Abstract
Genes encoding l-sorbose metabolism of Lactobacillus casei ATCC 393 have been identified on a 6.8-kb chromosomal DNA fragment. Sequence analysis revealed seven complete genes and a partial open reading frame transcribed as two units. The deduced amino acid sequences of the first transcriptional unit (sorRE) showed high similarity to the transcriptional regulator and the l-sorbose-1-phosphate reductase of the sorbose (sor) operon from Klebsiella pneumoniae. The other genes are transcribed as one unit (sorFABCDG) in opposite direction to sorRE. The deduced peptide sequence of sorF showed homology with the d-sorbitol-6-phosphate dehydrogenase encoded in the sor operon from K. pneumoniae and sorABCD to components of the mannose phosphotransferase system (PTS) family but especially to domains EIIA, EIIB, EIIC and EIID of the phosphoenolpyruvate-dependent l-sorbose PTS from K. pneumoniae. Finally, the deduced amino acid sequence of a truncated gene (sorG) located downstream of sorD presented high similarity with ketose-1,6-bisphosphate aldolases. Results of studies on enzyme activities and transcriptional analysis revealed that the two gene clusters, sorRE and sorFABCDG, are induced by l-sorbose and subject to catabolite repression by d-glucose. Data indicating that the catabolite repression is mediated by components of the PTS elements and by CcpA, are presented. Results of sugar uptake assays in L. casei wild-type and sorBC mutant strains indicated that l-sorbose is taken up by l-sorbose-specific enzyme II and that L. casei contains an inducible d-fructose-specific PTS. Results of growth analysis of those strains and a man sorBC double mutant suggested that l-sorbose is probably also transported by the d-mannose PTS. We also present evidence, from studies on a sorR mutant, suggesting that the sorR gene encodes a positive regulator of the two sor operons. Sequence alignment of SorR, SorC (K. pneumoniae), and DeoR (Bacillus subtilis) revealed that they might constitute a new group of transcriptional regulators.
In the enteric bacteria Klebsiella pneumoniae and Escherichia coli, the ketose l-sorbose is transported and phosphorylated through a phosphoenolpyruvate-dependent l-sorbose-specific phosphotransferase system (PTS) (36, 46, 47). The metabolism of l-sorbose involves the formation of d-fructose-6-phosphate, which enters the glycolysis pathway (Fig. 1A). During uptake of a PTS carbohydrate, the common PTS proteins, enzyme I (EI) and HPr, transfer a phosphoryl group from phosphoenolpyruvate to the substrate-specific EII complexes. The l-sorbose PTS EII consists of two membrane-bound proteins, EIICSor and EIIDSor, and two soluble components, EIIASor and EIIBSor (47). Sequence analysis revealed that EIISor was homologous to d-mannose PTS elements of E. coli (10), EIILev of Bacillus subtilis (21), and the putative EIIAga of E. coli (28). These four PTSs constitute the so-called mannose PTS class. In this class, HPr phosphorylates the EIIA domain at a conserved histidine, and the phosphoryl group is transferred in the next step to another histidyl residue in the EIIB domain. Sugar translocation occurs through EIIC and EIID, integrated in the membrane, and EIIB carries out the concomitant sugar phosphorylation (26, 29). The K. pneumoniae EIISor could also transport d-fructose, and the E. coli EIIABMan domain could complement the lack of the two soluble proteins EIIASor and EIIBSor in K. pneumoniae (47). Similarly, EIICLev and EIIDLev elements of the levanase operon of B. subtilis were able to transport d-mannose in addition to d-fructose. Moreover, these two domains could substitute in E. coli for the function of EIICMan and EIIDMan for d-mannose uptake and act as λ bacteriophage receptors (23).
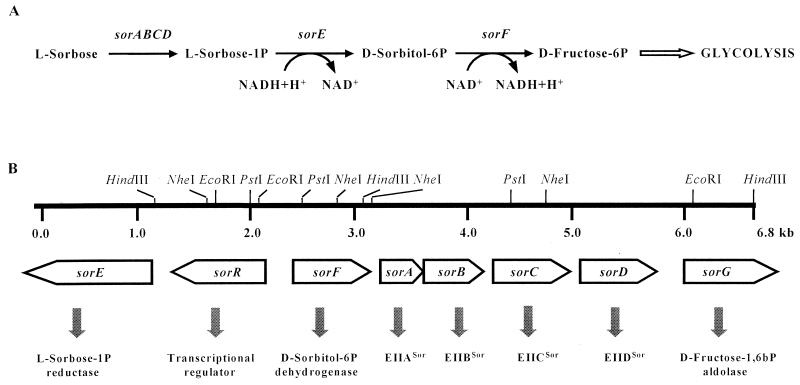
FIG. 1.
(A) Schematic representation of the transport and metabolism of l-sorbose. (B) Simplified restriction map and genetic organization of the sorbose operons from L. casei.
PTS elements are involved in sugar uptake and also in the regulation of chemotaxis, carbon metabolism, and modulation of gene expression (9, 20, 27). Many catabolic operons in bacteria are subject to carbon catabolite repression (CR) by rapidly metabolizable carbon sources, especially glucose (31, 37). The main CR mechanism identified in E. coli can be summarized as follows. EIIABGlc acts as the sensor of extracellular glucose inducing adenylate cyclase. This increases the cyclic AMP pool, which acts as an effector of the true repressor, the catabolite-activating protein, that binds to the promoter region of the genes under CR (26). However, in gram-positive bacteria with low GC content, CR follows a completely different pattern. Besides the phosphorylation by EI-P on HPr residue histidine 15, HPr(His-P), required for PTS sugar transport, HPr can also be phosphorylated at the serine 46 residue, HPr(Ser-P), by a specific ATP-dependent protein kinase (7). Then, HPr(Ser-P) interacts with CcpA, which binds to a conserved sequence in the promoter regions, the catabolite-responsive element (cre), and negatively controls the expression of several carbohydrate catabolic enzymes (8, 16, 22). However, recent studies showed that there could be other mechanisms for the control of gene expression in the presence of readily metabolizable carbon sources. A family of regulator proteins that contain consensus PTS-regulated domains (PRD), such as the positive regulator LicR from B. subtilis (40), and antiterminators like BglG from E. coli (11), LicT, GlcT and SacY from B. subtilis (see reviews in references 37 and 41), and LacT from Lactobacillus casei (13), has been described. These regulators can activate the transcription of certain genes in the presence of the inducing molecule by dephosphorylation of a PRD, but their activity also can be modulated by HPr(His-P) phosphorylation of specific histidine residues in a second PRD in such a way that in the presence of rapidly metabolizable carbon sources, the regulators would be inactive. This mechanism has been considered an alternative form of CR mediated by HPr. A major interest of our laboratory includes carbohydrate-specific regulation of gene expression in L. casei, a facultative heterofermentative lactic acid bacterium frequently isolated from fermented food products and from human and other mammalian intestines. L. casei contains a d-mannose-specific PTS for glucose and mannose uptake. It was shown that the lack of a functional EIIMan leads to derepression of lac and rbs genes. In this strain, glucose can also enter the cell by a proton motive force-driven permease (44). Likewise, the chromosomal ccpA gene from L. casei was isolated and sequenced, and a mutant deficient in this gene was constructed (24).
Results of an investigation of the components of the EIIMan family in L. casei ATCC 393 cured of plasmid pLZ15 (strain BL23) prompted a study of the sor operons, the results of which are presented here. Sequence analysis revealed eight open reading frames (ORFs) whose products are involved in l-sorbose transport and its conversion to d-fructose-6-phosphate. These genes are organized in two clusters, sorRE and sorFABCDG, which are induced by l-sorbose and repressed by d-glucose. These findings constitute the first data on the genetics and metabolism of l-sorbose in gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. casei wild-type strain BL23, man mutant BL23D (44), ccpA mutant BL71 (24), man ccpA double mutant BL72 (13), and ptsI mutant BL126 (R. Viana, V. Monedero, and G. Pérez-Martínez, unpublished data) were routinely grown at 37°C under static conditions on MRS medium (Oxoid), MRS basal medium (44), or MRS fermentation medium (Adsa-Micro) supplemented with sugars at the concentrations indicated. E. coli DH5α, E. coli DH10B, and E. coli XL1-Blue (Stratagene, La Jolla, Calif.) were used as hosts in cloning procedures. These strains were grown in Luria-Bertani medium at 37°C with vigorous shaking. Agar plates containing the same media were prepared. E. coli transformants were selected with ampicillin (50 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (40 μg/ml); L. casei transformants were selected with erythromycin (5 μg/ml).
Plasmids and transformation.
The L. casei library (13) was constructed in pJDC9 (6). Plasmids pUC18 and pBluescript II SK+ (Stratagene) were used for subcloning and sequencing experiments. The integrative vector pRV300 (19) was used for insertional inactivation of genes in the L. casei chromosome. Transformation of L. casei and E. coli strains was performed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) as previously described (25) and as recommended by the manufacturer, respectively.
Cloning and DNA sequencing.
Total DNA was isolated from L. casei as described before (25) and used as the template in a standard PCR with two primers, man11 (5′-GGAGGSASSSCCATATAAYGC) and man2 (5′-AGCYTGRCCATGAATYAA), designed from the sequence of the L. curvatus mannose PTS genes (43). An amplified DNA fragment of the expected size was cloned in pUC18 and checked by DNA sequencing; then new internal primers were designed and used to screen by PCR a genetic library of L. casei in E. coli DH5α (13). Isolation of plasmid DNA from E. coli, restriction endonuclease analysis, ligations, inverse PCR experiments, and construction of subclones in pBluescript II KS+ were performed by standard procedures (32). DNA sequencing was carried out by using an ABI PRISM dRhodamine Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase and an automatic ABI 310 DNA sequencer (Perkin-Elmer Corp.). M13 universal and reverse primers or primers that hybridize within the cloned DNA were used for this purpose. Alignment of sequences from different clones and analysis of the sequence for ORFs were carried out with version 8.1 of the Genetics Computer Group (Madison, Wis.) package. Sequence similarities were analyzed with the BLAST and FASTA programs.
Northern blot analysis.
RNA was isolated from L. casei cells grown in MRS fermentation medium with 0.5% appropriate sugars to an optical density at 550 nm (OD550) of 0.8. Cells were collected by centrifugation, washed with 50 mM EDTA (pH 8.0), resuspended in 1 ml of TRIZOL reagent (Gibco BRL, Grand Island, N.Y.), and then mechanically disrupted with glass beads in a cell disruptor (Savant Instruments, Holbrook, N.Y.). Total RNA was isolated according to the protocol of the TRIZOL manufacturer. Sample preparation, denaturing agarose gel electrophoresis, and RNA transfer were performed by standard methods (32). DNA probes for sorE, sorR, sorF, sorC, and sorG genes were synthesized in PCRs using L. casei chromosomal DNA as the template, deoxynucleoside triphosphate mix with digoxigenin-dUTP from the Boehringer digoxigenin-labeling kit, and TaqPlus Precision (Stratagene). Each probe was obtained by using two primers that hybridized within the appropriate gene: sorE probe with primers sorE1 (5′-CCATAATCCAGCAGTACTTG) and sorE2 (5′-CTTTCTTCATTGGGCCTGCG), sorR probe with sorR1 (5′-AAGGCGTTGTTTCAATTGCC) and sorR3 (5′-ACACGTGTCCACAGGCTAAG), sorF probe with sorF1 (5′-AGCAGAGAAGATTAATGGC) and manR21 (5′-TAGGATAATTGGTGCGCTAAAGG), sorC probe with sorSalI (5′-CAATGTCGACACATCGACCTTGC) and sorKpnI (5′-GCATTAGGTACCTAACACTCATC), and sorG probe with ald3 (5′-TGCGCCTCATCCGCATATG) and ald4 (5′-CGAGAGGCAGCACTAACCG).
Primer extension analysis.
The conditions for growth of L. casei cultures in 0.5% l-sorbose and the method used to isolate total RNA were as described above. Primer extension experiments were performed with avian myeloblastosis virus reverse transcriptase (Amersham) and primers sorF3 (5′-AATACCTGATGAACCGCCTG), 63 bp downstream of the sorF ATG start codon, and sorR4 (5′-GTATCAGACCGTGCATGTAG), 79 bp downstream of the sorR ATG start codon. Oligonucleotides were 5′ labeled and used in primer extension reactions with 15 μg of RNA. Reverse transcription products were resolved on 6% urea-containing polyacrylamide gels, together with DNA sequencing reactions performed with the same primers according to standard methods (32).
Chromosomal inactivation of the sorbose-specific PTS genes.
A DNA fragment containing 200 bp of the 3′ end of sorA and 204 bp of the 5′ end of sorB was amplified by PCR with primers man11m (5′-CGTCAGCAGAATTAGTTTTG) and manR22 (5′-TGCCGTTAACGAAAACCATGTAG). Another internal DNA fragment containing 543 bp of sorC was amplified by PCR with primers sorSalI (5′-CAATGTCGACACATCGACCTTGC) and sorKpnI (5′-GCATTAGGTACCTAACACTCATC). To create SalI and KpnI restriction sites (in italics), the original sequence was modified. Both DNA fragments were cloned in the appropriate orientation in the integrative vector pRV300. The resulting plasmid, pSB301, was then used to transform L. casei, and the transformants were selected on agar plates with erythromycin. Chromosomal integration of plasmid pSB301 was confirmed by PCR analysis. Subsequently, one of the integrants was selected and used to inoculate fresh MRS medium without antibiotic. After growth for approximately 200 generations, appropriate dilutions were replica plated onto agar plates with and without erythromycin. Strains that had undergone the second recombination and therefore were cured of plasmid could be selected as erythromycin sensitive. Chromosomal DNA of these strains was isolated and subjected to PCR analysis. Mutant BL23S, with the sorB carboxy-terminal region and the sorC amino-terminal region deleted, was selected. Plasmid pSB301 was also used to inactivate the sorbose-specific genes in the man mutant chromosome (BL23D) (44). The integrants resistant to erythromycin were analyzed by PCR, and one of them, named BL23DS, was selected.
Chromosomal inactivation of the sorR gene.
The 400-bp PstI-NheI sorR fragment was isolated from the original clone in pJDC9 and subcloned in the integrative vector pRV300. The resulting plasmid, pSR302, was then electroporated into L. casei, and the transformants were selected on agar plates with erythromycin. Chromosomal integration of the plasmid pSR302 was checked by PCR analysis. The sugar fermentation pattern of the integrants was analyzed, and one of them, designated BL23R, was selected.
Fructose uptake assays.
Fructose uptake assays were carried out as described previously (4, 5). L. casei wild-type and sorBC mutant strains were grown in 50 ml of MRS fermentation medium supplemented with 0.5% fructose or 0.5% sorbose to an OD550 of 0.6 to 0.8. The PTS EI-deficient mutant (Viana et al., unpublished data) was grown in the same medium supplemented with 0.5% ribose plus 0.5% fructose or 0.5% ribose plus 0.5% sorbose. Cells were collected by centrifugation, washed twice with 10 mM potassium phosphate buffer (pH 7.4) containing 1 mM MgCl2, and resuspended in the same buffer (0.1 mg [dry weight]/ml). The cells were incubated at 37°C, and a mix of labeled d-[14C]fructose (0.3 mCi/mmol) and nonlabeled fructose (final concentrations of 25, 75, 125, and 250 μM) was added. At intervals of 0, 15, 30, 60 and 120 s, samples (1 ml) were withdrawn and filtered through Millipore membranes. The filters were washed, and radioactivity was quantified by scintillation counting.
Enzyme assays.
For determination of enzymatic activities in L. casei, cultures were grown on MRS fermentation medium supplemented with 0.5% appropriate sugar to an OD550 of 0.8. Crude extracts were prepared by disrupting cells with glass beads in a cell disruptor (Savant Instruments) for four periods of 1 min with intervals of 1 min on ice. l-Sorbose-1-phosphate reductase (Sor-PR) activity was determined as described by Wöhrl and Lengeler (49). d-Frutose-1-phosphate was used as the substrate at 2.5 mM; the buffer contained 12.5 mM morpholinepropanesulfonic acid (pH 6.2), 0.1 mM NADH, and 0.2 mM MnCl2. d-Sorbitol-6-phosphate dehydrogenase (Stol-PDh) activity was determined in 10 mM Tris buffer (pH 7.5)–2 mM NAD+, and d-glucitol-6-phosphate (d-sorbitol-6-phosphate) was used as the substrate at 2 mM. The rate of NADH oxidation for Sor-PR and the rate of NAD+ reduction for Stol-PDh were determined by measuring the rate of absorbance change at 340 nm. Specific enzyme activities are given in nanomoles per minute per milligram of protein. Protein concentrations were determined by the method of Bradford (2).
Nucleotide sequence accession number.
The nucleotide sequence data reported here have been submitted to the GenBank database under accession no. AF129168.
RESULTS
Cloning and sequence analysis of the sor genes.
PCR amplification of L. casei DNA with two primers synthesized from the sequence of the L. curvatus d-mannose-specific PTS genes (manA and manB) (43) yielded a DNA fragment with the expected size (280 bp). This fragment was sequenced and used to screen by PCR a genetic library of L. casei in E. coli DH5α (13), and a positive clone containing a 3,639-bp insert in pJDC9 was obtained. Different restriction endonuclease fragments derived from this DNA insert were subcloned into pBlueScript II SK+ and sequenced. Sequence analysis revealed four ORFs. The highest homology was detected with Sor-PR (58% identical residues), the sorbose operon regulator (34% identical residues), and Stol-PDh (61% identical residues) of the sor operon of K. pneumoniae (45). The fourth ORF showed similarity with the EIIA component of the l-sorbose-specific PTS of K. pneumoniae (30% identical residues), the d-mannose-specific PTS of E. coli (35% identical residues), and the d-fructose-specific PTS of the lev operon from B. subtilis (34% identical residues) (10, 21, 45).
Several attempts to obtain clones containing the upstream and downstream regions of the cloned DNA insert were unsuccessful. Reverse PCR experiments allowed us to sequence a 3,196-bp fragment downstream of the 3′ end and a 300-bp fragment upstream of the 5′ end. Sequence analysis revealed four additional ORFs. They showed high similarity to PTS proteins such as EIIB, EIIC, and EIID of the l-sorbose-specific PTS from K. pneumoniae (65, 70, and 72% identical residues, respectively) (45), EIIAB, EIIC, and EIID of the d-mannose-specific PTS from E. coli (54, 61, and 57% identical residues, respectively) (10), and EIIB, EIIC, and EIID of the lev operon from B. subtilis (42, 51, and 54% identical residues, respectively) (21). The last ORF showed similarity to known ketose-bisphosphate aldolases, notably to the d-fructose-bisphosphate aldolase of B. subtilis (42% identical residues) (42).
We have named the sor genes of L. casei according to sequence homology of the proteins that they encode. Hence the gene for the transcriptional regulator is sorR, those encoding the sorbose-specific PTS elements are sorA, sorB, sorC, and sorD, and then the metabolic genes, named in alphabetical order, are sorE, sorF, and sorG, for the genes encoding Sor-PR, Stol-PDh, and d-fructose-1,6-bisphosphate aldolase, respectively. Schematic representation of the l-sorbose pathway and a restriction endonuclease map of the l-sorbose operons of L. casei are illustrated in Fig. 1. A putative promoter region was identified in each DNA strand of the intergenic region between sorR and sorF (Fig. 2). Both regions are AT rich and have potential −10 and −35 boxes. A cre-like sequence consisting of a perfect palindrome of 14 bp was also found in that region (17).
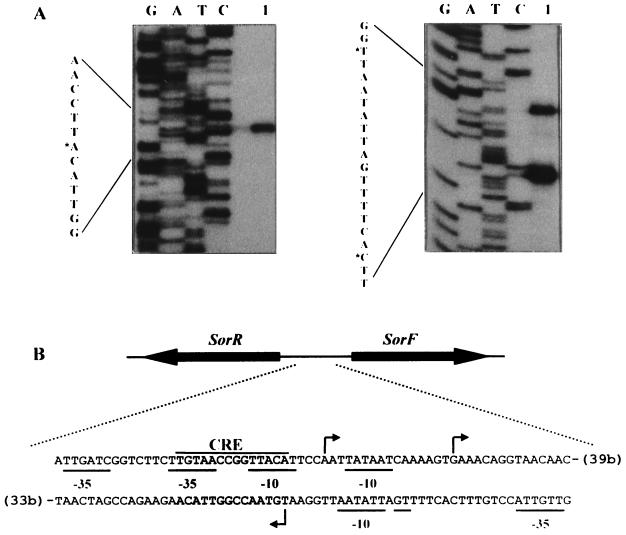
FIG. 2.
(A) Primer extension signals for sorRE (left) and sorFABCDG (right) promoters. Lanes G, A, T, and C contain the DNA sequencing reactions; lane 1 contains the products of primer extension. The DNA sequence shown is the complementary strand of the mRNA. Asterisks indicate nucleotides corresponding to the main extension products. (B) Sequence of the sor promoter region. The transcriptional start sites of the sorRE and sorFABCDG promoters are indicated by arrows. The putative promoter sequences, −35 and −10, are underlined. The cre-like sequence is also shown. Numbers in parentheses indicate distances in nucleotides from the sorR and sorF ATG start codons.
sorE (412 amino acids) has two potential ATG start codons separated by four codons, but only one putative ribosome-binding site (35) was detected in front of the second ATG. Downstream of this gene, a potential rho-independent terminator was found. The second ORF, sorR, was found upstream of sorE. The deduced amino acid sequence corresponded to a protein of 318 amino acids (Mr, 34,913). An intergenic region of 136 bp separates the third ORF, sorF, which is transcribed in the direction opposite that of sorR and sorE and encodes a protein of 266 amino acids (Mr, 28,459). sorA encodes the first protein of the l-sorbose-specific PTS system, EIIASor, with 138 amino acids (Mr, 14,985). The stop codon of sorA overlapped in one base with the start codon of sorB. This ORF was predicted to encode EIIBSor (164 amino acids; Mr, 18,304). EIIASor and EIIBSor contain histidyl residues at positions 10 and 14, respectively, which align with the phosphorylation sites of the homologous IIA and IIB proteins. Thirteen base pairs from the sorB we found the start codon of sorC. Its predicted amino acid sequence corresponds to a protein, EIICSor, of 277 amino acids (Mr, 29,002). Downstream we found sorD, the deduced product of the ORF corresponds to a 282-amino-acid protein, EIIDSor (Mr, 30,788). After a stretch of 191 bp, an incomplete ORF, sorG, truncated at amino acid 276, was found. Ribosome-binding sites were found always between 4 and 10 bp upstream of the start codons of all ORFs.
Nature of SorR.
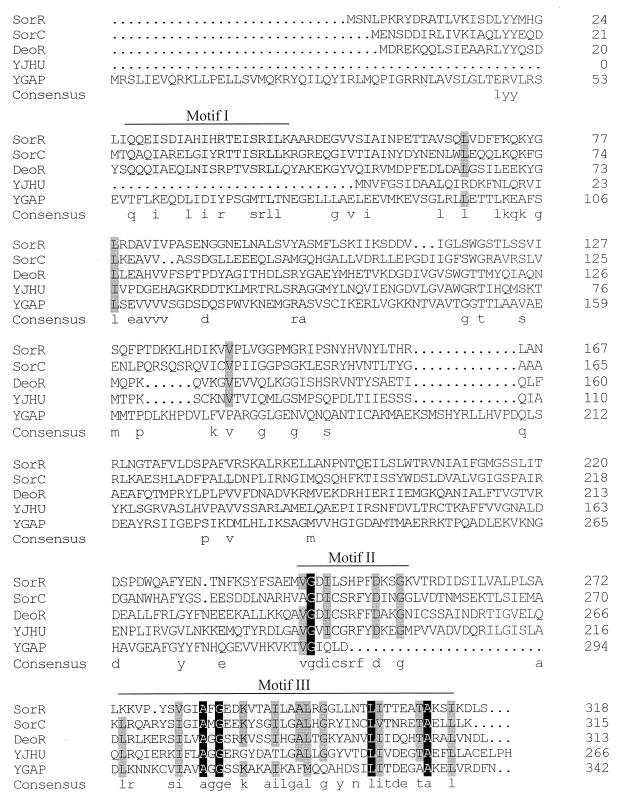
When the predicted amino acid sequence of SorR was used in a Blast search, the most homologous proteins were the sor operon regulator (SorC) from K. pneumoniae (45), the deoxyribonucleoside regulator (DeoR) from B. subtilis (33), the hypothetical transcriptional regulator in the fecI-fimB intergenic region (YJHU) from E. coli (3), and the hypothetical transcriptional regulator of the gap operon (YGAP) from Bacillus megaterium (34). Alignment of the SorR sequence with those sequences is shown in Fig. 3. The amino-terminal region contains the hypothetical DNA-binding helix-turn-helix motif, except for the YJHU sequence, which suggests that it could be a sugar kinase rather than a transcriptional regulator (39). The carboxy-terminal region reveals two highly conserved motifs, motifs II and III, of uncertain function. However, motif III showed homology with the carboxy-terminal region of the ROK family, which includes transcriptional repressors, ORFs of unknown function, and sugar kinases (39), suggesting that it could be involved in sugar binding. Based on the unique features just mentioned, L. casei SorR, K. pneumoniae SorC, and B. subtilis DeoR could be representative of a new group of transcriptional regulators.
FIG. 3.
Multiple amino acid sequence alignment of SorR from L. casei (this work), SorC from K. pneumoniae (accession no. P37078), DeoR from B. subtilis (accession no. P39140), YJHU from E. coli (accession no. P39356), and YGAP from B. megaterium (accession no. P35168). The residue number of each protein is indicated at the right. The consensus sequence (at least three residues conserved) is shown in lowercase letters. Residues conserved in all proteins are shown against a dark background. Residues that are similar in four sequences appear against a shaded background. Motif I indicates the position of the putative DNA-binding sites; motifs II and III indicate highly conserved regions.
Growth pattern on d-glucose and l-sorbose of L. casei strains.
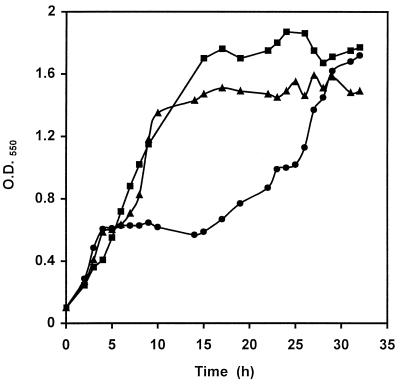
When the wild type was grown on a basal MRS medium supplemented with 0.1% d-glucose and 0.2% l-sorbose, growth stopped for about 10 h, possibly after d-glucose was consumed (Fig. 4). However, the man mutant, BL23D, displayed no diauxic growth, and in the ccpA mutant, BL71, the plateau of diauxic growth was reduced to only 3 h. These results indicated that the sor operon is repressed by glucose and that the repression is mediated by PTS elements and by the CcpA protein.
FIG. 4.
Growth of L. casei on MRS basal medium containing 0.1% glucose plus 0.2% sorbose. ●, wild type (BL23); ▴, ccpA mutant (BL71); ■, man mutant (BL23D).
Transcription analysis of the sor genes.
Northern blot experiments were performed with internal DNA fragments of the coding regions of sorE, sorR, sorF, sorC, and sorG as probes. The results with the sorE and sorF probes are shown in Fig. 5. The same data were obtained with the other probes assayed: a strong signal was found with RNA isolated from L. casei cells grown in the presence of l-sorbose, weaker bands were detected when d-glucose was present in the growth medium, and weak or no signals were observed when d-ribose was used as the carbon source. These results suggested that transcription of the sor genes was induced by the addition of l-sorbose in the culture and repressed by d-glucose.
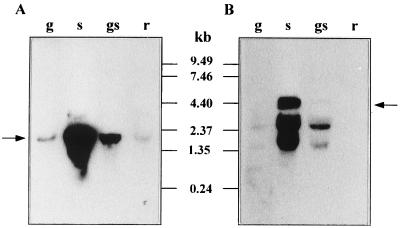
FIG. 5.
Northern blot analysis of sor mRNA, using an internal DNA fragment of sorE (A) or sorF (B) as the probe. RNA was isolated from L. casei grown with 0.5% glucose (g), 0.5% sorbose (s), 0.5% glucose plus 0.5% sorbose (gs), or 0.5% ribose (r). Arrows indicate transcripts of about 2,200 and 4,400 nucleotides; positions of size standards are marked in the center.
A signal corresponding to a transcript of about 2,200 nucleotides was obtained when sorE and sorR fragments were used as probes. This mRNA size could correspond to the size of sorR plus sorE. A transcript of about 4,400 nucleotides was detected with sorF and sorC probes. This size suggested that sorFABCDG are transcribed as one unit. These probes also detected two smaller bands that possibly are an artifact due to the position of rRNA, as has been reported by others (15, 38).
The transcriptional start sites of the sor operons were determined by primer extension experiments (Fig. 2). Two primer extension products were obtained when we performed the analysis with primer sorF3, which anneals at the 5′ end of sorF. The initiation sites correspond to guanine and adenine residues, 54 and 70 bp upstream of the sorF ATG start codon, respectively. A transcription initiation site, 62 bp upstream of the sorR ATG start codon, was identified when we used primer sorR4, which anneals at the 5′ end of sorR (Fig. 2).
Disruption of the L. casei l-sorbose-specific PTS genes.
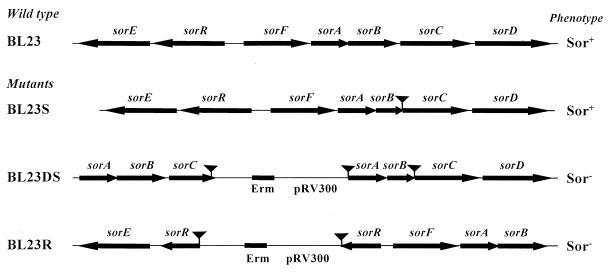
To test the biological function of the l-sorbose-specific PTS genes, a sorBC mutant was constructed as described in Materials and Methods. This strain, named BL23S, had a deletion of 117 amino acids at the carboxy-terminal region of sorB and 28 amino acids at the amino-terminal region of sorC. This deletion also introduced a frameshift mutation at the fusion point between sorB and sorC (Fig. 6). The mutant exhibited a sugar fermentation pattern identical to that of the wild type (BL23), with the difference that when grown on MRS basal medium with 0.5% l-sorbose as the carbon source, the mutant's doubling time (206 min) was greater than that of the wild type (157 min). When the same culture medium contained 0.1 or 0.05% l-sorbose, the mutant did not grow. A ptsI mutant did not ferment l-sorbose (Viana et al., unpublished data), suggesting that l-sorbose was probably transported by another PTS with less efficiency. To determine if l-sorbose was also taken up by the d-mannose-specific PTS, we disrupted the l-sorbose-specific PTS genes in the man mutant chromosome (Fig. 6). The resulting strain, named BL23DS, barely grew on MRS fermentation medium with 0.5% sorbose, presenting a very high doubling time of 301 min (the man mutant has a doubling time on sorbose of 168 min). These experiments indicated that with high l-sorbose concentrations, this sugar could also be taken up through the EIIMan elements.
FIG. 6.
Schematic representation of the genetic organization of the sorbose operons in wild-type and mutant strains of L. casei. The strategy for disrupting the sor genes is described in Materials and Methods. Arrows indicate ORFs and their orientations. Deleted DNA fragments are represented by triangular symbols. The erythromycin gene (Erm) and DNA from plasmid pRV300 are also indicated. Phenotypes were tested by streaking single colonies on MRS fermentation medium with 0.5% l-sorbose. + and − indicate fermentation and no fermentation, respectively.
Insertional inactivation of sorR.
To provide evidence for the functional importance of SorR for l-sorbose metabolism in L. casei, plasmid pRV300, containing a DNA fragment internal to sorR, was integrated into the chromosome of L. casei (Fig. 6). The integrants obtained were analyzed for the ability to ferment l-sorbose on MRS fermentation medium. All integrants exhibited an l-sorbose-negative phenotype; we picked one, named BL23R, for further analysis.
Fructose transport activity in wild-type L. casei and the sorBC mutant.
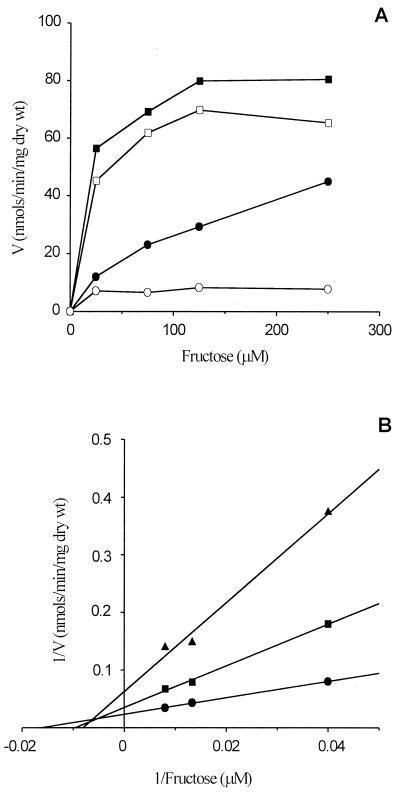
Since EIISor components have been shown to transport the analogue d-fructose (47), we tried to determine the uptake rate of d-fructose in L. casei wild-type and sorBC and ptsI mutant strains pregrown on d-fructose or l-sorbose. There have been no previous reports on d-fructose transport in L. casei. The ptsI mutant (Viana et al., unpublished data) showed no d-fructose uptake (data not shown). The wild-type and sorBC mutant strains pregrown on d-fructose can efficiently incorporate d-fructose (apparent Kms of 12 and 16 μM, respectively). The sorBC mutant pregrown on l-sorbose showed very little d-fructose uptake; however, the wild type was able to transport d-fructose at a lower rate (apparent Km of 61 μM) than when it was pregrown on d-fructose (Fig. 7A). These data suggest that the d-fructose transport activity detected in the d-fructose-pregrown sorBC mutant was due to a fructose-inducible PTS, possibly a d-fructose-specific PTS. The existence of an inducible EIIFru would explain the difference between the transport activity of L. casei wild-type cells pregrown on d-fructose and l-sorbose. The remaining activity of the L. casei wild type pregrown on l-sorbose would account for d-fructose translocation through EIISor. To confirm that d-fructose is indeed transported through EIISor, d-[14C]fructose uptake was measured with increasing amounts of l-sorbose (Fig. 7B). When the L. casei wild type was pregrown on l-sorbose, there was a competitive inhibition of d-fructose uptake (estimated Ki of 104 μM) by as little as 10 μM l-sorbose. However, no inhibition of d-fructose uptake by l-sorbose was detected in the sorBC mutant pregrown on d-fructose (data not shown). These results suggested that the sorBC mutant was impaired in EIISor.
FIG. 7.
(A) d-[14C]fructose uptake rate in L. casei wild-type (BL23) and sorBC mutant strains. Cells were pregrown on MRS fermentation medium with 0.5% fructose (■, wild type; □, sorBC mutant) or 0.5% sorbose (●, wild type; ○, sorBC mutant). (B) Lineweaver-Burk plot of d-[14C]fructose uptake rate without sorbose (●) or with sorbose (■, 10 μM; ▴, 100 μM) by the wild type pregrown on MRS fermentation medium with 0.5% sorbose.
Enzymatic activities in L. casei wild-type and sor mutant strains.
When the wild-type and mutant strains were grown on d-ribose as the sole carbon source, Sor-PR and Stol-PDh activities were below the detection level. The presence of l-sorbose in the culture medium induced both activities (Table 1). Activities in the sorBC mutant, BL23S, were similar to those in the wild type. However, the PTS transporter double mutant, BL23DS, had very low activities. These results confirmed that l-sorbose could also be taken up by the d-mannose-specific PTS, hence inducing the operon. Neither Sor-PR nor Stol-PDh activity was detected in sorR::pRV300 mutant BL23R crude extracts. Finally, both activities in the man mutant were higher than in the wild type, and Stol-PDh activity was higher than in the sorBC mutant. These activity levels are comparable to those in the wild type grown on l-sorbose (without d-ribose) (Table 2). Differences in activities due to the presence of ribose were consistently observed in other experiments, but an explanation could not be found.
TABLE 1.
Enzyme activities in L. casei wild-type and man, sorBC, and sorR mutant strains
| Strainb | Activity (nmol/min/mg of protein)a
|
|||
|---|---|---|---|---|
|
l-Sorbose-1-P reductase
|
d-Sorbitol-6-P dehydrogenase
|
|||
| Rib | Rib + Sor | Rib | Rib + Sor | |
| BL23 (wild type) | <10 | 760 ± 68 | <10 | 775 ± 23 |
| BL23S (sorBC) | <10 | 669 ± 106 | <10 | 1,083 ± 66 |
| BL23R (sorR) | <10 | <10 | <10 | <10 |
| BL23D (man) | <10 | 1,263 ± 68 | <10 | 992 ± 129 |
| BL23DS (man sorBC) | <10 | 26 ± 25 | <10 | 138 ± 19 |
Data represent averages ± standard deviations from at least three experiments.
L. casei strains were grown on MRS fermentation medium supplemented with 0.5% d-ribose (Rib) or with 0.5% d-ribose plus 0.5% l-sorbose (Rib + Sor).
TABLE 2.
Enzyme activities in L. casei wild-type and regulatory mutant strains
| Strainb | Activity (nmol/min/mg of protein)a
|
|||||
|---|---|---|---|---|---|---|
|
l-Sorbose-1-P reductase
|
d-Sorbitol-6-P dehydrogenase
|
|||||
| Glu | Sor | Glu + Sor | Glu | Sor | Glu + Sor | |
| BL23 (wild type) | <10 | 1,275 ± 117 | <10 | <10 | 1,135 ± 105 | <10 |
| BL23D (man) | <10 | 1,359 ± 221 | 223 ± 20 | <10 | 1,182 ± 66 | 229 ± 33 |
| BL71 (ccpA) | <10 | 1,070 ± 55 | 27 ± 3 | <10 | 1,956 ± 258 | 46 ± 10 |
| BL72 (man ccpA) | <10 | 696 ± 96 | 378 ± 41 | <10 | 1,481 ± 100 | 952 ± 169 |
Data represent averages ± standard deviations from at least three experiments.
L. casei strains were grown on MRS fermentation medium supplemented with 0.5% d-glucose (Glu), with 0.5% l-sorbose (Sor), or with 0.5% d-glucose plus 0.5% l-sorbose (Glu + Sor).
Influence of EIIMan and CcpA on sorE and sorF expression.
Glucose repression exerted on Sor-PR and Stol-PDh was tested in the wild type (BL23), in the man mutant (BL23D), in the ccpA mutant (BL71), and in the man ccpA double mutant (BL72) (Table 2). The wild type showed the strongest repression with the addition of glucose, followed by the ccpA mutant, with only a 2% residual activity. The man mutant in a glucose-plus-sorbose mix exhibited a 16 and 19% residual activities of Sor-PR and Stol-PDh, respectively. This partial loss of glucose repression explains the lack of diauxic growth of this strain (Fig. 4). The major loss of glucose repression was observed in the man ccpA double mutant, which showed 54 and 64% residual activities of Sor-PR and Stol-PDh, respectively. However, the activity levels of the fully induced wild type were not recovered. This degree of glucose repression suggests that an additional mechanism of CR could still exist.
DISCUSSION
The genes encoding PTS proteins and enzymes involved in l-sorbose metabolism in L. casei have been sequenced, and some of them have been characterized by inactivation through Campbell-like recombination. d-Fructose was used as analog of l-sorbose in uptake experiments. It was transported by EIISor, as this activity was missing in the sorBC mutant (BL23S), and evidence for an inducible d-fructose-specific PTS was found. Growth experiments with the wild type and a man mutant indicated that although l-sorbose is mainly taken up via EIISor, EIIMan also could play a secondary role in l-sorbose transport. In fact, this multivalent sugar specificity could be expected from members of the mannose PTS family (26). Assays of the metabolic enzymes showed that in the sorBC mutant (BL23S), Sor-PR and Stol-PDh activities were induced by l-sorbose as in the wild type. The sorBC::pRV300 double mutant man (BL23DS) showed almost no Sor-PR activity and a low, but significantly higher, Stol-PDh activity (Table 1), suggesting that a PTS element (EIIMan or EIISor) or the translocation product, l-sorbose-1-phosphate, is an essential effector for the SorR-mediated induction of the sor operons.
In K. pneumoniae, all sor genes are expressed from a single promoter, for which SorC would act as a repressor in its normal, uninduced state and as an inducible activator in the presence of l-sorbose (48). In L. casei, insertional inactivation of sorR led to no expression of sorF, indicating that sorR encodes a positive regulator. Its inactivation also abolished the expression of sorE; however, this might be due to a polar effect of the mutation, as sorR and sorE were cotranscribed. Alignment of the deduced amino acid sequence of SorR with sequences of the most similar proteins found in database revealed three highly conserved motifs, a putative DNA-binding helix-turn-helix at the amino terminus (motif I), and two other conserved areas (motifs II and III) for which no clear function could be deduced. However, homology of motif III with a region in proteins of the ROK family (39) suggested a sugar-binding function. The conclusions drawn from the alignments must be considered carefully because although SorC from K. pneumoniae was included in the alignment (Fig. 3), other authors reported that SorC showed homology to other sugar regulators (14) which are not shared by L. casei SorR or the other proteins shown in Fig. 3.
Many observations indicated that expression of the sor operons in L. casei was subject to CR. As mentioned earlier, adenylate cyclase, cyclic AMP, and a catabolite-activating protein-like protein are not related to CR in AT-rich gram-positive bacteria; instead, this effect depends on the phosphorylation of the HPr Ser46 residue and the association of HPr(Ser-P) with CcpA to promote its binding to the cre sequences. In this respect, a cre site has been found in the promoter region of the sor operons which matches the consensus sequence for gram-positive bacteria (17). It overlaps the putative −10 region of the first likely promoter, as well as the −35 region of the second promoter of the sorFABCDG cluster and the −10 region of the sorRE promoter (Fig. 2). Indirect evidence suggested that CcpA and elements of the d-mannose-specific PTS are directly or indirectly related to the CR of the sor operons. The diauxic growth exhibited by the wild type in l-sorbose plus d-glucose was abolished in the man mutant, which is consistent with the partial relief of glucose repression on Sor-PR and Stol-PDh in this strain. The activities of the sorbose catabolic enzymes were less sensitive to the presence of glucose in the man ccpA double mutant (showing only 1.6- to 2-fold repression). However, the ccpA mutant still displayed a remarkable CR of these activities. These data also indicate that yet another CR mechanism may operate in regulation of the sor operon, reminiscent of results of earlier studies on regulation of the L. casei lac operon (1, 12). In future work, the roles of SorR and other elements in this residual CR will be investigated. Additionally, when the L. casei strains were grown in the presence of l-sorbose, the level of Stol-PDh activity was significantly higher in the ccpA mutant than in the wild type. This may indicate that a metabolite derived from l-sorbose such as fructose-1,6-bisphosphate can stimulate CcpA, thus exerting some CR effect over the sorFABCDG promoter. A similar self-repressing effect has also been found for lactose in the lac operon of L. casei (12).
Processes such as inducer exclusion or inducer expulsion are very important and could indeed occur in this system. However, because the experiments in this work were designed to study the genetic induction or CR of the genes in the sor operons, sugar uptake and l-sorbose catabolic activities (Sor-PR and Stol-PDh) were always quantified in fully induced or repressed cells, for which inducer expulsion or exclusion effects could not be studied.
Among the genes characterized, a distal gene (sorG) possibly encoding a d-fructose-1,6-bisphosphate aldolase was found. After d-fructose-1,6-bisphosphate is formed by the activity of Sor-PR and Stol-PDh (18, 36) (Fig. 1), it seems likely that a phosphofructokinase yields d-fructose-1,6-bisphosphate, which then is hydrolyzed to dihydroxyacetone phosphate and glyceraldehyde phosphate by a d-fructose-1,6-bisphosphate aldolase. If SorG is confirmed to be an aldolase and since sorG is cotranscribed and coordinately regulated with sorFABCD genes, it would exclusively function during l-sorbose metabolism in L. casei. This would be expected in a highly specialized pathway when genes normally used for sugar catabolism are not fully induced. Previous studies showed that in the heterofermentative Lactobacillus brevis, the genes encoding a complete PTS, a d-fructose-1-phosphate kinase, and a d-fructose-1,6-bisphosphate aldolase are inducible by d-fructose (30). The presence of this pathway in L. casei may be related to its ecophysiology in the intestinal tract, as it would provide this bacterium an excellent machinery for scavenging vegetable cells and cell wall hydrolysates.
In this study, transport and metabolism of l-sorbose were analyzed for the first time in a gram-positive bacterium. The genetic organization of these operons is significantly different from those in K. pneumoniae and E. coli, where the gene encoding the transcriptional regulator (sorC) precedes the rest of the genes and all are expressed from a single promoter (46, 47). In L. casei, however, sorRE and sorFABCDG are divergently transcribed, and their expression is subject to SorR-mediated l-sorbose induction, with a strong CR, occurring primarily through the PTS/CcpA signal transduction pathway.
ACKNOWLEDGMENTS
This work was supported by the Biotech Programme of the European Community (contract BIO4-CT96-0380) and by funds from the Spanish Government (ALI95-0035). A. Veyrat was supported by a grant from the Consellería de Cultura, Educación y Ciencia de la Generalitat Valenciana, and M. Santos had an HCM Senior Scientist Training Contract of the E.U. (CHBG-CT93-0459).
REFERENCES
- 1.Alpert C-A, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the BglG family of transcriptional antiterminators. J Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Burland V, Plunkett III G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chassy B M, Thompson J. Regulation of lactose-phosphoenolpyruvate-dependent phosphotransferase system and β-d-phosphogalactosidase galactohydrolase activities in Lactobacillus casei. J Bacteriol. 1983;154:1195–1203. doi: 10.1128/jb.154.3.1195-1203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassy B M, Thompson J. Regulation and characterization of the galactose phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J Bacteriol. 1983;154:1204–1214. doi: 10.1128/jb.154.3.1204-1214.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J-D, Morisson D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher J, Saier M H., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1984;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolitic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 9.Deutscher J, Fischer C, Charrier V, Galinier A, Lindner C, Darbon E, Dossonnet V. Regulation of carbon metabolism in Gram-positive bacteria by protein phosphorylation. Folia Microbiol. 1997;42:171–178. doi: 10.1007/BF02818974. [DOI] [PubMed] [Google Scholar]
- 10.Erni B, Zanolari B, Kocher H P. The mannose-permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage λ DNA. J Biol Chem. 1987;262:5238–5247. [PubMed] [Google Scholar]
- 11.Görke B, Rak B. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 1999;18:3370–3379. doi: 10.1093/emboj/18.12.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosalbes M J, Monedero V, Pérez-Martínez G. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J Bacteriol. 1999;181:3928–3934. doi: 10.1128/jb.181.13.3928-3934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosalbes M J, Monedero V, Alpert C-A, Pérez-Martínez G. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol Lett. 1997;148:83–89. doi: 10.1111/j.1574-6968.1997.tb10271.x. [DOI] [PubMed] [Google Scholar]
- 14.Heuel H, Shakeri-Garakani A, Turgut S, Lengeler J W. Genes for d-arabitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology. 1998;144:1631–1639. doi: 10.1099/00221287-144-6-1631. [DOI] [PubMed] [Google Scholar]
- 15.Huang F G, Coppola G, Calhoun D H. Multiple transcripts encoded by the ilvGMEDA gene cluster of Escherichia coli K-12. J Bacteriol. 1992;174:4871–4877. doi: 10.1128/jb.174.15.4871-4877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 17.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 18.Kelker N E, Simkins R A, Anderson R L. Pathway of l-sorbose metabolism in Aerobacter aerogenes. J Biol Chem. 1972;247:1479–1483. [PubMed] [Google Scholar]
- 19.Leloup L, Ehrlich S D, Zagorec M, Morel-Deville F. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol. 1997;63:2117–2123. doi: 10.1128/aem.63.6.2117-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lux R, Jahreis K, Bettenbrock K, Parkinson J S, Lengeler J W. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. The levanase operon of Bacillus subtilis includes a fructose-specific PTS regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin-Verstraete I, Michel V, Charbit A. The levanase operon of Bacillus subtilis expressed in Escherichia coli can substitute for the mannose permease in mannose uptake and bacteriophage lambda infection. J Bacteriol. 1996;178:7112–7119. doi: 10.1128/jb.178.24.7112-7119.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monedero V, Gosalbes M J, Pérez-Martínez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postma M, Leer R J, van Luijk N, van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reizer J, Romano A H, Deutscher J. The role of phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, in the regulation of carbon metabolism in Gram-positive bacteria. J Cell Biochem. 1993;51:19–24. doi: 10.1002/jcb.240510105. [DOI] [PubMed] [Google Scholar]
- 28.Reizer J, Ramseier T M, Reizer A, Charbit A, Saier M H., Jr Novel phosphotransferase genes revealed by bacterial genome sequencing: a gene cluster encoding a putative N-acetylgalactosamine metabolic pathway in Escherichia coli. Microbiology. 1996;142:231–250. doi: 10.1099/13500872-142-2-231. [DOI] [PubMed] [Google Scholar]
- 29.Saier M H, Jr, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 30.Saier M H, Jr, Ye J-J, Klinke S, Nino E. Identification of an anaerobically induced phosphoenolpyruvate-dependent fructose-specific phosphotransferase system and evidence for the Embden-Meyerhof glycolytic pathway in the heterofermentative bacterium Lactobacillus brevis. J Bacteriol. 1996;178:314–316. doi: 10.1128/jb.178.1.314-316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J-J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Saxild H H, Andersen L N, Hammer K. dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J Bacteriol. 1996;178:424–434. doi: 10.1128/jb.178.2.424-434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlapfer B S, Zuber H. Cloning and sequencing of the genes encoding glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase and triosephosphate isomerase (gap operon) from mesophilic Bacillus megaterium: comparison with corresponding sequences from thermophilic Bacillus stearothermophilus. Gene. 1992;122:53–62. doi: 10.1016/0378-1119(92)90031-j. [DOI] [PubMed] [Google Scholar]
- 35.Shine J, Dalgarno L. The 3′ terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprenger G A, Lengeler J W. l-Sorbose metabolism in Klebsiella pneumoniae and Sor+ derivatives of Escherichia coli K-12 and chemotaxis toward sorbose. J Bacteriol. 1984;157:39–45. doi: 10.1128/jb.157.1.39-45.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 38.Sulavik M C, Clewel D B. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J Bacteriol. 1996;178:5826–5830. doi: 10.1128/jb.178.19.5826-5830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Titgemeyer F, Reizer J, Reizer A, Saier M H., Jr Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994;140:2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 40.Tobisch S, Stülke J, Hecker M. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J Bacteriol. 1999;181:4995–5003. doi: 10.1128/jb.181.16.4995-5003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tortosa P, Aymerich S, Lindner C, Saier M H, Jr, Reizer J, Le Coq D. Multiple phosphorylation of SacY, a Bacillus subtilis antiterminator negatively controlled by the phosphotransferase system. J Biol Chem. 1997;272:17230–17237. doi: 10.1074/jbc.272.27.17230. [DOI] [PubMed] [Google Scholar]
- 42.Trach K, Chapman J W, Piggot P, LeCoq D, Hoch J A. Complete sequence and transcriptional analysis of the spoOF region of the Bacillus subtilis chromosome. J Bacteriol. 1988;170:4194–4208. doi: 10.1128/jb.170.9.4194-4208.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veyrat A, Gosalbes M J, Pérez-Martínez G. Lactobacillus curvatus has a glucose transport system homologous to the mannose family of phosphoenolpyruvate-dependent phosphostransferase systems. Microbiology. 1996;142:3469–3477. doi: 10.1099/13500872-142-12-3469. [DOI] [PubMed] [Google Scholar]
- 44.Veyrat A, Monedero V, Pérez-Martínez G. Glucose transport by the phosphoenolpyruvate:mannose phosphotransferase system in Lactobacillus casei ATCC 393 and its role in carbon catabolite repression. Microbiology. 1994;140:1141–1149. doi: 10.1099/13500872-140-5-1141. [DOI] [PubMed] [Google Scholar]
- 45.Wehmeier U F, Lengeler J W. Sequence of the sor-operon for l-sorbose utilization from Klebsiella pneumoniae KAY2026. Biochim Biophys Acta. 1994;1208:348–351. doi: 10.1016/0167-4838(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 46.Wehmeier U F, Wöhrl B M, Lengeler J W. Molecular analysis of the phosphoenolpyruvate-dependent l-sorbose:phosphotransferase system from Klebsiella pneumoniae and of its multidomain structure. Mol Gen Genet. 1995;246:610–618. doi: 10.1007/BF00298968. [DOI] [PubMed] [Google Scholar]
- 47.Wehmeier U F, Nobelmann B, Lengeler J W. Cloning of the Escherichia coli sor genes for l-sorbose transport and metabolism and physical mapping of the genes near metH and iclR. J Bacteriol. 1992;174:7784–7790. doi: 10.1128/jb.174.23.7784-7790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wöhrl B M, Wehmeier U F, Lengeler J W. Positive and negative regulation of expression of the l-sorbose (sor) operon by SorC in Klebsiella pneumoniae. Mol Gen Genet. 1990;224:193–200. doi: 10.1007/BF00271552. [DOI] [PubMed] [Google Scholar]
- 49.Wöhrl B M, Lengeler J W. Cloning and physical mapping of the sor genes for l-sorbose transport and metabolism from Klebsiella pneumoniae. Mol Microbiol. 1990;4:1557–1565. doi: 10.1111/j.1365-2958.1990.tb02067.x. [DOI] [PubMed] [Google Scholar]