Abstract
In Escherichia coli, nine essential cell division proteins are known to localize to the division septum. FtsL is a 13-kDa bitopic membrane protein with a short cytoplasmic N-terminal domain, a membrane-spanning segment, and a periplasmic domain that has a repeated heptad motif characteristic of leucine zippers. Here, we identify the requirements for FtsL septal localization and function. We used green fluorescent protein fusions to FtsL proteins where domains of FtsL had been exchanged with analogous domains from either its Haemophilus influenzae homologue or the unrelated MalF protein to show that both the membrane-spanning segment and the periplasmic domain of FtsL are required for localization to the division site. Mutagenesis of the periplasmic heptad repeat motif severely impaired both localization and function as well as the ability of FtsL to drive the formation of sodium dodecyl sulfate-resistant multimers in vitro. These results are consistent with the predicted propensity of the FtsL periplasmic domain to adopt a coiled-coiled structure. This coiled-coil motif is conserved in all gram-negative and gram-positive FtsL homologues identified so far. Our data suggest that most of the FtsL molecule is a helical coiled coil involved in FtsL multimerization.
Cell division in bacteria culminates with the formation of the midcell septum followed by cell separation. In Escherichia coli, at least nine gene products, FtsZ, FtsA, ZipA, FtsQ, FtsL, FtsI, FtsK, FtsW, and FtsN, have been identified as being essential for septation, and the septal localization of all of them has been reported previously (1–3, 5, 8, 12, 20, 29, 30, 31). The nature of the mechanism leading to the selection of the midcell division site is still unknown and poses one of the most fundamental question in cell biology. One approach to this issue is to determine the nature of the septal localization signal that targets cell division proteins to the division site.
Recent progress indicates that only portions of certain Fts proteins are necessary for septal localization. In the case of FtsK, the N-terminal-15% portion of this protein (comprising the membrane-spanning segments) is sufficient for localization to the division site but not for complementation of an ftsK defect (16). For FtsN, the periplasmic domain promotes targeting of this protein to the septum (29). Studies with replacements of the cytoplasmic and transmembrane domain of the bitopic membrane protein FtsQ suggest that the cytoplasmic domain and the membrane-spanning segment have little or no role in FtsQ targeting and function at the division site (3, 5). In contrast, for FtsI, a bitopic membrane protein with a large periplasmic enzymatic domain involved in peptidoglycan biosynthesis, replacements of the cytoplasmic or transmembrane domain of FtsI failed to complement and localize at the division site. These results, along with the fact that no common targeting motif can be identified in these proteins, suggest that each cell division protein carries a distinct and specific localization signal.
In this paper, we identify the requirements for septal localization and function of the cell division protein FtsL. Although the function of FtsL is still unknown, its sequence is conserved in gram-negative and gram-positive bacteria, and it is an essential cell division protein in E. coli and Bacillus subtilis (7, 9). FtsL is a bitopic cytoplasmic membrane protein whose structure includes a small amino-terminal cytoplasmic domain (37 residues), a single transmembrane segment (20 residues), and a small periplasmic domain (64 residues). The sequence of the periplasmic domain of FtsL exhibits a stretch of five leucines separated from each other by seven residues which has features of an α-helical leucine zipper. The presence of this motif raises the possibility that FtsL interacts with itself or other proteins to form homo- or heterodimers. Studies in which the three domains of FtsL were exchanged with those of other bitopic membrane proteins showed that all three domains of FtsL are required for its function (11). Different possible roles can be attributed to FtsL domains, including that of signaling localization to the cell septum. Thus, one possible consequence of exchanging FtsL domains might be to interfere with its localization.
We recently used fluorescence microscopy to demonstrate that E. coli FtsL (FtsLEcol) is localized to the division site and that FtsL localization depends on the prior recruitment of FtsZ, FtsA, ZipA, and FtsQ but not FtsI and FtsN. These results, along with similar experiments done with FtsI and FtsQ, led us to propose that FtsL is recruited after FtsZ, ZipA, FtsA, and FtsQ and before FtsI and FtsN (8).
In this paper, we use green fluorescent protein (GFP) fusion technology to analyze the septal localization and function of FtsL derivatives in which the cytoplasmic domain and/or membrane-spanning segment of FtsL is replaced with analogous parts of either MalF, a maltose transport protein with no role in cell division, or FtsLHinf, the Haemophilus influenzae FtsLEcol homologue. Our main finding is that both the membrane-spanning segment and the periplasmic domain of FtsLEcol, but not the cytoplasmic domain, are required for localization to the division site. Further, we found that mutagenesis of the leucine repeat heptad motif in the periplasmic domain of FtsL severely impaired both localization and function. Finally, we report that the FtsL periplasmic domain can drive formation of sodium dodecyl sulfate (SDS)-resistant multimers in vitro. These findings are consistent with the likely existence of a coiled-coil structure predicted for the periplasmic domain of all FtsL homologues, which may be involved in a homo- or heterodimerization process important in the mechanism of cell division.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
Bacterial strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. Strain construction was done by generalized transduction with P1 (24) or specialized transduction with λInCh. λInCh picks up plasmid-borne genes by homologous recombination. In this case, the genes were gfp-ftsL or gfp-ftsL derivatives, bla, and lacIq from pDSW207-based plasmids (31). Chromosomal insertions made with λInCh were subsequently stabilized by selecting for a deletion that removed most of the phage, including its killing functions (D. Boyd et al., unpublished data; see also http://beck1.med.harvard.edu/LambdaInCh.html).
TABLE 1.
Bacterial strains used
| Strain | Relevant genetic markers or features | Constructiona | Source or reference |
|---|---|---|---|
| Strains without gfp fusions | |||
| KS272 | F− ΔlacX74 galE galK thi rpsL ΔphoA(PvuII) | Laboratory collection | |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | Laboratory collection | |
| JP313 | MC4100 Δara714 | J. Pogliano | |
| LMG145 | KS272 ftsL::TnphoAL81ΔIS50R(Kanr)/pLMG180 | 9 | |
| JMG47 | KS272 ftsL::TnphoAL81ΔIS50R(Kanr)/pLD45 | P1 on LMG145 × KS272 pLD45, selecting for Kmr | This study |
| DHB4 | F− Δlacpro lacIq/Δ(malF) ΔphoA (PvuII) phoR ΔlacX74 Δ(ara leu)7697 araD139 galE (or U) galK Strr | D. Boyd | |
| DHB9 | DHB4 recA1 srl::Tn10 | D. Boyd | |
| JMG48 | KS272 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG64 | P1 on LMG145 × KS272 pJMG64, selecting for Kmr | This study |
| JMG33 | KS272 ftsL::TnphoAL81ΔIS50R(Kanr) recA1 srl::Tn10/pJMG64 | P1 on DHB9 × JMG48, selecting for Tetr | This study |
| DHB6521 | SM551 λInCh (Kanr) | D. Boyd et al., unpublished data | |
| Depletion backgrounds | |||
| JMG343 | JMG33/pAM238 | This study | |
| JMG344 | JMG33/pJMG40 | This study | |
| JMG100 | JMG48/pJMG473 | This study | |
| JMG348 | JMG33/pJMG38 | This study | |
| JMG347 | JMG33/pJMG39 | This study | |
| JMG359 | JMG33/pJMG444 | This study | |
| JMG350 | JMG33/pJMG157 | This study | |
| JMG351 | JMG33/pJMG158 | This study | |
| JMG354 | JMG33/pJMG162 | This study | |
| JMG355 | JMG33/pJMG163 | This study | |
| JMG356 | JMG33/pJMG164 | This study | |
| JMG353 | JMG33/pJMG161 | This study | |
| JMG352 | JMG33/pJMG159 | This study | |
| gfp fusion in wild-type background | |||
| EC452 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp | 30 | |
| EC438 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-ftsL | 8 | |
| EC531 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-ftsL | 8 | |
| JMG287 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLL | λInCh using pDWS207 derivative pJMG453 | This study |
| JMG441 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-LLL | λInCh using pDWS207 derivative pJMG453 | This study |
| JMG377 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-FLL | λInCh using pDWS207 derivative pJMG447 | This study |
| JMG469 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-FLL | λInCh using pDWS207 derivative pJMG447 | This study |
| JMG289 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LFL | λInCh using pDWS207 derivative pJMG448 | This study |
| JMG292 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-FFL | λInCh using pDWS207 derivative pJMG449 | This study |
| JMG290 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLF | λInCh using pDWS207 derivative pJMG441 | This study |
| JMG291 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-FFF | λInCh using pDWS207 derivative pJMG450 | This study |
| JMG295 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLI | λInCh using pDWS207 derivative pJMG474 | This study |
| JMG299 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLΔL | λInCh using pDWS207 derivative pJMG465 | This study |
| JMG300 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLHinfL | λInCh using pDWS207 derivative pJMG461 | This study |
| JMG371 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-HLL | λInCh using pDWS207 derivative pJMG455 | This study |
| JMG372 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-LHL | λInCh using pDWS207 derivative pJMG475 | This study |
| JMG374 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-HHL | λInCh using pDWS207 derivative pJMG456 | This study |
| JMG373 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-LHH | λInCh using pDWS207 derivative pJMG458 | This study |
| JMG370 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-LLH | λInCh using pDWS207 derivative pJMG459 | This study |
| JMG375 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-HLH | λInCh using pDWS207 derivative pJMG457 | This study |
| JMG376 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-FtsLHinf | λInCh using pDWS207 derivative pJMG454 | This study |
| JMG303 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLL(L70H) | λInCh using pDWS207 derivative pJMG466 | This study |
| JMG304 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLL(L84D) | λInCh using pDWS207 derivative pJMG467 | This study |
| JMG301 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLGCN4L | λInCh using pDWS207 derivative pJMG462 | This study |
| JMG302 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-LLC/EBPL | λInCh using pDWS207 derivative pJMG463 | This study |
| gfp fusions in depletion backgrounds | |||
| JMG366 | JMG531 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | 8 | |
| JMG367 | JMG441 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG441, selecting for Kmr | This study |
| JMG420 | JMG377 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG377, selecting for Kmr | This study |
| JMG416 | JMG371 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG371, selecting for Kmr | This study |
| JMG418 | JMG373 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG373, selecting for Kmr | This study |
| JMG415 | JMG370 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG370, selecting for Kmr | This study |
| JMG419 | JMG375 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG375, selecting for Kmr | This study |
| JMG422 | JMG376 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG376, selecting for Kmr | This study |
| JMG417 | JMG372 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG372, selecting for Kmr | This study |
| JMG442 | JMG374 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | P1 on LMG145 × JMG374, selecting for Kmr | This study |
P1 indicates P1 transduction. For example, JMG367 was constructed by infecting JMG441 with P1 lysate made from LMG145.
TABLE 2.
Plasmids used
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pBAD18 | PBAD arabinose regulation, Ampr | 10 |
| pBAD39 | Arabinose regulation, IPTG depa, lacIq StrepS Ampr | L. M. Guzman, unpublished data |
| pAM238 | Plac regulation, Specr | J. P. Bouché, unpublished data |
| pGHIIA44 | H. influenzae ATCC clone, ftsLHinf Ampr | ATCC |
| pGB017 | C/EBP Ampr | 4 |
| pY88 | GCN4 Ampr | 24 |
| pDSW207 | P207-gfp, Ampr | 30 |
| pDSW236 | pDSW207-ftsL, Ampr | 8 |
| pJMG453 | pDSW207-LLL, Ampr | This study |
| pJMG447 | pDSW207-FLL, Ampr | This study |
| pJMG448 | pDSW207-LFL, Ampr | This study |
| pJMG449 | pDSW207-FFL, Ampr | This study |
| pJMG450 | pDSW207-FFF, Ampr | This study |
| pJMG451 | pDSW207-LLF, Ampr | This study |
| pJMG474 | pDSW207-LLI, Ampr | This study |
| pJMG454 | pDSW207-ftsLHinf, Ampr | This study |
| pJMG455 | pDSW207-HLL, Ampr | This study |
| pJMG456 | pDSW207-HHL, Ampr | This study |
| pJMG475 | pDSW207-LHL, Ampr | This study |
| pJMG457 | pDSW207-HLH, Ampr | This study |
| pJMG458 | pDSW207-LHH, Ampr | This study |
| pJMG459 | pDSW207-LLH, Ampr | This study |
| pJMG461 | pDSW207-LLHinfL, Ampr | This study |
| pJMG462 | pDSW207-LLGCN4, Ampr | This study |
| pJMG463 | pDSW207-LLC/EBPL, Ampr | This study |
| pJMG465 | pDSW207-LLΔL, Ampr | This study |
| pJMG466 | pDSW207-LLL(L70H), Ampr | This study |
| pJMG467 | pDSW207-LLL(L84D), Ampr | This study |
| pJMG197 | ftsL Cmr | 8 |
| pJMG64 | ftsL, IPTG dep, lacIq StrepS Ampr | This study |
| pJMG180 | pBAD18-ftsL, Ampr | 10 |
| pJMG460 | pBAD18-LLxL, Ampr | This study |
| pLD45 | pBAD18-LLL, Ampr | 10 |
| pLD63 | pBAD18-FLL, Ampr | 10 |
| pLD94 | pBAD18-LFL, Ampr | 10 |
| pLD90 | pBAD18-FFL, Ampr | 10 |
| pLD47 | pBAD18-FFF, Ampr | 10 |
| pLD116 | pBAD18-LLI, Ampr | 10 |
| pJMG427 | pBAD18-LLHinfL, Ampr | This study |
| pJMG429 | pBAD18-HLL, Ampr | This study |
| pJMG446 | pBAD18-HLH, Ampr | This study |
| pJMG433 | pBAD18-LHH, Ampr | This study |
| pJMG434 | pBAD18-LLH, Ampr | This study |
| pJMG430 | pBAD18-LHL, Ampr | This study |
| pJMG431 | pBAD18-HHL, Ampr | This study |
| pJMG452 | pBAD18 ftsLHinf, Ampr | This study |
| pJMG464 | pBAD18-LLΔL, Ampr | This study |
| pJMG438 | pBAD18-LLL(L70H), Ampr | This study |
| pJMG439 | pBAD18-LLL(L84D), Ampr | This study |
| pJMG428 | pBAD18-LLGCN4L, Ampr | This study |
| pJMG440 | pBAD18-LLC/EBPL, Ampr | This study |
| pJMG40 | pAM238-LLL, Specr | This study |
| pJMG473 | pAM238-LLxL, Specr | This study |
| pJMG38 | pAM238-FLL, Specr | This study |
| pJMG39 | pAM238-LFL, Specr | This study |
| pJMG444 | pAM238-FFL, Specr | This study |
| pJMG158 | pAM238-LLHinfL, Specr | This study |
| pJMG162 | pAM238-ftsLHinf, Specr | This study |
| pJMG157 | pAM238-LLΔL, Specr | This study |
| pJMG155 | pAM238-LLLΔ, Specr | This study |
| pJMG163 | pAM238-LLL(L70H), Specr | This study |
| pJMG164 | pAM238-LLL(L84D), Specr | This study |
| pJMG161 | pAM238-LLGCN4L, Specr | This study |
| pJMG159 | pAM238-LLC/EBPL, Specr | This study |
IPTG dep, IPTG-dependent replication.
Media.
Media were NZY or Luria-Bertani (LB) medium (9, 27). l-Arabinose or d-glucose was added as indicated to modulate expression of genes under control of the PBAD promoter (10). For localization studies, isopropyl-β-d-thiogalactoside (IPTG) was used to modulate expression of gfp fusions under the control of modified trc promoters: 2.5 μM for P207-gfp-ftsL derivatives (8, 31). The IPTG-dependent plasmid pJMG64 was maintained with 5 mM IPTG. Antibiotics were used for selection at the following concentrations: ampicillin (AMP) at 200 μg/ml for plasmids and 25 μg/ml for chromosomal alleles, kanamycin (KAN) at 40 μg/ml, chloramphenicol at 30 μg/ml, tetracycline (TET) at 15 μg/ml, and spectinomycin (SPEC) at 25 μg/ml. In complementation experiments, pJMG64 was counterselected with streptomycin used at 1.5 mg/ml.
Molecular biological procedures.
Standard procedures were used for cloning and analysis of DNA, PCR, electroporation, and transformation (27). Enzymes used to manipulate DNA were from New England Biolabs (Beverly, Mass.). Oligonucleotides were from Gibco BRL (Gaithersburg, Md.). DNA sequencing was performed by the Micro Core Facility in the Department of Microbiology and Molecular Genetics at Harvard Medical School.
Sequence analysis.
Amino acid sequence comparisons were performed with the Clustal package program (14). Homology searches were performed with Blast 2.0 at the National Center for Biotechnology Information website (http: //www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html). Hydropathy plots and DNA sequence manipulations were done with DNAStrider 1.3 (22). Coiled-coil propensity was determined with Coiled 2.2 (19) on the ISREC website (http: //www.isrec.isb-sib.ch/software/COILS_form.html).
Construction of gfp fusions. (i) gfp-ftsL and gfp-LLL.
Plasmids for making gene fusions to gfp were based on pDSW207 (31). pDSW236 is described in reference 8. To make pDSW207-LLL, LLL was amplified by PCR with pLD45 (11) as template and oligonucleotides L1 (CGAGAATTCAACAACAACATGATCAGCAGACTGACAGAA) and L2 (TCGAAGCTTTTTATTTTTGCACTACGATATTTTC) as primers. The amplified DNA was digested with EcoRI and HindIII (sites underlined) and ligated into the same sites of pDSW207 to create pJMG453. The fusion protein encoded by pJMG453 has the linker sequence YKEFNNNMI, where YK are the last residues of GFP and MI are the first two residues of FtsL.
(ii) Fusions to FtsL-MalF swap proteins.
To make gfp-FLL, gfp-LFL, and gfp-FFL, the FLL, LFL, and FFL genes were amplified by PCR with pLD63, pLD94, and pLD90 (11), respectively, as template and oligonucleotides F1 (CG AGAATTCAACAACAACATGGATGTCATTAAAAAGAAACATTGGTG GC) (FLL and FFL) or L1 (LFL) and pBADrev (ACCGCTTCTGCGTTCTGATT) as primers. The amplified DNA was digested with EcoRI (site underlined) and HindIII (site present downstream of the constructs in pLD63, pLD94, and pLD90) and ligated into the same sites of pDSW207 to create pJMG447, pJMG448, and pJMG449. The fusion protein encoded by pJMG448 has the same linker sequence as that in GFP-LLL. The fusion proteins encoded by pJMG447 and pJMG449 have the linker sequence YKEFNNNMD, where YK are the last two residues of GFP and MD are the first two residues of FLL and FFL, whose amino terminus is derived from malF.
To make gfp-FFF, the FFF gene was amplified by PCR with pLD47 (11) as template and oligonucleotides F1 and F2 (TCGAAGCTTTTAATCAAACTTCATTCGCGTGGC) as primers. The amplified DNA was digested with EcoRI and HindIII (site underlined) and ligated into the same sites of pDSW207 to create pJMG450. The fusion protein encoded by pJMG450 has the same linker sequence as that in GFP-FLL.
To make gfp-LLF, the MscI-HindIII fragment of pLD47, corresponding to the MalF region beyond the first transmembrane segment, was introduced into pJMG453 (pDSW207-LLL) with its MscI-HindIII fragment deleted to create pJMG451. The fusion protein encoded by pJMG451 has the same linker sequence as that in GFP-FLL.
(iii) Fusions to FtsL-FtsI swap proteins.
To make gfp-LLI, the LLI gene was amplified by PCR with pLD116 (11) as template and oligonucleotides L1 and pBADrev. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pDSW207 to create pJMG474. The fusion protein encoded by pJMG474 has the same linker sequence as that of pDSW236.
(iv) Fusions to FtsLEcol-FtsLHinf swap proteins.
ftsLHinf was amplified by PCR with pGHIIA44 (American Type Culture Collection [ATCC], Manassas, Va.) as template and oligonucleotides HiL-5 (CCCCGAATTCAGAGAGGACGAATGCATGTCTGAAAATAATAAGCCTCG and HiL-3 (CCCCAAGCTTTTATCCTTATTCTCTAATTTCAACTTCTTGC) as primers. In HiL-5, the sequence located downstream of the EcoRI site corresponds to the region placed upstream of E. coli FtsL (the ribosome binding site is in boldface) followed by the H. influenzae coding sequence (italicized). The amplified DNA was digested with EcoRI and HindIII (sites underlined) and ligated into the same sites of pBAD18 to create pJMG452.
To make gfp fusions to FtsLEcol-FtsLHinf swap proteins, the swap constructs were first made in pBAD18 derivatives. To make HLL, the DNA fragment encoding the cytoplasmic domain of FtsLHinf was amplified by PCR with pJMG452 as template and oligonucleotides pBADfw (CTGACGCTTTTTATCGCAAC) and Hieag-3 (GGGCGGCCGGAAGAAAATAAATCTTCG) as primers. The amplified DNA was digested with EcoRI (site present upstream of the constructs in pJMG452) and EagI (site underlined) and ligated into the same sites of pLD45 to create pJMG429.
To make LHL, the DNA fragment encoding the membrane-spanning domain of FtsLHinf was amplified by PCR with pJMG452 as template and oligonucleotides Hieag-5 (CCCCGGCCGGTTAGTGGTGTTGCTGTTAATAGGG) and Himsc-3 (GGGTGGCCAATCCAAATCGTCCCCATTGCAGAAACTA) as primers. The amplified DNA was digested with EagI (site underlined) and MscI (site underlined) and ligated into the same sites of pLD45 to create pJMG430.
To make LLH, the DNA fragment encoding the periplasmic domain of FtsLHinf was amplified by PCR with pJMG452 as template and oligonucleotides Himsc-5 (CCCTGGCCATAAAACTCGCCAATTAATTTC) and HiL-3 as primers. The amplified DNA was digested with MscI (site underlined) and HindIII and ligated into the same sites of pLD45 to create pJMG434.
To make HHL, the EagI-HindIII DNA fragment of pJMG430 was inserted into the same sites of pJMG429 to create pJMG431.
To make gfp-LHH, the MscI-HindIII DNA fragment of pJMG434 was inserted into the same sites of pJMG430 to create pJMG433.
To make HLH, the EagI-HindIII DNA fragment of pJMG434 was inserted into the same sites of pJMG432 to create pJMG446.
Plasmids for making gene fusions to gfp were all based on pDSW207. ftsLHinf and HLH were amplified by PCR with, respectively, pJMG452 and pJMG446 as templates and oligonucleotides Haeinf1 (CCCGAATTCAACAACAACATGTCTGAAAATAATAAGCCTCG) and pBADfw as primers. The amplified DNA was digested with EcoRI (site underlined) and HindIII and ligated into the same sites of pDSW207 to create pJMG454 and pJMG457. The fusion proteins encoded by pJMG454 and pJMG457 have the linker sequence YKEFNNNMS, where YK are the last residues of GFP and MS are the first two residues of FtsL. HLL and HHL were amplified by PCR with pJMG429 and pJMG431 as templates and oligonucleotide HiL-1 and pBADrev as primers. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pDSW207 to create pJMG455 and pJMG456, respectively. LHH and LLH were amplified by PCR with pJMG433 and pJMG434 as templates and oligonucleotides L1 and pBADrev as primers. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pDSW207 to create pJMG458 and pJMG459, respectively.
(v) Fusions to FtsLEcol periplasmic domain mutants.
LLHinfL, LLGCN4L, and LLCEB/PL were constructed as follows. First, a silent mutation was introduced in the DNA region encoding the periplasmic domain of LLL by the megaprimer methods (28). The megaprimer was generated with pLD45 as template and oligonucleotides Lxho3mp (GAATGGCGCAACCTGATCCTCGAGGAGAATGC) and pBADfw as primers. LLxL was then amplified with pLD45 as template and the megaprimer and pBADrev as primers. The mutation modified the 86th and 87th codons of LLL (CTT GAA [Leu Glu]) to (CTCGAG [Leu Glu]) and introduced a unique XhoI site in the LLL sequence to create pJMG460.
The partial exchange of the periplasmic domain of FtsLEcol with the homologous region of ftsLHinf (LLHinfL) (see Fig. 3) was achieved as follows: a 104-bp DNA fragment corresponding to residues 45 to 72 of the ftsLHinf periplasmic domain, flanked by MscI and XhoI restriction sites, was amplified by PCR with pJMG452 as template and oligonucleotides LzHinf-5 (CCCCTGGCCATAAAACTCGCCAATTAATTTC) and LzHinf-3 (GGGGCTCGAGTTGTAAATACGGTATTC) as primers. The amplified DNA was digested with MscI and XhoI (sites underlined) and ligated into the same sites of pJMG460 to create pJMG427 (see Fig. 3).
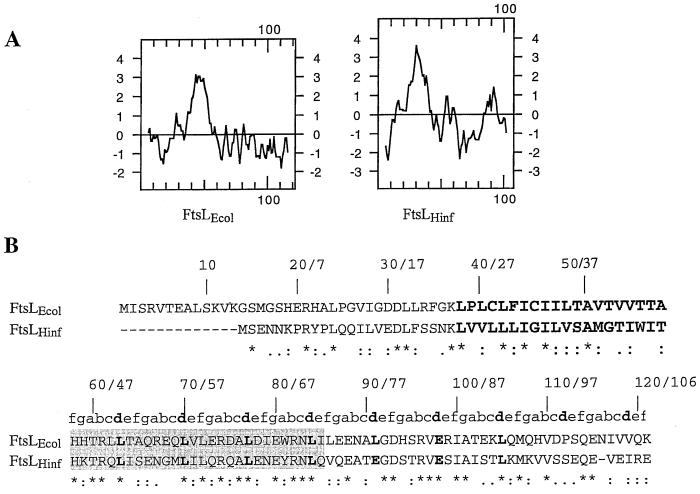
FIG. 3.
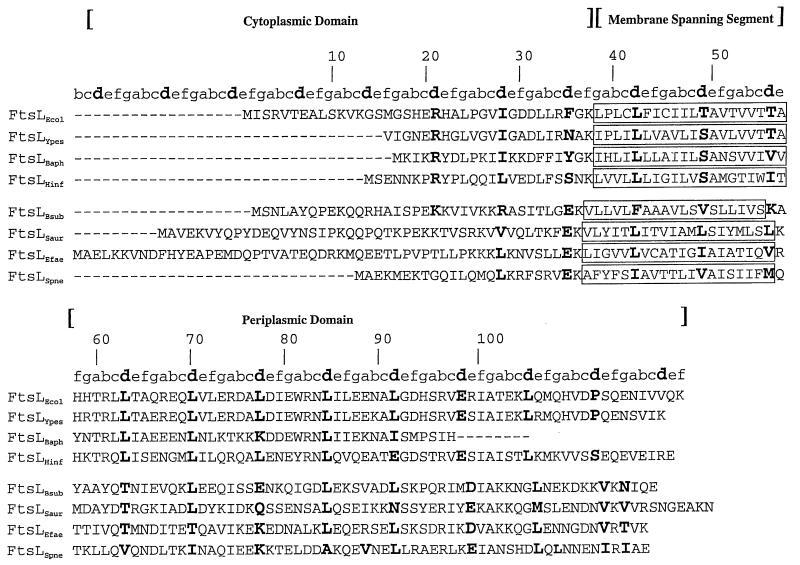
Comparison between E. coli and H. influenzae FtsL. (A) Kyte and Doolittle hydropathy plot comparison between FtsLEcol and FtsLHinf. (B) Amino acid sequence comparison between FtsLEcol and FtsLHinf. The first and second numbers indicated above the sequences correspond to amino acid positions in FtsLEcol and FtsLHinf, respectively. FtsLEcol and FtsLHinf membrane-spanning segments are in oversized boldface. The shaded area corresponds to the region exchanged in the LLHinfL swap (see text). The a to f positions of the heptad repeat of the potential coiled-coil domain are indicated above the sequence of the periplasmic domain of FtsLEcol and FtsLHinf. The d position is indicated in boldface. Conserved residues are indicated with a star below the sequence. Conservative substitutions are indicated with dots.
The partial exchange of the periplasmic domain of FtsLEcol with the analogous region of the yeast transcription factor GCN4 was realized as follows: a 104-bp DNA fragment corresponding to residues 248 to 275 of GCN4, flanked by MscI and XhoI restriction sites, was amplified by PCR with pY88 (25) as template and oligonucleotides LzGCN4-5 (CCCCTGGCCAAAGAATGAAACAACTTGAAGAC) and LzGCN4-3 (GGGGCTCGAGCTTTAATCTGGCAACCTCATT) as primers. The amplified DNA was digested with MscI and XhoI (sites underlined) and ligated into the same sites of pJMG460 to create pJMG428 (see also Table 5).
TABLE 5.
Localization frequencies of and complementation by GFP-FtsL mutants
| Construct | Structurea | Localization frequencyb
|
Complementatione
|
||
|---|---|---|---|---|---|
| No. of cells scored | Band at midcell (%) | Swap not fused to GFPc | Swap fused to GFPd | ||
| GFP-LLL |  |
342 | 69 | +++ | +++ |
| GFP-LLΔL | 355 | − | ND | ||
| GFP-LLL(L70H) | 310 | 17 | + | ND | |
| GFP-LLL(L84D) | 380 | 28 | + | ND | |
| GFP-LLGCN4L | 232 | <1 | − | ND | |
| GFP-LLC/EBPL | 218 | <1 | − | ND | |
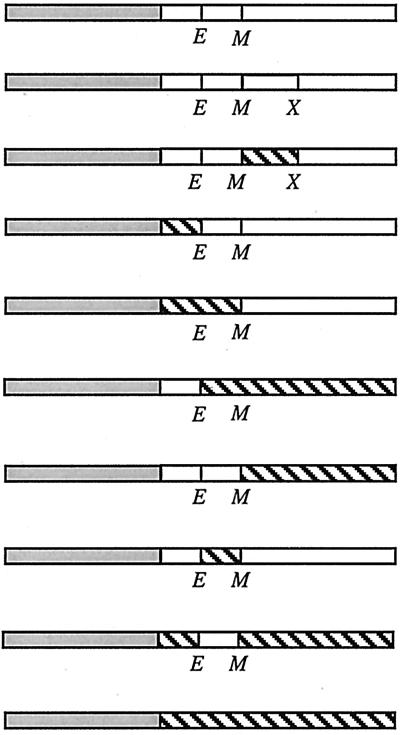
Open boxes represent domains of E. coli FtsL (FtsLEcol). Shaded boxes denote GFP. Hatched boxes represent GCN4 leucine zipper. The checkered box represents C/EBP leucine zipper. E and M indicate borders of the transmembrane domain where restriction sites for EagI and MscI, respectively, have been introduced. X indicates the position of the XhoI site introduced to allow partial swapping inside the region encoding the periplasmic domain of FtsL. In LLL, the former changes a Lys to an Arg, and the latter changes an Ala to a Gly. The cytoplasmic and transmembrane domains of LLL contain 37 and 20 amino acids, respectively. The periplasmic domain of LLL contains 64 amino acids.
Strains used: GFP-LLL, JMG287; GFP-LLΔL, JMG299; GFP-LLL(L70H), JMG303; GFP-LLL(L84D), JMG304; GFP-LLGCN4L, JMG301; GFP-LLC/EBPL, JMG302.
Strains used: LLL, JMG344; LLΔL, JMG350; LLL(L70H), JMG355; LLL(L84D), JMG356; LLGCN4L, JMG353; LLC/EBPL, JMG352.
Strain used: GFP-LLL, JMG367. ND, not determined.
For complementation test, plus signs indicate colony size and minus signs indicate no growth.
The partial exchange of the periplasmic domain of FtsLEcol with the analogous region of the mammalian transcription factor C/EBP was achieved as follows: a 104-bp DNA fragment corresponding to residues 312 to 339 of the rat liver C/EBP, flanked by MscI and XhoI restriction sites, was amplified by PCR with pGB017 (4) as template and oligonucleotides LzCEBP-5 (CCCCTGGCCAGAAGGTGTTGGAGTTGACCAG) and LzCEBP-3 (GGGGCTCGAGCCGCAGCGTGTCCAGTTCACGG) as primers. The amplified DNA was digested with MscI and XhoI (sites underlined) and ligated into the same sites of pJMG460 to create pJMG440 (see also Table 5).
Plasmids for making gene fusions to gfp were based on pDSW207. pDSW207-LLHinfL, pDSW207-LLGCN4L, and pDSW207-LLCEB/PL were made by PCR with, respectively, pJMG427, pJMG428, and pJMG440 as template and oligonucleotides L1 and L2 as primers. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pDSW207 to create pJMG61, pJMG462, and pJMG463. The fusion proteins encoded by pJMG461, pJMG462, and pJMG463 have the same linker as pJMG236.
LLΔL was made as follows: the 86-bp MscI-XhoI fragment of LLxL was deleted from pJMG460. The resulting deleted plasmid was treated with mung bean nuclease, removing four extra hanging bases from the XhoI site. The resulting construct was self-ligated, creating a 30-residue in-frame deletion in the periplasmic domain of LLL (LLΔL) in pJMG464. pDSW207-LLΔL (pJMG465) was made as described above with pJMG464 as template and L1 and L2 as primers.
To create LLL(L70H), we mutagenized LLL by incorporation of a phosphorylated oligonucleotide during PCR amplification (23). We used pLD45 as template and oligonucleotide pBADfw, phosphorylated oligonucleotide MnL2 (GCTCAGCGCGAACAACACGTGCTGGAGCGAGATGC), and pBADrev as primers. MnL2 introduced an L-to-H mutation in the 70th codon of FtsL. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pBAD18 to create pJMG438.
To create LLL(L84D), we mutagenized LLL by incorporation of a phosphorylated oligonucleotide during PCR amplification (23). We used pLD45 as template and oligonucleotide pBADfw, phosphorylated oligonucleotide MnL4 (GACATTGAATGGCGCAACGATATCCTTGAAGAGAATGC), and pBADrev as primers. MnL4 introduced an L-to-D mutation in the 84th codon of FtsL. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pBAD18 to create pJMG439. pDSW207-LLL(L70H) (pJMG466) and pDSW207-LLL(L84D) (pJMG467) were made as described above with pJMG438 and pJMG439 as template and L1 and L2 as primers.
Other constructs.
pAM238 derivative plasmids described in Table 2 were created as follows: pBAD18-based constructs were amplified by PCR with oligonucleotides pBADfw and pBADrev as primers. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pAM238. LLLΔ was constructed as follows: a stop linker (LzStop1 [TCGACCAAAAATAATACGTAA] and LzStop2 [ACGTTTACGTATTATTTTTGG]) was inserted at the XhoI site of LLxL in pJMG473. The resulting plasmid (pJMG155) presents a deletion of residues 87 to 119 in the FtsL coding sequence. For pJMG64, the SacI-HindIII fragment of pLMG180 was introduced into pBAD39 linearized with the same enzymes to create pJMG64.
Construct verification.
All constructs were checked by PCR with specific and vector-based primers. Then each construct was sequenced in the pBAD18 or pAM238 context. For GFP fusions, the gene coding for the protein fused to GFP was sequenced from a PCR product obtained by amplification of chromosomal DNA. The sequence was checked up to the GFP-swap junction point. The constructs FFF and LLF were sequenced up to the junction point with the end of the region coding for the first membrane-spanning segment. The integrity of the rest of the malF portion was then checked by testing the ability of LLF (pJMG451) to complement a malF mutation in DHB4 on a MacConkey maltose plate.
Growth conditions. (i) GFP-fusion protein expression. (a) Wild-type background.
Cells expressing GFP fusions (“gfp fusion in wild-type background” section of Table 1) were grown overnight at 30°C in NZY medium plus AMP, diluted 1:200 in fresh NZY medium (without AMP) containing 2.5 to 10 μM IPTG, and grown on a roller at 30°C until the optical density at 600 nm (OD600) reached 0.25.
(b) Depletion experiments.
To deplete FtsL from strains indicated in the “gfp fusion in depletion background” section of Table 1, strains were grown overnight in NZY with AMP, KAN, chloramphenicol, and 0.2% arabinose (which allowed high-level expression of FtsL from plasmid pJMG197). Cultures were diluted into fresh medium containing chloramphenicol and 0.02% arabinose and grown to an OD600 of 0.5. Cells were washed and resuspended in NZY medium and then diluted 1:200 into NZY with 0.2% glucose (which shut off synthesis of FtsL from pJMG197) or NZY with 0.2% arabinose plus 2.5 to 5 μM IPTG. After about 3 to 5 h, an OD600 of 0.25 was reached and cells had become filamentous in the glucose culture. Samples were harvested and prepared for microscopy and Western blot analysis.
(ii) FtsL derivative protein expression from pBAD18-based constructs.
pBAD18-based FtsL derivatives were introduced into KS272, grown overnight at 37°C in LB medium plus AMP, diluted 1:200 in fresh LB medium, and grown on a roller at 37°C until the OD600 reached 0.25. Then, arabinose or glucose (0.2%) was added and cultures were grown for 1 to 2 more h until the OD600 reached 0.6 to 1.
Complementation of an ftsL null mutation. (i) Complementation of FtsL derivative as GFP fusions inserted on the E. coli chromosome.
GFP-FtsL fusions were tested for complementation as follows. Strains corresponding to the “gfp fusion in depletion background” section in Table 1 were grown on low-AMP (25 μg/ml)-KAN-chloramphenicol-arabinose plates and were tested on plates containing low AMP, KAN, and 2.5 μM IPTG, supplemented with 0.2% glucose. Under these conditions, where the pJMG197-borne copy of ftsL is repressed, only strains expressing a functional GFP-FtsL derivative could grow.
(ii) Complementation of null mutant by counterselection.
The functionality of FtsL derivatives in the pAM238 context was tested as follows. FtsL variants made in pBAD18 were subcloned in pAM238 (a plasmid with a pSC101 origin of replication) under plac promoter control and introduced in strain JMG33 containing pJMG64 (Table 1 and Table 2). pJMG64 is a pBAD39-derived plasmid expressing a wild-type allele of ftsL, whose induction (with arabinose) is required to complement the chromosomal ftsL-phoA null mutation. pBAD39 is a counterselectable pBAD18-derived plasmid, carrying a lacI gene, whose replication is IPTG dependent and which expresses rpsL, conferring a dominant sensitivity to streptomycin (1.5 mg/ml). The complete features of pBAD39 will be described in detail elsewhere (L. M. Guzman, unpublished data). The rationale of the counterselection-based complementation test is the following: without IPTG and AMP and in the presence of streptomycin, cells carrying pJMG64 are counterselected. The expression of genes carried by the pAM238-based plasmid is then constitutive in JMG33 deprived of pJMG64, as there is no longer a lacI gene in the cells. Any functional FtsL derivatives would then support the growth of JMG33 in the absence of pJMG64.
Strains corresponding to the “depletion background” section in Table 1 were grown on plates containing IPTG, AMP, KAN (Tn5 associated with the chromosomal ftsL null allele), TET (Tn10 associated with the chromosomal recA allele), SPEC, and arabinose (0.2%). Under counterselection conditions without IPTG and AMP but with SPEC, KAN, TET, streptomycin, and glucose (0.2%), where the pJMG64-borne copy of ftsL is counterselected, only strains expressing a functional FtsL derivative could grow.
Microscopy and image analysis.
Cells were grown and processed for protein localization experiments as described in reference 8. Microscopy and image analysis were performed as described in reference 8.
Western blotting.
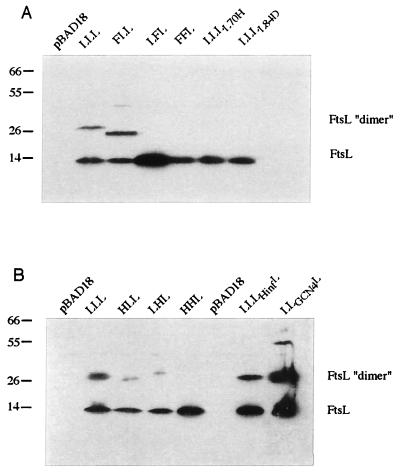
Western blotting was performed by standard techniques as described in reference 8. To determine the steady-state levels of the GFP fusion proteins, we harvested cells from the same cultures as those used for fluorescence microscopy. Cells were pelleted by centrifugation and resuspended with 10 μl of SDS sample buffer per OD600 unit of 0.1. Samples were boiled for 3 min, and 10 μl of sample was loaded onto an SDS-polyacrylamide gel. To determine the multimerization state of the FtsL derivatives, cells were pelleted by centrifugation and resuspended with 10 μl of SDS sample buffer per OD600 unit of 0.1. In most of the experiments, samples were not boiled and 10 μl of sample was loaded onto a 10% (GFP fusion) or 13% (FtsL swaps) SDS-polyacrylamide gel.
RESULTS
The membrane-spanning segment and the periplasmic domain of FtsL are both required for septal localization.
In previous complementation tests, we used swap proteins where the three domains of FtsL were exchanged with topologically equivalent domains of MalF, FtsI, and FtsQ. This analysis indicated that all three domains were required for FtsL function (11). One simple interpretation of these data is that all three domains participate in septal localization. To test this hypothesis, we fused a bright variant of GFP (6) at the amino terminus (cytoplasmic domain) of several swap proteins: LLL, FLL, FFL, LFL, LLF, LLI, and FFF (Table 3). The three-letter names indicate the source of each domain in the swap proteins: the first letter indicates that of the cytoplasmic region, the second indicates that of the transmembrane segment, and the third indicates that of the periplasmic domain. For example, LFL refers to a protein in which the cytoplasmic and periplasmic domains are derived from FtsL and the membrane-spanning domain is derived from the first transmembrane segment from MalF. In LLI, I stands for the periplasmic domain of FtsI, another bitopic membrane protein involved in cell division. LLL and FFF constructs differ from wild-type FtsL and MalF in that two and four amino acids at the borders of the membrane-spanning segment have been altered due to introduction of restriction sites in corresponding regions of the ftsL and malF genes, respectively (11). We integrated the genes coding for GFP fused to the N terminus of LLL, FLL, FFL, LFL, LLF, LLI, and FFF to generate merodiploid strains similar to those used for localization of GFP-FtsL (5, 8, 31). Cells expressing GFP fusions to the FtsL-MalF swap proteins were fixed and examined by fluorescence microscopy. GFP-LLL localized as well as the control protein in which GFP was fused to the wild-type FtsL, while GFP-FFF did not localize to the septum. The GFP-FLL protein was the only swap protein to localize to the septum with a frequency similar to that of GFP-LLL (Table 3 and Fig. 1A).
TABLE 3.
Localization frequencies of and complementation by GFP-FtsL and GFP-FtsLEcol-MalF swap proteins
| Construct | Structurea | Localization frequencyb
|
Complementatione
|
||
|---|---|---|---|---|---|
| No. of cells scored | Band at midcell (%) | Swap not fused to GFPc | Swap fused to GFPd | ||
| GFP-FtsL |
 |
342 | 72 | +++ | +++ |
| GFP-LLL | 310 | 69 | +++ | +++ | |
| GFP-FLL | 307 | 70 | − | − | |
| GFP-FFL | 295 | <1 | − | ND | |
| GFP-LFL | 301 | <1 | − | ND | |
| GFP-LLF | 293 | <1 | ND | ND | |
| GFP-LLI | 320 | <1 | − | ND | |
| GFP-FFF | 311 | <1 | − | ND | |
Open boxes represent domains of FtsL. Shaded boxes denote GFP. Hatched boxes represent domains derived from MalF. The checkered box represents the periplasmic domain of FtsI. E and M indicate borders of the transmembrane domain where restriction sites for EagI and MscI, respectively, have been introduced. In FtsL, the former changes a Lys to an Arg and the latter changes an Ala to Gly. Diagrams of fusion proteins are not to scale. The cytoplasmic and transmembrane domains of FtsL contain 37 and 20 amino acids, respectively, while those from MalF contain 16 and 25 amino acids, respectively. The periplasmic domain of FtsL contains 64 amino acids. The periplasmic domain of FtsI contains 537 amino acids. The region past the first transmembrane domain of MalF contains 478 amino acids.
Strains used: FtsL, EC438; LLL, JMG287; FLL, JMG469; FFL, JMG292; LFL, JMG289; LLF, JMG290; LLI, JMG295; FFF, JMG291.
Strains used: FtsL, strain from reference 10; LLL, JMG344; FLL, JMG348; FFL, JMG359; LFL, JMG347; LLI, strain from reference 10; FFF, strain from reference 10.
Strains used: GFP-FtsL, JMG366 (8); GFP-LLL, 367; GFP-FLL, JMG420.
For complementation test, plus signs indicate colony size and minus signs indicate no growth. ND, not determined.
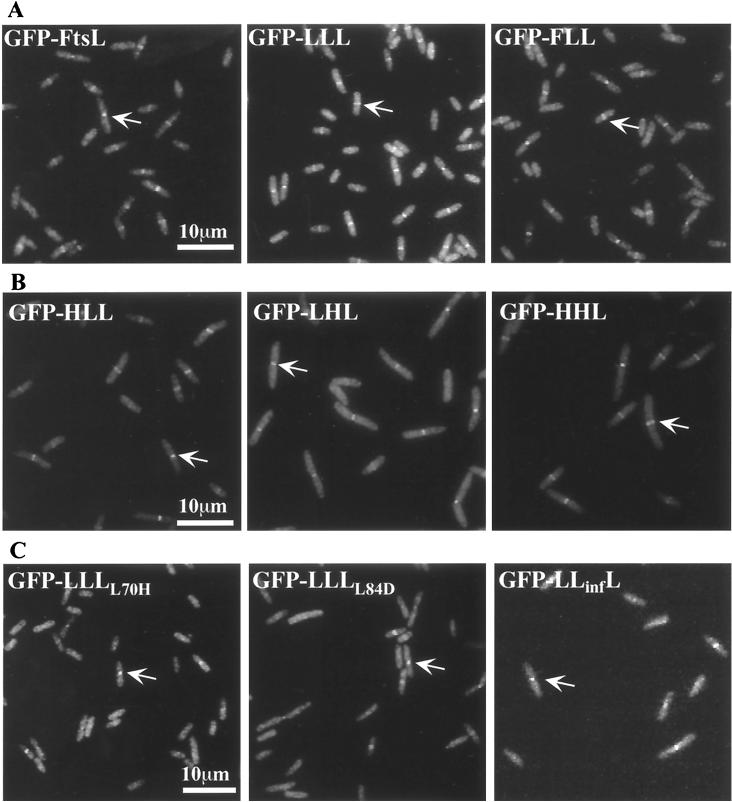
FIG. 1.
Subcellular location of GFP-FtsL swap proteins in cells fixed during exponential growth. Arrows indicate rings at division sites. (A) Septal localization of FtsLEcol-MalF swap proteins fused to GFP. Strains used were as follows: GFP-FtsL, EC438; GFP-LLL, JMG287; and GFP-FLL, JMG469. (B) Septal localization of FtsLEcol-FtsLHinf swap proteins fused to GFP. Strains used were as follows: GFP-HLL, JMG371; GFP-LHL, JMG372; and GFP-HHL, JMG374. (C) Septal localization of FtsL periplasmic mutants fused to GFP. Strains used were as follows: GFP-LLL(L70H), JMG303; GFP-LLL(L84D), JMG304; and GFP-LLHinfL, JMG300.
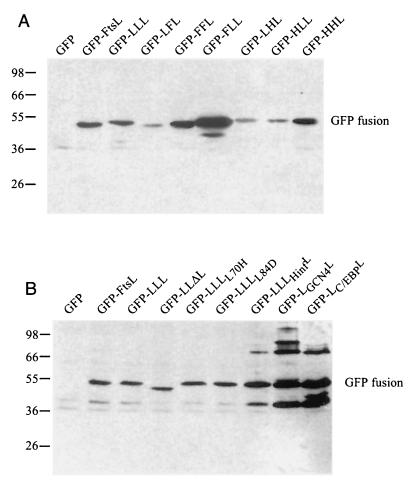
The lack of localization of GFP-LFL, GFP-FFL, GFP-LLF, and GFP-LLI is consistent with the inability of these constructs, without GFP attached, to complement an ftsL null mutant. The efficient septal localization frequency of GFP-FLL contrasted with the lack of functionality of the FLL protein in the cell division process (11) (Table 3). This finding led us to test the complementation of a null mutation in ftsL by GFP-FLL. The gfp-LLL and gfp-FLL fusions were tested for complementation in the FtsL depletion strain. Cells expressing GFP-LLL grew well in the presence of glucose (FtsL depletion) while cells expressing GFP-FLL did not. These complementation results are consistent with those obtained with the LLL and FLL proteins when not fused to GFP (Table 3). Differences in localization did not result from differences in expression; Western blotting was performed and indicated that, although the stability and abundance of each swap protein vary, they are all expressed at a significant level (Fig. 2). The fact that GFP-FFF and GFP-LLF complement a malF mutation (Materials and Methods) suggests that these proteins are also expressed at appropriate levels. The amount of GFP-LLI was determined with anti-FtsI antibodies (data not shown).
FIG. 2.
Steady-state levels of GFP-FtsL-MalF swap proteins as determined by Western blotting with an anti-FtsL antibody. Molecular mass standards (kilodaltons) are indicated to the left of the blot, while the positions of GFP-FtsL fusions are shown to the right. The GFP fusion protein produced is indicated above each lane. Strains used were as follows: (A) GFP, EC452; GFP-FtsL, EC438; GFP-LLL, JMG287; GFP-LFL, JMG289; GFP-FFL, JMG292; GFP-FLL, JMG469; GFP-LHL, JMG372; GFP-HLL, JMG371; and GFP-HHL, JMG374; (B) GFP, EC452; GFP-FtsL, EC438; GFP-LLL, JMG287; GFP-LLΔL, JMG299; GFP-LLL(L70H), JMG303; GFP-LLL(L84D), JMG304; GFP-LLHinfL, JMG300; GFP-LLGCN4L, JMG301; and GFP-LLC/EBPL, JMG302.
The results in Table 3 suggest that the FtsL septal localization signal is carried by a combination of the membrane-spanning segment and the periplasmic domain but that localization of these two domains to the division site is not sufficient to complement for FtsL function.
The H. influenzae FtsL homologue neither complements nor localizes in E. coli.
To further investigate the essential features of the FtsL localization signal, we asked whether a homologue of E. coli FtsL from another bacterial species could complement for FtsL function. Such an approach has proven to be informative in studies of the function of the cell division proteins FtsA and FtsZ (21). We chose the H. influenzae FtsL (FtsLHinf) as a good candidate to test for complementation and localization in E. coli. The FtsLHinf amino acid sequence is 40% identical to that of the FtsL of E. coli, and the chromosomal position and locus genetic organization of ftsLHinf are very similar to those of ftsLEcol. The homology between the two proteins is distributed evenly across the sequence except for the N terminus of FtsLHinf, which is shorter by 14 residues. FtsLEcol and FtsLHinf present the same predicted bitopic topology and have a conserved heptad motif in their periplasmic domain (Fig. 3). We cloned the ftsLHinf gene from the H. influenzae ATCC cosmid library and expressed it under the pBAD arabinose-inducible promoter with the ftsLEcol translation signals. Complementation tests showed that ftsLHinf did not complement an ftsLEcol null mutant. To verify the expression of FtsLHinf under these conditions, we generated fusions of FtsLHinf to alkaline phosphatase, where alkaline phosphatase is fused after the last residue of the FtsLHinf periplasmic domain. In this construct, the promoter and translation signals are the same as in constructs expressing FtsLEcol and FtsLHinf. PhoA fusions have been used to verify expression and proper topology of FtsLEcol (11). The hybrid protein FtsLHinf-PhoA gave high levels of alkaline phosphatase expression, as observed on indicator plates containing 5-bromo-3-chloro-indolylphosphate (data not shown), indicating that FtsLHinf, as FtsLHinf-PhoA, was expressed and assumed the predicted topology and arrangement in the membrane. Therefore, the lack of functionality of FtsLHinf does not appear to be due to lack of expression or proper insertion in the envelope.
Based on sequence homologies and similarity of genetic organization of the ftsLHinf chromosomal region, it is likely that FtsLHinf fulfills in H. influenzae the role played by FtsLEcol in E. coli. Our results suggest that FtsLHinf in E. coli is deficient for a key functional feature required to support cell division. Whether this deficiency relates to an inability to localize at the division site is analyzed below.
Portions of FtsLHinf and FtsLEcol are functionally interchangeable.
To test whether portions of the FtsLHinf protein could functionally substitute for the homologous regions of FtsLEcol, we replaced the first half of the periplasmic domain of the swap protein LLL (residues 58 to 85 [Fig. 3]) with its corresponding region from FtsLHinf (residues 45 to 72). This new protein, named LLHinfL, is able to complement an ftsL null mutant, although less efficiently than LLL or wild-type FtsL (Table 4).
TABLE 4.
Localization frequencies of and complementation by GFP-FtsLEcol-FtsLHinf swap proteins
| Construct | Structurea | Localization frequencyb
|
Complementatione
|
||
|---|---|---|---|---|---|
| No. of cells scored | Band at midcell (%) | Swap not fused to GFPc | Swap fused to GFPd | ||
| GFP-LLL |
 |
310 | 75 | +++ | +++ |
| GFP-LLxL | NDf | ND | +++ | ND | |
| GFP-LLHinfL | 255 | 40 | + | ND | |
| GFP-HLL | 278 | 82 | ND | +++ | |
| GFP-HHL | 310 | 76 | ND | − | |
| GFP-LHH | 268 | <1 | ND | − | |
| GFP-LLH | 319 | <1 | ND | − | |
| GFP-LHL | 308 | 80 | ND | − | |
| GFP-HLH | 242 | <1 | ND | − | |
| GFP-FtsLHinf | 273 | <1 | − | − | |
Open boxes represent domains of E. coli FtsL (FtsLEcol). Shaded boxes denote GFP. Hatched boxes represent domains derived from wild-type H. influenzae FtsL (FtsLHinf). E and M indicate borders of the transmembrane domain where restriction sites for EagI and MscI, respectively, have been introduced. X indicates the position of the XhoI site introduced to allow partial swapping inside the region encoding the periplasmic domain of FtsL. In FtsLEcol, the former changes a Lys to an Arg and the latter changes an Ala to a Gly. In FtsLHinf, the former changes an Asn and a Lys to a Gly and an Arg, respectively, and the latter changes a Thr to a Gly. Diagrams of fusion proteins are not to scale. The cytoplasmic and transmembrane domains of FtsLEcol contain 37 and 20 amino acids, respectively, while those from FtsLHinf contain 24 and 20 amino acids, respectively. The periplasmic domain of FtsLEcol contains 64 amino acids. The periplasmic domain of FtsLHinf contains 63 amino acids.
Strains used: GFP-LLL, JMG441; GFP-LLHinfL, JMG300; GFP-HLL, JMG371; GFP-HHL, JMG374; GFP-LHH, JMG373; GFP-LLH, JMG370; GFP-LHL, JMG372; GFP-HLH, JMG375; GFP-FtsLHinf, JMG376.
Strains used: LLL, JMG344; LLxL, JMG100; LLHinfL, JMG351; FtsLHinf, JMG354.
Strains used: GFP-LLL, JMG367; GFP-HLL, JMG416; GFP-HHL, JMG442; GFP-LHH, JMG418; GFP-LLH, JMG415; GFP-LHL, JMG417; GFP-HLH, JMG419; GFP-FtsLHinf, JMG422.
For complementation test, plus signs reflect colony size and minus signs indicate no growth.
ND, not determined.
To test whether FtsLHinf and LLHinfL localize to the division site, we fused GFP to the N terminus of FtsLHinf and LLHinfL. As expected from complementation studies, the GFP-FtsLHinf protein did not localize to the division site but rather showed a uniform fluorescent background. In contrast, GFP-LLHinfL localized in 40% of the cells, compared to 70% septal localization for GFP-LLL (Fig. 1C and Table 4). The level of localization was consistent with the complementation results: weaker complementation correlates with less frequent localization (Table 4). These results indicated that FtsLHinf is nonfunctional in E. coli but that part of its periplasmic domain can be functionally exchanged with the homologous region of the FtsLEcol domain.
We then constructed several swap proteins where the three domains of FtsLEcol were exchanged with topologically equivalent domains of FtsLHinf: HLL, HHL, LHH, LLH, and LHL, where, for example, LHL refers to a protein in which the cytoplasmic and periplasmic domain are derived from FtsLEcol and the transmembrane domain is from FtsLHinf. To test for localization of the swap proteins, cells expressing GFP fusions to the N terminus of the swap genes were fixed and examined by fluorescence microscopy. As shown in Fig. 1B and Table 4, all fusions containing the periplasmic domain of FtsLEcol showed efficient localization to the division site. In contrast with the localization results obtain with GFP-LLHinfL, the whole periplasmic domain of FtsLHinf is not sufficient to direct the fusion protein to the division site and plausibly accounts for the lack of function of FtsLHinf in E. coli.
We then tested the functionality of these GFP fusion proteins in the FtsL depletion strain. Complementation results are presented in Table 4 and show that (i) all functional fusions (GFP-LLL and GFP-HLL) also localize to the division site, (ii) function is strictly associated with the simultaneous presence of the membrane-spanning and periplasmic domains of FtsLEcol, (iii) the cytoplasmic domain of FtsLHinf is interchangeable with that of FtsLEcol, and (iv) FtsLHinf and FtsLEcol membrane-spanning segments are not fully equivalent. Exchange between them has no effect on localization but prevents complementation. These results are consistent with those obtained with the FtsL-MalF swaps and further demonstrate that the correlation between function and localization is not reciprocal. They confirm the role of the periplasmic domain of FtsLEcol in the cell septum localization and suggest that the membrane-spanning segment of FtsLEcol may have a role beyond signaling cell localization, since the FtsLHinf transmembrane segment is not functional in constructs that nevertheless localize to the division site.
The integrity of the FtsL periplasmic domain is required for localization.
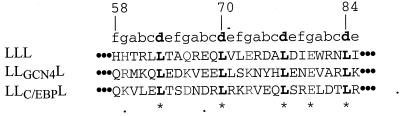
Our data suggested that the periplasmic domain of the E. coli FtsL contains information essential for localization to the division site. This domain displays features of a potential leucine zipper with regular heptad repeats with Leu residues in most of the d position of the heptad (Fig. 3). This motif may promote the formation of a helical structure that encompasses at least the entire periplasmic domain (64 residues). We constructed FtsL mutants to test whether this potential structure is part of the septal localization signal. We first deleted portions of the FtsL periplasmic domain corresponding to the first three heptads: LLΔL had 27 residues (residues 58 to 85 were removed [Table 5]). This construct was not functional, nor did it localize as a GFP fusion. In order to determine the effect of the disruption of the potential leucine zipper motif, we mutagenized the second [Leu70 by a His in LLL(L70H)] and fourth [Leu84 to an Asp in LLL(L84D)] leucines located at the d positions of the heptad in the interaction face of the potential structural motif. The ability of these FtsL variants to complement for FtsL function was tested: LLL(L70H) and LLL(L84D) complemented weakly compared to the positive control LLL (Table 5). The poor functionality of LLL(L70H) and LLL(L84D) correlated with a reduced frequency of localization of GFP-LLL(L70H) and GFP-LLL(L84D) where GFP was fused to the N terminus of these proteins. In these cases, only about 20% of the cells displayed a septal localization signal (Table 5 and Fig. 1C). The consequences of the mutagenesis of leucines 70 and 84 on FtsL are consistent with a contribution of these leucines to a structural leucine zipper-type motif. To determine whether it would be sufficient for FtsL function to simply replace this region with a known leucine zipper, we exchanged the first three repeats of the periplasmic domain of FtsL (residues 58 to 85) with two well-characterized leucine zippers: the yeast GCN4 transcription activator and the mammalian C/EBP transcription activator (LLGCN4L and LLC/EBPL, respectively). We took great care in these constructs to keep unchanged the spacing of the leucines (Fig. 4). Neither construct complemented an FtsL defect or allowed localization to the division site (Table 5). Western blotting indicated that the stability and abundance of each FtsL variant were similar to or higher than that of FtsL or LLL (Fig. 2; see also Fig. 6).
FIG. 4.
Sequence alignment of the regions that are not common to LLL, LLGCN4L, and LLC/EBPL swap proteins. Dotted lines represent flanking FtsL sequence which is the same in the three proteins. Leu70 and Leu84 are mutated in LLL(L70H) and LLL(L84D).
FIG. 6.
Western blot of FtsL SDS-resistant multimers. The figure shows the multimerization state of FtsL swap proteins as determined by Western blotting with an anti-FtsL antibody. Molecular mass standards (kilodaltons) are indicated to the left of the blot, while the positions of FtsL monomers and dimers are shown to the right. The FtsL swap protein produced is indicated above each lane. Background strain used was KS272. Plasmids used were as follows: (A) control, pBAD18; LLL, pLD45; FLL, pLD63; LFL, pLD94; FFL, pLD90; LLL(L70H), pJMG438; and LLL(L84D), pJMG439; (B) control, pBAD18; LLL, pLD45; HLL, pJMG432; LHL, pJMG430; HHL, pJMG431; LLHinfL, pJMG427; and LLGCN4L, pJMG428.
Taken together, these results indicate that the integrity of the periplasmic domain of FtsL is required for accurate septal localization. None of the swap proteins in which a portion of this domain was removed or exchanged yielded a fully functional and correctly localized protein. The fact that the two leucine mutants GFP-LLL(L70H) and GFP-LLL(L84D) showed a deficient localization pattern suggests that the heptad motif could be instrumental in the localization process; however, merely exchanging this motif with a well-characterized leucine zipper does not allow localization in GFP-LLGCN4L and GFP-LLC/EBPL.
The heptad motif of the periplasmic domain of E. coli FtsL is conserved in all identified FtsL homologues and has the characteristics of a coiled-coil domain.
The results presented above suggested that the heptad repeats of the periplasmic domain of FtsL may be required for FtsL function and that they may be indicative of structural features of its periplasmic domain. Heptad repeat motifs such as those found in FtsLEcol and FtsLHinf have been reported to have a high propensity to form α-helical multimeric coiled coils. Coiled coils are helical bundles of two to five helices that have a distinctive packing of amino acid side chains at helix-helix interfaces (18). This packing results in every seventh residue occupying an equivalent position on the helix surface. The seven positions are named a to g: a and d are usually occupied by hydrophobic residues and form the helix interface (core). Oppositely, charged residues in positions e and g flank the core and stabilize the protein through ionic interactions. The outer positions, b, c, and f, are not directly involved in the formation of the helical bundle and are free to interact with surrounding molecules. A wide variety of proteins involved in maintaining cell shape, in organizing the cytoplasm, and in cell movement and membrane fusion are coiled coils of two or more α-helices (17).
In order to estimate the degree of conservation of the heptad motif among FtsL homologues, we carried out a multiple alignment between several previously identified as well as newly described FtsL homologues. FtsL homologues so identified were found in B. subtilis, Streptococcus aureus, Streptococcus pneumoniae, Enterococcus faecalis, Yersinia pestis, and Buchnera aphidicola. These homologues were identified by us and others in both gram-negative and gram-positive bacteria in complete and incomplete genome databases based on (i) their homologies with either FtsLEcol or FtsLHinf, (ii) the similarity of their genetic locus position (upstream of ftsI in a cell division operon encoding other fts gene homologues), and (iii) their predicted bitopic membrane protein topology. Multiple alignment of the FtsL homologues so identified is presented in Fig. 5, where the a to g positions of the residues in the heptads are indicated above the alignment. In gram-negative bacterial FtsL homologues, the d position is mostly occupied by hydrophobic (Leu) and, to a lesser extent, by charged residues (E). Interestingly, in a typical coiled-coil core, the a position is often occupied by charged residues instead of hydrophobic residues. The g position is mainly occupied by charged residues, especially in the gram-negative bacterial homologues. Gram-positive bacterial FtsL proteins conform more to a canonical coiled coil, where a and d hydrophobic positions define the apolar stripe between helices.
FIG. 5.
Multiple alignment of FtsL homologues. Predicted membrane-spanning segments are boxed. The a to f positions of the heptad repeat of the potential coiled-coil domain are indicated above the sequence. The d position is indicated in boldface. The numbers above the alignment refer to the E. coli FtsL amino acid position. FtsLEcol, E. coli FtsL; FtsLYpes, Y. pestis FtsL; FtsLBaph, B. aphidicola FtsL; FtsLHinf, H. influenzae FtsL; FtsLBsub, B. subtilis FtsL; FtsLSaur, S. aureus FtsL; FtsLEfae, E. faecalis FtsL; FtsLSpne, S. pneumoniae FtsL.
In order to determine the propensity of FtsLEcol to form coiled coils, we submitted the FtsLEcol amino acid sequence to a widely used coiled-coil prediction algorithm, COILS 2.2 (18, 19). This analysis revealed that FtsLEcol has a low propensity (5% depending on the window length [Table 6]) to form a coiled-coil structure in the periplasmic domain. Strikingly, when we performed the same analysis with FtsL from H. influenzae, B. subtilis, S. aureus, S. pneumoniae, and E. faecalis, the periplasmic domain of these FtsLs showed a maximum propensity score of 100%. As noted by Lupas and coworkers (18), one of the limits of the method is that the absence of a peak is not as conclusive as the presence of a peak; there are many explanations of why the program might fail to predict a coiled-coil structure. On the other hand, a propensity of 100% is a very strong indication of a coiled-coil structure. This predicted coiled-coil propensity is very high in most of the FtsL homologues except for E. coli FtsL and its closest homologue, Y. pestis FtsL, where a low coiled-coil propensity is predicted (5% for Y. pestis and 80% for B. aphidicola [Table 6]).
TABLE 6.
Coiled-coil-forming potential in FtsL gram-negative and gram-positive homologues
| Organism | Predicted coiled coila | Avg scoreb (%) |
|---|---|---|
| Escherichia coli | 60–90 | 5 |
| Escherichia coli (W81 to Y) | 60–90 | 90c |
| Yersinia pestis | 40–80 | 5 |
| Yersinia pestis (W81 to Y) | 40–80 | 90c |
| Buchnera aphidicola | 35–70 | 80 |
| Haemophilus influenzae | 40–80 | 100 |
| Bacillus subtilis | 60–100 | 100 |
| Streptococcus aureus | 60–120 | 95 |
| Streptococcus pneumoniae | 40–110 | 100 |
| Enterococcus faecalis | 75–115 | 100 |
Approximate location of the strongest prediction for coiled coil in FtsL amino acid sequence.
Scores were calculated according to the work of Lupas et al. (18) with the COILS program on the ISREC web server (http://www.isrec.isb-sib.ch/software /COILS_form.html). Window length, 21; matrix, MTIDK weighted. For E. coli and Y. pestis FtsL, the value reported is the average obtained with all four matrices used.
Coiled-coil propensity prediction: consequence of the replacement of W81 (1) by Y (2) in E. coli FtsL and of W66 (3) by Y (4) in Y. pestis FtsL.
Interestingly, the coiled-coil propensity of E. coli and Y. pestis FtsL increases up to almost 100% if only one residue is changed in their amino acid sequence (Y81 instead of W81 for E. coli FtsL and Y66 instead of W66 for Y. pestis FtsL). Apparently, the tryptophan instead of tyrosine residue at position 81 (or 66 for Y. pestis FtsL) greatly reduces the coiled-coil propensity probability (Table 6). This result may be due to the fact that the scoring matrix for the COILS program is derived exclusively from long, parallel, two-stranded coiled coils where W is highly disfavored at the a position. Indeed, certain peculiarities of two-stranded coiled coils, such as a restricted amount of space in packing the core layers, will be used to judge coiled-coil sequences that do not have these problems by virtue of the fact that they form three-, four-, or even five-stranded coiled coils, which have increasingly more packing space in their core. This signifies that if the FtsL coiled-coil is not two stranded, then it may well be easy to accommodate the W in position a of the core, even though this is very difficult in a two-stranded coiled coil (A. Lupas, personal communication). In B. aphidicola FtsL, which is one-third shorter than E. coli FtsL, the disruptive effect of the tryptophan W81 located at the end of the putative coiled coil is predicted to be smaller than in the case of E. coli or Y. pestis FtsL, where it is located in the middle of the coiled coil.
Besides the striking conservation of the heptad repeats in the periplasmic domain, the FtsL multiple alignment also shows that the helical structure may extend through the membrane-spanning segment and the cytoplasmic domain (Fig. 4).
The periplasmic domain of E. coli FtsL is a multimerization domain.
The data presented above suggest that FtsL may adopt a coiled-coil structure that could encompass both the transmembrane and the periplasmic domain. Coiled-coil structures have been involved in the formation of multimers where two to five helices interact to form a superhelical bundle. The stability of the coiled-coil interactions is variable but may be high and in some cases thermostable and SDS resistant (13). To test whether potential FtsL multimers were stable enough to resist denaturation by SDS, we carried out an SDS-polyacrylamide gel electrophoresis analysis of a protein extract from a strain where FtsL was expressed from a multicopy plasmid under an inducible promoter. On SDS-polyacrylamide gels, two forms could be detected with anti-FtsL antibodies: a lower form that ran at the expected monomeric FtsL molecular mass (about 14 kDa) and an upper form running at 30 kDa that may correspond to an FtsL homodimer (Fig. 6). In some experiments, higher-order multimers could also be seen, although less consistently than the apparent dimer (data not shown). The abundance of the dimeric form was considerably reduced in boiled samples (data not shown). In order to determine whether the formation of this putative dimer is dependent on the presence of the periplasmic domain, cell extracts from strains expressing LLL, FLL, LFL, and FFL swap proteins as well as LLL(L70H) and LLL(L84D) were submitted to the same analysis. Samples from cells expressing LLL and FLL, but not LFL or FFL, displayed the putative dimer bands (Fig. 6A). Samples from strains expressing the two leucine mutants displayed a much-reduced ability to dimerize that could not be observed consistently (not shown in Fig. 6 and noted as +/− in Table 7). These results demonstrate that only constructs carrying the membrane-spanning segment and periplasmic domain of FtsL multimerize and that mutagenesis of two d positions [LLL(L70H) and LLL(L84D)] of the putative coiled-coil motif reduces the dimerization ability. To carry further this analysis, we analyzed cell extracts from strains expressing LLL, HLL, LHL, and HHL swap proteins as well as LLHinfL and LLGCN4L. Figure 6B shows that LLL, HLL, LHL, and LLHinfL dimerize while HHL dimers could not consistently be observed (Fig. 6 and noted as +/− in Table 7). The levels of dimerization of HLL and LHL were found to be consistently lower than the one observed with FLL, although HLL and LHL were stable enough at the steady-state level. Samples from strains expressing LLHinfL displayed about the same ability to dimerize as LLL. The introduction of a well-characterized dimerization motif in LLGCN4L led to the efficient formation of dimers and even higher-order multimers (tetramers?) (Fig. 6). These results suggest that the FtsLHinf membrane-spanning segment cannot fully replace its E. coli homologue with regard to the multimerization phenotype although it fully complements the localization phenotype. It also shows that multimerization alone is not sufficient to fulfill the FtsL role in cell division.
TABLE 7.
Localization frequencies, complementation, and dimerization phenotypes of some of the FtsL swap constructs described in this study
| Construct | Localization frequency (%) | Complementationa | SDS-resistant dimersb |
|---|---|---|---|
| Controls | |||
| FtsL | 72 | +++ | ++++ |
| LLL | 69 | +++ | +++ |
| MalF-E. coli FtsL swaps | |||
| FLL | 70 | − | +++ |
| LFL | <1 | − | − |
| FFL | <1 | − | − |
| H. influenzae-E. coli FtsL swaps | |||
| LLHinfL | 40 | +/− | ++ |
| HLL | 82 | + | + |
| LHL | 80 | − | +/− |
| HHL | 76 | − | +/− |
| HLH | <1 | − | − |
| LHH | <1 | − | − |
| E. coli FtsL mutants | |||
| LLL(L70H) | 17 | +/− | +/− |
| LLL(L84D) | 28 | +/− | +/− |
| E. coli FtsL leucine zipper swap | |||
| LLGCN4L | <1 | − | ++++ |
For the complementation test, plus signs reflect colony size, minus signs indicate no growth, and +/− indicates poor growth.
The plus signs reflect the propensity to form SDS-resistant dimers. +/−, dimers were not consistently observed in all the experiments.
Taken together, these results are consistent with the hypothesis that both the transmembrane segment and the periplasmic domain of FtsL form a coiled-coil structure that could lead to multimerization. Since poorly dimerizing constructs still localize efficiently, multimerization may be a functionally critical event that could take place after FtsL localization to the division site.
DISCUSSION
In this paper, we have analyzed the structure-function relationship of the essential cell division protein, FtsL. This analysis is facilitated by the simple domain structure of FtsL, a bitopic membrane protein composed of an N-terminal cytoplasmic domain, a transmembrane segment, and a periplasmic domain. We have studied the role of these domains by swapping them or portions of them with other cell division proteins or with an unrelated protein. In these studies, we have examined the role of these different domains in three properties of FtsL. These properties are (i) the ability to form dimers or even multimers on SDS gels, (ii) the ability to localize to the site of formation of the cell septum, and (iii) the ability to provide an essential function in the cell division process. The results of the swap studies (summarized in Table 7) show that the three functions of FtsL are separable.
Oligomerization of FtsL.
We have shown that, on SDS gels, a substantial proportion of FtsL is found in a band corresponding to dimers. We believe that this oligomerization is made possible by the apparent coiled-coil structure that the periplasmic domain, with its heptad repeats of leucines, may assume, perhaps along with the two other domains of the protein (see below). We find that dimerization is correlated with the presence of the intact E. coli FtsL periplasmic and transmembrane domains; the presence of the periplasmic domain alone is not sufficient. Furthermore, mutations in either of two leucines of the heptad repeats in the periplasmic domain reduce the efficiency of dimerization. However, dimerization per se is not sufficient for the two other properties of FtsL: localization to the septum and function in cell division. This we showed by providing an alternative dimerization signal in the periplasmic domain, the leucine zipper region of the Saccharomyces cerevisiae GCN4 protein. In this case, the protein still forms dimers on gels but is nonfunctional for the other properties of FtsL. Thus, specific sequences in the periplasmic domain are required for the localization process, beyond those required for dimerization.
Localization to the septum.
Localization of FtsL to the septal region of the E. coli cells also appears to be dependent on the presence of its periplasmic domain and on a transmembrane segment either from E. coli FtsL or from the H. influenzae homologue. The HLL, HHL, and LHL swap constructs all localize efficiently to the septum. Furthermore, the FLL construct, in which a cytoplasmic domain of the unrelated membrane protein, MalF, replaces the normal FtsL cytoplasmic domain, also localizes to the septum with a frequency comparable to that of the wild type. Thus, the cytoplasmic domain is needed neither for dimerization nor for localization. However, since the localization studies were done in a background that expresses wild-type FtsL from the chromosome, we cannot distinguish between the possibility that FLL localizes on its own to the septum and the possibility that it localizes via an interaction of the dimerization domain with an already localized wild-type FtsL.
The mutations altering the conserved leucines in the periplasmic domain reduce the efficiency of localization, as does the replacement of the heptad repeat region of FtsL with the comparable region from the H. influenzae FtsL. While the studies of dimerization suggested that not just any dimerization domain will allow proper localization, the results for localization indicate that not even a close relative of FtsL from H. influenzae provides enough of the specific sequence to the periplasmic domain that is required for efficient localization. The degree of specificity required in this case suggests that the periplasmic domain of FtsL may interact with another cell division protein that is not strongly conserved in sequence between bacterial species.
Function in cell division.
Finally, even certain swap constructs that localize very efficiently to the cell septum do not provide the complete set of functions required to complement an FtsL null mutation. The constructs FLL, HHL, and LHL all localize to the septum with the same efficiency as wild-type FtsL, but none of them complement. The finding with the FLL swap protein indicates that the cytoplasmic domain of FtsL, which is not essential for dimerization or septal localization, is, nevertheless, required for function. Complementation by the HLL construct indicates that the H. influenzae cytoplasmic domain of FtsL is sufficiently close in structure to that of the E. coli protein, despite a low degree of sequence conservation, that it can function properly. This is somewhat surprising since the FtsLHinf cytoplasmic domain is 14 residues shorter than the one of FtsLEcol. Thus, the essential region of the cytoplasmic domain of FtsLEcol may lie in the sequence that starts after residue 15 (Fig. 3). Two other gram-negative bacterial FtsL homologues, identified in Y. pestis and B. aphidicola, also display short cytoplasmic domains. Multiple alignments show that each of the three homologues has two conserved charged residues in common (R21 and D31 in FtsLEcol [Fig. 5]) that may be good mutagenesis targets for identifying critical residues in the FtsL cytoplasmic domain.
In sum, all three domains of FtsL are required for its function in cell division, but they play different roles. The cytoplasmic domain appears to be needed for some step late in the functioning of FtsL, after it has arrived at the cell septum. The periplasmic domain is involved in dimerization, a step that may be required after localization can take place or a step that may be required to maintain a stably localized protein. The transmembrane segment of FtsL is required for complementation also; even the related membrane-spanning segment from the H. influenzae FtsL will not suffice for function. Again, such a finding suggests that the interactions of E. coli FtsL with other components of the cell division machinery require specific sequences of both the transmembrane and periplasmic domains.
Interestingly, B. subtilis DivIC protein stability was reported to depend on B. subtilis FtsL synthesis (7). The DivIC product is required for formation of the vegetative and sporulation septum (15) and bears a striking structural resemblance to E. coli FtsL, including a predicted coiled-coil periplasmic domain. Although there is no obvious DivIC homologue in E. coli, an attractive possibility would be that FtsL interacts in a complex containing a DivIC homologue.
A potentially unbroken transmembrane and periplasmic coiled-coil motif is involved in localization to the division site.
Localization and complementation analysis as well as coiled-coil predictions is compatible with a helical structure prolonged throughout the membrane-spanning segment and the cytoplasmic domain. This raises the possibility that the FtsL coiled coil could run essentially unbroken through the membrane and be involved in the formation of multimers. FtsL SDS-resistant multimers of higher order than dimers can be observed in some conditions, suggesting that, at least in vitro, interaction of more than two molecules may take place. Whereas the presence in vivo of such multimers remains to be demonstrated, preliminary data involving in vivo cross-linking between cysteines introduced in the periplasmic domain (Leu63 to Cys and Leu70 to Cys) suggest that the observed SDS-resistant multimers are likely to be homomultimers (J. M. Ghigo, unpublished results).
What is the function of FtsL? Coiled-coil domains are found in transcription factors and fibrous and structural proteins as well as membrane fusion proteins. As pointed out before (8), based on its small size and the absence of recognizable catalytic domain, it seems unlikely that FtsL fulfills a biosynthetic function of its own. Rather, the results presented in this study are consistent with the hypothesis of a structural role for FtsL. Although there is limited information on the roles of proteins involved in septation in E. coli, so far, they often fall into two hypothetical categories: cytokinesis (i.e., FtsZ) and cell envelope biogenesis (i.e., FtsI). Since FtsL homologues are known in gram-positive bacteria, it is unlikely that FtsL functions in outer membrane biosynthesis or interaction. Interestingly, most cell division proteins known to act before FtsL, i.e., FtsZ, ZipA, FtsA, and FtsQ (8), have known homologues in gram-positive bacteria or have been shown to interact with FtsZ. An attractive possibility would then be that FtsL participates in a process that takes place in both gram-positive and gram-negative bacteria and that is related to cytokinetic aspects of septation.
FtsL, through the formation of densely packed multimers, may be involved in linking together the cytokinetic elements or in providing a membrane anchor to the FtsZ contractile ring. Such an anchor may help to resist the shearing forces developed during the contraction of the cytokinetic ring. Alternatively, since coiled-coil domains have been shown elsewhere to be involved in membrane interactions (26), an attractive hypothesis would be that the FtsL coiled coil could be playing a similar role in septation.
ACKNOWLEDGMENTS
We are grateful to L. M. Guzman for the gift of the unpublished plasmid pBAD39. We thank D. S. Weiss, J. C. Chen, N. Buddelmeijer, A. Lupas, and J. M. Betton for their helpful comments and discussions.
This work was supported by grants from the American Cancer Society and the National Institutes of Health (GM 38922) to J.B. J.-M.G. was supported by the Institut Pasteur, Paris, France. J.B. is an American Cancer Society Research Professor.
REFERENCES
- 1.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 2.Addinall S G, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 3.Buddelmeijer N, Aarsman M E, Kolk A H, Vicente M, Nanninga N. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J Bacteriol. 1998;180:6107–6116. doi: 10.1128/jb.180.23.6107-6116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustos S A, Schleif R F. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J C, Weiss D S, Ghigo J M, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Daniel R A, Harry E J, Katis V L, Wake R G, Errington J. Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- 8.Ghigo J M, Weiss D S, Chen J C, Yarrow J C, Beckwith J. Localization of FtsL to the Escherichia coli septal ring. Mol Microbiol. 1999;31:725–737. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- 9.Guzman L M, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L M, Weiss D S, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale C A, de Boer P A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 13.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 14.Levin P A, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G, Draper G C, Donachie W D. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 16.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 17.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 18.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 19.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Sun Q, Wang R, Singh G, Jonietz E L, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marck C. ‘DNA Strider‘: a ‘C‘ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael S F. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques. 1994;16:410–412. [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Ottemann K M, Mekalanos J J. Analysis of Vibrio cholerae ToxR function by construction of novel fusion proteins. Mol Microbiol. 1995;15:719–731. doi: 10.1111/j.1365-2958.1995.tb02380.x. [DOI] [PubMed] [Google Scholar]
- 25.Rizo J, Sudhof T C. Mechanics of membrane fusion. Nat Struct Biol. 1998;5:839–842. doi: 10.1038/2280. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 28.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 30.Weiss D S, Chen J C, Ghigo J M, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss D S, et al. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]