Abstract
Background & Aims
MUC1 is abnormally expressed in colorectal cancer, including colitis-associated colorectal cancer (CAC), but its role in tumorigenesis is unclear. This study investigated MUC1’s effects in murine models of colitis and CAC and elucidated mechanisms of action.
Methods
Colitis and CAC were induced in mice by exposure to dextran sodium sulfate or azoxymethane plus dextran sodium sulphate. Clinical parameters, immune cell infiltration, and tumor development were monitored throughout disease progression. Experiments in knockout mice and bone marrow chimeras were combined with an exploration of immune cell abundance and function.
Results
Deficiency of Muc1 suppressed inflammation, inhibited tumor progression, increased abundance of CD8+ T lymphocytes, and reduced abundance of macrophages in colon tumors. Bone marrow chimeras showed promotion of CAC was primarily mediated by Muc1-expressing hematopoietic cells, and that MUC1 promoted a pro-tumoral immunosuppressive macrophage phenotype within tumors. Mechanistic studies revealed that Muc1 deficiency remarkably reduced interleukin-6 levels in the colonic tissues and tumors that was mainly produced by infiltrating macrophages at day 21, 42, and 85. In bone marrow-derived macrophages, MUC1 promoted responsiveness to chemoattractant and promoted activation into a phenotype with high Il6 and Ido1 expression, secreting factors which inhibited CD8+ T cell proliferation. MUC1 potently drives macrophages to produce interleukin-6, which in turn drives a pro-tumorigenic activation of signal transducer and activator of transcription 3 in colon epithelial tumor and stromal cells, ultimately increasing the occurrence and development of CAC.
Conclusions
Our findings provide cellular and molecular mechanisms for the pro-tumorigenic functions of MUC1 in the inflamed colon. Therapeutic strategies to inhibit MUC1 signal transduction warrant consideration for the prevention or therapy of CAC.
Keywords: Colitis-associated Colorectal Cancer, IL-6, Inflammation, Macrophages, MUC1
Abbreviations used in this paper: AOM, azoxymethane; BM, bone marrow; BMDM, bone marrow-derived macrophage; BMMC, bone marrow-derived mononuclear cell; BSA, bovine serum albumin; CAC, colitis-associated cancer; CRC, colorectal cancer; DAI, disease activity index scores; DSS, dextran sodium sulphate; H&E, hematoxylin and eosin; IBD, inflammatory bowel disease; IFN, interferon; IHC, immunohistochemical; IL, interleukin; LPS, lipopolysaccharide; Mɸ, macrophages; NFĸB, nuclear factor kappa B; PBS, phosphate buffered saline; PCR, polymerase chain reaction; STAT3, signal transducer and activator of transcription 3; TAMs, tumor-associated macrophages; TCM, tumor-conditioned medium; UC, ulcerative colitis; WT, wild type
Graphical abstract
Summary.
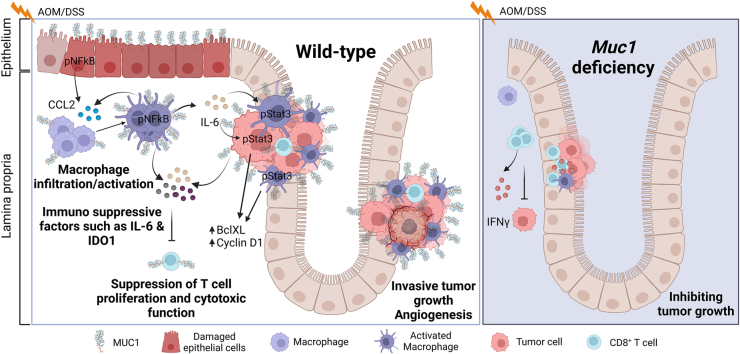
MUC1 cell surface mucin plays a pivotal role in colonic inflammation-induced cancer development by facilitating CCL2-mediated macrophage infiltration and modulating macrophage function, promoting interleukin-6 production and signal transducer and activator of transcription 3 activation and suppressing anti-tumor immunity, thereby promoting colitis and colitis-associated cancer.
Chronic inflammation is a significant risk factor for cancer initiation and progression in the colon.1 Patients with ulcerative colitis (UC) with long-term uncontrolled inflammation are at an increased risk of developing colorectal cancer (CRC), known as colitis-associated cancer (CAC).2 3 The microenvironment in the inflamed intestine leading to CAC comprises a complex network of multiple cell types communicating through the autocrine and paracrine production of cytokines, chemokines, and other factors to control the growth, survival, and progression of mutated epithelial cells. This network controls tumorigenesis and progression, both intrinsically and extrinsically. Intrinsically, cytokines like interleukin (IL)-6 and tumor necrosis factor-α, promote tumor cell proliferation and survival by activating transcription factors such as signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappa B (NFĸB).4 Extrinsically, chemokines and cytokines also regulate the recruitment, activation, and tumor-promoting/suppressing activities of immune and stromal cells, which promote tumor growth through tissue remodeling and angiogenesis.5 However, not all chronic inflammatory diseases increase cancer risk, and some, such as psoriasis, may even reduce cancer risk.5,6 It is not clear how inflammatory factors drive tumorigenesis in the colon.5
Immune cells have complex, heterogeneous, and, in some cases, counteracting functions in the colonic tumor microenvironment. The most abundant immune cells within the tumor microenvironment are the tumor-associated macrophages (TAMs) and T cells.5 In recent years, increasing attention has focused on TAMs. Macrophages (Mɸs) are hematopoietic cells that can undergo a spectrum of activation states respond to different stimuli. These can be typified as M1 (induced by interferon [IFN]γ and lipopolysaccharide [LPS]) and M2 (induced by IL-4, IL-10, and IL-13), often exploited in in vitro experiments. However, these extreme Mɸ phenotypes do not fully recapitulate the plasticity and range of differentiation states of TAMs in vivo, where Mɸs are polarized to diverse and more complex phenotypes.7 Indeed, tumor cells can manipulate TAMs’ polarization by releasing cytokines, chemokines, and extracellular matrix components that give rise to a large range of pro-tumoral macrophages.7 During tumor evolution, most TAMs are considered to have an M2 phenotype that promotes tumor angiogenesis,8,9 and facilitates tumor cell invasion and metastasis formation.8,10 However, most confirmed tumor-promoting cytokines are M1 cytokines such as IL-6.11 Like Mɸs, T cells can exert both tumor-suppressive and tumor-promoting effects, depending on their differentiation state and effector functions.12 Increased T cell numbers in the tumor microenvironment, specifically activated CD8+ cytotoxic T cells and CD4+ helper T 1 cells, correlate with better survival in invasive colon cancer.5,13 In addition, in experimental models, cytotoxic T cells have been shown to protect against CAC tumorigenesis.14
Genome-wide association studies have vastly increased our knowledge of inflammatory bowel disease (IBD) genetics. MUC1 is a cell surface mucin present on intestinal epithelial cells and some leukocytes. In health, MUC1 is expressed at relatively low levels on the apical surface of ductal epithelial cells in the colonic epithelium that provide protection against toxic substances and harmful infection, playing a protective role in maintaining intestinal integrity.15,16 Repeated injury and regeneration of intestinal epithelial cells caused by chronic inflammation or other factors can lead to overexpression of MUC1.16,17
MUC1 polymorphisms have been linked to Crohn's disease18 and are associated with developing gastritis and gastric cancer following H. pylori infection.19 Aberrant expression of cell surface mucins occurs in many cancers and is linked to the initiation, progression, and poor prognosis of multiple types of adenocarcinoma.20,21 High MUC1 expression represented a marker of poor prognosis in CRC and was associated with advanced TNM stage, greater depth of invasion, and lymph node metastasis.22 However, few studies have explored a functional role for MUC1 in driving CAC. When assessed using the dextran sulfate sodium (DSS) model of chemically induced colitis, Muc1-/- mice are protected from inflammation as they demonstrate a thickened mucus layer with fewer infiltrating T cells.23 In contrast, Muc1-/- mice have exacerbated chronic inflammation in both Th1- and Th2- mediated colitis models.24 These diverse results highlight the complexity of MUC1 physiology in IBD.
Recent studies suggest that MUC1 expression is upregulated in patients with both UC and CRC and that targeting the MUC1 cytoplasmic domain attenuates inflammation in human MUC1-transgenic mouse models of CAC.25,26 These observations raise the intriguing possibility that blockade of MUC1 function can dampen the ongoing mucosal inflammation in UC, and reduce the risk of CAC. However, a single study conducted in wild-type (WT) mice transplanted with Muc1-/- bone marrow showed that MUC1 inhibited rather than promoted the formation and growth of tumors, which was attributed to less infiltrating CD11b+ GR+ myeloid-derived suppressive cells following exposure to carcinogens and inflammation,27 suggesting an anti-inflammatory/anti-tumorigenic role for MUC1. In contrast, human MUC1-transgenic mice have activated inflammatory pathways, enhanced colitis severity, and more aggressive CACs.26,28 An improved understanding of the role of MUC1 in inflammation and especially in the microenvironment of CAC will provide novel opportunities for prevention and therapeutic intervention. Therefore, the current study focused on providing definitive evidence of the role of MUC1 in regulating the tumor microenvironment in CAC and the underlying cellular and molecular mechanisms of MUC1 action.
We found that Muc1-/- mice are protected against DSS-induced colitis and azoxymethane (AOM)-DSS induced CAC. Somewhat unexpectedly, we found that promotion of CAC is primarily mediated by Muc1-expressing bone marrow-derived cells, with MUC1 promoting infiltration and activation of Mɸs. Mechanistic studies revealed that Muc1 deficiency remarkably reduced proinflammatory cytokine IL-6 levels in the colonic tissues and tumors that was mainly produced by infiltrating macrophages. Greater infiltration of macrophages and increased levels of IL-6 were also observed in the colonic mucosa of WT mice at days 21 and 42 in the early stages of CAC before tumors arise. MUC1-dependant IL-6 production elicited STAT3 activation in neoplastic cells, further promoting tumor growth. Together, our results highlight a pro-neoplastic role for leukocyte Muc1 in regulating Mɸ accumulation and activation. Therefore, MUC1 represents a promising target for therapeutic intervention.
Results
Loss of Muc1 Protects Against Colitis-associated Tumorigenesis and Progression
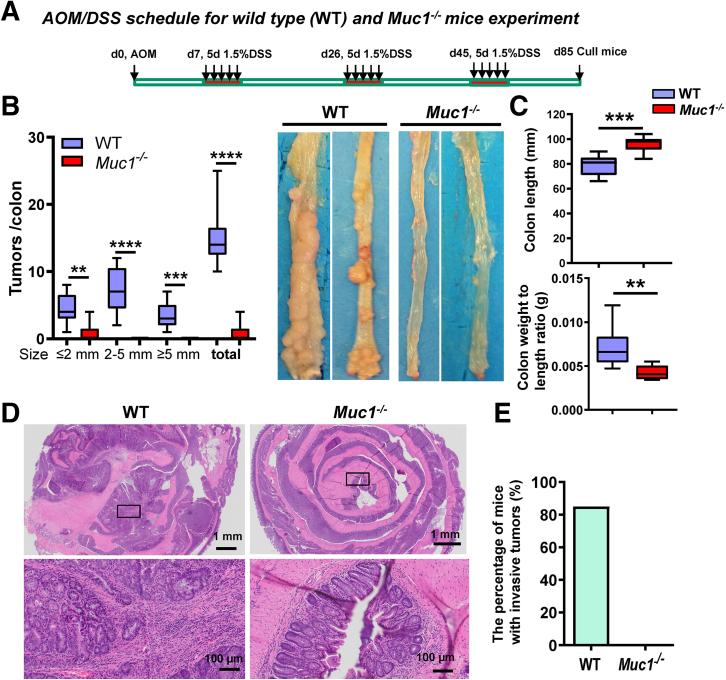
To ascertain the roles of MUC1 in inflammation-associated tumorigenesis, we used an established mouse model of CAC induced by the carcinogen AOM followed by 3 cycles of DSS (Figure 1, A).29, 30, 31 Muc1-/- mice developed significantly fewer and smaller tumors in the colon compared with WT mice. Macroscopically visible tumors formed in only 5 of 13 Muc1-/- mice (0.85 ± 0.37 tumors per colon) compared with 13 of 13 WT mice (14.85 ± 1.08 tumors per colon) (Figure 1, B), and these mice had lower colon weight and longer colon length than WT mice (Figure 1, C). No submucosal invasion was observed in tumors of the Muc1-/- mice, whereas 85% of the WT mice had invasive tumors (WT vs Muc1-/-, 85% vs 0%) (Figure 1, D and E). indicating that, in the absence of Muc1, tumors arise less frequently and do not progress to invasive tumors. These data underline a role of MUC1 not only in colitis-associated tumor initiation but also in the progression of dysplasia and invasive behavior of tumors.
Figure 1.
MUC1 promotes development and progression of CAC. (A) Scheme showing the induction procedure for the AOM/DSS model of CAC. At sacrifice on day 85: (B) Number of colon tumors (left panel, n ≥ 13); and representative macroscopic images of the colonic mucosa (right panel). (C) Colon length and weight (n ≥ 13). (D) Representative H&E–stained sections of colonic tissue. (E) The percentage of mice with submucosal invasive tumors in the WT and Muc1-/- mice (n ≥ 13). Statistics: box plots show median, quartiles, and range (B–C); nonparametric Mann-Whitney U test: n ≥ 13; ∗Muc1-/- vs WT; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. The data are representative of 3 independent experiments.
Muc1 Deficiency Reduces the Innate Immune Response and Colitis Severity Following DSS Treatment
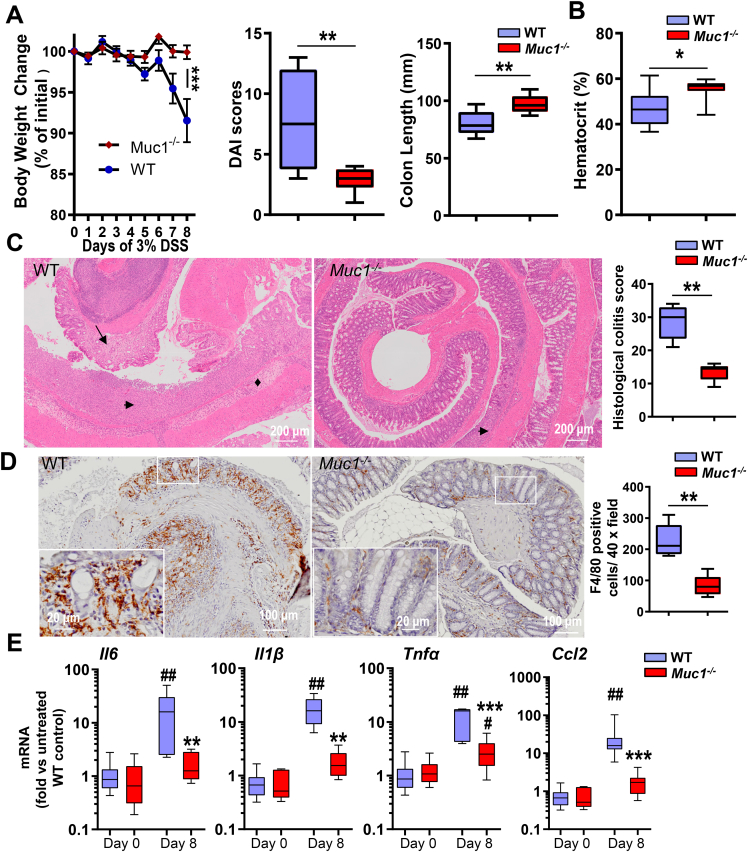
Given the above, we further investigated and characterized the development of DSS-induced colitis in Muc1-/- mice. With an acute 8-day exposure to DSS, Muc1-/- mice exhibited less severe colitis than WT mice, based on clinical signs and histopathological examination (Figure 2). Weight loss and daily activity index (DAI) scores (DAI score of rectal bleeding, diarrhea, body weight change) were significant lower in DSS-treated Muc1-/- mice when compared with WT mice (Figure 2, A). Furthermore, WT mice had a significantly shorter colon length and lower hematocrit when compared with Muc1-/- mice after 8 days of DSS treatment (Figure 2, A–B). This suggests that Muc1-/- mice have reduced DSS-induced colonic damage and intestinal bleeding. Hematoxylin and eosin (H&E) staining of WT colons showed the entire colon was severely inflamed with extensive crypt destruction and ulceration and disappearance of goblet cells, together with mucosal edema, whereas Muc1-/- mice had significantly less disturbed morphology (Figure 2, C).
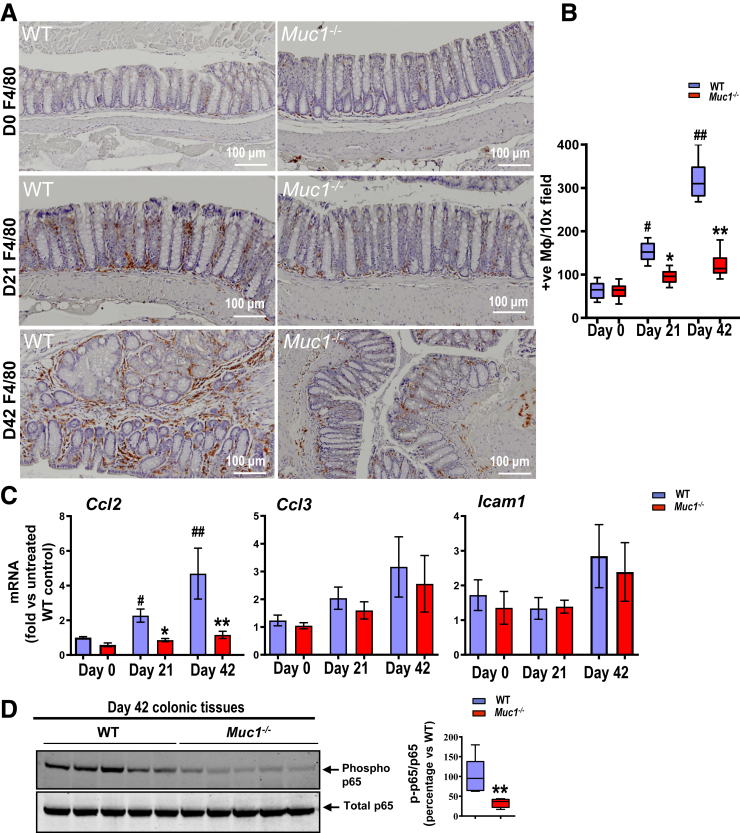
Figure 2.
Muc1-deficient mice are protected against DSS-induced colitis. (A–E) Assessments of WT and Muc1-/- mice given 3% DSS in drinking water for 8 days. (A) Percent body weight loss, the total clinical DAI (combined scores of rectal bleeding, diarrhea, and loss of body weight), and colon length (n ≥ 8). (B) Hematocrit (n ≥ 8). (C) H&E-stained colon sections from representative WT and Muc1-/- mice treated with DSS (left panels); scale bars, 200 μm. Histological colitis scores grading severity of inflammation and tissue damage shown on right (n ≥ 8). Crypt destruction and ulceration (arrow heads), disappearance of goblet cells (arrow), and mucosal edema (diamonds) are highlighted. (D) Representative sections showing F4/80-stained macrophages in the colon of WT and Muc1-/- mice exposed to DSS (left panels); scale bars are shown. Quantitative assessment of macrophage density in the colon shown on the right (n ≥ 8). (E) Colonic mRNA expression of genes encoding inflammatory cytokines and chemokines (n ≥ 8). Statistics: mean ± standard error of the mean (A), box plots show median, quartiles, and range (A–E); Mann-Whitney U test, # vs WT Day 0, ∗ vs WT Day 8, ∗∗ or ## P < .01. ∗∗∗P < .001. The data are from 1 of 3 independent experiments.
Quantification of immunohistochemical (IHC) staining for F4/80 showed significantly lower number of Mɸs infiltrating the colons of Muc1-/- mice compared with WT mice (Figure 2, D). In line with these observations, mRNA expression of inflammatory cytokines (Il1β, Il6 and Tnfα) and the chemokine Ccl2 were also significantly lower in the DSS-treated colons of the Muc1-/- mice compared with WT mice (Figure 2, E).
Hematopoietic Muc1 is Primarily Responsible for CAC Promotion
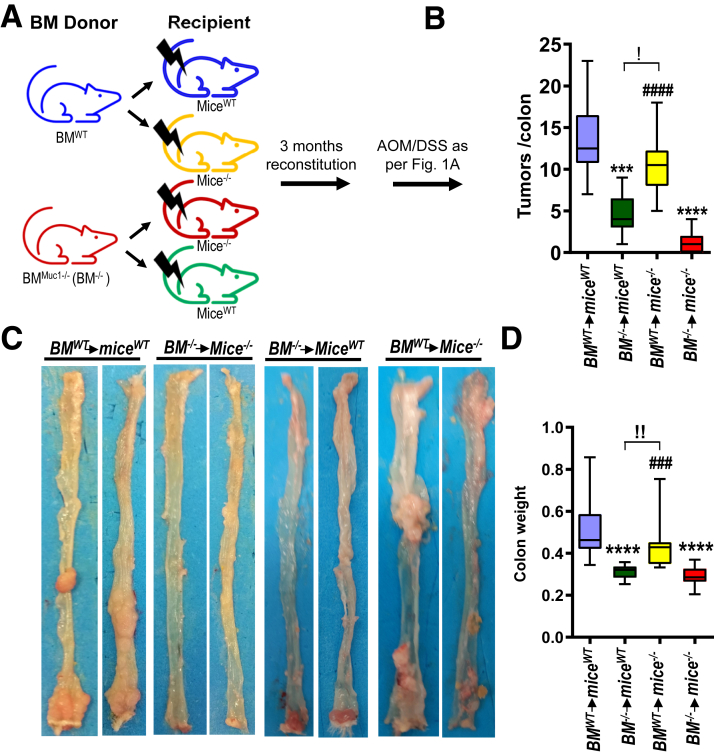
Considering that MUC1 is expressed in both colonic epithelial and immune cells, we examined the contribution of each compartment in promoting CAC using bone marrow (BM) chimeras (Figure 3, A). As expected, WT mice (miceWT) receiving WT BM (BMWT) developed more tumors than Muc1-/- mice (mice-/-) receiving Muc1-/- BM (BM-/-) (Figure 3, B–C). Muc1-/- mice transplanted with WT-derived BM cells (BMWTmice-/-) had significantly higher tumor burden and heavier colon weight than that in either WT or Muc1-/- mice transplanted with Muc1-/--derived BM cells (Figure 3, B–D). These observations indicate that Muc1 expression in hematopoietic cells play a major role in promoting CAC.
Figure 3.
Promotion of inflammation-associated colon cancer by MUC1 is mediated primarily by hematopoietic cells. Four groups of BM chimeric mice were generated as shown; 3 months following BM transplantation, mice were treated with AOM followed by DSS as per Figure 1, A and sampled after 85 days. (A) Scheme of treatment. (B) Colon tumor counts. (C) Representative macroscopic images of colons. (D) Colon weight. Statistics: box plots show median, quartiles, and range; Mann-Whitney U test, n ≥ 13; ∗ vs BMWTmiceWT, # vs BM-/-mice-/-, ! vs BM-/-miceWT, !P < .05; !!P < .01; ∗∗∗or ###P < .001; ∗∗∗∗ or ####P < .0001. The data are representative of 2 independent experiments.
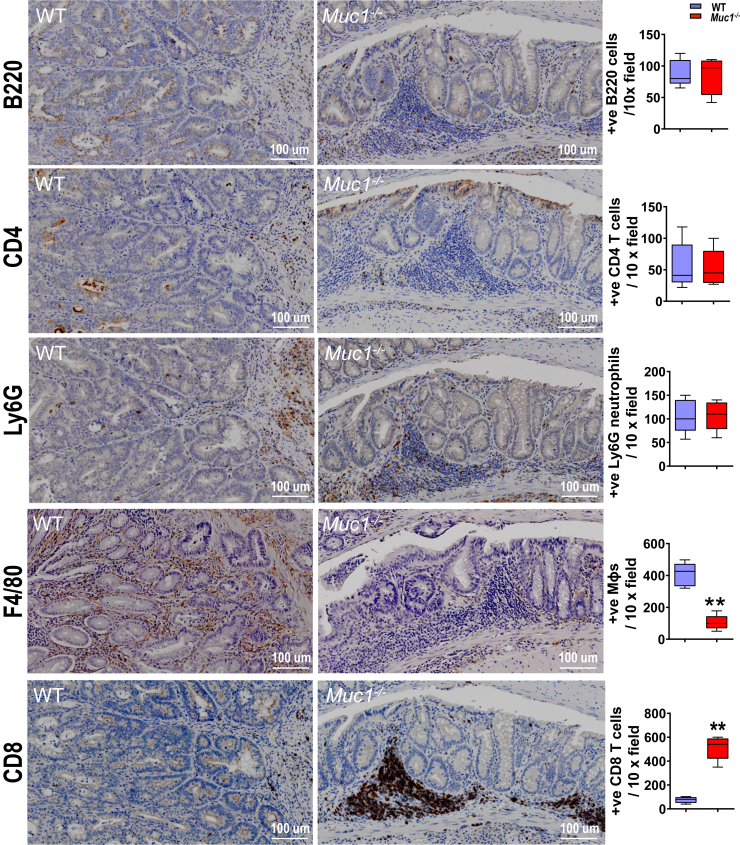
Muc1 Deficiency is Associated With Decreased Recruitment of Macrophages and Increased Abundance of CD8+ T Cells into the Tumor-containing Tissue
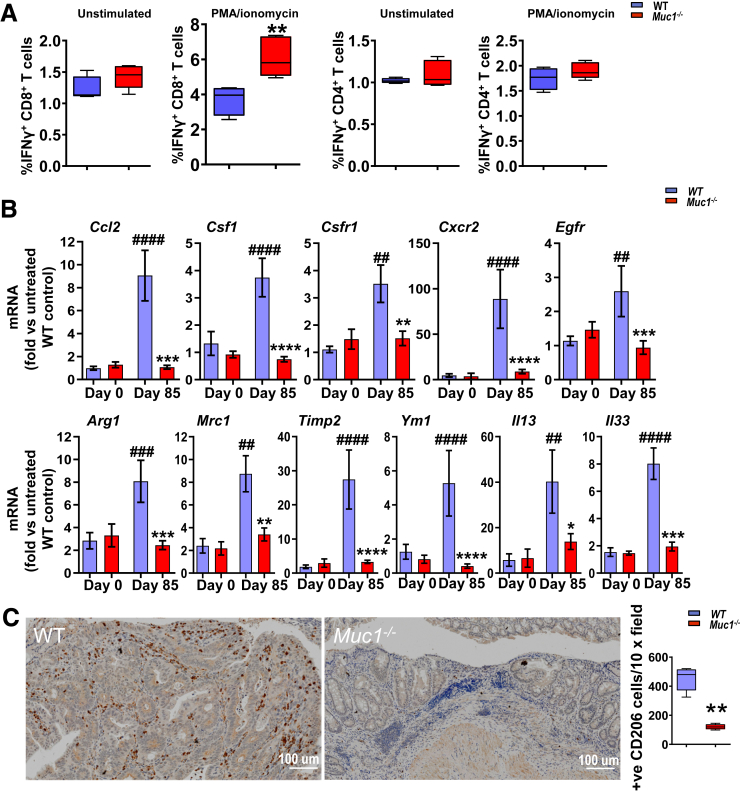
To explore the hematopoietic basis for the tumor-promoting role of Muc1, we investigated the recruitment of specific leukocyte subsets to tumor stroma in WT and Muc1-/- mice. IHC staining was used to measure tumor infiltration of Mɸs (F4/80), neutrophils (Ly6G), B cells (B220), CD4+ T cells (CD4), and CD8α+ T cells (CD8). Although the abundance of CD4+ T cells, B cells or neutrophils did not significantly differ between Muc1-/- and WT mice at day 85 in AOM/DSS-treated tumor-containing tissues (Figure 4), we detected a marked reduction (∼4-fold) in Mɸs and increased (∼5-fold) CD8+ T cells in colonic tissues of Muc1-/- mice compared with WT mice (Figure 4). Importantly, cytotoxic CD8+ T cells isolated from mesenteric lymph nodes of Muc1-/- mice produced more IFNγ when compared with CD8+ T cells from mesenteric lymph nodes of WT mice (Figure 5, A), consistent with greater potential antitumor immunity.
Figure 4.
Reduced infiltration of Mɸs and increased number of CD8+T cells in the colons of Muc1-/-mice treated with AOM/DSS. Mice were treated with AOM followed by DSS as per Figure 1, A and sampled after 85 days. (A) Representative IHC pictures of colonic sections take from WT and Muc1-/- mice on day 85 and stained with B220, CD4, Ly6G, F4/80, and CD8 on the left, and quantification of positive cells counts in colon section on the right (n ≥ 8). Statistics: box plots show median, quartiles, and range; Mann-Whitney U test; ∗vs WT day 85, ∗∗P < .01. The data are representative of 3 independent experiments.
Figure 5.
Muc1 deficiency in CAC correlates with increased activation of CD8+T cells in tumor-draining lymph nodes and a reduced gene signature from alternatively activated Mɸs in tumors. Mice were treated with AOM followed by DSS as per Figure 1, A and sampled after 85 days. (A) Quantification of IFNγ+ CD4 or CD8 T cell of mesenteric node lymphocytes with AOM/DSS treatment based on flow cytometry (n ≥ 8). (B) Real-time quantitative PCR gene expression analysis of alternative Mɸs markers from WT and Muc1-/- tumors on day 85 of the CAC model (n ≥ 13). (C) Representative micrographs of colonic sections from WT and Muc1-/- mice on day 85 and stained with CD206 by IHC on the left, and quantification of positive cells counts in colon section on the right (n ≥ 8). Statistics: box plots show median, quartiles, and range (A and C), mean ± standard error of the mean (B); Mann-Whitney U test, # vs WT day 0, ∗vs WT day 85, ∗P < .05; ∗∗or ##P < .01; ∗∗∗ or ###P < .001; ∗∗∗∗ or ####P < .0001. The data are representative of 3 independent experiments.
Gene expression analysis showed reduced inflammatory and M2 Mɸ gene signatures (Timp2, Ym1) in tumors of Muc1-/- mice (Figure 5, B). This finding, accompanied by the reduction in CD206+ Mɸ (Figure 5C) in Muc1-/- tumors, suggests that loss of Muc1 reduces the accumulation of alternatively activated macrophages into tumors. Given that we did not detect any difference in gene expression in the colons of healthy WT and Muc1-/- mice (Figure 5, B), the recruitment and activation of Mɸ in response to signals from the inflammatory tumor microenvironment appears substantially influenced by MUC1 on hematopoietic cells.
Loss of Muc1 Reduces Macrophage Infiltration via CCL2 in the Early Stages of CAC Development
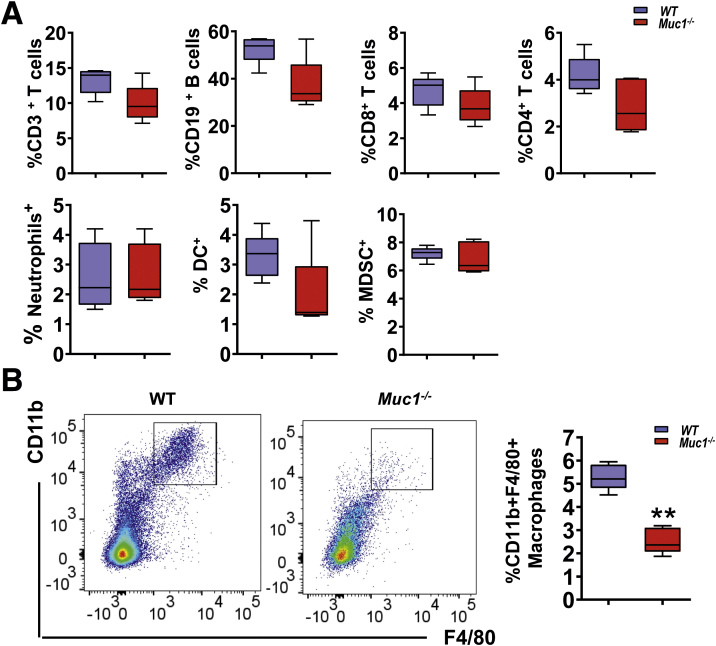
Chronic inflammation contributes to tumor initiation in CAC, and infiltration of immune cells, specifically myeloid cells, plays a central role in tumorigenesis.5,11 To determine if Muc1 is playing a role in immune cell infiltration, we assessed an early stage of CAC development prior to the development of visual tumors. To this end, immune cell profiles in the colonic lamina propria at day 42 of AOM/DSS treatment (a time just before tumors develop) were analyzed by flow cytometry (Figure 6). No differences in the abundance of CD3+, CD4+, or CD8+ T lymphocytes, B lymphocytes, neutrophils, dendritic cells, or myeloid-derived suppressor cells were detected (Figure 7, A). However, Muc1-/- mice had significantly reduced numbers of Mɸs (CD11b+F4/80+) compared with WT mice (Figure 7, B). Colonic tissue staining confirmed a reduction in Mɸ abundance at day 21 and 42 of AOM/DSS treatment compared with WT mice (Figure 8, A–B). To further investigate this, gene expression of chemokines Ccl2, Ccl3, and the adhesion molecule Icam1 were assessed. Increasing Ccl2 mRNA expression was evident at day 21 and 42 in WT mice; however, this was not evident in the Muc1-/- mice (Figure 8, C). No differences were observed in the expression of Ccl3 or Icam1 between WT and Muc1- deficient colonic tissues at day 21 and 42 (Figure 8, C). CCL 2 plays an important role in recruitment of monocytes/Mɸs into tumors and their subsequent polarization TAMs into a tumor-permissive tissue microenvironment.32,33
Figure 6.
Flow cytometry plots showing the gating strategy used to identify T cells, B cells, Mɸs, myeloid-derived suppressive cells, and dendritic cells in lamina propria mononuclear cell preparations.
Figure 7.
Leukocyte infiltration in the colon during the pre-malignant stages of CAC. Lamina propria mononuclear cells (LPMCs) were prepared by enzymatic digestion from AOM/DSS treated mice at day 42 and analyzed by flow cytometry. (A) The abundance of T cells, B cells, myeloid-derived suppressive cells, dendritic cells, and neutrophils in lived LPMC of WT and Muc1-/- at day 42 (n ≥ 6). (B) Representative plots for CD11b vs F4/80 within a Ly6G-gate for Mɸs in LPMCs. Right, percentage of CD11b+F4/80+Ly6G- Mɸs from LPMCs (n ≥ 6). Statistics: box plots show median, quartiles and range; Mann-Whitney U test, n ≥ 6. ∗ vs WT, ∗∗P < .01. The data are representative of 3 independent experiments.
Figure 8.
MUC1 influences colonic Mɸ infiltration via CCL2 production in premalignant stages of CAC. (A) Representative IHC staining for F4/80 at day 0, 21, and 42 of AOM/DSS- treated mice. (B) Quantification of Mɸ based on F4/80 staining positive cells count per 10× field in colon tissues at day 0, 21, and 42 of AOM/DSS-treated mice (n ≥ 6). (C) Relative expression of Ccl2, Ccl3, and Icam1 normalized to β-actin from WT and Muc1-/- colon tissue at day 0, 21, and42 of AOM/DSS (n ≥ 6). (D) Immunoblot analysis of phosphorylated and total NFĸBp65. Each lane represents colonic tissue from an individual mouse (n = 5). Densitometry analysis of level of proteins corrected for total NFĸB is shown. Statistics: box plots show median, quartiles and range (B and D), mean ± standard error of the mean (C); Mann-Whitney U test, # vs WT at day 0, ∗ vs WT at same day, ∗ or #P < .05, ∗∗ or ##P < .01. The data are representative of 3 independent experiments.
CCL2 can be secreted by tumor, stromal, and immune cells and is regulated by NF-ĸB.34 To study whether loss of Muc1 alters NFĸB activity during CAC, colon tissues were assessed by Western blot. We found lower levels of phosphorylated NFĸB p65 protein in colonic tissues of Muc1-/- mice compared with WT mice at day 42 and day 85 (Figure 8, D; Figure 9, A).
Figure 9.
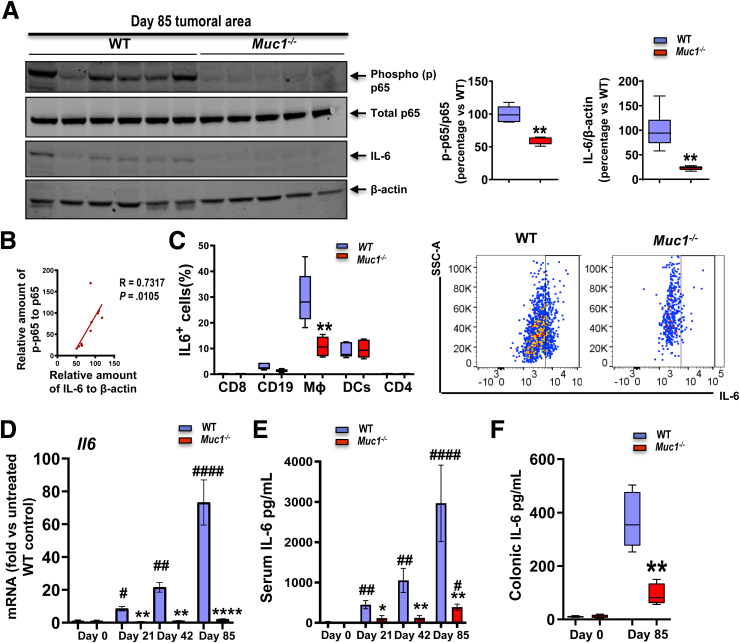
Reduced activation of NFĸB and induction of IL-6 in the tumors of Muc1-/-mice treated with AOM/DSS. Mice were treated with AOM followed by DSS as per Figure 1, A and sampled after 85 days. (A) Immunoblot analysis of NFĸB and its downstream tumor promoting gene IL-6. Each lane represents a pool of all tumors from an individual mouse (n ≥ 5). Densitometry analysis of level of proteins corrected for β-actin is shown. (B) Correlation plot of p-p65 and IL-6 in the tissues from (A). (C) Left, Quantification of IL6+ CD8, CD4, CD19, Mɸs, and dendritic cells of lamina propria mononuclear cells (LPMCs) at Day 42 AOM/DSS treatment based on flow cytometry (n ≥ 6). Right, Representative plots for IL-6 within a CD11b+F4/80+Ly6G-gate for Mɸs in LPMCs. (D) Real-time quantitative PCR analysis of Il6 expression from WT and Muc1-/- colon tissues on days 0, 21, 42, and 85 of the CAC model (n ≥ 6). (E) Concentration of IL-6 in serum of WT and Muc1-/- mice on day 0, 21, 42, and 85 (n ≥ 6). (F) Concentration of IL-6 in colonic tissue (50 mg tissue/mL assayed) of WT and Muc1-/- mice on day 0 and day 85 (n ≥ 6). Statistics: box plots show median, quartiles and range (A, C, and F), mean ± standard error of the mean (D and E); Mann-Whitney U test, # vs WT at day 0, ∗ vs WT at same time point, ∗ or #P < .05; ∗∗ or ##P < .01; ∗∗∗∗ or ####P < .0001. The data are representative of 3 independent experiments.
MUC1 Promotes Colorectal Tumorigenesis via Enhanced NFĸB and IL-6/STAT3 Activation
NF-κB also regulates the production of the pleiotropic cytokine IL-6, which is often elevated in CAC. Analysis of IL-6 protein by Western blotting demonstrated greater IL-6 in WT colonic tissues that correlated strongly with NFĸBp65 activation (Figure 9, A–B), further strengthening the potential functional link between these proteins in vivo. By using flow cytometry to characterize the cellular source of IL-6 in leukocytes from WT and Muc1-/- mice, we showed that Mɸs were not only the major producers of IL-6, but also a higher percentage of Mɸs from colonic tissue of WT mice expressed IL-6 (median, 28%) than from Muc1-/- mice (median, 12%) at day 42 of AOM/DSS treatment (Figure 9, C). Although a smaller proportion of IL-6 was produced in dendritic cells and B cells, there was no difference in IL-6 production between WT and Muc1-/- mice in these cells (Figure 9, C).
Because IL-6 plays a crucial role in colon tumorigenesis,35 we measured IL-6 at early, mid, and late timepoints in colonic tissue and serum. Increasing Il6 mRNA expression was evident over the course of tumor development in WT mice. Strikingly however, this was not evident in the Muc1-/- mice (Figure 9, D). This also correlated with increasing IL-6 protein in serum over time in the WT mice vs much lower increases of protein over time in Muc1-/- mice (Figure 9, E). Enzyme-linked immunoassay measurements of IL-6 protein in colonic tissue at D85 were consistent with lower Il6 mRNA levels in Muc1-/- tumors after AOM/DSS compared with WT tumors (Figure 9, F).
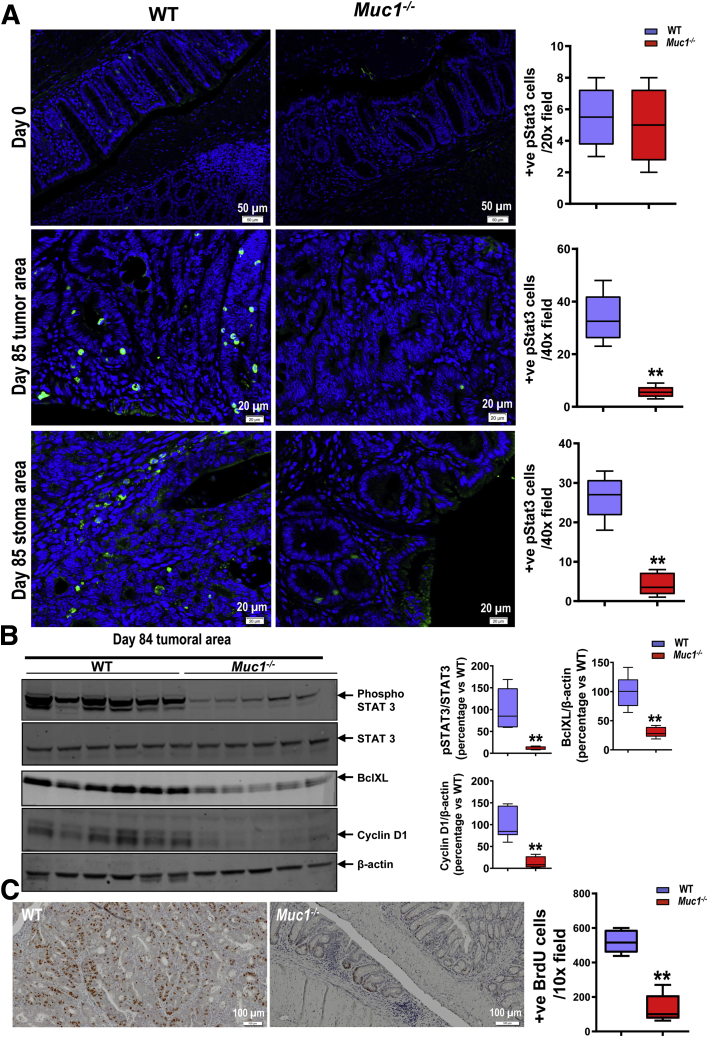
IL-6 activity is mediated through STAT3,36 a critical modulator of chronic inflammation and malignant progression.37 Intriguingly, MUC1 has also been shown to activate STAT3 in human breast38 and lung cancer.39 Therefore, we quantified the amount of phosphorylated STAT3 at a late stage of tumor development by immunofluorescence. Fewer phospho-STAT3 positive cells were observed in Muc1-/- colons compared with WT mice (Figure 10, A). Cells exhibiting strong nuclear staining for p-STAT3 were seen in tumors and the adjacent epithelium and stroma in WT colons (Figure 10, A), but these were mostly absent in the Muc1-/- tissue. We confirmed reduced levels of activated STAT3 (phospho-STAT3) in Muc1-/- colon tissue by Western blotting (Figure 10, B). Finally, we assessed target protein activity downstream of STAT3 and found lower levels of cyclin D1 (driver of cell cycle G1/S-phase transition) and BclXL (critical anti-apoptotic gene in colon cancer40) in Muc1-/- tumor tissue than WT tumor tissue (Figure 10, B). Consistent with this finding, less BrdU was incorporated into Muc1-/- tumors (Figure 10, C), demonstrating their reduced proliferative activity. Taken together, these results suggest that loss of Muc1 inhibits tumor cell proliferation through STAT3 and its downstream targets.
Figure 10.
Reduced STAT3 activation in Muc1-/-mice. Mice were treated with AOM followed by DSS as per Figure 1, A and sampled after 85 days. (A) Representative immunofluorescence pictures of colonic sections take from WT and Muc1-/- mice on day 0 and 85 (n ≥ 6). (B) Immunoblot analysis of STAT3 activation and its downstream genes in the tumor as indicated. Each lane represents a pool of all tumors from an individual mouse (n ≥ 5). (C) IHC of BrdU staining in colon sections at day 85 of AOM/DSS (n ≥ 8). Statistics: box plots show median, quartiles and range; Mann-Whitney U test, ∗ vs WT, ∗∗P < .01. The data are representative of 3 independent experiments.
MUC1 Skews Macrophages to a Pro-tumoral Phenotype
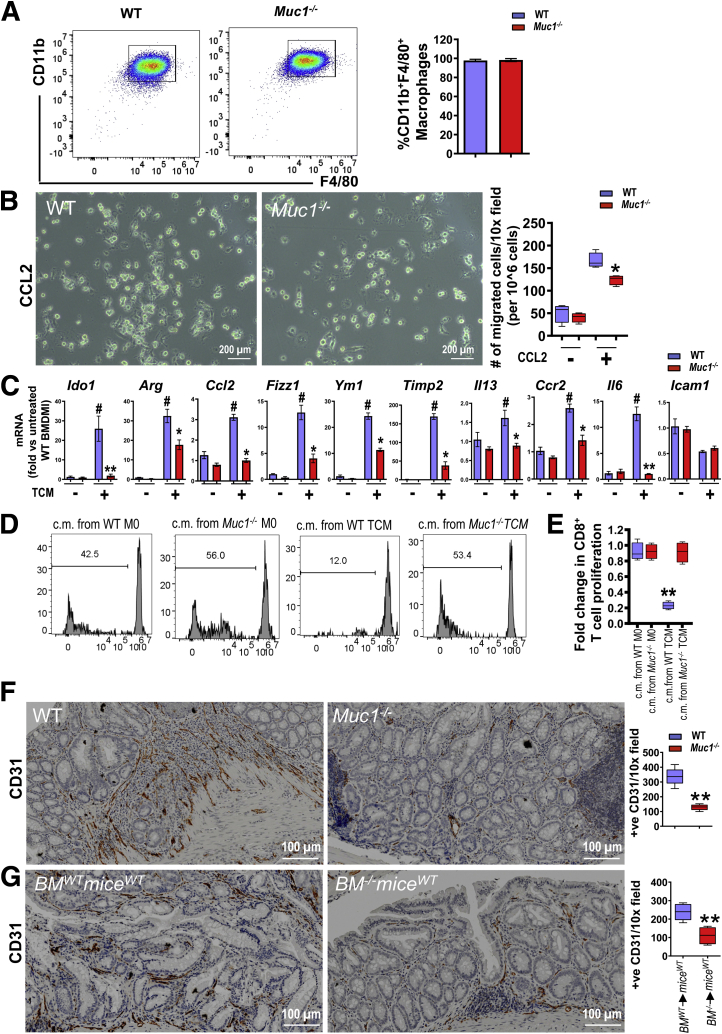
We next determined whether MUC1 intrinsically regulates macrophage recruitment or acts through other leukocytes to impact recruitment. BM-derived macrophage (BMDM) cultures were generated from WT and Muc1-/- mice. BM progenitors differentiated into Mɸ with comparable efficiency in both strains (Figure 11, A). The migration of BMDM towards CCL2 was assessed. Muc1-/- BMDM showed a significant 2-fold reduction in cell migration in response to CCL2 compared with WT BMDM (Figure 11, B).
Figure 11.
MUC1 specifies a pro-tumoral phenotype of Mɸs. (A) BM-derived cells differentiated into Mɸ (BMDM) with comparable efficiency in both the WT and Muc1-/- after stimulation with MCSF for 7 days, with ∼98% CD11b+F4/80+ cells (n = 5). (B) 106 BMDM were seeded on the upper layer of an 8-μm transwell chamber and medium with or without CCL2 at 100 ng/mL was placed below the chamber. After 24-hour incubation, migrated cells were counted (n = 5). (C) mRNA expression of indicated genes in WT and Muc1-/- BMDM cultured in the presence or absence of colon TCM (n = 5). D–E, Flow cytometry analysis (D) and quantification (E) showing carboxy fluorescein succinimidyl ester (CFSE) proliferation assays performed on CD8+ T cells isolated from mouse spleens after exposing to Mɸs conditioned media (c.m.) (n = 5). Mɸs were polarized with stimuli (TCM) or without stimuli (M0) for 48 hours, then media was washed out and replaced. c.m. for the experiment was collected after 24 hours. (F) CD31 IHC staining of colon sections from WT and Muc1-/- mice on day 85 (n ≥ 8). (G) CD31 IHC staining of colon sections from BMWTmiceWT and BM-/-miceWT mice on day 85 as described in Figure 3 (n ≥ 8). Statistics: mean ± standard error of the mean (A and C) box plots show median, quartiles and range (B, E–G); Mann-Whitney U test, P value is shown or # vs WT without TCM ∗ vs WT with TCM (C and E), ∗ vs WT (F and G), ∗ or #P < .05; ∗∗P < .01; ∗∗∗P < .001. The data are representative of 3 independent experiments except for (G).
Because we observed a reduction in macrophage-associate gene expression in tumor tissue in the Muc1-/- mice, we next investigated whether loss of Muc1 altered BMDM responses to tumor-conditioned medium (TCM) generated from the MC38 murine colon adenocarcinoma cell line. In WT BMDM, TCM induced expression of a complex set of genes, including pro-inflammatory Il6 and immunosuppressive Arg1 and Ido1 (Figure 11, C), supporting the notion that the tumor microenvironment induces a complex response from BMDM. In contrast, the response of the Muc1-/- BMDM to the TCM was attenuated; most notably, the upregulation of Ido1 and Il6 was completely abrogated (Figure 11, C). IDO1 is a potent enzyme working in dendritic cells and macrophages to limit the extracellular availability of tryptophan, essential for T cell proliferation.41,42 Blocking IDO1 activity increases T cell proliferation, survival, and function.43,44 We hypothesized that the low level of Ido1 expression in Muc1-/- BMDM in response to TCM would translate to increased T cell proliferation. To test this, we generated conditioned media by incubating BMDM with TCM for 48 hours followed by 24 hours rest. The conditioned media was added to CD8+ T cells, and their proliferation was assessed in a suppression assay (after 5 days). As expected, TCM-exposed WT BMDM-conditioned media reduced CD8+ T cell proliferation. In contrast, TCM-exposed Muc1-/- BMDM-conditioned media induced as much proliferation as conditioned media from unstimulated BMDM (Figure 11, D–E). Taken together, these experiments show that MUC1 promotes macrophage recruitment and regulates functional responses relevant to the tumor microenvironment.
High numbers of TAMs are positively associated with tumor angiogenesis and facilitate cell invasion and metastasis.45 As we have observed a reduced number of Mɸ in the tumors of Muc1-/- mice, we assessed the vascularization of the tumors in our model. The endothelial marker CD31 was significantly reduced in tumors in Muc1-/- and in the bone marrow-specific Muc1-deficient mice tumors compared with their corresponding controls (Figure 11, F), suggesting a role for MUC1 in hematopoietic cells in the tumor microenvironment that promotes tumor angiogenesis.
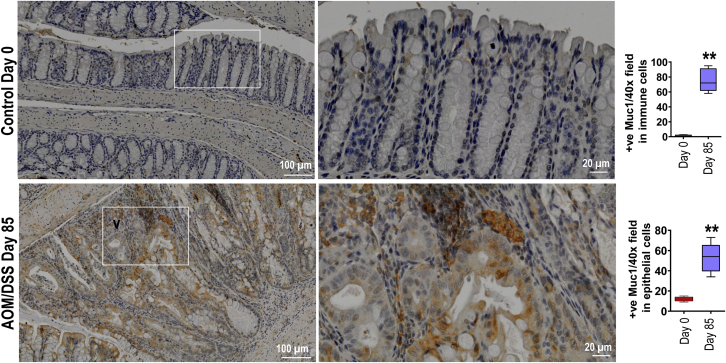
We also compared MUC1 expression in our AOM/DSS CAC model and observed that MUC1 is significantly upregulated in both epithelial and immune cells by IHC than untreated control (Figure 12).
Figure 12.
MUC1 is upregulated in both tumor and immune cells in repose to AOM/DSS. MUC1 IHC staining colon section from WT mice ether treated with AOM followed by DSS as per Figure 1, A at day 85 or untreated at day 0 (n ≥ 6). Statistics: box plots show median, quartiles and range. Mann-Whitney U test. ∗ vs Day 0, ∗∗P < .01. The data are representative of 3 independent experiments.
Discussion
Taken together, our findings support the notion that MUC1 promotes colonic epithelial tumor development and that MUC1 in immune cells rather than epithelial cells is predominantly responsible. Although overexpression of MUC1 in epithelial tumor cells has been shown to be crucial to the transformation process, MUC1’s role in immune cells during cancer development and its role in modulating interaction between tumor cells and the immune system has not been well-appreciated. We showed that MUC1 functions as a neoplastic factor that regulates Mɸ infiltration and function, and fosters a tumor-permissive microenvironment that influences tumor initiation and progression to invasive carcinoma in the AOM/DSS animal model of CAC.46 AOM treatment followed by repeated cycles of DSS resulted in chronic inflammation and the development of colon tumors in 100% of the treated WT mice. In the absence of Muc1, colonic inflammation was reduced, as was the tumor incidence, size, and invasiveness. Our experiments reveal multiple underlying mechanisms by which MUC1 promotes cancer development, demonstrating that MUC1 has a major role in immune cell signalling, thus promoting chronic inflammation-associated cancer development. In addition to previous reports identifying MUC1 as an epithelial cancer cell-intrinsic driver of aggressive behavior in colorectal, gastric, and non-small cell lung cancer,47 the current finding demonstrates that MUC1 in innate immune cells drives cancer progression, at least in CRC, adding to its promise as a therapeutic target.
MUC1 is a cell surface mucin that in healthy physiology promotes healing from acute injury by promoting cellular proliferation and survival. However, MUC1’s role in the development of cancer during chronic inflammation is not thoroughly studied, and the limited data to date has delivered some conflicting results. By using models of CAC in mice overexpressing transgenic human MUC1, MUC1 was shown to drive progression of colitis to CRC with the mechanism ascribed to inducing the expansion of intestinal stem cells26 and epithelial NFĸB activation.28 However, these studies did not investigate or consider the effect of MUC1 on the immune cells with regards to inflammatory signaling and contribution to CAC development. There is a previous study that examined the role of MUC1 in immune cells on CAC and that suggested that Muc1 expression on hematopoietic cells decreased tumor formation in the AOM/DSS model, while at the same time, increased colitis severity by inhibiting expansion of myeloid-derived suppressor cells.27 These results are completely opposite to our findings, which demonstrated that Muc1 expression on hematopoietic cells plays a major role in driving the progression of colitis to CRC. The opposing results are difficult to reconcile as both studies utilize a similar AOM/DSS protocol. However, while we used WT and Muc1-/- mice on a clean C57BL/6 background, the background of Muc1-/- mice was not made clear, and additionally, only irradiated mice were used in the previous study.
Our results indicate that one of the mechanisms driving MUC1-dependent promotion of colitis and CAC is the ability of MUC1 to stimulate Mɸ infiltration into the inflamed colonic tissues both before and after development of tumors. These data suggest that MUC1 may sustain an inflammatory circuit that promotes the growth of colon cancer during inflammation. Indeed, in the absence of Muc1, colonic inflammation was reduced, and Muc1 deficient mice were resistant to DSS-induced colitis and produced less inflammatory cytokines, such as Il6 and Tnfα, thought to be master regulators of tumor-associated inflammation and tumorigenesis in the colon.48 We also demonstrated that the majority of IL-6-producing cells were Mɸs, and Muc1-deficient bone marrow-derived Mɸs cultured in vitro produced significantly less IL-6 compared with WT Mɸs, which in turn, results in less MUC1-dependent IL-6/STAT3 activation in epithelial cells with the potential for transformation. Although several inflammatory molecules have been shown to modulate the different stages of CAC development, our data indicate that MUC1-induced IL-6 production may provide an important link between immune cell infiltration and activation of STAT3 pro-tumorigenesis pathways during CAC development.
Mɸs have emerged as key effector cells in the tumor microenvironment of many solid tumor malignancies, including CRC. CRC is associated with a significantly increased infiltration of Mɸs, particularly skewed toward a M2 differentiation state, which has previously been demonstrated to correlate with metastasis and poor prognosis of the patients.49 Our understanding of the factors that influence Mɸ and/or monocyte recruitment, polarization, and function continues to expand. We report here a strong association between MUC1 and an accumulation Mɸ during AOM/DSS treatment. Therefore, we investigated 2 notions to account for decreased tumor burdens in in this model: that Muc1-deficient mice lack key chemo-attractants for Mɸ, and that Muc1-deficient Mɸ have intrinsic defects in differentiation, activation, and migration. Both CCL2 and ICAM1 have been shown to act as mediators that recruit Mɸs50,51 and promote colitis-associated tumorigenesis.52 Our results suggested that Muc1-deficient mice are not able to upregulate colonic Ccl2 gene expression under inflammatory conditions. This low level of Ccl2 expression in the colonic mucosa of Muc1-deficient mice may contribute to low levels of Mɸ infiltration due to the lack of chemotactic signal upon inflammation. Because CCL2 can be expressed by epithelial cells,53 and high levels of expression of secreted form of MUC1-1 have been shown to be associated with upregulation of CCL2 in the tumor cells,54 it is possible that the deficiency of MUC1 in epithelial cells leads to lower epithelial CCL2 secretion. We also found reduced Ccl2 expression in Muc1-deficient BMDM activated in vitro. Thus, our data support that both epithelial and Mɸ MUC1 influences Mɸ infiltration by boosting CCL2 production, which is likely mediated by enhanced NFĸB activation. The intracellular mechanisms by which MUC1 enhances NFĸB inflammatory signaling shown previously include: (1) binding to and promoting activation of the transforming growth factor–β activated kinase 1 and the IĸB kinase complex, and (2) binding directly to the NFĸB p65 transcription factor to drive NFĸB target genes.55,56 The cell surface MUC1 extracellular domain expressed on leukocytes has also been demonstrated to mediate trans-endothelial cellular migration of Mɸs by binding to endothelial ICAM-1,57,58 partly by instigating Src-CrlL-Rac1/Cdc42 facilitated actin cytoskeletal protrusive motility.59,60 Although a lower level of ICAM-1 expression in the Muc1 deficient pancreas leads to reduction of infiltration of Mɸs in a coxsackie virus B3-infection model,61 we did not observe a difference of expression of Icam1 between WT and Muc1 deficient colonic tissues during AOM/DSS treatment.
We also found that Muc1-deficient Mɸs matured normally but had intrinsic defects in chemotactic migration. Decreased CCL2-induced migration as may be in part due to lower expression levels of Ccr2, which we found to be poorly induced after activation. Our findings are supported by a recent investigation showing Muc1-deficient BMDMs had reduced Ccr2 expression, resulting in decreased LPS-induced migration in a coxsackie virus B3-infection model.61 Thus, our results suggest that MUC1 contributes to colonic tissue Mɸ infiltration by enhancing both epithelial and Mɸ CCL2 production and by promoting Mɸ migration responses to CCL2.
TAMs are phenotypically plastic, and factors produced by the cancer cells and the tumor microenvironment can induce Mɸs to become tumor-promoting by producing various factors, such as Ido1, vascular endothelial growth factor, and IL-6, which can inhibit the activation and proliferation of effector T cells critical to anti-tumor immunity, and promote cancer cell survival, proliferation, and metastasis.1,11,42 In vitro studies of a sialyated tumor-associated glycoform of MUC1 have been shown that MUC1 can induce: (1) monocytes to secrete tumor progression factors; (2) Mɸs to display TAM-like phenotype, with increased expression of the immunosuppressive molecule, programmed death-ligand 1; and (3) differentiation of monocytes to TAM through binding of Siglec-9 expressed on myeloid cells.62,63 However, prior to the current study, it was unknown whether MUC1 expressed on Mɸs also contributes to the detrimental microenvironment promoting colon cancer progression. Our data shows increased expression of M2 Mɸs gene signatures in Muc1-expressing AOM/DSS tumors, together with enhanced angiogenesis. Our in vitro studies show that MUC1 influences the response of macrophages to tumor-derived factors, with a prominent influence on Ido1 and Il6 expression. Both IDO1 and IL-6 are key molecules produced by TAMs that promote colon cancer progression,11,64 indicating that MUC1 directly promotes the pro-tumoral function of Mɸs. The ability of MUC1 to trigger Ido1 and Il6 expression in Mɸs is most likely due to promoting activation of NFĸB, as MUC1 has been shown binding to transforming growth factor-β-activated kinase 1 directly and confer the transforming growth factor-β-activated kinase 1 linked with TRAF6, leading to activation of NFĸB in colon cancer cells.55
TAMs are known to inhibit T cell proliferation and cytotoxic activity and promote tumorigenesis in CRC.11 Consistent with this, we found fewer CD8+ T cells in AOM/DSS tumors when MUC1 was present in hematopoietic cells. The reduced abundance of CD8+ T cells most likely involves reduced proliferation of CD8+ T cells in the tumor microenvironment because: (1) we did not observe a difference in CD8+ T cell infiltration at day 42 of AOM/DSS treatment prior to tumor development, and (2) we demonstrated that only WT TCM-treated BMDM produced inhibitors of CD8+ T cell proliferation. However, the CD8+ T cell from tumor-draining lymph nodes of AOM/DSS tumor-bearing Muc1-/- mice expressed higher levels of IFNγ than CD8+ T cells derived from WT mice, suggesting effects also in the lymph node away from the tumor microenvironment, potentially explained by impaired antigen-presenting cell activation in the tumor microenvironment. Together, these findings highlight the dynamic roles of MUC1 in regulating immune cell function during inflammation and tumorigenesis.
We also showed that Muc1 expression is required for IL-6 production during intestinal inflammation and tumor formation after AOM/DSS administration. The significantly increased level of IL-6 in Muc1 proficient mice was associated with amplified activation of STAT3, both in the colonic epithelium and stroma at day 85 of AOM/DSS. Activation of the STAT3 pathway by IL-6, produced by TAMs, is required for CAC formation in AOM/DSS-dependent mouse models,37 and STAT3 activation has been demonstrated to have an important role in tumor progression by augmenting tumor survival and angiogenesis and suppressing antitumor immunity.5 Therefore, it is plausible that the reduced incidence and size of tumors seen in Muc1-deficient mice is in part dependent on the reduced activation of the IL-6/STAT3 axis due to reduced Mɸ recruitment and their altered activation. Moreover, STAT3 induces expression of genes important for proliferation (such as cyclin D) and suppression of apoptosis (BclXL),37,65 induction of the Cyclin D and BclXL protein, is a common observation during the mucosal response to the AOM/DSS insult 66 that was absent in Muc1-/- mice, which further supports our model of MUC1-dependent IL-6/STAT3 signalling as a promoter of tumors.
We chose to investigate the role of MUC1 in the AOM/DSS model because this model recapitulates the aberrant crypt foci-adenoma-carcinoma sequence29 that occurs in human CRC and gives large numbers of spontaneously arising colonic tumors,29 which presents similarities with human CRC pathogenesis. Because of its high reproducibility and potency, as well as affordable and simple model of application, the AOM/DSS has become a well-accepted model to interrogate the link between inflammation and cancer development in the colon.66
Our study proposed a schematic model of MUC1 function on CAC carcinogenesis as follows: intrinsically, administration of AOM and DSS leads to MUC1-enhanced NFĸB activation in epithelial cells and tissue resident macrophages, which in turn leads to CCL2 production that attracts Mɸ infiltration into the inflamed colonic tissues. Activation of these recruited Mɸs results in amplification of production of inflammatory chemokines, cytokines, and reactive oxygen species, thus creating a tumor initiation environment that is potentiated by Muc1 expression on the Mɸs. MUC1-enhanced IL-6 production further induces a strong STAT3 response in these tumor-initiating cells and immune cells via autocrine and paracrine pathways. STAT3 promotes hyperproliferation and survival of the cells through upregulation the expression of cyclin D1 (drivers of cell cycle G1/S-phase transition) and BclXL and thus accelerates tumor growth and progression. As tumors emerge, the MUC1-modulated tumor microenvironment is not permissive for the evolution of CD8 T cell-mediated anti-tumor immunity. We predict that production of both CCL2 and IL-6 by cells expressing MUC1 are important drivers of the increased tumor burden in the AOM/DSS model. Higher levels of CCL2 in the Muc1-expressing mice is likely to underlie the infiltration and activation of Mɸ in the colon, which occurred during the early stage of tumor development. Subsequently, high levels of IL-6 produced by Muc1-expressing Mɸ is likely to contribute to tumor survival, growth, and progression. Future experiments with CCL2 and IL-6 neutralizing antibodies treatment would provide insights into the relative importance of these factors in DSS-azoxymethane carcinogenesis. However, to dissect the importance of MUC1 in their generation, and to define the immune cell sources, these neutralizing experiments would be best combined with genetic models of immune cell-specific Muc1 deficiency.
Blockade of the IL-6/STAT3 axis has been proposed as a mechanism to treat diverse chronic inflammatory disorders, including IBD.67 Given the unacceptable side effects of systemic IL-6/STAT3 inhibition,68 MUC1 inhibitors are currently under active development, encouraged by an acceptable safety profile of a phase I trial of the MUC1 inhibitor-GO-203.25 Our genetic Muc1 knock out mice results agree with recent studies testing the efficacy of cytoplasmic MUC1 inhibition on regression of colon tumors in vivo.26 Our study, together with others, encourages the potential use of MUC1 inhibition for preventing and/or treating inflammation-associated malignancies, as well as the underlying inflammation.
Materials and Methods
Mice
Muc1+/+(WT) and Muc1-/- male mice on a C57BL/6 background were bred at Mater Research Institute, The University of Queensland, Translational Research Institute, Queensland, Australia. The experiments involving mice were conducted in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes 8th edition (2013) and were approved by University of Queensland Animal Experimentation Ethics Committee. Mice were sex- and age-matched within experiments (6–12 weeks of age; n = 8–15).
Induction of CAC
Colon tumors were induced using a AOM and DSS model (Figure 1, A) as previously described.30,31 Briefly, mice were injected intraperitoneally with 10 mg/kg AOM (A5486; Sigma). Seven days later, 1.5% DSS (molecular mass, 36–40 kDa) was given in the drinking water for 5 days followed by regular drinking water for 2 weeks. This cycle was repeated twice. Mice were sacrificed at day 85; grossly visible polyps were counted and measured with calipers by a trained researcher blinded to the genotype and experimental condition of the animals. The colons were bisected longitudinally, and one-half was snap frozen for tissue sample and the other one-half was Swiss rolled and fixed in 10% buffered formalin overnight and transferred to 70% ethanol for subsequent paraffin embedding and histological analysis.
Induction of Colitis and Assessment of Clinical Scores of Colitis
Induction of acute colitis was performed as previously described.69 In brief, acute colitis was induced through administration of 3% DSS (molecular mass, 36–40 kDa) via drinking water for 8 days. Symptoms of colitis were assessed by the daily DAI.69 The DAI is a score based on sum of subscores for diarrhea, rectal bleeding, and loss of body weight. In brief, stool scores were determined as follows: 0, well-formed pellets; 1, semi-formed stools; 2, soft stools; 3, liquid stools that adhered to the anus. Bleeding scores were determined as follows: 0, no blood; 1, small traces of blood in stool; 2, blood traces in stool clearly visible; 3, gross rectal bleeding. Weight loss scores were determined as follows; 0, less than 5% weight loss; 1, 5% to 15% weight loss; 2, 15% to 20% weight loss; 3, more than 20% weight loss.
Histological Analysis Scoring of Inflammation
Colons were examined using 4-μm sections and stained with H&E. Histological assessment of colonic mucosae was carried out in a blinded fashion using a protocol described previously.70,71
BM Chimeras
Male mice were subjected to two doses of 5.5 Gy irradiation separated by a 3-hour interval, then, the next day, transplanted intravenously with 5 × 106 BM cells harvested from femurs and tibias of female donor mice. Three months after BM reconstitution, mice were used for CAC experiments. The following 4 groups of chimeric mice were generated: BMWTmiceWT (WT BM was transplanted into WT mice), BM-/-miceWT (Muc1-/- BM was transplanted into WT mice), BMWTmice-/- (WT BM was transplanted into Muc1-/- mice), and BM-/-mice-/- (Muc1-/- BM was transplanted into Muc1-/- mice).
IHC and Immunofluorescence
Paraffin sections were affixed to adhesive slides and air-dried overnight at 37 °C. After dewaxing in xylol and rehydration through descending graduated ethanol, sections were subjected to antigen retrieval in either citrate buffer pH 6.1 (for CT2, CD31, BrdU, and p-STAT3) or pH 9 (for F4/80 and CD206) (Dako, Carpinteria, CA). For IHC, sections were treated with 3% hydrogen peroxide in phosphate buffered saline (PBS) for 10 minutes. Sections were blocked with blocking solution (10% goat serum, 2% bovine serum albumin [BSA] in PBS with 0.5% Tween 20 [Sigma-Aldrich, St. Louis, MO]) and then incubated overnight at 4 °C with antibody against F4/80-clone A3.1 (Abcam, cat. No. ab6640), CD31 – clone D8V9E (Cell Signaling Technology, cat. No. 77699), MUC1 cytoplasmic tail – clone MH1 (CT2) (Thermo Fisher Scientific, cat. No. MA5-11202), BrdU (Abcam, cat. No ab2284), CD206 – clone 15-2; (Abcam, cat. No. ab64693) All the sections were further stained with appropriate peroxidase-conjugated secondary antibody, detected with DAB, and counter stained with hematoxylin. For B220, Ly6G, CD4, and CD8, IHC stains were outsourced to a professional pathology laboratory with validated protocols (QIMR, Queensland, Australia).
For immunofluorescence p-STAT3 staining, sections were blocked as described above and incubated with anti-phosphorylated (p)-STAT3 (Tyr705) clone D3A7 (Cell Signaling Technology, cat. No. 9145) diluted to in 1:200 dilution to detect p-STAT3. The slides were then incubated with Alexa Fluor 488 goat anti-rabbit (Molecular Probes) prepared in PBS-T with 2% BSA. The slides were counterstained with DAPI (Molecular Probes) and imaged using the confocal laser-scanning microscope Olympus FV1200 microscopy.
Immunoblot Analysis
Protein concentrations were measured using the BCA Protein Assay Kit (Thermo Scientific, Waltham, MA) with BSA as a standard. Aliquots of the lysate containing 30 μg of total protein were mixed in SDS-PAGE Laemmli buffer (0.05M Tris-HCI, pH 6.8; 0.1% 2-mercaptoethanol; 1.0% SDS; 5% glycerol; and 0.15% bromophenol blue), boiled for 5 minutes, and resolved on 4% to 12% acrylamide gels. Resolved proteins were transferred to polyvinylidene difluoride membranes, probed with appropriate antibodies, and detected by chemiluminescence or dual-label infrared analysis.
Isolation of Cells from Organs
For isolating immunocytes of lamina propria, colonic tissues were carefully separated and cut into small pieces. Intraepithelial immunocytes were separated by incubating tissues in 0.15% dithioerythritol (Sigma) for 30 minutes at room temperature. After washing with media, the remaining tissue was incubated with 1 mm EDTA for 30 minutes at room temperature to remove the epithelium. After incubation, EDTA was washed, and the tissue was minced and digested in RPMI culture medium containing 0.25 μg/ml liberase TL (Sigma) and 10 U/mL RNase-free DNaseI (Sigma) in a bacterial incubator for 15 to 25 minutes at 37 °C. Single-cell suspensions were passed through a 70-mm mesh cell strainer. Discontinuous (30% and 70%) Percoll (GE) gradient centrifugation was used to enrich immunocytes.
BMs were collected from femurs by flushing. Mesenteric lymph nodes were dissected and disaggregated on a 70-μm cell strainer.
Flow Cytometry Analysis
Immune cells isolated from colonic lamina propria and mesenteric lymph nodes were stained using fluorescently labeled antibodies for 30 minutes on ice after blocking with Fc-block (Cd16/CD32, Biolegend). The following anti-mouse antibodies were used (most from Bio Legend unless specified): Live or dead BD Horizon Alexa Fluor 700 (cat no 564997, BD), CD3e BUV395 (Clone 145-2C11, BD), CD 19 PercP (clone1D3, cat no 115532), CD4 BV711 (clone RM4-5, cat no 100549), CD8α BV605 (clone 53-6.7, cat no 100743), Ly-6GBV510 (clone 1A8, cat no 127601), CD11cBv421(clone N418,cat no 117329), CD11bPecy7 (clone M1/70, cat no 101215), F4/80 APC (clone T45-2342, cat no 566787, BD), I-A/I-EFITC(MHCII, clone M5/114.15.2, cat no 107605), Gr1 APC Cy7 (clone RB6-8C5, cat no 108423), IL-6PE (clone MP5-20F3, cat no 504503), IFNγBV785 (clone XMG1.2, cat no 563773, BD) following the manufacturer’s instructions with greater than 106 events collected by a LSR Fortessa flow cytometer (BD Biosciences, Schwechat, Austria). Data were analyzed using the Flow Jo v10 software (Flow Jo, LLC, Ashland, OR).
Mouse BM-derived Mononuclear Cell (BMMC) Cultures
For this step, 1 × 106 BM cells per well were cultured in tissue culture-treated 10-cm plates in 10 mL of complete medium (RPMI 1640 supplemented with glutamine, penicillin, streptomycin, 2-mercaptoethanol [all from Invitrogen]), 10% heat-inactivated fetal calf serum (Source BioScience), M-CSF (20 ng/ml, Peprotech) to allow differentiation of BMMCs. One-half of the medium was removed at day 3, and new medium supplemented with M-CSF (20 ng/ml) and warmed at 37 °C was added. The BMMCs were used for experiments at day 6 or 7.
In Vitro Suppression Assay
In vitro suppression assays were performed in 96-well U-bottom plates. CD8+ cells were isolated from native splenocytes by using EasySep mouse CD8+ T cell isolation kit (STEMCELL Technologies, Vancouver, Canada) according to manufacturer’s instructions. Isolated cells were labeled with 5 μM CFSE (Molecular Probes) and activated with ani-CD3 and ani-CD28 antibodies (BD) according to the manufacturer’s instructions. Condition media of polarized macrophages was added to the culture. After 5 days, cells were acquired by a LSR Fortessa flow cytometer (BD Biosciences, Schwechat, Austria). Data were analyzed using the Flow Jo v10 software (Flow Jo, LLC, Ashland, OR).
RNA Preparation and Real-time Polymerase Chain Reaction (PCR)
Total RNA was prepared using the RNeasy Mini Kit (Qiagen, Valencia, CA). The quantity and quality of the RNA was determined by spectrophotometry (ND-1000; NanoDrop Technologies Inc, Wilmington, DE). Total RNA (1 μg) from each sample was used for first strand cDNA synthesis using SuperScriptTM III reverse transcriptase (Life Technologies) following the manufacturer's instructions. Real-time PCR was performed on a Rotor-Gene 3000 cycler (Qiagen) by using SYBR Green I fluorescence (Life Technologies) in the presence of Platinum Taq DNA-Polymerase (Life Technologies), 3 mM MgCl2, 2 μM of each of the primers, and 200 μM dNTPs. The cycling conditions were denaturation for 10 minutes at 95 °C, followed by 40 amplification cycles of 20 seconds of denaturation at 94 °C, 30 seconds of annealing at 60 °C, and 30 seconds of extension at 72 °C. To confirm the specificity of the amplified DNA, a melting curve was determined at the end of each run. The reaction efficiency was determined with a dilution series of cDNA containing the PCR products. Expression of the target genes were normalized to that of β-actin, and the results were presented as their ratios (arbitrary units).
The primers used for PCR were designed from Primer Bank (http://pga.mgh.harvard.edu/primebank/index.html) or using Oligoperfect Designer (Life Technologies), and their sequences to amplify the target genes are shown in Table 1.
Table 1.
Primer Set for Quantitative PCR to Detect the Indicated mRNA Levels
| Forward primers | Reverse primers | |
|---|---|---|
| β actin | 5'-CTTCTTGGGTATGGAATCCTGTG-3' | 5'-AGCACTGTGTTGGCATAGAGGTC-3' |
| Ccl2 | 5'-TTCTGGGCCTGCTGTTCAC-3' | 5’-GGCGTTAACTGCATCTGGCTG-3' |
| Csfr1 | 5'-GGACCTACCGTTGTACCGAG-3' | 5'-CAAGAGTGGGCCGGATCTTT-3' |
| Csf2 | 5'-GGCCTTGGAAGCATGTAGAGG-3' | 5'-GGAGAACTCGTTAGAGACGACTT-3' |
| Cxcr2 | 5'-GCCCTGCCCATCTTAATTCTAC-3' | 5'-ACCCTCAAACGGGATGTATTG-3' |
| Egfr | 5'-GCATCATGGGAGAGAACAACA-3' | 5'-CTGCCATTGAACGTACCCAGA-3' |
| Arg1 | 5'-CGTAGACCCTGGGGAACACTAT-3' | 5'-TCCATCACCTTGCCAATCCC-3' |
| Mrc1 | 5'-TTCAGCTATTGGACGCGAGG-3' | 5'-GAATCTGACACCCAGCGGAA-3' |
| Il6 | 5'-CCCCAATTTCCAATGCTCTCC-3' | 5'-GGATGGTCTTGGTCCTTAGCC-3' |
| Timp2 | 5'-GCAACTCGGACCTGGTCATAA-3' | 5'-CGGCCCGTGATGAGAAACT-3' |
| Ym1 | 5'-GAAGCTCTCCAGAAGCAATCCTG-3' | 5'-CAGAAGAATTGCCAGACCTGTGA-3' |
| IL13 | 5'-CTCCCTCTGACCCTTAAGGAG-3' | 5'-GAAGGGGCCGTGGCGAAACAG-3' |
| IL33 | 5'-CGGATCCACTTCACTTTTAACACAGTC-3' | 5'-GAGATCTTTAGATTTTCGAGAGCTTA-3' |
| Ccr2 | 5'-AGGAGCCATACCTGTAAATGCC-3' | 5'-TGTGGTGAATCCAATGCCCT-3' |
| Ido1 | 5'-CGACAAGGGCTTCTTCCTCGTC-3' | 5'-TGGGTCCACAAAGTCACGCATC-3' |
| Icam1 | 5'-AGCTCGGAGGATCACAAACG-3' | 5'-TCCAGCCGAGGACCATACAG-3' |
| Fizz1 | 5'-TCCCAGTGAATACTGATGAGA-3' | 5'-CCACTCTGGATCTCCCAAGA-3' |
PCR, Polymerase chain reaction.
Statistical Analyses
All statistical analyses were performed using Prism v5 (GraphPad Software). Sample sizes for experiments were determined by power analyses based on the variation shown in previous experiments and predicted effect sizes considered to be of biological relevance. No data were excluded from any analyses. The normal distribution of data was assessed by probability plots, and where a normal distribution could not be established, nonparametric testing was used. The statistical test used and the sample sizes for individual analyses are provided within the figure legends.
Acknowledgment
The authors thank the National Health and Medical Research Council, Mater Foundation, Gastroenterological Society of Australia Mostyn Family Grant, and Gastroenterological Society of Australia Project Grant for the funding support, technical assistance of the Translational Research Institute core facilities for histology, flow cytometry, and microscopy.
CRediT Authorship Contributions
Yong Hua Sheng, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Supporting; Investigation: Equal; Methodology: Lead; Project administration: Lead; Resources: Supporting; Supervision: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Lead)
Julie M. Davies, PhD (Data curation: Equal; Investigation: Equal; Project administration: Equal; Validation: Lead; Visualization: Equal; Writing – review & editing: Equal)
Ran Wang, PhD (Conceptualization: Supporting; Data curation: Equal; Formal analysis: Supporting; Validation: Equal)
Kuan Yau Wong, PhD (Data curation: Equal; Formal analysis: Equal; Validation: Equal)
Rabina Giri, PhD (Data curation: Equal; Formal analysis: Supporting; Validation: Supporting)
Yuanhao Yang, PhD (Data curation: Supporting; Software: Lead; Validation: Equal)
Jakob Begun, PhD, MD (Supervision: Supporting; Validation: Supporting; Visualization: Supporting)
Timothy H. Florin, PhD, MD (Conceptualization: Supporting; Writing – review & editing: Equal)
Sumaira Z. Hasnain, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Michael A. McGuckin, PhD (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Supervision: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Health and Medical Research Council project grants 1060698 and 1164141, and funding by the Mater Foundation. Yong Hua Sheng was partly supported by a Gastroenterological Society of Australia Mostyn Family Grant and Gastroenterological Society of Australia Project Grant. The Translational Research Institute is supported by a grant from the Australian Government.
Contributor Information
Yong H. Sheng, Email: yong.sheng@mater.uq.edu.au.
Michael A. McGuckin, Email: michael.mcguckin@unimelb.edu.au.
References
- 1.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein C.N., Blanchard J.F., Kliewer E., Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91:854–862. doi: 10.1002/1097-0142(20010215)91:4<854::aid-cncr1073>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Terzic J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Neurath M.F. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 5.Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickoloff B.J., Ben-Neriah Y., Pikarsky E. Inflammation and cancer: is the link as simple as we think? J Invest Dermatol. 2005;124:x–xiv. doi: 10.1111/j.0022-202X.2005.23724.x. [DOI] [PubMed] [Google Scholar]
- 7.Cassetta L., Fragkogianni S., Sims A.H., Swierczak A., Forrester L.M., Zhang H., Soong D.Y.H., Cotechini T., Anur P., Lin E.Y., Fidanza A., Lopez-Yrigoyen M., Millar M.R., Urman A., Ai Z., Spellman P.T., Hwang E.S., Dixon J.M., Wiechmann L., Coussens L.M., Smith H.O., Pollard J.W. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35:588–602.e10. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Ginderachter J.A., Movahedi K., Hassanzadeh Ghassabeh G., Meerschaut S., Beschin A., Raes G., De Baetselier P. Classical and alternative activation of mononuclear phagocytes: picking the best of both worlds for tumor promotion. Immunobiology. 2006;211:487–501. doi: 10.1016/j.imbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Pollard J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 10.Sica A., Schioppa T., Mantovani A., Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42:717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Erreni M., Mantovani A., Allavena P. Tumor-associated macrophages (TAM) and inflammation in colorectal cancer. Cancer Microenviron. 2011;4:141–154. doi: 10.1007/s12307-010-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth M.J., Dunn G.P., Schreiber R.D. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv Immunol. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 13.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoue F., Bruneval P., Cugnenc P.H., Trajanoski Z., Fridman W.H., Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 14.Olguin J.E., Medina-Andrade I., Molina E., Vazquez A., Pacheco-Fernandez T., Saavedra R., Perez-Plasencia C., Chirino Y.I., Vaca-Paniagua F., Arias-Romero L.E., Gutierrez-Cirlos E.B., Leon-Cabrera S.A., Rodriguez-Sosa M., Terrazas L.I. Early and partial reduction in CD4(+)Foxp3(+) regulatory T cells during colitis-associated colon cancer induces CD4(+) and CD8(+) T cell activation inhibiting tumorigenesis. J Cancer. 2018;9:239–249. doi: 10.7150/jca.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuley J.L., Linden S.K., Png C.W., King R.M., Pennington H.L., Gendler S.J., Florin T.H., Hill G.R., Korolik V., McGuckin M.A. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2324. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng Y.H., Hasnain S.Z., Florin T.H., McGuckin M.A. Mucins in inflammatory bowel diseases and colorectal cancer. J Gastroenterol Hepatol. 2012;27:28–38. doi: 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- 17.McGuckin M.A., Linden S.K., Sutton P., Florin T.H. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 18.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., Anderson C.A., Bis J.C., Bumpstead S., Ellinghaus D., Festen E.M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C.G., Montgomery G.W., Prescott N.J., Raychaudhuri S., Rotter J.I., Schumm P., Sharma Y., Simms L.A., Taylor K.D., Whiteman D., Wijmenga C., Baldassano R.N., Barclay M., Bayless T.M., Brand S., Buning C., Cohen A., Colombel J.F., Cottone M., Stronati L., Denson T., De Vos M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van Gossum A., Guthery S.L., Halfvarson J., Verspaget H.W., Hugot J.P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panes J., Phillips A., Proctor D.D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A.H., Stokkers P.C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S.R., Brant S.R., Rioux J.D., D'Amato M., Weersma R.K., Kugathasan S., Griffiths A.M., Mansfield J.C., Vermeire S., Duerr R.H., Silverberg M.S., Satsangi J., Schreiber S., Cho J.H., Annese V., Hakonarson H., Daly M.J., Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinall L.E., King M., Novelli M., Green C.A., Daniels G., Hilkens J., Sarner M., Swallow D.M. Altered expression and allelic association of the hypervariable membrane mucin MUC1 in Helicobacter pylori gastritis. Gastroenterology. 2002;123:41–49. doi: 10.1053/gast.2002.34157. [DOI] [PubMed] [Google Scholar]
- 20.Jonckheere N., Skrypek N., Van Seuningen I. Mucins and tumor resistance to chemotherapeutic drugs. Biochim Biophys Acta. 2014;1846:142–151. doi: 10.1016/j.bbcan.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Nath S., Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C., Liu T., Yin L., Zuo D., Lin Y., Wang L. Prognostic and clinicopathological value of MUC1 expression in colorectal cancer: a meta-analysis. Medicine (Baltimore) 2019;98 doi: 10.1097/MD.0000000000014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersson J., Schreiber O., Hansson G.C., Gendler S.J., Velcich A., Lundberg J.O., Roos S., Holm L., Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida A., Lau C.W., Zhang M., Andoh A., Shi H.N., Mizoguchi E., Mizoguchi A. The membrane-bound mucin Muc1 regulates T helper 17-cell responses and colitis in mice. Gastroenterology. 2012;142:865–874.e2. doi: 10.1053/j.gastro.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kufe D.W. MUC1-C in chronic inflammation and carcinogenesis; emergence as a target for cancer treatment. Carcinogenesis. 2020;41:1173–1183. doi: 10.1093/carcin/bgaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Zhang N., Jin C., Long M.D., Rajabi H., Yasumizu Y., Fushimi A., Yamashita N., Hagiwara M., Zheng R., Wang J., Kui L., Singh H., Kharbanda S., Hu Q., Liu S., Kufe D. MUC1-C drives stemness in progression of colitis to colorectal cancer. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poh T.W., Madsen C.S., Gorman J.E., Marler R.J., Leighton J.A., Cohen P.A., Gendler S.J. Downregulation of hematopoietic MUC1 during experimental colitis increases tumor-promoting myeloid-derived suppressor cells. Clin Cancer Res. 2013;19:5039–5052. doi: 10.1158/1078-0432.CCR-13-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascio S., Faylo J.L., Sciurba J.C., Xue J., Ranganathan S., Lohmueller J.J., Beatty P.L., Finn O.J. Abnormally glycosylated MUC1 establishes a positive feedback circuit of inflammatory cytokines, mediated by NF-kappaB p65 and EzH2, in colitis-associated cancer. Oncotarget. 2017;8:105284–105298. doi: 10.18632/oncotarget.22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Robertis M., Massi E., Poeta M.L., Carotti S., Morini S., Cecchetelli L., Signori E., Fazio V.M. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng Y.H., Wong K.Y., Seim I., Wang R., He Y., Wu A., Patrick M., Lourie R., Schreiber V., Giri R., Ng C.P., Popat A., Hooper J., Kijanka G., Florin T.H., Begun J., Radford K.J., Hasnain S., McGuckin M.A. MUC13 promotes the development of colitis-associated colorectal tumors via beta-catenin activity. Oncogene. 2019;38:7294–7310. doi: 10.1038/s41388-019-0951-y. [DOI] [PubMed] [Google Scholar]
- 31.Sheng Y.H., Giri R., Davies J., Schreiber V., Alabbas S., Movva R., He Y., Wu A., Hooper J., McWhinney B., Oancea I., Kijanka G., Hasnain S., Lucke A.J., Fairlie D.P., McGuckin M.A., Florin T.H., Begun J. A nucleotide analog prevents colitis-associated cancer via beta-catenin independently of inflammation and autophagy. Cell Mol Gastroenterol Hepatol. 2021;11:33–53. doi: 10.1016/j.jcmgh.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClellan J.L., Davis J.M., Steiner J.L., Enos R.T., Jung S.H., Carson J.A., Pena M.M., Carnevale K.A., Berger F.G., Murphy E.A. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: role of MCP-1. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1087–G1095. doi: 10.1152/ajpgi.00252.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Yao W., Yuan Y., Chen P., Li B., Li J., Chu R., Song H., Xie D., Jiang X., Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura T. The production of monocyte chemoattractant protein-1 (MCP-1)/CCL2 in tumor microenvironments. Cytokine. 2017;98:71–78. doi: 10.1016/j.cyto.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Hsu C.P., Chung Y.C. Influence of interleukin-6 on the invasiveness of human colorectal carcinoma. Anticancer Res. 2006;26:4607–4614. [PubMed] [Google Scholar]
- 36.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad R., Rajabi H., Kosugi M., Joshi M.D., Alam M., Vasir B., Kawano T., Kharbanda S., Kufe D. MUC1-C oncoprotein promotes STAT3 activation in an autoinductive regulatory loop. Sci Signal. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J., McConnell M.J., Yu B., Li J., Balko J.M., Black E.P., Johnson J.O., Lloyd M.C., Altiok S., Haura E.B. MUC1 is a downstream target of STAT3 and regulates lung cancer cell survival and invasion. Int J Oncol. 2009;35:337–345. [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng Y.H., He Y., Hasnain S.Z., Wang R., Tong H., Clarke D.T., Lourie R., Oancea I., Wong K.Y., Lumley J.W., Florin T.H., Sutton P., Hooper J.D., McMillan N.A., McGuckin M.A. MUC13 protects colorectal cancer cells from death by activating the NF-kappaB pathway and is a potential therapeutic target. Oncogene. 2017;36:700–713. doi: 10.1038/onc.2016.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terness P., Bauer T.M., Rose L., Dufter C., Watzlik A., Simon H., Opelz G. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J Exp Med. 2002;196:447–457. doi: 10.1084/jem.20020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prendergast G.C., Smith C., Thomas S., Mandik-Nayak L., Laury-Kleintop L., Metz R., Muller A.J. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol Immunother. 2014;63:721–735. doi: 10.1007/s00262-014-1549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munn D.H., Shafizadeh E., Attwood J.T., Bondarev I., Pashine A., Mellor A.L. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X., Shin N., Koblish H.K., Yang G., Wang Q., Wang K., Leffet L., Hansbury M.J., Thomas B., Rupar M., Waeltz P., Bowman K.J., Polam P., Sparks R.B., Yue E.W., Li Y., Wynn R., Fridman J.S., Burn T.C., Combs A.P., Newton R.C., Scherle P.A. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood. 2010;115:3520–3530. doi: 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- 45.Solinas G., Germano G., Mantovani A., Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol. 2009;86:1065–1073. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka T., Kohno H., Suzuki R., Yamada Y., Sugie S., Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94:965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu F., Liu F., Zhao H., An G., Feng G. Prognostic significance of mucin antigen MUC1 in various human epithelial cancers: a meta-analysis. Medicine (Baltimore) 2015;94:e2286. doi: 10.1097/MD.0000000000002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atreya R., Neurath M.F. Signaling molecules: the pathogenic role of the IL-6/STAT-3 trans signaling pathway in intestinal inflammation and in colonic cancer. Curr Drug Targets. 2008;9:369–374. doi: 10.2174/138945008784221116. [DOI] [PubMed] [Google Scholar]
- 49.Forssell J., Oberg A., Henriksson M.L., Stenling R., Jung A., Palmqvist R. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res. 2007;13:1472–1479. doi: 10.1158/1078-0432.CCR-06-2073. [DOI] [PubMed] [Google Scholar]
- 50.Liou G.Y., Doppler H., Necela B., Edenfield B., Zhang L., Dawson D.W., Storz P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5:52–63. doi: 10.1158/2159-8290.CD-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimoto H., Sangai T., Ishii G., Ikehara A., Nagashima T., Miyazaki M., Ochiai A. Stromal MCP-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int J Cancer. 2009;125:1276–1284. doi: 10.1002/ijc.24378. [DOI] [PubMed] [Google Scholar]
- 52.Popivanova B.K., Kostadinova F.I., Furuichi K., Shamekh M.M., Kondo T., Wada T., Egashira K., Mukaida N. Blockade of a chemokine, CCL2, reduces chronic colitis-associated carcinogenesis in mice. Cancer Res. 2009;69:7884–7892. doi: 10.1158/0008-5472.CAN-09-1451. [DOI] [PubMed] [Google Scholar]
- 53.Lazennec G., Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosso J.F., Herbert L.M., Owen J.L., Lopez D.M. MUC1/sec-expressing tumors are rejected in vivo by a T cell-dependent mechanism and secrete high levels of CCL2. J Immunol. 2004;173:1721–1730. doi: 10.4049/jimmunol.173.3.1721. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi H., Jin C., Rajabi H., Pitroda S., Alam M., Ahmad R., Raina D., Hasegawa M., Suzuki Y., Tagde A., Bronson R.T., Weichselbaum R., Kufe D. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad R., Raina D., Trivedi V., Ren J., Rajabi H., Kharbanda S., Kufe D. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernier A.J., Zhang J., Lillehoj E., Shaw A.R., Gunasekara N., Hugh J.C. Non-cysteine linked MUC1 cytoplasmic dimers are required for Src recruitment and ICAM-1 binding induced cell invasion. Mol Cancer. 2011;10:93. doi: 10.1186/1476-4598-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahn J.J., Chow J.W., Horne G.J., Mah B.K., Emerman J.T., Hoffman P., Hugh J.C. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin Exp Metastasis. 2005;22:475–483. doi: 10.1007/s10585-005-3098-x. [DOI] [PubMed] [Google Scholar]
- 59.Shen Q., Rahn J.J., Zhang J., Gunasekera N., Sun X., Shaw A.R., Hendzel M.J., Hoffman P., Bernier A., Hugh J.C. MUC1 initiates Src-CrkL-Rac1/Cdc42-mediated actin cytoskeletal protrusive motility after ligating intercellular adhesion molecule-1. Mol Cancer Res. 2008;6:555–567. doi: 10.1158/1541-7786.MCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 60.Haddon L., Hugh J. MUC1-mediated motility in breast cancer: a review highlighting the role of the MUC1/ICAM-1/Src signaling triad. Clin Exp Metastasis. 2015;32:393–403. doi: 10.1007/s10585-015-9711-8. [DOI] [PubMed] [Google Scholar]
- 61.Liu X., Clemens D.L., Grunkemeyer J.A., Price J.D., O'Connell K., Chapman N.M., Storz P., Wen H., Cox J.L., Reid W.L., Hollingsworth M.A., Thayer S. Mucin-1 is required for Coxsackie virus B3-induced inflammation in pancreatitis. Sci Rep. 2019;9 doi: 10.1038/s41598-019-46933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beatson R., Tajadura-Ortega V., Achkova D., Picco G., Tsourouktsoglou T.D., Klausing S., Hillier M., Maher J., Noll T., Crocker P.R., Taylor-Papadimitriou J., Burchell J.M. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol. 2016;17:1273–1281. doi: 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beatson R., Graham R., Grundland Freile F., Cozzetto D., Kannambath S., Pfeifer E., Woodman N., Owen J., Nuamah R., Mandel U., Pinder S., Gillett C., Noll T., Bouybayoune I., Taylor-Papadimitriou J., Burchell J.M. Cancer-associated hypersialylated MUC1 drives the differentiation of human monocytes into macrophages with a pathogenic phenotype. Commun Biol. 2020;3:644. doi: 10.1038/s42003-020-01359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prendergast G.C., Malachowski W.P., DuHadaway J.B., Muller A.J. Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klampfer L. The role of signal transducers and activators of transcription in colon cancer. Front Biosci. 2008;13:2888–2899. doi: 10.2741/2893. [DOI] [PubMed] [Google Scholar]
- 66.Greten F.R., Eckmann L., Greten T.F., Park J.M., Li Z.W., Egan L.J., Kagnoff M.F., Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber S., Aden K., Bernardes J.P., Conrad C., Tran F., Hoper H., Volk V., Mishra N., Blase J.I., Nikolaus S., Bethge J., Kuhbacher T., Rocken C., Chen M., Cottingham I., Petri N., Rasmussen B.B., Lokau J., Lenk L., Garbers C., Feuerhake F., Rose-John S., Waetzig G.H., Rosenstiel P. Therapeutic interleukin-6 trans-signaling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology. 2021;160:2354–2366.e11. doi: 10.1053/j.gastro.2021.02.062. [DOI] [PubMed] [Google Scholar]
- 68.Yang L., Lin S., Xu L., Lin J., Zhao C., Huang X. Novel activators and small-molecule inhibitors of STAT3 in cancer. Cytokine Growth Factor Rev. 2019;49:10–22. doi: 10.1016/j.cytogfr.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Sheng Y.H., Lourie R., Linden S.K., Jeffery P.L., Roche D., Tran T.V., Png C.W., Waterhouse N., Sutton P., Florin T.H., McGuckin M.A. The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis. Gut. 2011;60:1661–1670. doi: 10.1136/gut.2011.239194. [DOI] [PubMed] [Google Scholar]
- 70.Tamaki H., Nakamura H., Nishio A., Nakase H., Ueno S., Uza N., Kido M., Inoue S., Mikami S., Asada M., Kiriya K., Kitamura H., Ohashi S., Fukui T., Kawasaki K., Matsuura M., Ishii Y., Okazaki K., Yodoi J., Chiba T. Human thioredoxin-1 ameliorates experimental murine colitis in association with suppressed macrophage inhibitory factor production. Gastroenterology. 2006;131:1110–1121. doi: 10.1053/j.gastro.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 71.Obermeier F., Kojouharoff G., Hans W., Scholmerich J., Gross V., Falk W. Interferon-gamma (IFN-gamma)- and tumour necrosis factor (TNF)-induced nitric oxide as toxic effector molecule in chronic dextran sulphate sodium (DSS)-induced colitis in mice. Clin Exp Immunol. 1999;116:238–245. doi: 10.1046/j.1365-2249.1999.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]