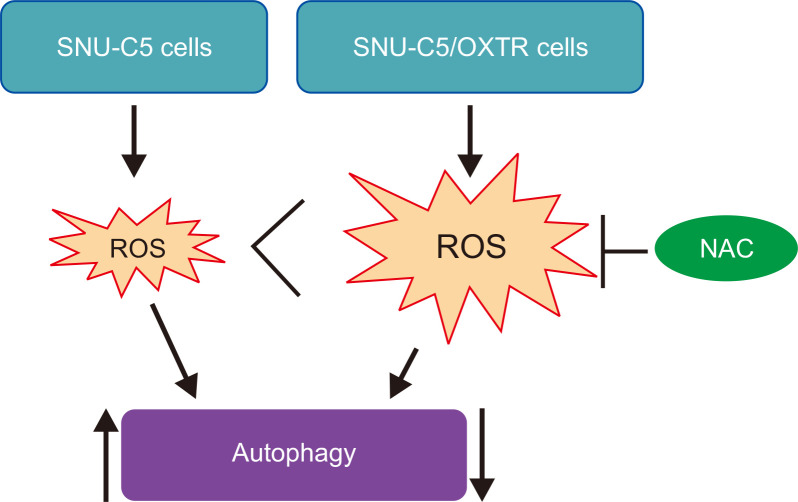
ROS, a by-product of oxygen metabolism, comprises highly reactive ions and molecules. Their role has been described in various pathophysiological conditions. Cancer cells contain increased ROS levels compared with those in normal cells. Elevated ROS levels in cancer cells contribute to the biochemical and molecular changes required for cancer initiation and progression, and the development of resistance to chemotherapy (Galadari et al., 2017). Nrf2 is a major transcription factor that promotes cellular defense against oxidative stress. Nrf2 controls the transcription of many genes responsible for antioxidative and detoxification, such as SOD1 and HO-1. It has been demonstrated that the autophagic survival of colon cancer cells is promoted by activating the Nrf2/HO-1 pathway (Cernigliaro et al., 2019). Regulation of autophagy is determined by the intensity and duration of ROS exposure and the cell type (Ornatowski et al., 2020). ROS are associated with both chemosensitization and chemoresistance (Galadari et al., 2017). Our data showed that the level of ROS as well as that of Nrf2, Cu/Zn SOD, and HO-1 was higher in SNU-C5/OXTR cells than in SNU-C5 cells. The ROS scavenger NAC significantly increased the level of AVO in SNU-C5/OXTR cells. We previously investigated the reduction of autophagy in 5-fluorouracil resistant SNU-C5 colon cancer cells (SNU-C5/5-FUR) (Yao et al., 2017). We found that the amount of ROS produced was higher and the occurrence of autophagy was lower in SNU-C5/5-FUR cells than in wild-type SNU-C5 cells. However, the ROS scavenger NAC failed to restore the occurrence of autophagy in SNU-C5/5-FUR cells. Therefore, at the time, we thought that ROS was not necessarily a key regulator of reduced autophagic activity in SNU-C5/5-FUR cells. However, in the present study, treatment of NAC in SNU-C5/OXTR cells restored autophagy. This may be owing to the use of different drug-resistant cells, degree of drug resistance, amount of ROS produced in the cells, and levels of the antioxidant system. Under normal circumstances, the production and removal of intracellular ROS are in a state of dynamic equilibrium. When the accumulation of ROS is excessive, this balance is disturbed. We suggest that SNU-C5/OXTR cells were exposed to a high level of ROS to maintain a drug-resistant environment for a long time. To maintain homeostasis, SNU-C5/OXTR cells activate the antioxidant system to scavenge ROS. In the present study, the amount of ROS increased in SNU-C5/OXTR cells compared to that in SNU-C5 cells were decreased autophagy formation; however, NAC recovered it. These findings suggest that ROS may be a critical regulator of autophagy in SNU-C5/OXTR cells (Fig. 5).
Fig. 5.
Schematic illustration of the mechanism of action of autophagy in SNU-C5 colon cancer cells and SNU-C5/OXTR colon cancer cells. Large amounts of ROS are produced in SNU-C5/OXTR cells compared to that in SNU-C5 cells. Enhanced autophagy is observed in SNU-C5 cells, which is significantly reduced in SNU-C5/OXTR cells; however, treatment with the ROS scavenger NAC significantly restored the reduced autophagy in SNU-C5/OXTR cells.
In conclusion, this study showed that low levels of autophagy are related to OXT resistance in SNU-C5/OXTR cells. The ability of SNU-C5/OXTR cells to undergo autophagy may be restored by restoring the normal physiological levels of ROS, thereby reducing the resistance and increasing the sensitivity of colon cancer cells to OXT. The findings demonstrate the role of autophagy in the mechanism of chemoresistance development and can serve as a basis for developing a novel strategy to overcome chemoresistance. More extensive and systematic research should be conducted in the future to gain better awareness in this field.
ACKNOWLEDGMENTS
This work was supported by a research grant from Jeju National University Hospital in 2020.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Alcindor T., Beauger N. Oxaliplatin: a review in the era of molecularly targeted therapy. Curr. Oncol. 2011;18:18–25. doi: 10.3747/co.v18i1.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclin E., Zaanan A., Vernerey D., Douard R., Gallois C., Laurent-Puig P., Bonnetain F., Taieb J. Subgroups and prognostication in stage III colon cancer: future perspectives for adjuvant therapy. Ann. Oncol. 2017;28:958–968. doi: 10.1093/annonc/mdx030. [DOI] [PubMed] [Google Scholar]
- Cernigliaro C., D'Anneo A., Carlisi D., Giuliano M., Gammazza A. M., Barone R., Longhitano L., Cappello F., Emanuele S., Distefano A., Campanella C., Calvaruso G., Lauricella M. Ethanol-mediated stress promotes autophagic survival and aggressiveness of colon cancer cells via activation of Nrf2/HO-1 pathway. Cancers (Basel) 2019;11:505. doi: 10.3390/cancers11040505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Chen Y. H., Huang T. Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 2019;26:80. doi: 10.1186/s12929-019-0569-y.d7f188eff41045d1a04b744e06a52c8d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Wang W., Zhao B., Zhang S., Miao J. Chloroquine inhibits cell growth and induces cell death in A549 lung cancer cells. Bioorg. Med. Chem. 2006;14:3218–3222. doi: 10.1016/j.bmc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Galadari S., Rahman A., Pallichankandy S., Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic. Biol. Med. 2017;104:144–164. doi: 10.1016/j.freeradbiomed.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zhou L., Zhang Y., Tian H., Li A., Han X. Effects of PP2A/Nrf2 on experimental diabetes mellitus-related cardiomyopathy by regulation of autophagy and apoptosis through ROS dependent pathway. Cell. Signal. 2019;62:109339. doi: 10.1016/j.cellsig.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Holohan C., Van Schaeybroeck S., Longley D. B., Johnston P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Huang H., Aladelokun O., Ideta T., Giardina C., Ellis L. M., Rosenberg D. W. Inhibition of PGE2/EP4 receptor signaling enhances oxaliplatin efficacy in resistant colon cancer cells through modulation of oxidative stress. Sci. Rep. 2019;9:4954. doi: 10.1038/s41598-019-40848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Kim D. Y., Kang S. H., Yun H. K., Kim J. L., Kim B. R., Park S. H., Na Y. J., Jo M. J., Jeong Y. A., Kim B. G., Lee D. H., Oh S. C. Docosahexaenoic acid enhances oxaliplatin-induced autophagic cell death via the ER stress/Sesn2 pathway in colorectal cancer. Cancers (Basel) 2019;11:982. doi: 10.3390/cancers11070982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. R., Kim K. J., Choi C. H., Lee T. B., Han S. I., Han H. K., Lim S. C. Effect of betulinic acid on anticancer drug-resistant colon cancer cells. Basic. Clin. Pharmacol. Toxicol. 2007;101:277–285. doi: 10.1111/j.1742-7843.2007.00115.x. [DOI] [PubMed] [Google Scholar]
- Kang K. A., Piao M. J., Ryu Y. S., Kang H. K., Chang W. Y., Keum Y. S., Hyun J. W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget. 2016;7:40594–40620. doi: 10.18632/oncotarget.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann M., Fakler H., Butzer U., Beger H. G., Link K. H. Oxaliplatin exerts potent in vitro cytotoxicity in colorectal and pancreatic cancer cell lines and liver metastases. Anticancer Res. 2000;20:3259–3264. [PubMed] [Google Scholar]
- Koustas E., Sarantis P., Theoharis S., Saetta A. A., Chatziandreou I., Kyriakopoulou G., Giannopoulou I., Michelli M., Schizas D., Papavassiliou A. G., Karamouzis M. V. Autophagy-related proteins as a prognostic factor of patients with colorectal cancer. Am. J. Clin. Oncol. 2019;42:767–776. doi: 10.1097/COC.0000000000000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku J. L., Park J. G. Biology of SNU cell lines. Cancer Res. Treat. 2005;37:1–19. doi: 10.4143/crt.2005.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Yoon Y. M., Han Y. S., Yun C. W., Lee S. H. Melatonin promotes apoptosis of oxaliplatin-resistant colorectal cancer cells through inhibition of cellular prion protein. Anticancer Res. 2018;38:1993–2000. doi: 10.21873/anticanres.12437. [DOI] [PubMed] [Google Scholar]
- Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4.04726baefac043eb9bf39450c8a8316d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Li X., Pan C., Lin W., Shao R., Liu Y., Zhang J., Luo Y., Qian K., Shi M., Bin J., Liao Y., Liao W. ATXN2L upregulated by epidermal growth factor promotes gastric cancer cell invasiveness and oxaliplatin resistance. Cell Death Dis. 2019;10:173. doi: 10.1038/s41419-019-1362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balibrea E., Martínez-Cardús A., Ginés A., Ruiz, de Porras V., Moutinho C., Layos L., Manzano J. L., Bugés C., Bystrup S., Esteller M., Abad A. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol. Cancer Ther. 2015;14:1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- Ornatowski W., Lu Q., Yegambaram M., Garcia A. E., Zemskov E. A., Maltepe E., Fineman J. R., Wang T., Black S. M. Complex interplay between autophagy and oxidative stress in the development of pulmonary disease. Redox Biol. 2020;36:101679. doi: 10.1016/j.redox.2020.101679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao M. J., Kang K. A., Zhen A. X., Fernando P. D. S. M., Ahn M. J., Koh Y. S., Kang H. K., Yi J. M., Choi Y. H., Hyun J. W. Particulate matter 2.5 mediates cutaneous cellular injury by inducing mitochondria-associated endoplasmic reticulum stress: protective effects of ginsenoside Rb1. Antioxidants. 2019;8:383. doi: 10.3390/antiox8090383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand A., Glenn K. S., Alvares C. P., White M. B., Thibodeau S. M., Karnes W. E., Jr. p53 functional loss in a colon cancer cell line with two missense mutations (218leu and 248trp) on separate alleles. Cancer Lett. 1996;98:183–191. doi: 10.1016/S0304-3835(06)80030-3. [DOI] [PubMed] [Google Scholar]
- Ray B., Gupta B., Mehrotra R. Binding of platinum derivative, oxaliplatin to deoxyribonucleic acid: structural insight into antitumor action. J. Biomol. Struct. Dyn. 2019;37:3838–3847. doi: 10.1080/07391102.2018.1531059. [DOI] [PubMed] [Google Scholar]
- Shiragami R., Murata S., Kosugi C., Tezuka T., Yamazaki M., Hirano A., Yoshimura Y., Suzuki M., Shuto K., Koda K. Enhanced antitumor activity of cerulenin combined with oxaliplatin in human colon cancer cells. Int. J. Oncol. 2013;43:431–438. doi: 10.3892/ijo.2013.1978. [DOI] [PubMed] [Google Scholar]
- Steinmetz T. D., Schlötzer-Schrehardt U., Hearne A., Schuh W., Wittner J., Schulz S. R., Winkler T. H., Jäck H. M., Mielenz D. TFG is required for autophagy flux and to prevent endoplasmic reticulum stress in CH12 B lymphoma cells. Autophagy. 2021;17:2238–2256. doi: 10.1080/15548627.2020.1821546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Li Y., Fang F. MiR-138 suppresses the PDK1 expression to decrease the oxaliplatin resistance of colorectal cancer. Onco. Targets Ther. 2020;13:3607–3618. doi: 10.2147/OTT.S242929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Liu Y., Li X. The interaction mechanism between autophagy and apoptosis in colon cancer. Transl. Oncol. 2020;12:100871. doi: 10.1016/j.tranon.2020.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D. F., Pan S. T., Huang G., Qiu J. X. ROS enhances the cytotoxicity of cisplatin by inducing apoptosis and autophagy in tongue squamous cell carcinoma cells. Int. J. Biochem. Cell Biol. 2020;122:105732. doi: 10.1016/j.biocel.2020.105732. [DOI] [PubMed] [Google Scholar]
- Yao C. W., Kang K. A., Piao M. J., Ryu Y. S., Fernando P. M. D. J., Oh M. C., Park J. E., Shilnikova K., Na S. Y., Jeong S. U., Boo S. J., Hyun J. W. Reduced autophagy in 5-fluorouracil resistant colon cancer cells. Biomol. Ther. (Seoul) 2017;25:315–320. doi: 10.4062/biomolther.2016.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim W. W., Mizushima N. Lysosome biology in autophagy. Cell Discov. 2020;6:6. doi: 10.1038/s41421-020-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C. W., Lee S. H. The roles of autophagy in cancer. Int. J. Mol. Sci. 2018;19:3466. doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]