Abstract
Xenorhabdus is a major insect pathogen symbiotically associated with nematodes of the family Steinernematidae. This motile bacterium displays swarming behavior on suitable media, but a spontaneous loss of motility is observed as part of a phenomenon designated phase variation which involves the loss of stationary-phase products active as antibiotics and potential virulence factors. To investigate the role of one of the transcriptional activators of flagellar genes, FlhDC, in motility and virulence, the Xenorhabdus nematophilus flhDC locus was identified by functional complementation of an Escherichia coli flhD null mutant and DNA sequencing. Construction of X. nematophilus flhD null mutants confirmed that the flhDC operon controls flagellin expression but also revealed that lipolytic and extracellular hemolysin activity is flhDC dependent. We also showed that the flhD null mutant displayed a slightly attenuated virulence phenotype in Spodoptera littoralis compared to that of the wild-type strain. Thus, these data indicated that motility, lipase, hemolysin, or unknown functions controlled by the flhDC operon are involved in the infectious process in insects. Our investigation expands the view of the flagellar regulon as a checkpoint coupled to a major network involving bacterial physiological aspects as well as motility.
The genus Xenorhabdus (Enterobacteriaceae) consists of the specific bacterial symbionts of the entomopathogenic nematodes of the family Steinernematidae (57) and was separated from the genus Photorhabdus (10) containing the symbionts of the entomopathogenic nematodes of the family Heterorhabditidae. Both genera are entomopathogenic gram-negative bacteria belonging to the family Enterobacteriaceae. The nematodes of the Steinernematidae carry their bacterial symbionts monoxenically in a special vesicle in the intestines of the infective stage (L3 juveniles), while the nematodes of the Heterorhabditidae carry their bacterial symbionts throughout their intestines (22). These bacteria are transported by their nematode hosts into the hemocoel of the insect prey which is killed, probably via a combination of toxin action and septicemia. The bacterial symbionts also contribute to the symbiotic relationship by establishing and maintaining suitable conditions for nematode reproduction (46). Recently, a novel toxin complex with both oral and injectable activities against a wide range of insects was identified in Photorhabdus luminescens (11).
The form of the bacterium that is normally isolated from symbiotic infective-stage nematodes is referred to as phase I. During in vitro culture or mass rearing of nematodes, Xenorhabdus and Photorhabdus strains spontaneously produce colonial variants which have been called phase II variants (9). The two variants of the bacteria have generally been shown to be equally pathogenic for the larvae of the greater wax moth, Galleria mellonella (4). Recently, Volgyi et al. (58) described for the first time a phase II variant that displayed reduced virulence in the Manduca sexta virulence assay.
The two variants of Xenorhabdus can be distinguished by several characteristics, which may be involved in insect virulence or association with nematodes. During the stationary period, phase I variants of Xenorhabdus adsorb dyes on agar plates, produce outer membrane protein OpnB (39), have protease, lipase, or lecithinase activity (56), secrete chemical antibiotics (3), synthesize mannose-resistant pili (43), and have protoplasmic paracrystalline inclusions (15). These properties are either apparently absent or greatly reduced in phase II variants. It is clearly demonstrated that the phase shift occurs during the stationary period of growth and that the phase variation phenomenon is highly variable, unpredictable, and reversible (26). It was suggested that phase variation is controlled by a putative master switch which affects a number of other regulatory systems differently that in turn control one or more phase variant characteristics.
Recently, we showed that swarming and swimming motility in different Xenorhabdus nematophilus strains were impaired by phase variation (28). In strain F1, the phase II variant was nonmotile and unable to synthesize flagellar filaments. Moreover, the flagellin-encoding gene, fliC, and the hook-associated protein 2 gene, fliD, were switched off at the transcriptional level in the phase II variants (29). These results suggested that the expression of a gene earlier in the transcriptional hierarchy of the flagellar regulon was impaired. Approximately 50 genes are involved in the biogenesis and function of a flagellum of Escherichia coli or Salmonella enterica serovar Typhimurium (for a review, see reference 42). These genes are transcriptionally regulated as a cascade and are coordinated with the flagellar hierarchy (35). At the top of the hierarchy is the class I operon, flhDC, whose products are required for expression of all other flagellar genes (7, 36). The E. coli FlhD and FlhC proteins act as an activator for class II operons including most of the structural genes for the flagellar hook-basal body complexes plus the alternative sigma factor fliA (41). The product of the fliA gene, ς28, directs the transcription of class III genes which encode the filament protein, hook-associated proteins, motor proteins, and various chemotaxis proteins (44). The central channel is believed to work as a passage not only for flagellar component proteins but also for flagellar regulatory protein FlgM, an anti-sigma factor (33, 36). Accumulation of FlgM in the cell by preventing its export blocks the transcription of class III genes, including flagellin.
FlhD is involved in processes other than flagellar expression that occur when cells enter the stationary phase (48). Recently, it was demonstrated that E. coli flhD mutants were unable to sense the depletion of serine from the medium that signals wild-type cells to reduce their cell division (48). Acetyl phosphate and phosphorylation of OmpR would mediate this effect (47, 54). Moreover, Givskov et al. (30) and more recently Young et al. (60) have shown that the flhDC operon controls phospholipase expression and secretion in Serratia liquefaciens and Yersinia enterocolitica. It was proposed that type III protein secretion by the flagellar apparatus may be a general mechanism for transport of proteins involved in virulence (60).
The simultaneous loss of motility and extracellular stationary-phase products during phase shift in Xenorhabdus led us to investigate whether global regulators, equivalent to the master operon flhDC of E. coli, control flagellar synthesis and other phase-variable properties of these entomopathogenic bacteria. The role for Xenorhabdus flhDC in bacterium-insect interaction was also studied.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used in this study are listed in Table 1. X. nematophilus F1 (phase I variant) was isolated from the nematode Steinernema carpocapsae Plougastel from Brittany, France. The phase II variants of X. nematophilus F1 were selected from in vitro cultures of the phase I variants. Phases I and II of F1 are indicated by addition of the suffixes /1 and /2 to the strain name, respectively, as previously described (9). At each subculture, phase status was identified by the differential adsorption of dye when grown on NBTA (nutrient agar supplemented with 25 mg of bromothymol blue per liter and 40 mg of triphenyltetrazolium chloride per liter) and by measuring activity against Micrococcus luteus (from the culture collection of the Institut Pasteur, Paris, France).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α MCR | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 (lacZYA-argF)U169 endA1 recA1 deoR thi-1 supE44 λ− gyrA96 relA1 | Gibco-BRL (Life Technologies) |
| HB101 | F−mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (strR) supE44 λ− | 12 |
| S17.1 | pro r− n+ Tpr Smr RP4-2-Tc::Mu::Tn7 recA thi | 55 |
| MC1000 flhD::Tn10 | F−araD139 (ara-leu)7697 galE15 galK16 Δ(lac)X74 rpsL(Str) hsdR2(rK− mK+) mcrA mcrB1 relA1 flhD::Tn10 | P. Matsumara |
| YK4323 | YK410 flhC::lacZ Apr | P. Matsumara |
| YK410 F−araD139 ΔlacU169 rpsL thi nalA thyA pyrC46 his | ||
| X. nematophilus | ||
| F1/1 | Wild type, phase I variant | Laboratory collection |
| F1/2 | Phase II variant | 28 |
| ΩIA | F1/1 flhD::ΩCm | This work |
| Plasmids | ||
| pUC19 | Apr cloning vehicle | Biolabs |
| pACYC184 | Tcr Cmr cloning vehicle | 14 |
| pRK404 | Tcr mobilizable plasmid | 18 |
| pRK2013 | Kmr (Tn903), ColE1 replicon, tra+ (RK2); helper plasmid | 25 |
| pPM61 | pMK2004 with the E. coli flhDC operon, Kmr Apr | 7 |
| pRK620 | 1.5-kb DraI-HpaI fragment from pPM61 in pRK404 (contains E. coli flhDC operon) | This work |
| pHP45-ΩCm | Apr Cmr ΩCm | 24 |
| pJQ200SK | GmrsacRB mob oriV (p15A replicon) | S. Forst |
| pAGE1 | 10-kb DNA EcoRI fragment from F1/1 in EcoRI site of pUC19 (contains flhDC operon) | This work |
| pAGE101 | 1.5-kb HindIII fragment from pAGE1 in pUC19 (contains flhD gene and 5′ flhC region) | This work |
| pAGE121 | Same as pAGE101 but in pRK404 | This work |
| pAGE105 | 3-kb EcoRI-ScaI fragment from pAGE1 in pUC19 | This work |
| pAGE135 | Same as pAGE1051 but in pACYC184 | This work |
| pAGE1051 | 1.25 kb DraI fragment from pAGE105 in pUC19 (contains flhDC genes without the CRP-cAMP consensus sequence) | This work |
| pAGE1351 | Same as pAGE1051 but in pACYC184 | This work |
| pAGE1052 | 0.15-kb DraI-SspI deletion from pAGE1051 insert (contains flhDC genes without the CRP-cAMP consensus sequence) | This work |
| pAGE1352 | Same as pAGE1052 but in pACYC184 | This work |
| pAGE1054 | 1.9-kb HindIII partial fragment from pAGE105 in pUC19 (contains flhDC operon) | This work |
| pAGE1254 | Same as pAGE1054 but in pRK404 | This work |
| pAGE1354 | Same as pAGE1054 but in pACYC184 | This work |
| pAGE1054-ΩCm | 3.5-kb BamHI fragment from pHP45-ΩCm in HpaI site of pAGE1054 (contains flhD::ΩCm) | This work |
| pAGEQ2 | 5.5-kb BamHI-PstI fragment from pAGE1054-ΩCm in pJQ200 (contains flhD::ΩCm) | This work |
Swimming motility was observed in mot agar (mot broth [1% tryptone, 0.5% NaCl, 10 mM MgSO4] with 0.35% agar [Difco]) at 28°C.
The final concentrations (in milligrams per liter) of antibiotics used for selection were as follows: ampicillin, 100 for E. coli strains and 50 for X. nematophilus; gentamicin, 30; kanamycin, 20; chloramphenicol, 20 for E. coli strains and 15 for X. nematophilus; and tetracycline, 10 for E. coli strains and 7.5 for X. nematophilus.
Molecular genetic techniques, Southern blotting, and Northern analysis.
Purification of genomic DNA from Xenorhabdus and recombinant techniques have been described elsewhere (13, 51). Double-stranded DNA from pAGE101 and pAGE105 (Table 1) were used as templates for sequencing reactions. Sequencing was performed with the long DNA sequencer (model 1377; Applied Biosystems). DNA sequence analysis and Southern blotting experiments were performed as previously described (29). In order to detect a locus homologous to E. coli flhD, X. nematophilus DNA digests were hybridized to the 1.5-kb DraI-HpaI fragment from pPM61 (Table 1) used as a probe. To compare the gross structure of the flhDC locus between phase variants I and II, a large fragment (7.5-kb EcoRI-BamHI fragment from pAGE1) containing chromosomal regions around X. nematophilus flhDC was used as a probe. To control the allelic exchange in the flhD null mutant, the 1.25-kb DraI fragment of pAGE105 was used as a probe. Total RNA from a log-phase culture (optical density at 540 nm of 0.5) of F1 strain phase variants grown in mot broth was purified per the manufacturer's instructions (5 Prime-3 Prime Perfect RNA total isolation kit). Equal loading in lanes was confirmed by absorbance at 260 nm. The ClaI-PstI fragment from pAGE1051 was used as a flhDC probe. Digoxigenin-labeled probes were synthesized, hybridized, and detected by the manufacturer's procedure (Boehringer Mannheim).
Gene disruption of flhD.
A chloramphenicol-resistant omega cassette (24) with transcriptional and translational terminators was inserted into the unique HpaI site within the X. nematophilus flhD gene inserted into the pJQ200SK plasmid to yield pAGEQ2 (Table 1). pJQ200SK plasmid is a derivative of pACYC184 carrying the sacB gene and the mob site from RP4. Preliminary experiments using exconjugant harboring plasmid (pRK404)-borne sacB gene showed that X. nematophilus cells were susceptible to 1% sucrose (A. Givaudan, unpublished data). Luria-Bertani (LB) medium without NaCl containing 2% sucrose was used throughout this study. The pAGEQ2 plasmid (a sacB-negative selection plasmid) (Table 1) carrying a chloramphenicol resistance-encoding interposon inserted in the flhD gene was transformed into E. coli S17.1 and introduced into X. nematophilus F1/1 by mating. Cmr and Sacr exconjugants were selected, and omega insertion was confirmed by Southern blot analysis. The resulting clone was designated ΩIA.
Complementation of flhD mutants.
Complementation was done by means of mating experiments. A low-copy-number mobilizable plasmid pRK404 (derivative of RK2) (Table 1) was used to transfer flhD (pAGE1254) or flhD (pAGE121) genes from variant I into flhD mutants as recipients. pAGE1254 and pAGE121 were constructed in the following way: the 1.9-kb BamHI-PstI fragment from pAGE1054 (Fig. 1) was ligated into the BamHI-PstI digest of pRK404 to yield pAGE1254; the 1.5-kb HindIII fragment from pAGE101 was ligated into the HindIII digest of pRK404 to yield pAGE121. Both plasmids were transformed into E. coli HB101 and MC1000 flhD::Tn10 as a control of motility restoration. E. coli HB101 carrying pAGE1254 or pAGE121 was conjugally mated to the mutant by a triparental cross on nitrocellulose with HB101 (pRK2013 was used as a helper plasmid [Table 1]). Exconjugants were selected for tetracycline and chloramphenicol resistance and screened for kanamycin sensitivity (absence of pRK2013).
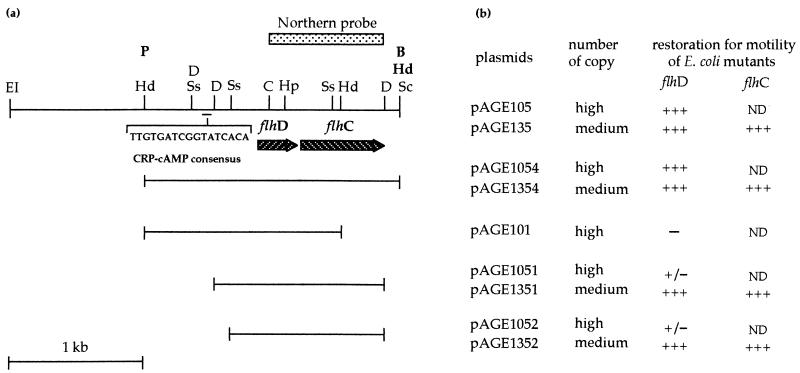
FIG. 1.
(a) Physical and genetic map of the X. nematophilus F1/1 flhDC locus. Selected restriction sites are shown as follows: B, BamHI; C, ClaI; D, DraI; EI, EcoRI; Hd, HindIII; Hp, HpaI; P, PstI; Sc, ScaI; Ss, SspI. pUC19 cloning sites are indicated in bold type. Hatched arrows indicate flhDC ORFs and the direction of gene transcription. The stippled box above the map represents the DNA probe used for the Northern blot. Solid bars represent DNA fragments inserted in pUC19 (high-copy-number plasmid) and/or pACYC184 (medium-copy-number plasmid). (b) Complementation analysis. The effects of various deletions on the restoration of motility of E. coli mutants MC1000 flhD::Tn10 (for flhD mutant) and YK4323 (for flhC mutant) are shown. +++, large spreading area in mot agar (diameter up to 20 mm); +/−, weak spreading area (5 mm); −, no spreading area; ND, not done.
Electron microscopy and phase-contrast microscopy.
Early-exponential-phase bacteria were washed in phosphate buffer (0.1 M; pH 8). Carbon-coated copper grids (400 mesh) were floated onto a drop of washed bacteria, rinsed in ultrapure grade water, and negatively stained with 0.5% (wt/vol) phosphotungstic acid (5 to 10 s). Electron microscopy was performed with a JEOL 1200 X transmission electron microscope. Swimming motility and paracrystalline inclusions were observed with an Olympus light microscope. Early-exponential-phase bacteria were observed for motility, while paracrystalline inclusions were more detectable in 48-h-old bacteria grown on nutrient agar.
Immunoblotting.
Lysates of whole cells were processed as follows. Whole-cell lysates from an exponential-phase culture in LB (for flagellin staining) and from 48-h-old cultures grown on nutrient agar (bioMérieux, Craponne, France) (for pilin staining) were prepared by boiling cells in Laemmli buffer (38). Proteins partitioned in sodium dodecyl sulfate gels were transferred to nitrocellulose membranes (BAS85; Schleicher & Schuell). Membranes were incubated in a 1:300 dilution of rabbit antiserum directed against the denatured 36-kDa flagellin (28) and in a 1:200 dilution of antiserum directed against the denatured 16-kDa pilin (43). Protein bands were detected as previously described (28).
Phenotypic characterization of flhD mutants.
Antibiotic, lecithinase, and DNase activities were tested by previously described methods (3, 8). Extracellular lipase was indicated by a halo of precipitated material surrounding the colony cultured on Tween 20, 40, and 60 agar as previously described (56). Hemolytic activity was determined by using blood agar plates and liquid hemolytic assay (50). (i) Bacteria were grown on Trypticase soy (bioMérieux) with 5% (vol/vol) defibrinated sheep blood (bioMérieux); hemolysis was determined by the presence of a clearing surrounding bacteria grown on standard sheep blood agar plates. (ii) Determination of the hemolytic activities in bacterial supernatant or in intracellular protein extracts (prepared as previously described [28]) was achieved by the liquid hemolytic assay (50). Briefly, bacterial cells were harvested during growth for 5 days. After centrifugation and ultrafiltration (0.2-μm-pore-size filters; Millipore), the extracts were mixed with a suspension (25 μl) of phosphate-buffered saline-washed sheep erythrocytes (bioMérieux) to a final concentration of 5% (vol/vol). The mixture was incubated at 37°C for 45 min. After centrifugation to remove unlysed cells and cell membranes, the hemoglobin released present in the samples was determined by measuring the optical density at 540 nm. The hemolytic unit (HU) (release of 100% of total hemoglobin) is defined as follows: (A540 for the sample with hemolysin − A540 for the control without hemolysin)/(A540 for the complete lysis caused by mixing ultrapure grade water).
The agglutination assays were performed as previously described (43) with bacterial suspensions (109 cells per ml) and sheep erythrocytes (bioMérieux). The titer was defined as the reciprocal of the highest dilution of the samples causing visible agglutination.
Standard biochemical traits were assayed by using Biotype 100 strips (bioMérieux) to study the metabolism of carbon sources (carbohydrates, amino acids, and organic acids) in assimilation tests, API 20E strips to detect enzymes involved in the metabolism and fermentation of a few carbohydrates and API 50CH strips to study the fermentation of carbohydrates.
In vivo pathogenicity assays.
The common cutworm, Spodoptera littoralis, was reared with a photoperiod of 12 h on an artificial diet at 24°C. Fourth-instar larvae were selected and surface sterilized with 70% (vol/vol) ethanol prior to intrahemocoelic injection. Then, with a Hamilton syringe, groups of 20 larvae were injected with 20 μl of bacterial culture. To make the experiments reproducible, bacteria were grown to late logarithmic phase in LB broth, washed, and diluted in phosphate-buffered saline. Treated larvae were individually incubated for up to 96 h, and the time at which insects died was recorded. Bacterial concentrations were determined by CFU by plating dilutions onto nutrient agar. Statistical analysis were performed by comparing survival experiments. The rank test (Wilcoxon test) was used to compare mortality patterns.
Nucleotide sequence accession number.
The sequence for the X. nematophilus flhDC operon has been assigned EMBL accession no. AJ012828.
RESULTS
Cloning, DNA sequence analysis, and expression of the flhDC genes from X. nematophilus.
A Southern blot prepared with X. nematophilus F1 genomic DNA cleaved with EcoRI was hybridized with an E. coli labeled flhDC probe. One hybridizing 10-kb fragment was detected (data not shown). We therefore enriched for EcoRI fragments in the size range of 8 to 12 kb for cloning into the EcoRI site of pUC19. From 600 recombinant colonies, one colony was able to restore E. coli MC1000 flhD::Tn10 motility in mot agar. Plasmid DNA (pAGE1) purified from this recombinant colony contained the expected 10-kb insert. Subcloning steps yielded plasmid pAGE1054 with the 1.9-kb HindIII insert which still complemented MC1000 flhD::Tn10 for full motility (Fig. 1). This insert was characterized by DNA sequence analysis. Two open reading frames (ORFs) in the same orientation were found within the 1,912 nucleotides. Both have distinct start and stop codons but Xenorhabdus flhD lacks an obvious ribosome-binding sequence (RBS) consensus, as previously reported for E. coli (7, 41). A good match to the RBS consensus upstream of the second ORF, flhC, was found. A stem-loop structure (with a minimum free energy of 31.3 kcal/mol) was found downstream of flhC; however, this structure is not followed by the series of uridine residues characteristic of E. coli rho-independent terminator (16). As expected, the predicted primary amino acid sequences are 72 and 78% identical to E. coli FlhD and FlhC, respectively. The strongest homology for X. nematophilus FlhDC is to FlhD (27) from Proteus mirabilis (91% similarity; 85% identity) and to FlhC (61) from Y. enterocolitica (88% similarity; 86% identity).
There is a putative cyclic AMP (cAMP)-receptor protein (CRP) binding site upstream of flhD (Fig. 1) which matches the CRP binding site consensus sequence (5′-AANTGTGA-N6-NCANATT-3′) (17). To investigate the influence of this latter sequence on restoration of motility to E. coli mutants, cloned fragments containing flhDC genes from Xenorhabdus carried on a high-copy-number plasmid (pUC19 derivatives) or a medium-copy-number plasmid (pACYC184) (Table 1) were transformed into E. coli MC1000 flhD::Tn10 strain and YK4323 (a flhC::Mu E. coli mutant). The transformants were examined for motility restoration in mot agar (Fig. 1) and under light microscope. Medium-copy-number plasmids containing flhDC with or without the CRP consensus sequence restore full motility to mutants, while the pUC19 derivatives with the same inserts without the CRP consensus sequence restored motility weakly in the MC1000 background (Fig. 1). Although a gene dosage effect could account for this latter result, this region may have some effects on restoration of E. coli mutant motility. Catabolite repression has been reported to repress E. coli flagellar production in the presence of glucose (1) at the level of the flhDC operon. Increase of glucose concentration in mot agar to 55 mM showed no inhibitory effect on all E. coli transformants previously obtained (see above). Moreover, Xenorhabdus motility was not impaired by adding glucose (up to 55 mM). Furthermore, complementation experiments demonstrated that an insert with flhD alone is not able to complement MC1000 flhD::Tn10 for motility, owing to the polar effect of the Tn10 insertion (Fig. 1). Southern blot hybridizations were used to compare the gross structure of the flhDC-flanking region in both X. nematophilus variants. Genomic DNA from F1/1 and F1/2 digested with four restriction enzymes (DraI, EcoRI, HindIII, and SspI) were Southern blotted and hybridized to a 7.3-kb EcoRI-BamHI fragment from pAGE1 used as a flhDC probe. The phase I and phase II variants showed the same hybridization patterns with all restriction enzymes used (data not shown). This suggests that the gross structure of the flhDC locus and its surrounding region are identical in both phase variants. Last, expression of flhDC genes was investigated by Northern blot analysis. Total cellular RNA from a log-phase culture of F1/1 and F1/2 was purified and hybridized with a flhDC probe (Fig. 1). A single mRNA band signal with an approximate size of 1.25 kb was labeled with variant I and variant II RNA (data not shown). It is likely that flhDC are transcribed as an operon. Moreover, the amount of the message was identical in both variants grown under the same conditions (data not shown).
X. nematophilus flhDC operon controls swimming and swarming motility and flagellar synthesis.
Previous studies have shown that X. nematophilus F1/1 displayed swarming and swimming motility and that the filament of the flagella is composed of one type of flagellin (the FliC protein) with an apparent molecular mass of 36.5 kDa (28). To assess whether the master flhDC operon controls motility in Xenorhabdus, a flhD chromosomal null mutant (ΩIA) was constructed via allelic exchange. Electron microscopy showed that ΩIA cells are nonmotile and nonflagellated, and Western blotting using antiflagellin antiserum confirmed that ΩIA was unable to synthesize 36.5-kDa flagellin subunits (data not shown). This aflagellate flhD mutant was also unable to migrate over the surface of a solid medium (0.8% agar) (data not shown) that allowed Xenorhabdus swarming behavior (28). Complementation experiments with low-copy-number mobilizable plasmid derivatives from pRK404 containing flhDC restored both swarming and swimming motility similar to that of wild-type strain F1/1 (Fig. 2A).
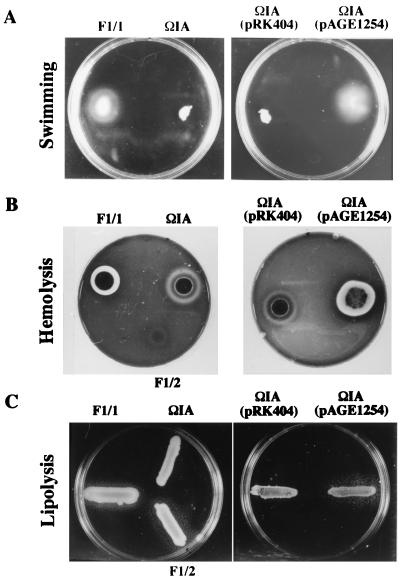
FIG. 2.
Phenotypic analysis of X. nematophilus phase variants, the flhD null mutant (ΩIA) and the complemented mutant [ΩIA(pAGE1254)]. In order to maintain the plasmid, ΩIA with the cloned X. nematophilus flhDC (pAGE1254) was inoculated into assay plates containing tetracycline (7.5 mg/liter). ΩIA(pRK404) was used as a control. (A) Swimming motility assays. Motility agar plates (mot broth with 0.35% agar) were point inoculated with the indicated strains and incubated for 12 h at 28°C. (B) Hemolysis activity on sheep blood agar. Portions (10 μl) of cultures of the indicated strains grown overnight were deposited on plates and incubated at 28°C for 72 h. (C) Lipolysis activity on Tween 20 agar. Samples of cultures of the indicated strains grown overnight were streaked as a line on the agar plates and incubated at 28°C for 72 h.
Phenotypic analysis of flhD null mutants.
First, 160 standard traits were assayed by using Biotype 100 strips, API 20E strips, and API 50CH strips to compare the wild type and flhD null mutant for the metabolism of carbon sources (assimilation and fermentation tests) and the presence of enzymes involved in the metabolism. In all tests, no difference was observed between F1/1 and ΩIA. The morphology of F1/1 and ΩIA cells grown in LB liquid media and nutrient agar was examined by light microscopy. Exponential-phase cells are mainly rods, although they become increasingly pleiomorphic with apparent spheroplasts during the stationary period. F1/1 and ΩIA (48-h-old) cells grown on solid media harbored one or two paracrystalline inclusions. The only morphological difference between the wild-type strain and flhD mutant was the presence of large amorphous cells in 3-day-grown cultures of ΩIA cells (representing 1% of population) (data not shown).
As previously described (28, 58), the swimming changes in X. nematophilus F1 during phase shift are associated with the loss of several phenotypic traits (Table 2). ΩIA exhibited three phenotypes different from its parental strain F1/1 (Table 2). As described above, this latter mutant is unable to swim in mot agar (Fig. 2A), but surprisingly, only a partial hemolysis zone on blood agar (Fig. 2B) and no halo on Tween agar (Fig. 2C) were observed. F1/1 colonies (48 h old) produced total hemolysis, while the flhD null mutant under the same conditions displayed only a faint halo of partial hemolysis. As shown in Fig. 2B, 72-h-old cultures of the mutant produce an unusual type of hemolysis previously described by Farmer et al. (23). This type of hemolysis, namely, partial hemolysis immediately around the colony and a thin line of complete hemolysis at some distance from the colony (Fig. 2B), has been designated annular hemolysis.
TABLE 2.
Phenotypic characteristics of phase variants and flhD null mutants of X. nematophilus F1a
| Strain | Description | Chemotaxis | Sheep blood hemolysisb | Lipolysisc of Tween 20, 40, and 60 | Lecithinased | Antibiotic productione | DNase | Btb adsorptionf | Ha titerg |
|---|---|---|---|---|---|---|---|---|---|
| F1/1 | Parental strain | + | T | W+ | + | + | + | B | 64 |
| ΩIA | F1/1 (flhD::ΩCm) | − | AH | − | + | + | + | B | 64 |
| ΩIA(pAGE1254)h | pAGE1254 carries flhDC operon | + | T | + | +W | +W | + | BG | 64 |
| ΩIA(pAGE121)h | pAGE121 carries only flhD | − | AH | − | + | + | + | BG | ND |
| F1/2 | Phase II variant | − | − | + | − | W+ | + | R | <2 |
All plates were cultured for 2 days at 28°C before assays were interpreted unless indicated otherwise. +, positive, −, negative.
T, total hemolysis; AH, annular hemolysis observed with 72-h-old cultures.
Tween plates were observed with 72-h-old-cultures. +, halo of precipitated material up to 6 mm in diameter; W+, perceptible halo less than 6 mm.
+, halo up to 8 mm in diameter; +W, perceptible halo with variable diameters depending on the experiment but always less than 8 mm.
Zones of growth inhibition of Micrococcus luteus or E. coli HB101(pRK404) (for assays using tetracycline plates, see footnote h). +, up to 40 mm in diameter; +W, perceptible inhibition zones with variable values depending on the experiment but always less than 40 mm; W+, zones less than 20 mm.
Btb, bromothymol blue; B, dark blue colonies; BG, blue-green colonies; R, red colonies.
Ha, sheep erythrocyte agglutination (see Materials and Methods for titer calculation). ND, not determined.
Assays were performed with addition of tetracycline (for plasmid maintenance) and compared with the same strain containing pRK404 without insert.
In order to estimate the production of extracellular hemolysin, liquid hemolysis assays were performed. Cell-free supernatants collected during cell growth of F1/1 and ΩIA were incubated with erythrocytes, and the hemoglobin released from erythrocytes was spectrophotometrically measured as the indicator of cytolysis (see Materials and Methods). Hemolytic activity (0.5 HU) appears during the stationary phase of the wild-type strain, while no activity was detected in ΩIA culture supernatant from cells grown for 5 days. No hemolytic activity against sheep erythrocytes was detected in ΩIA intracellular protein extracts. Intracellular proteins from F1/1 grown in the same culture conditions, used as a control, exhibited cytolytic activity (0.2 HU). Moreover, no Tween lipase was detected in F1/1 supernatants containing active extracellular hemolysin. The other characteristics tested (lecithinase-like activity, antibiotic production, DNase activity, hemagglutination of sheep erythrocytes, and pilin synthesis) (Table 2) were comparable to those observed in the parental strain. Table 2 also showed that phenotypes of the flhD mutant were different than those of the phase II variant.
Complementation experiments with low-copy-number mobilizable plasmid containing flhDC (see above) restored motility (Fig. 2A), hemolysis (Fig. 2B), and Tween lipase activities for ΩIA (Fig. 2C) (Table 2). The flhD null mutant (ΩIA) carrying only the flhD gene in trans had phenotypes similar to those of the noncomplemented mutant (Table 2). Although lecithinase and antibiotic phenotypes were not altered in ΩIA mutants, a variable reduction (20 to 50%) in halo size from that of F1/1 was observed when flhDC were placed in trans in this mutant (Table 2). In order to confirm this latter observation, when pRK620 containing the E. coli flhDC genes (Table 1) and pAGE1254 were transferred into this mutant or in the wild-type strain F1/1, respectively, a significant reduction of lecithinase and antibiotic production was again obtained in these exconjugants (data not shown).
Virulence of flhD mutant and phase variants for Spodopera littoralis.
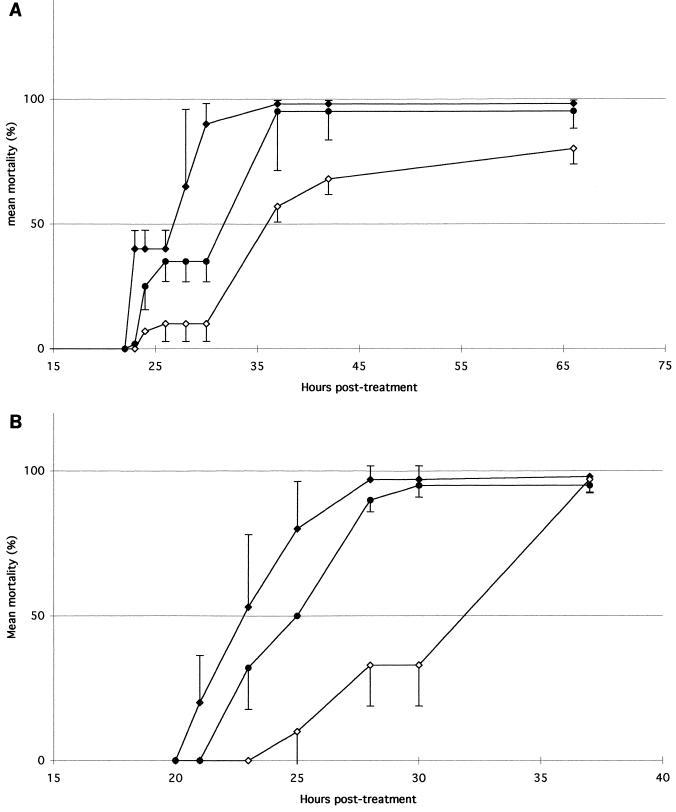
The fact that the flhD mutant lost three potential virulence factors prompted us to question the in vivo relevance of these phenotypes. Living cells of wild-type X. nematophilus are highly virulent when injected into hemolymph of insects; the 50% lethal dose (LD50) for the lepidopteran Galleria mellonella was less than five cells (4). To assess the effects of flhD mutations on virulence in insects, two bacterial doses of F1/1, F1/2, and ΩIA were injected into the hemocoel of another lepidopteran, Spodoptera littoralis. At both doses used, almost all injected larvae were dead within about 35 h (except for ΩIA) and septicemia was observed in every case. To check that the flhD mutant and the phase II variant remain nonmotile in an insect environment, hemolymph samples were collected during septicemia and motility was observed under a light microscope. As expected, only F1/1 displayed a motile phenotype. The LD50s calculated for the three strains (F1/1, ΩIA, and F1/2) were very low, less than 20 bacteria. When mortality was monitored over a 3-day period postinjection, mortality patterns of F1/1- and ΩIA-injected larvae were significantly different (P < 0.001) with both doses of bacteria (Fig. 3). By 30 h postinjection, 90% of F1/1-injected larvae were dead, while only 10% ΩIA-injected larvae were dead (Fig. 3A). The calculated time to 50% lethality (TL50) in both experiments (Fig. 3) was longer for ΩIA (36 and 32 h, respectively) than for the wild type (26 and 22 h, respectively). The TL50 of F1/1 was significantly (P < 0.001) shorter than that for F1/2 in both experiments (Fig. 3).
FIG. 3.
Mortality of Spodoptera littoralis infected with two doses of bacterial cells. End-exponential-phase bacteria were injected in fourth-instar larvae. (A) Means (± standard errors of the means [SEMs]) of bacterial cells injected with F1/1, F1/2, and ΩIA are 52 (±13), 47 (±4), and 47 (±13), respectively. (B) Means (± SEMs) of bacterial cells injected with F1/1, F1/2, and ΩIA are 250 (±10), 265 (±47), and 275 (±45), respectively. Mortality values are based on injections into 20 larvae. Results expressed as percentages represent the averages of mortality from three independent experiments. Errors bars indicate standard deviations. Symbols: ⧫, F1/1; ●, F1/2; ◊, ΩIA.
DISCUSSION
The flhDC operon controls flagellar synthesis, motility, and swarming in Xenorhabdus.
The DNA sequence analysis revealed extensive homology of the coding regions for FlhD and FlhC with their respective homologues from other members of the family Enterobacteriaceae. Insertional inactivation of the chromosomal flhD locus produced nonswarmer, nonmotile Xenorhabdus cells unable to synthesize flagellin. Complementation studies using the Xenorhabdus flhDC locus in trans restored swimming and a swarming behavior to the flhD null mutants. These data demonstrated that the master operon, flhDC, controls flagellar synthesis, motility, and swarming behavior as previously reported in other bacteria (Serratia, Proteus, and Yersinia) (20, 27, 61).
We previously showed that phase I cells of strain F1 were motile and able to swarm on appropriate medium, while the phase II variants were nonmotile cells which did not synthesize flagellin (28). It was also shown that the fliC gene encoding flagellin, which belongs to the class III flagellar genes, was expressed in phase I variant but not in phase II variant cells (29). This latter result suggested that a gene higher in the transcriptional hierarchy of the flagellar regulon is switched off in phase II variants. This study showed that flhDC gene structure and expression in phase II variants are not altered, suggesting that this locus is not responsible for flagellar variation phenomenon. In E. coli and S. enterica serovar Typhimurium, the class III flagellar genes are under the control of FliA (ς28). However, it was demonstrated that the promoter region of certain Salmonella class III genes including fliD also contains class II promoters (FlhDC dependent) (32, 37). Indeed, it was previously shown that X. nematophilus phase II variants were able to produce a small amount of fliD mRNA (29). This may indicate that the weak transcription of phase II fliD gene is dependent on FlhDC and that fliA expression in phase II variant is impaired. We are presently studying the fliA-flgM regulatory system in Xenorhabdus.
Lipolysis and extracellular hemolysin activity are flhDC dependent.
Examination of X. nematophilus flhD mutant phenotypes revealed the surprising result that the flagellar master operon is required for expression of at least two nonflagellar products involved in lipolysis and hemolysis. Even if these properties are also phase-characteristic phenotypes like motility (Table 2), it is clear that the complete extinction of the flagellar regulon gives a mutant with phenotypes quite different from that of the phase II variant. However, examination of about a hundred phase-independent phenotypes revealed no difference between the flhD mutant and the wild-type strain. The results presented here indicate that at least two separate genetic networks, the flagellar regulon and an unidentified phase shift-dependent network, differentially control the expression of these three functions in X. nematophilus.
Gene disruption and complementation experiments illustrate that expression of flagellar proteins and secreted products involved in hemolysis and lipolysis are clearly dependent on the presence of both flhD and flhC genes (Table 2). In E. coli, FlhD alone, not the FlhDC heterotetramer, is involved in cell division (48). In contrast, the intact operon is required to control phospholipase A-encoding genes in Serratia liquefaciens and Y. enterocolitica (30, 60). The Serratia phospholipase pA promoter exhibits similarity to FliA-controlled promoter (31). Moreover, it was clearly demonstrated in both genera that the phospholipase operon belongs to the class III flagellar genes (30, 60). In Xenorhabdus, it was reported that molecules involved in lecithinase and broad lipase activity are distinct (56). The ability to produce lecithinase in the Xenorhabdus flhD mutant lacking Tween lipase and hemolysin (Table 2) also suggests that the Xenorhabdus flhDC operon controls genes quite different than the phospholipase A-encoding genes controlled by the flhDC operon in Serratia and Yersinia (31, 52).
It was also recently proposed that the type III export apparatus of the flagellar system transports the virulence-associated phospholipase A and several unknown nonflagellar secreted proteins in Y. enterocolitica (60). To date, no report has described the direct involvement of the master flagellar operon in expression and secretion of other biologically active molecules. However, HpmA hemolysin in Proteus mirabilis is coinduced with flagellin during swarm cell differentiation (6). The molecules or genes involved in lipolysis or hemolytic activity in Xenorhabdus are not known. Nevertheless, the absence of intracellular hemolytic activity in the flhD mutant suggests that the FlhDC control of both transcription and export should be considered in Xenorhabdus.
The β-hydroxybutanoyl homoserine lactone autoinducer increased Tween lipase activity in transpositional mutants of X. nematophilus (19) and can restore virulence to avirulent mutants (19). Thus, Tween lipase activity in X. nematophilus could be controlled by both flhD and quorum sensing. This link between both global regulators has already been described during formation of swarming colonies in S. liquefaciens (21, 40).
Hemolysis in Photorhabdus, formerly called Xenorhabdus luminescens, was first described by Farmer et al. (23), who observed an unusual reaction on a sheep blood plate, which was designated annular hemolysis (5). This reaction was at first considered to be a marker in recognizing P. luminescens strains isolated from clinical specimens (23) but also occurred in other P. luminescens isolated from nematodes (5). The Xenorhabdus flhD mutant produces annular ring hemolysis reaction on blood agar (Fig. 2B). Unlike the wild-type strain, the lack of extracellular hemolysis production in the flhD mutant should allow for this observation. This particular phenotype of the flhD mutant strongly suggests the production of two types of hemolysin in X. nematophilus F1, (i) one flhDC-independent hemolysin giving the annular ring phenotype on blood agar and (ii) one flhDC-dependent extracellular hemolysin with cytolytic activity against sheep erythrocytes. The X. nematophilus extracellular hemolysin may be involved in cytotoxic effects observed on insect immunocompetent cells. New insect hemocyte cytotoxic factors recently identified from in vitro incubation of the nematode-bacterium complex provided from the bacterial symbiont X. nematophilus were shown to be distinct from lipopolysaccharide (49).
flhDC-mediated properties are involved in virulence in insects.
A series of data illustrates relationships between flagellum-mediated motility and bacterial virulence in hosts (45). A few examples at the molecular level showed that regulation of flagellar synthesis, not motility, is indeed coupled to virulence. In S. enterica serovar Typhimurium, motility per se is not required for pathogenesis, but appropriate flagellar gene expression appears necessary for full virulence (53). In Bordetella bronchiseptica, the genetic network that couples virulence gene regulation to motility has been identified. Motility is repressed by the virulence control system through an analogue of flhDC, and negative control is crucial for in vivo virulence (2). X. nematophilus is highly pathogenic to insects, with a LD50 of less than 20 bacteria to kill Galleria (4) or Spodoptera (this study). However, virulence factors are generally unknown in this genus. The transposition mutants are often useful to identify virulence-associated genes. However, in X. nematophilus, multiple Tn5 insertions were found (34). Even if the Tn5 mutants were pleiotropic, all five avirulent mutants were nonmotile and partially impaired in blood hemolysis (59). These data prompt us to study virulence of flhD mutants that displayed similar phenotypes. Surprisingly, flhD mutants remain virulent and are only attenuated, showing significant increases in TL50 compared to that of the wild-type strain. All these data taken together suggest that one or more flhDC-mediated properties are involved in infection but are not necessary for this process. It is likely that pathogenicity of Xenorhabdus towards insect hosts is determined by a large number of factors, and the loss of several of them may not change virulence.
It is clear from this study that the Xenorhabdus flhDC operon is an important global regulator affecting stationary phase-expressed virulence factors like extracellular hemolysin and Tween lipase besides flagellum-mediated motility. These data illustrate, for the first time in insects, the relationship between the flagellar regulon and virulence. This view of flhDC as a major checkpoint of genetic regulation argues for the presence of multiple flhDC-dependent genes outside the flagellar regulon involved in biogenesis of flagellum.
ACKNOWLEDGMENTS
We thank Jean Luciani for valuable assistance, Steve Forst (University of Wisconsin) and Noel Boemare for useful discussions, Sylvaine Artero for statistical analysis, and Alan Kirk (USDA, Montpellier, France) for help with English.
This work was supported in part by a grant from Institut National de Recherche Agronomique (grant no. AIP 188).
REFERENCES
- 1.Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 3.Akhurst R J. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- 4.Akhurst R J. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes, Neoaplectana and Heterorhabditis. J Gen Microbiol. 1980;121:303–309. [Google Scholar]
- 5.Akhurst R J, Mourant R G, Baud L, Boemare N E. Phenotypic and DNA relatedness between nematode symbionts and clinical strains of the genus Photorhabdus (Enterobacteriaceae) Int J Syst Bacteriol. 1996;46:1034–1041. doi: 10.1099/00207713-46-4-1034. [DOI] [PubMed] [Google Scholar]
- 6.Allison C, Lai H C, Hughes C. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol Microbiol. 1992;6:1583–1591. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boemare N, Thaler J-O, Lanois A. Simple bacteriological tests for phenotypic characterization of Xenorhabdus and Photorhabdus phase variants. Symbiosis. 1997;22:167–175. [Google Scholar]
- 9.Boemare N E, Akhurst R J. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae) J Gen Microbiol. 1988;134:1835–1845. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- 10.Boemare N E, Akhurst R J, Mourant R G. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43:249–255. [Google Scholar]
- 11.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, Ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 12.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 13.Brenner D J, McWhorter A C, Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S, Cohen S N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 15.Couche G A, Gregson R P. Protein inclusions produced by the entomopathogenic bacterium Xenorhabdus nematophilus subsp. nematophilus. J Bacteriol. 1987;169:5279–5288. doi: 10.1128/jb.169.11.5279-5288.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 17.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 18.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 19.Dunphy G, Miyamoto C, Meighen E. A homoserine lactone autoinducer regulates virulence of an insect-pathogenic bacterium, Xenorhabdus nematophilus (Enterobacteriaceae) J Bacteriol. 1997;179:5288–5291. doi: 10.1128/jb.179.17.5288-5291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberl L, Winson M K, Sternberg C, Stewart G S, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-L-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 22.Endo B Y, Nickle W R. Ultrastructure of the intestinal epithelium, lumen and associated bacteria in Heterorhabditis bacteriophora. J Helminthol. 1991;58:202–212. [Google Scholar]
- 23.Farmer J J D, Jorgensen J H, Grimont P A, Akhurst R J, Poinar G O, Jr, Ageron E, Pierce G V, Smith J A, Carter G P, Wilson K L, Hickman-Brenner F W. Xenorhabdus luminescens (DNA hybridization group 5) from human clinical specimens. J Clin Microbiol. 1989;27:1594–1600. doi: 10.1128/jcm.27.7.1594-1600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 25.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 27.Furness R B, Fraser G M, Hay N A, Hughes C. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J Bacteriol. 1997;179:5585–5588. doi: 10.1128/jb.179.17.5585-5588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Givaudan A, Baghdiguian S, Lanois A, Boemare N. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol. 1995;61:1408–1413. doi: 10.1128/aem.61.4.1408-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Givaudan A, Lanois A, Boemare N. Cloning and nucleotide sequence of a flagellin encoding genetic locus from Xenorhabdus nematophilus: phase variation leads to differential transcription of two flagellar genes (fliCD) Gene. 1996;183:243–253. doi: 10.1016/s0378-1119(96)00452-0. [DOI] [PubMed] [Google Scholar]
- 30.Givskov M, Eberl L, Christiansen G, Benedik M J, Molin S. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 31.Givskov M, Molin S. Expression of extracellular phospholipase from Serratia liquefaciens is growth-phase-dependent, catabolite-repressed and regulated by anaerobiosis. Mol Microbiol. 1992;6:1363–1374. doi: 10.1111/j.1365-2958.1992.tb00857.x. [DOI] [PubMed] [Google Scholar]
- 32.Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 34.Hurlbert R E. Investigations into the pathogenic mechanisms of the bacterium-nematode complex: the search for virulence determinants of Xenorhabdus nematophilus ATCC 19061 could lead to agriculturally useful products. ASM News. 1994;60:473–478. [Google Scholar]
- 35.Komeda Y. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J Bacteriol. 1982;150:16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 37.Kutsukake K, Ide N. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol Gen Genet. 1995;247:275–281. doi: 10.1007/BF00293195. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Leisman G B, Waukau J, Forst S A. Characterization and environmental regulation of outer membrane proteins in Xenorhabdus nematophilus. Appl Environ Microbiol. 1995;61:200–204. doi: 10.1128/aem.61.1.200-204.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindum P W, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 43.Moureaux N, Karjalainen T, Givaudan A, Bourlioux P, Boemare N. Biochemical characterization and agglutinating properties of Xenorhabdus nematophilus F1 fimbriae. Appl Environ Microbiol. 1995;61:2707–2712. doi: 10.1128/aem.61.7.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 45.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 46.Poinar G O, Thomas G M. Significance of Achromobacter nematophilus Poinar et Thomas (Achromobacteriaceae, Eubacteriales) in the development of the nematode DD 136. Parasitology. 1966;56:385–390. doi: 10.1017/s0031182000070980. [DOI] [PubMed] [Google Scholar]
- 47.Pruss B M. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch Microbiol. 1998;170:141–146. doi: 10.1007/s002030050626. [DOI] [PubMed] [Google Scholar]
- 48.Pruss B M, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribeiro C, Duvic B, Oliveira P, Givaudan A, Palha F, Simões N, Brehélin M. Insect immunity—effects of factors produced by a nematobacterial complex on immunocompetent cells. J Insect Physiol. 1999;45:677–685. doi: 10.1016/s0022-1910(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 50.Rowe G E, Welch R A. Assays of hemolytic toxins. Methods Enzymol. 1994;235:657–667. doi: 10.1016/0076-6879(94)35179-1. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt C K, Darnell S C, O'Brien A D. The attenuated phenotype of a Salmonella typhimurium flgM mutant is related to expression of FliC flagellin. J Bacteriol. 1996;178:2911–2915. doi: 10.1128/jb.178.10.2911-2915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 56.Thaler J O, Duvic B, Givaudan A, Boemare N. Isolation and entomotoxic properties of the Xenorhabdus nematophilus F1 lecithinase. Appl Environ Microbiol. 1998;64:2367–2373. doi: 10.1128/aem.64.7.2367-2373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas G M, Poinar G O. Xenorhabdus gen. nov., a genus of entomopathogenic and nematophilic bacteria of the family Enterobacteriaceae. Int J Syst Bacteriol. 1979;29:352–360. [Google Scholar]
- 58.Volgyi A, Fodor A, Szentirmai A, Forst S. Phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol. 1998;64:1188–1193. doi: 10.1128/aem.64.4.1188-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu J M, Olsen M E, Kahn M L, Hurlbert R E. Characterization of Tn5-induced mutants of Xenorhabdus nematophilus ATCC 19061. Appl Environ Microbiol. 1991;57:1173–1180. doi: 10.1128/aem.57.4.1173-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young G M, Smith M J, Minnich S A, Miller V L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]