Abstract
Pseudomonas putida KT2442 is a root-colonizing strain which can use proline, one of the major components in root exudates, as its sole carbon and nitrogen source. A P. putida mutant unable to grow with proline as the sole carbon and nitrogen source was isolated after random mini-Tn5–Km mutagenesis. The mini-Tn5 insertion was located at the putA gene, which is adjacent to and divergent from the putP gene. The putA gene codes for a protein of 1,315 amino acid residues which is homologous to the PutA protein of Escherichia coli, Salmonella enterica serovar Typhimurium, Rhodobacter capsulatus, and several Rhizobium strains. The central part of P. putida PutA showed homology to the proline dehydrogenase of Saccharomyces cerevisiae and Drosophila melanogaster, whereas the C-terminal end was homologous to the pyrroline-5-carboxylate dehydrogenase of S. cerevisiae and a number of aldehyde dehydrogenases. This suggests that in P. putida, both enzymatic steps for proline conversion to glutamic acid are catalyzed by a single polypeptide. The putP gene was homologous to the putP genes of several prokaryotic microorganisms, and its gene product is an integral inner-membrane protein involved in the uptake of proline. The expression of both genes was induced by proline added in the culture medium and was regulated by PutA. In a P. putida putA-deficient background, expression of both putA and putP genes was maximal and proline independent. Corn root exudates collected during 7 days also strongly induced the P. putida put genes, as determined by using fusions of the put promoters to ′lacZ. The induction ratio for the putA promoter (about 20-fold) was 6-fold higher than the induction ratio for the putP promoter.
Pseudomonas putida KT2442 is an efficient root colonizer in a number of agriculturally important plants. In field assays, the root colonization of corn and broad bean by this P. putida strain ranged from about 105 to 107 CFU per g of soil, depending on the year and the season (38, 39). However, in soils without plants, the number of viable cells never surpassed 103 CFU per g of soil (39) and frequently remained at a level below 102 CFU per g of soil. Little is known about the nature of the nutrient source available for this strain during root colonization. Amino acids present in plant exudates may help satisfy the energy demands of rhizobacteria (25). Our group and others have identified the amino acids present in the root exudates of corn plants. Almost all of the 20 amino acids most frequently present in the proteins can be detected, with proline one of the most abundant (4, 8, 29, 41, 56; C. Ramos and L. Molina, unpublished results). These observations raise the possibility that, at least in the corn root rhizosphere, proline catabolism may play a relevant role in supporting root colonization. Nevertheless, information regarding proline catabolism by Pseudomonas strains is scarce (34, 35).
The first step for proline catabolism requires the entry of this amino acid into the cells (60). In enteric bacteria, proline is taken up by several transport systems that differ in their Vmax and affinity for proline. The PutP protein represents the major proline uptake system in Escherichia coli and Salmonella spp., with a Km of about 2 μM (61). The uptake of proline via PutP is coupled to the entry of sodium ions (7, 10, 26, 47, 60).
Proline is converted into glutamate in a two-step process carried out by proline dehydrogenase (PDH) (EC 1.5.99.8) and pyrroline-5-carboxylate dehydrogenase (P5CDH) (EC 1.5.1.12) (21, 33, 59). In eukaryotes, PDH and P5CDH are encoded by two different genes (30, 58), while in enteric bacteria (2, 31, 63), Rhodobacter capsulatus (27), Rhizobium meliloti (25), and Bradyrhizobium japonicum (53), both steps for proline utilization are catalyzed by a single polypeptide encoded by the putA gene. In addition to these enzymatic activities, the PutA protein, at least in enterobacteria, is involved in the transcriptional control of the put genes. It seems that PutA functions as a repressor, inhibiting expression from the divergent put genes (33, 44).
In the present study, we isolated a P. putida KT2442 mutant unable to use proline as its sole C and N source. The mutation was complemented by using a P. putida cosmid library, and we rescued and analyzed the complete nucleotide sequence of the P. putida put genes. We also show that put gene expression in this strain is inducible by proline present in root exudates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
P. putida KT2442 was described in an earlier publication (18). It can use benzoate as its sole C source and exhibits resistance to rifampin, chloramphenicol, and ampicillin. Strain S14D2 is a KT2242 mutant unable to use proline as its sole C and N source (Table 1). The E. coli strains used in this study are shown in Table 1.
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2442 | Rifr Apr Cmr; prototroph | 18 |
| S14D2 | Rifr KmrputA::mini-Tn5 luxAB-Km | This study |
| E. coli | ||
| HB101 | SmrrecA | 5 |
| DH5αF′ | recA | 62 |
| CC118λpir | Rifr; λ-pir lysogen | 22 |
| RM2 | Δ(putA putP) | 20 |
| Plasmids | ||
| pCK220 | Apr Kmr mini-Tn5::′luxAB | 52 |
| pCRR831 | Tcr; chimeric cosmid of P. putida library bearing the proline utilization operon | C. Ramos and L. Molina |
| pMIS5 | TcrPputA::′lacZ oriRK2 | This study |
| pMIS12 | TcrPputP::′lacZ oriRK2 | This study |
| pMP220 | Tcr ′lacZ oriRK2 | 50 |
| pPC6 | Apr; 6-kb AatII-PvuII fragment from the serovar Typhimurium chromosome in pBR322 | 20 |
| pRK600 | Cmrmob+ tra+ | 22 |
| pUC18/19 | Apr; cloning vector | 57 |
Apr, Rifr, Kmr, Cmr, Smr, and Tcr indicate resistance to ampicillin, rifampin, kanamycin, chloramphenicol, streptomycin, and tetracycline, respectively.
Bacterial cells were grown in Luria-Bertani medium or minimal M9 medium with succinate (20 mM) and/or proline (20 mM) as a C source (1). When proline (20 mM) was used as the sole C and N source, M9 depleted of ammonium, called M8, was used. When necessary, ampicillin, chloramphenicol, kanamycin, rifampin, and tetracycline were added to final concentrations of 100, 30, 25, 10, and 10 μg/ml, respectively.
DNA techniques.
Plasmid DNA was isolated by the alkaline lysis method with the QIAprep spin plasmid minipreps kit (Qiagen catalog no. 27104). Total DNA was isolated by modifying the method of Kado and Liu as described by Ramos-González et al. (46), except that the 30-min incubation step at 55°C was omitted. DNA digestions with restriction enzymes, ligations, and transformations were performed by standard procedures (48).
DNA in both strands was sequenced with the dideoxy sequencing method, using the ABI Prism dRhodamine terminator kit (reference no. 403042; Perkin-Elmer).
Southern hybridization and DNA labeling.
DNA fragments were separated in agarose gels and transferred onto nylon membranes by capillary blotting as previously described (48). Specific probes for hybridization were recovered from agarose gels with an agarose gel DNA extraction kit (reference no. 1696505; Boehringer Mannheim). All probes were labeled with digoxigenin by Klenow random primer extension according to the recommended procedure (3). Blotted filters were prehybridized, hybridized, washed, and immunologically developed according to the supplier's instructions. High-stringency conditions (50% [vol/vol] formamide at 42°C) were used.
Mutagenesis of P. putida by the mini-Tn5 luxAB-Km transposon.
Triparental matings involving P. putida KT2442 as the recipient, E. coli CC118λpir(pCK220) as the transposon donor strain (52), and E. coli HB101(pRK600) as the helper strain were carried out as described by de Lorenzo and Timmis (16). Transconjugants of P. putida were selected on M9 minimal medium plates with 5 mM benzoic acid as the sole C source and supplemented with kanamycin and rifampin. About 5,000 independent clones were tested for their ability to grow on M8 minimal medium with proline as the sole C and N source. Four mutants unable to produce colonies on minimal medium with proline were kept for further studies.
Complementation assays.
The pCRR831 cosmid (Table 1) (C. Ramos and L. Molina, unpublished results) selected from a P. putida KT2442 gene bank (M. I. Ramos-González, unpublished data) was used for complementation studies. pCRR831 was transferred by conjugation by the filter-mating technique (16) to the P. putida S14D2 mutant unable to grow with proline as the sole C and N source. Filters with a mixture of donor [E. coli HB101(pCRR831)], recipient (P. putida S14D2), and helper [E. coli HB101(pRK600)] strains at a ratio of 1:5:1 were incubated for 4 h at room temperature on Luria-Bertani plates. The cells were suspended in 1 ml of M9 minimal medium, and 100 μl was plated on selective minimal medium (M9 minimal medium with 10 mM benzoic acid, 10 μg of rifampin per ml, and 10 μg of tetracycline per ml). The transconjugants obtained were tested for their ability to grow on proline as the sole C and N source.
Enzyme assay.
P. putida cells were grown on succinate, proline, or succinate plus proline as the sole C source. Cells were harvested by centrifugation, resuspended in a Tris buffer (pH 7.0; 100 mM), and permeabilized with toluene by vortexing. PDH activity was measured at 30°C in a 7-ml reaction mixture that contained 100 μmol of Tris buffer (pH 7.0), 45 μmol of proline, and 4.5 μmol of o-aminobenzaldehyde. The Δ1-pyrroline-5′-carboxylic acid (P5C) that formed reacted with o-aminobenzaldehyde to produce a complex that exhibited maximal absorbance at 443 nm (17). The absorbance was corrected with a blank consisting of the same reaction mixture with water instead of proline. PDH activity was expressed as the number of nanomoles of P5C formed per milligram of protein.
Protein concentration in the cell extracts was determined with the Bradford reagent (Bio-Rad reference no. 500.0006; Bio-Rad, Madrid, Spain) with bovine serum albumin as the standard.
Collection of corn root exudates.
Seeds were germinated on a sterile petri dish with water-agar. Seedlings were transferred to a grid, and the hair root was submerged into a sterile solution of M9 medium without ammonium. After 7 days, the seeds were removed, and the solution was filtered through a 0.2-μm sterile nitrocellulose filter and stored at −20°C until use. Proline concentrations in these exudates ranged between 50 and 100 μM.
Construction of PputA::lacZ and PputP::lacZ fusions.
The divergent putA and putP promoter region was amplified by PCR from total chromosomal DNA of P. putida KT2442 with primers 5′-TTACGAATTCCGATGTAGATCACGAAGG-3′ and 5′-TTACGGAATTCTGCTTTGAGTCGCTCACGG-3′, which are provided with a restriction site for EcoRI. Upon amplification, as recommended by Ausubel et al. (3), DNA was restricted with EcoRI and ligated to plasmid pMP220 digested with EcoRI, so that transcriptional fusions of the putA or putP promoters to a promoterless ′lacZ gene were generated. The nature of the fusion can be distinguished by PCR amplification with an oligonucleotide primer based on the lacZ sequence and on putA- or putP-based primers, which result in a 0.8-kb fragment. The plasmid bearing the PputA::′lacZ fusion was named pMIS5, and the one bearing the PputP::′lacZ fusion was called pMIS12. The fusions were further confirmed by sequencing the whole promoter region and the first 20 codons of the ′lacZ gene.
β-Galactosidase activity was measured in P. putida KT2440 and in P. putida S14D2 bearing pMIS5 or pMIS12 and grown on M9 minimal medium with 20 mM succinic acid in the absence or the presence of 20 mM proline. Activity was determined according to Gallegos et al. (19), and activity was given in Miller units (36).
RESULTS
Growth of P. putida KT2442 on proline as the sole C and N source and isolation of mutants unable to metabolize proline.
We first tested whether P. putida KT2442 was able to use proline as the sole source of C, N, or both nutrients. This strain was grown on M9 minimal medium with succinate as the sole C source. The culture was diluted 100-fold into M8 minimal medium with 20 mM proline and 10 mM NH4Cl (proline as the sole C source), 20 mM succinate and 20 mM proline (proline as the sole N source), and 20 mM proline (proline as the C and N source). The strain grew exponentially with generation times of 1.70, 1.44, and 2.27 h when proline was used as the sole C, N, and C plus N source, respectively.
We then mated P. putida KT2442 with E. coli CC118λpir(pCK220) as described in Materials and Methods, and four mutants defective in proline utilization, called S14D2, S14D11, S15D3, and S16D2, were found.
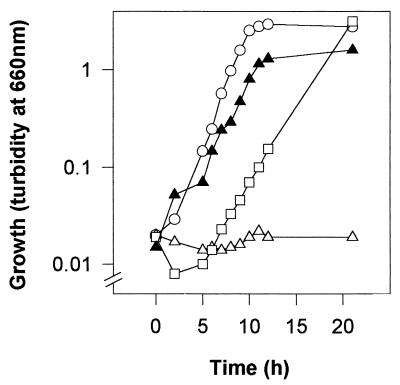
To further confirm this initial selection, growth of the strains was tested in liquid M8 minimal medium with proline as the sole C and N source. Mutant S14D2 did not grow on minimal medium after prolonged incubation (Fig. 1), whereas the other three mutants did grow, although they had a very long lag period before growth started. See Fig. 1 for mutant S14D11. We measured the PDH activity of the wild-type and the mutant strains growing on M9 with succinate or succinate plus proline. The results obtained are shown in Table 2. Neither the wild-type nor the mutant strains exhibited any statistically significant activity when grown on succinate alone, but the wild-type had high activity levels when it grew in the presence of proline. Mutants S14D11, S15D3, and S16D2 also had high levels of PDH activity when grown in the presence of proline (results not shown). In contrast, mutant S14D2 showed no activity when cells were grown on M9 with succinate and proline (Table 2). On the basis of these results, we considered S14D2 a true proline utilization-deficient strain, and it was retained for further studies. The other three mutants (S14D11, S15D3, and S16D2) were discarded.
FIG. 1.
Growth on minimal medium with proline as the sole C and N source of the wild-type P. putida KT2442 and its mutant strains deficient in proline utilization. Growth was monitored as an increase in turbidity of the culture. ○, P. putida KT2442; ▵, P. putida S14D2; □, P. putida S14D11; ▴, P. putida S14D2(pCRR831).
TABLE 2.
PDH activity of P. putida KT2442 and its mini-Tn5 transposon derivativesa
| Strain | PDH activity (%)
|
|
|---|---|---|
| −Pro | +Pro | |
| KT2442 | 5 | 100 |
| S14D2 | 1 | 5 |
| S14D2(pCCR831) | 3 | 72 |
Cells were grown on M9 minimal medium with succinate as the sole C source in the absence (−Pro) and the presence (+Pro) of 20 mM proline. PDH activity was determined as described in Materials and Methods. One hundred percent of activity corresponded to 190 nmol of P5C produced per milligrams of protein per minute.
Complementation of mutant S14D2 by pCRR831, cloning, and sequencing of the put genes.
A P. putida KT2442 gene bank constructed in the broad-host-range pLAFR3 cosmid (M. I. Ramos-González, unpublished data) was used to complement E. coli RM2 (Table 1), a mutant unable to grow on proline because of a deletion of the putA and putP genes (20). A plasmid called pCRR831 was found to restore the ability to use proline as the sole C and N source to the E. coli mutant strain (C. Ramos and L. Molina, unpublished results). We transferred the Tcr pCRR831 plasmid to P. putida S14D2 and selected Kmr Tcr transconjugants able to grow on M8 minimal medium with proline as the sole C source. The frequency of appearance of transconjugants was 10−5 per recipient, and 100% of the transconjugants were able to grow on M8 liquid medium with proline as the sole C and N source. Figure 1 shows the growth of one random P. putida S14D2(pCRR831) clone, compared with the growth of the wild type and the mutant S14D2. This finding suggests that pCRR831 carries the proline degradation genes. To corroborate this finding, we determined the PDH activity of P. putida S14D2(pCRR831) growing on succinate or succinate plus proline. As expected, pCRR831 restored this activity in the mutant strain to levels similar to those found in the wild-type strain, when cells grew in the presence of proline (Table 2).
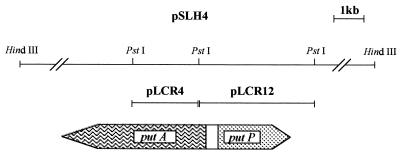
To locate the put genes in pCRR831, cosmid DNA was digested with PstI and hybridized against the 4.2-kb MluI fragment of plasmid pPC6 (20), which carries the putA putP genes of Salmonella enterica serovar Typhimurium. The P. putida put genes were located within two PstI fragments of 4.3 and 2.0 kb, which were subcloned in pUC19 to yield plasmids pLCR12 and pLCR4, respectively (Fig. 2). The DNA in both PstI fragments was sequenced on both strands. The DNA sequences were compared with those deposited in the GenBank database, and the analysis revealed that the 4.3-kb DNA fragment bore the whole putP gene (1,479 bp), part of the ′putA′ gene (450 bp), and the intergenic region between putP and putA (355 bp). These genes were transcribed divergently. Plasmid pLCR4, bearing a 2-kb insert of the P. putida genome, also contained part of the putA gene; however, the translated DNA sequence did not exhibit a stop codon, nor did it account for the expected size of the PutA protein when compared with the PutA sequences deposited in GenBank. To complete the putA gene, a 12-kb HindIII fragment of pCRR831 was subcloned in pUC19 to yield pSLH4 (Fig. 2). DNA was sequenced with specific 20-mer primers, based on available P. putida putA sequences, until the complete putA gene sequence was obtained (3,948 bp). In all, the putA and putP genes and the intergenic region covered 5,757 bp. The DNA sequence is available from GenBank under accession no. AF153207. Downstream of both coding sequences, stem-loop transcription terminator sequences were found, which suggests that each gene makes a monocistronic mRNA.
FIG. 2.
Localization of put genes of P. putida KT2442 in vectors pLCR4, pLCR12, and pSLH4. The pLCR4 plasmid contains 2 kb of the putA gene, and pLCR12 contains 450 bp of the putA gene, all the putP genes, and the intervening regulatory region between the two genes that are transcribed divergently. A 12-kb insert in plasmid pSLH4 bears the complete proline utilization operon.
The insertion of the mini-Tn5 ′luxAB-Km transposon in the genome of P. putida S14D2 was first located within the putA gene, based on hybridization assays. The region surrounding the mini-Tn5 was PCR amplified and the insertion was specifically identified at nucleotide 1635 of the putA gene sequence.
Analysis of putA and putP gene products.
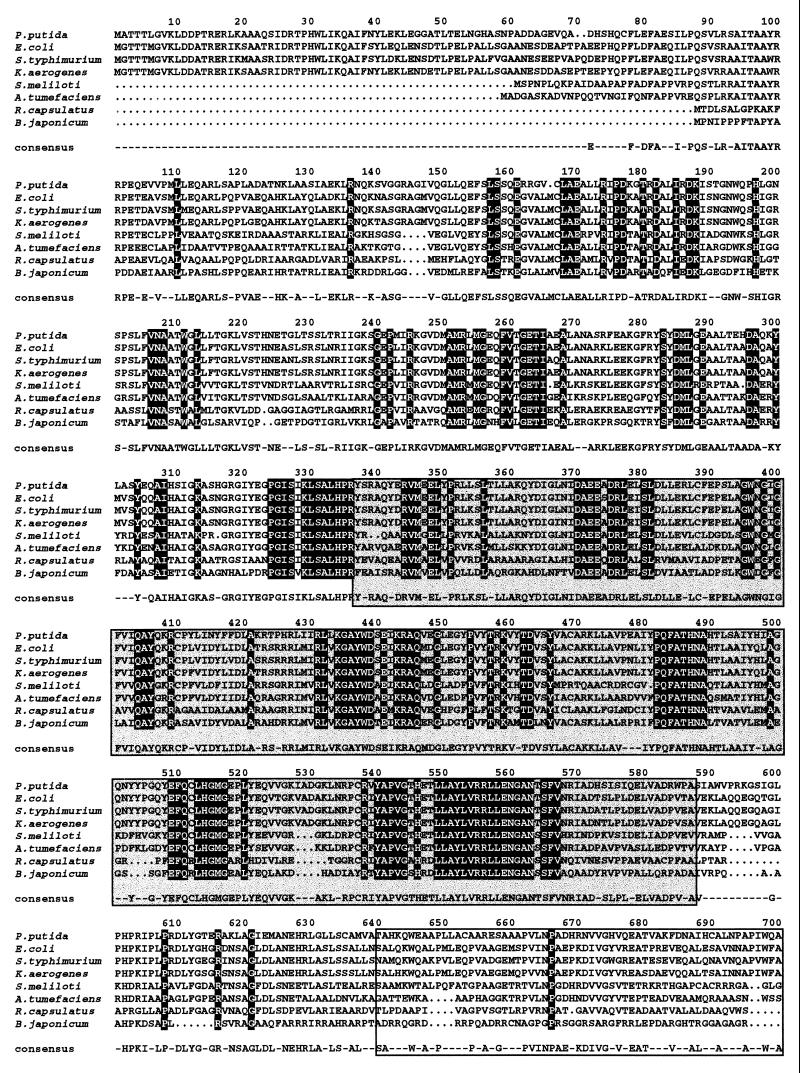
The putA gene yielded the predicted PutA protein, which is 1,315 amino acids long and shows homology to PutA from different organisms such as Klebsiella aerogenes (71% identity) (54), Salmonella serovar Typhimurium (69% identity) (2), E. coli (69% identity) (31), R. meliloti (54% identity) (25), and B. japonicum (42% identity) (53). The highest homology was the domain involved in PDH activity (amino acids 337 to 588 in the P. putida PutA protein) (Fig. 3). Within this domain, a flavin adenine dinucleotide-binding pocket (residues 312 to 354) was identified. This domain exhibited homology with PDHs from Saccharomyces cerevisiae and Drosophila melanogaster and therefore seems to be involved in the conversion of proline to P5C, which equilibrates in solution with glutamic acid semialdehyde.
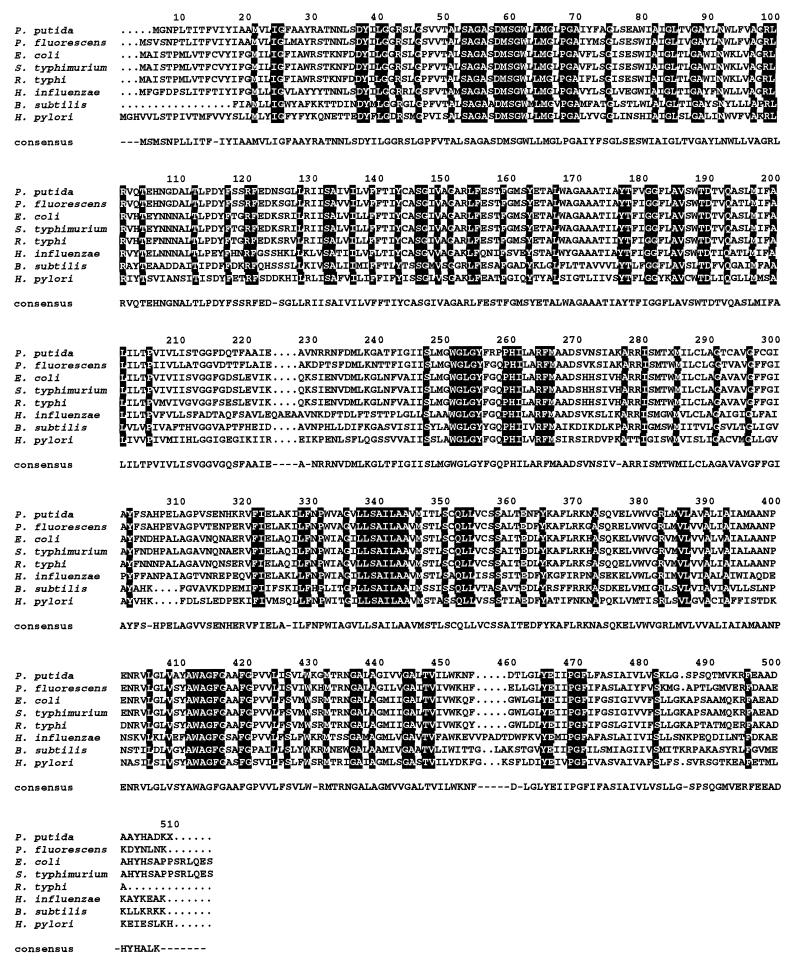
FIG. 3.
Sequence alignment of PutA proteins of prokaryotic origin. The strains and sources of the protein sequences were as follows: P. putida (this study); E. coli (31); Salmonella serovar Typhimurium (2); K. aerogenses (54); Agrobacterium tumefaciens (14); R. capsulatus (27); S. meliloti (25); B. japonicum (53). The ALIGN program was used. If the residue is identical in all the aligned proteins, it appears printed on a black background. If the residue is identical in 50% of the aligned proteins, it appears on a gray background. The amino acid chosen for the consensus was present at the given position in at least 50% of the aligned sequences. The PDH domain, residues 337 to 588, is shown in a grey box, and the P5CDH domain, residues 641 to 1074, is also boxed.
According to Ling et al. (31), amino acids 641 to 1074 are required for P5CDH activity. An NADPH pocket (residues 850 to 857) with the sequence FTGSTEVG was found within this region (31), which is highly similar to the corresponding PutA region in E. coli and Salmonella serovar Typhimurium (Fig. 3). This domain exhibited homology with aldehyde dehydrogenases, i.e., methylmalonate dehydrogenase, betaine dehydrogenase, and 2-hydroxymuconic acid semialdehyde dehydrogenase (9, 11, 13, 42, 45, 51). This finding suggests that the real substrate of this activity of PutA is glutamic acid semialdehyde.
A third region with high homology between PutA proteins but of unknown function is located between amino acids 78 and 190. In E. coli, the PutA protein is able to associate with the cell membranes. Three hydrophobic segments between residues 158 and 167, 767 and 817, and 1205 to 1220 may be important for such interactions. These segments are present in the P. putida PutA protein. In general, the interdomains were less conserved (Fig. 3).
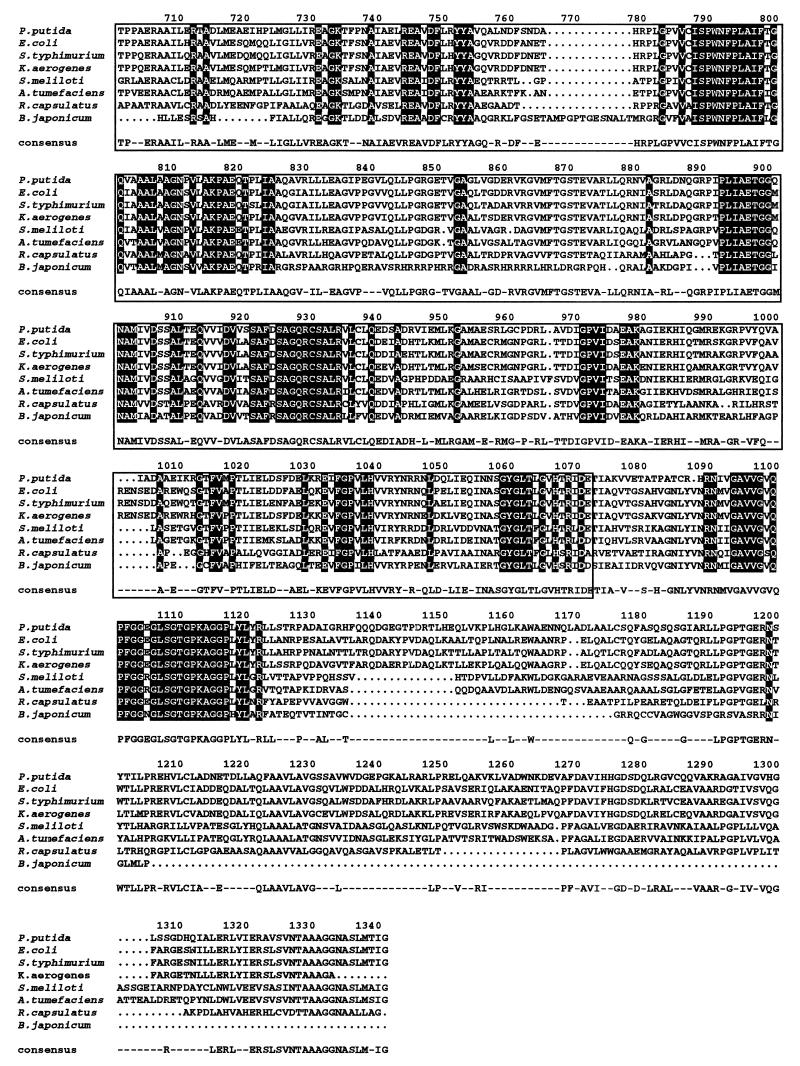
The P. putida PutP protein is 493 amino acids long and exhibits 85% similarity with PutP from Pseudomonas fluorescens, 76% with Salmonella serovar Typhimurium, and 78% with E. coli. The Scamprosite program predicted 12 transmembrane segments for the P. putida PutP protein, and multiple alignments revealed extended homology with PutP from other sources that corresponded to transmembrane segments (Fig. 4), whereas cytoplasmic and periplasmic loops were less well conserved. In addition, PutP presents homology to transport systems that are involved in the uptake of chemicals related to sodium entry, i.e., E. coli porter systems for inositol, phenylacetic acid, and pantothenate (7, 15, 49, 55, 58).
FIG. 4.
Sequence alignment of PutP proteins. Strains and sources of the sequences were as follows: P. putida (this study); Bacillus subtilis (64); P. fluorescens (23); E. coli (40); Salmonella serovar Typhimurium (37); Rickettsia typhi (40); and Haemophilus influenzae and Helicobacter pylori (55). The ALIGN program was used. Other details are as in the legend for Fig. 3.
Expression from the putA and putP gene promoters.
To determine the expression of the put genes, the divergent put promoter region was fused in a broad-host-range vector to ′lacZ as described in Materials and Methods to generate transcriptional fusions yielding pMIS5 and pMIS12. These plasmids were transferred to the wild-type P. putida KT2442 and to its mutant P. putida S14D2. β-Galactosidase (LacZ) activity in P. putida KT2442 with one of these plasmids was measured in cells growing on minimal medium with succinate and succinate plus proline under highly aerated conditions. In wild-type cells growing on succinate, basal activity from PputP (700 Miller units) was twofold higher than for PputA (350 Miller units) (Table 3). In the presence of proline, the increase in activity was 4- and 20-fold for the PputP fusion and the PputA fusion, respectively (Table 3). These results suggest that the genes for proline catabolism are inducible.
TABLE 3.
Expression from the put promoters in P. putidaa
| Strain and fusion | Growth conditions | β-Galactosidase activity |
|---|---|---|
| Wild type | ||
| PputP::′lacZ | Succinate | 700 ± 50 |
| Succinate plus proline | 2,800 ± 100 | |
| PputA::′lacZ | Succinate | 350 ± 30 |
| Succinate plus proline | 6,950 ± 100 | |
| Mutant S14D2 | ||
| PputP::′lacZ | Succinate | 2,700 ± 200 |
| Succinate plus proline | 2,600 ± 150 | |
| PputA::′lacZ | Succinate | 7,900 ± 200 |
| Succinate | 8,600 ± 250 |
P. putida KT2442 and P. putida S14D2 bearing pMIS5 (PputA::′lacZ) or pMIS12 (PputP::′lacZ) were grown on M9 minimal medium with succinate or succinate plus proline. β-Galactosidase activity (in Miller units) was determined in permeabilized whole cells according to the method of Gallegos et al. (19). Data are the average of four independent assays.
Expression of the putA and putP genes was also measured in the S14D2 mutant strain bearing pMIS5 or pMIS12 in cells growing on M9 minimal medium with succinate or with proline. In both the absence and the presence of proline, high levels of expression were found, about 2,700 Miller units for the PputP::′lacZ fusion and about 8,000 Miller units for the PputA fusion. These results suggest that the PutA protein is involved in the control of expression from the putA and putP gene promoters.
Induction of the Pput promoters by corn root exudates.
P. putida KT2442 bearing plasmid pMIS5 or pMIS12 was grown on minimal medium with succinate as the sole C source until the mid-exponential growth phase was reached. Cells were then either harvested and suspended in M8 minimal medium without a C source or suspended in 7-day-old root exudates. The suspensions were incubated at room temperature without agitation for 30 min to follow induction from the put promoters. The level of β-galactosidase activity from PputA and PputP when cells were incubated in the presence of corn root exudates was around 20- and 4-fold higher than the basal level (Table 4). This suggests that proline present in root exudates was able to promote expression of the P. putida put catabolic genes.
TABLE 4.
Induction of the put promoters in the presence of corn root exudatesa
| Fusion | Incubation conditions | β-Galactosidase activity |
|---|---|---|
| PputA::′lacZ | Corn exudate | 425 ± 50 |
| M8 | 20 ± 10 | |
| PputP::′lacZ | Corn exudate | 270 ± 50 |
| M8 | 60 ± 10 |
P. putida KT2442 cells bearing pMIS5 (PputA::′lacZ) or pMIS12 (PputP::′lacZ) grown on M9 minimal medium with succinate were harvested and suspended in M8 minimal medium (M8) or in the same medium enriched for 7 days with corn root exudates (corn exudate) and incubated at room temperature without agitation. β-Galactosidase activity was determined as described in the footnote to Table 3.
DISCUSSION
Recent studies have focused their attention on the possible role of amino acids as carbon substrates to support growth of microorganisms in the rhizosphere of plants (24, 28, 63, 65). Proline has been found to be a major compound in the corn root exudates; therefore, this amino acid could be an important energy source for bacteria during the first stages of colonization of the roots of plants. How deficiency in the utilization of proline or other amino acids affects rhizosphere colonization has not yet been studied in detail, although an R. meliloti mutant altered in proline catabolism exhibited reduced ability to colonize the alfalfa root (25).
In this work we have approached the study of proline utilization in P. putida, for which we isolated mutants unable to use proline as their C or N source. P. putida S14D2 was considered a true proline utilization-deficient mutant because it did not grow with proline, in contrast with other mutants isolated in this study that showed retarded growth on proline. We found that in the S14D2 mutant strain, the mini-Tn5 transposon was inserted in the chromosome within a gene involved in proline catabolism (putA). Analysis of the P. putida putA gene product revealed a domain structure similar to that of enteric bacteria such as R. capsulatus and B. japonicum in which the two steps for proline degradation to glutamate are catalyzed by a single bifunctional dehydrogenase enzyme (2, 25, 27, 31, 51, 53). Analysis of the P. putida PutP protein suggests that it is an integral inner-membrane protein that belongs to the family of Na+ substrate symporters (15, 49, 58, 60). We showed that the putA gene is adjacent to the putP gene and that these genes are transcribed divergently, as is the case for enteric bacteria.
In P. putida, the putA and putP genes seem to be regulated at the transcriptional level, with proline—either supplied in culture medium or in root exudates—acting as an inducer, as the expression from the putA and putP gene promoters increased by about 20- and 4-fold, respectively, in the presence of proline. In a putA mutant background, high levels of expression from these genes occurred, suggesting that the P. putida PutA protein acts as a repressor of putA and putP gene expression, as also described for enteric bacteria (11, 44). The fact that proline metabolism in the soil bacterium P. putida is regulated by a mechanism similar in principle to that of enteric bacteria is rather surprising in the light of the differences in the ecological habitats of these organisms. These similarities in the regulation of the put genes in enteric bacteria and in Pseudomonas prompted us to compare the intergenic regions between putA and putP in these microorganisms. Figure 5 shows an alignment of the intergenic region between putA and putP of Salmonella serovar Typhimurium, E. coli, K. aerogenes and P. putida, from which it can be seen that this region is 63 to 65 bp longer in enteric bacteria than in P. putida, with a very large gap (28 nt) being observed near the ATG start codon of the putP gene. In all four microorganisms, putA and putP genes are transcribed divergently, although differences in the location of promoters are known, with overlapping promoters in Salmonella serovar Typhimurium and well-separated transcription starts in K. aerogenes and P. putida (12, 44; S. Vílchez and J. L. Ramos, unpublished results). In Salmonella serovar Typhimurium, the intergenic putA-putP DNA is intrinsically curved and it has been found that up to five segments (marked in Fig. 5 by a line above the sequence) could be bound by purified PutA protein. In enteric bacteria, it has been suggested that the integration host factor plays a role in the expression from putA and putP, and two sites (positions 1 to 13 and 330 to 344) (Fig. 5) in the Salmonella serovar Typhimurium promoter region were found (6, 43, 44). Those sites are not well conserved in the corresponding aligned sequence in P. putida, and at present, we cannot predict whether or not integration host factor plays a role in the transcription of the put genes in the soil bacterium P. putida.
FIG. 5.
Alignment of the putA and putP intergenic regions of enteric bacteria and P. putida. The alignment includes the region between the start codons of putA and putP. Gaps were introduced to allow maximal scoring in the alignment with identical positions being shown in boldface. The overlined bases indicate putative PutA binding sites.
Therefore, we can conclude that although the pattern of gene control of the putA and putP genes is similar in enteric bacteria and in the soil-borne P. putida KT2440, the molecular mechanisms of control may be very distinct.
ACKNOWLEDGMENTS
Susana Vílchez and Lázaro Molina contributed equally to the experimental work.
Part of this study was supported by a grant from the European Commission (BIO4-CT98-0283).
REFERENCES
- 1.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range or the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen S W, Sentis Willis A, Maloy S R. DNA sequence of the putA gene from Salmonella typhimurium: a bifunctional membrane-associated dehydrogenase that binds DNA. Nucleic Acids Res. 1993;21:1676. doi: 10.1093/nar/21.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 4.Barber D A, Martin J K. The release of organic substances by cereal roots into the soil. New Phytol. 1976;76:68–80. [Google Scholar]
- 5.Boyer H W, Roullard-Dussoix D. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Brown E D, Wood J M. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- 7.Cairney J, Higgins F C, Booth I R. Proline uptake through the major transport system of Salmonella typhimurium is coupled to sodium ions. J Bacteriol. 1984;160:22–27. doi: 10.1128/jb.160.1.22-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaboud A. Isolation, purification and chemical composition of maize root cap slime. Plant Soil. 1983;73:395–402. [Google Scholar]
- 9.Chang C, Yoshida A. Cloning and characterization of the gene encoding mouse mitochondrial aldehyde dehydrogenase. Gene. 1994;148:331–336. doi: 10.1016/0378-1119(94)90708-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen C C, Tsuchiya T, Yamane Y, Wood J M, Wilson J H. Na+(Li+)-proline transport in Escherichia coli. J Membr Biol. 1985;84:157–164. doi: 10.1007/BF01872213. [DOI] [PubMed] [Google Scholar]
- 11.Chen C S, Yoshida A. Enzymatic properties of the proline dehydrogenase encoded by newly cloned human alcohol dehydrogenase ADH6 gene. Biochem Biophys Res Commun. 1991;181:743–747. doi: 10.1016/0006-291x(91)91253-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen L-M, Maloy S. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J Bacteriol. 1991;173:783–790. doi: 10.1128/jb.173.2.783-790.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Achkar C, Gudas L J. Enzymatic conversion of retinaldehyde to retinoic acid by cloned murine cytosolic and mitochondrial aldehyde dehydrogenases. Mol Pharmacol. 1994;46:88–96. [PubMed] [Google Scholar]
- 14.Cho K, Fuqua C, Martin B S, Winans S C. Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J Bacteriol. 1996;178:1872–1880. doi: 10.1128/jb.178.7.1872-1880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379:458–460. doi: 10.1038/379458a0. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 17.Dendinger S, Brill W J. Regulation of proline degradation in Salmonella typhimurium. J Bacteriol. 1970;103:144–152. doi: 10.1128/jb.103.1.144-152.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin F G H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallegos M T, Williams P A, Ramos J L. Transcriptional control of the multiple catabolic pathways encoded on the TOL plasmid pWW53 of Pseudomonas putida MT53. J Bacteriol. 1997;179:5024–5029. doi: 10.1128/jb.179.16.5024-5029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn D R, Myers R S, Kent C R, Maloy S R. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon, and DNA sequence of the put control region. Mol Gen Genet. 1988;213:125–133. doi: 10.1007/BF00333408. [DOI] [PubMed] [Google Scholar]
- 21.Hayward D C, Delaney S J, Campbell H D, Ghysen A, Benzer S, Kasprzak A B, Cotsell J N, Young I G, Gabor Miklos G L. The sluggish-A gene of Drosophila melanogaster is expressed in the nervous system and encodes proline oxidase, a mitochondrial enzyme involved in glutamate biosynthesis. Proc Natl Acad Sci USA. 1993;90:2979–2983. doi: 10.1073/pnas.90.7.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoya H, Nakamura K. DNA sequence of proline permease gene from Pseudomonas fluorescens and predicted structure of proline permease. Biosci Biotechnol Biochem. 1994;5:2099–2101. doi: 10.1271/bbb.58.2099. [DOI] [PubMed] [Google Scholar]
- 24.Jiménez-Zurdo J I, van Dillewijn P, Soto M J, de Felipe M R, Olivares J, Toro N. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol Plant-Microbe Interact. 1995;8:492–498. doi: 10.1094/mpmi-8-0492. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez-Zurdo J I, Garcia-Rodriguez F M, Toro N. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol Microbiol. 1997;23:85–93. doi: 10.1046/j.1365-2958.1997.1861555.x. [DOI] [PubMed] [Google Scholar]
- 26.Kayama Y, Kawasaki T. Stimulatory effect of lithium ion on proline transport by whole cells of Escherichia coli. J Bacteriol. 1976;128:157–164. doi: 10.1128/jb.128.1.157-164.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keuntje B, Masepohl B, Klipp W. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J Bacteriol. 1995;177:6432–6439. doi: 10.1128/jb.177.22.6432-6439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl D H, Schubert K R, Carter M B, Hagedam C H, Shearer G. Proline metabolism in N2-fixing root nodules: energy transfer and regulation of purine synthesis. Proc Natl Acad Sci USA. 1988;85:2036–2040. doi: 10.1073/pnas.85.7.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohl D H, Straub P, Shearer G. Does proline play a special role in bacterial metabolism? Plant Cell Environ. 1994;17:1257–1262. [Google Scholar]
- 30.Krywicki K A, Brandriss M C. Primary structure of the nuclear PUT2 gene involved in the mitochondrial pathway for proline utilization in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2837–2842. doi: 10.1128/mcb.4.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling M, Allen S W, Wood J M. Sequence analysis identifies the proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J Mol Biol. 1994;243:950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- 32.Maloy S R. The proline utilization operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1513–1519. [Google Scholar]
- 33.Maloy S, Stewart V. Autogenous regulation of gene expression. J Bacteriol. 1993;175:307–316. doi: 10.1128/jb.175.2.307-316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meile L, Leisinger T. Purification and properties of the bifunctional proline dehydrogenase/1-pyrroline-5-carboxylate dehydrogenase from Pseudomonas aeruginosa. Eur J Biochem. 1982;129:67–75. doi: 10.1111/j.1432-1033.1982.tb07021.x. [DOI] [PubMed] [Google Scholar]
- 35.Meile L, Soldati L, Leisinger T. Regulation of proline catabolism in Pseudomonas aeruginosa PAO. Arch Microbiol. 1982;132:189–193. doi: 10.1007/BF00508729. [DOI] [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Miller K, Maloy S. DNA sequence of the putP gene from Salmonella typhimurium and predicted structure of proline fermease. Nucleic Acids Res. 1990;18:3057. doi: 10.1093/nar/18.10.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. Garcia, L. Wyke, and J. L. Ramos. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem., in press.
- 39.Molina L, Ramos C, Ronchel M C, Molin S, Ramos J L. Construction of an efficient biologically contained Pseudomonas putida strain and its survival in outdoor assays. Appl Environ Microbiol. 1998;64:2072–2078. doi: 10.1128/aem.64.6.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson K, Selander R K. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman E I. The rhizosphere; carbon sources and microbial populations. In: Fritter A R, editor. Ecological interactions in soil. Boston, Mass: Blackwell Scientific Publications, Ltd.; 1985. pp. 107–121. [Google Scholar]
- 42.Norlund I, Shingler V. Nucleotide sequence of the meta-cleavage pathway enzymes 2-hydroxymuconic semialdehyde dehydrogenase and 2-hydroxymuconic semialdehyde hydrolase from Pseudomonas CF600. Biochim Biophys Acta. 1990;2:227–230. doi: 10.1016/0167-4781(90)90046-5. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien K, Deno G, Ostrovsky de Spicer P, Gardner J F, Maloy S R. Integration host factor facilitates repression of the put operon in Salmonella typhimurium. Gene. 1992;118:13–19. doi: 10.1016/0378-1119(92)90243-i. [DOI] [PubMed] [Google Scholar]
- 44.Ostrovsky de Spicer P, O'Brien K, Maloy S. Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J Bacteriol. 1991;173:211–219. doi: 10.1128/jb.173.1.211-219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purdue P E, Lumb M J, Danpure C J. Molecular evolution of alanine/glycoxylate aminotransferase 1 intracellular targeting: analysis of the marmoset and rabbit genes. Eur J Biochem. 1992;207:757–766. doi: 10.1111/j.1432-1033.1992.tb17106.x. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-González M I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratzkin B, Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978;133:744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Smanik P A, Liu Q, Furminger T L, Ryu K, Xing S, Mazzaferri E L, Jhiang S M. Cloning of the human sodium ion symporter. Biochem Biophys Res Commun. 1996;226:339–345. doi: 10.1006/bbrc.1996.1358. [DOI] [PubMed] [Google Scholar]
- 50.Spaink H, Okker R, Wijffelman C, Pees E, Lugtenberg B. Promoters in the nodulation region of the Rhizobium leguminosarum sym. plasmid pRL1J1. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 51.Steele M I, Lorenz D, Hatter K, Park A, Sokatch J R. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J Biol Chem. 1992;267:13585–13592. [PubMed] [Google Scholar]
- 52.Sternberg C, Eberl L, Poulsen L K, Molin S. Detection of bioluminescence from individual bacterial cells: a comparison of two different low-light imaging systems. J Biochem Biolumin. 1997;12:7–13. doi: 10.1002/(SICI)1099-1271(199701/02)12:1<7::AID-BIO427>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Straub P F, Reynolds P H S, Althomsons S, Mett V, Zhu Y, Shearer G, Kohl D H. Isolation, DNA sequence analysis, and mutagenesis of a proline dehydrogenase gene (putA) from Bradyrhizobium japonicum. Appl Environ Microbiol. 1996;62:221–229. doi: 10.1128/aem.62.1.221-229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Surber, M. W., and S. Maloy. 1997. GenBank accession number AFO38838.
- 55.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Petterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. [DOI] [PubMed]
- 56.Vancura V. Plant metabolites in soil. In: Kunc F, Vancura V, editors. Soil microbial associations: control of structures and functions. Amsterdam, The Netherlands: Elsevier; 1988. pp. 156–165. [Google Scholar]
- 57.Vieira J, Messing J. The pUC plasmid: an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 58.von Blohn C, Kempf B, Kappes R M, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma β. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 59.Wang S-S, Brandriss M C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987;7:4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wengender P A, Miller K J. Identification of a putP proline permease gene homolog from Staphylococcus aureus by expression cloning of the high-affinity proline transport system in Escherichia coli. Appl Environ Microbiol. 1995;61:252–259. doi: 10.1128/aem.61.1.252-259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood J M, Zadworny D. Characterization of an inducible porter required for l-proline catabolism by Escherichia coli K12. Can J Biochem. 1979;57:1191–1199. doi: 10.1139/o79-155. [DOI] [PubMed] [Google Scholar]
- 62.Woodcock D M. Quantitative evolution of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia M, Zhu Y, Cao X, You L, Chen Z. Cloning, sequencing and analysis of a gene encoding Escherichia coli proline dehydrogenase. FEMS Microbiol Lett. 1996;127:235–242. doi: 10.1111/j.1574-6968.1995.tb07479.x. [DOI] [PubMed] [Google Scholar]
- 64.Yamane K, Kumeno H, Kurita K. The 25 degrees-36 degrees region of the Bacillus subtilis chromosome: the germination of the sequence of a 146-kb segment and identification of 113 genes. Microbiology. 1995;142:3047–3056. doi: 10.1099/13500872-142-11-3047. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y-X, Shearer G, Kohl D H. Proline fed to intact soybean plants influences acetylene reducing activity, and content and metabolism of proline in bacteroids. Plant Physiol. 1992;98:1020–1028. doi: 10.1104/pp.98.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]