Key Points
Question
Can a machine learning model predict treatment success of the initial antiseizure medication?
Findings
With the use of routinely collected clinical information, this cohort study developed a deep learning model on a pooled cohort of 1798 adults with newly diagnosed epilepsy seen in 5 centers in 4 countries. The model showed potential in predicting treatment success on the first prescribed antiseizure medication.
Meaning
This study’s findings demonstrate the potential feasibility of personalized prediction of treatment response in patients with newly diagnosed epilepsy.
Abstract
Importance
Selection of antiseizure medications (ASMs) for epilepsy remains largely a trial-and-error approach. Under this approach, many patients have to endure sequential trials of ineffective treatments until the “right drugs” are prescribed.
Objective
To develop and validate a deep learning model using readily available clinical information to predict treatment success with the first ASM for individual patients.
Design, Setting, and Participants
This cohort study developed and validated a prognostic model. Patients were treated between 1982 and 2020. All patients were followed up for a minimum of 1 year or until failure of the first ASM. A total of 2404 adults with epilepsy newly treated at specialist clinics in Scotland, Malaysia, Australia, and China between 1982 and 2020 were considered for inclusion, of whom 606 (25.2%) were excluded from the final cohort because of missing information in 1 or more variables.
Exposures
One of 7 antiseizure medications.
Main Outcomes and Measures
With the use of the transformer model architecture on 16 clinical factors and ASM information, this cohort study first pooled all cohorts for model training and testing. The model was trained again using the largest cohort and externally validated on the other 4 cohorts. The area under the receiver operating characteristic curve (AUROC), weighted balanced accuracy, sensitivity, and specificity of the model were all assessed for predicting treatment success based on the optimal probability cutoff. Treatment success was defined as complete seizure freedom for the first year of treatment while taking the first ASM. Performance of the transformer model was compared with other machine learning models.
Results
The final pooled cohort included 1798 adults (54.5% female; median age, 34 years [IQR, 24-50 years]). The transformer model that was trained using the pooled cohort had an AUROC of 0.65 (95% CI, 0.63-0.67) and a weighted balanced accuracy of 0.62 (95% CI, 0.60-0.64) on the test set. The model that was trained using the largest cohort only had AUROCs ranging from 0.52 to 0.60 and a weighted balanced accuracy ranging from 0.51 to 0.62 in the external validation cohorts. Number of pretreatment seizures, presence of psychiatric disorders, electroencephalography, and brain imaging findings were the most important clinical variables for predicted outcomes in both models. The transformer model that was developed using the pooled cohort outperformed 2 of the 5 other models tested in terms of AUROC.
Conclusions and Relevance
In this cohort study, a deep learning model showed the feasibility of personalized prediction of response to ASMs based on clinical information. With improvement of performance, such as by incorporating genetic and imaging data, this model may potentially assist clinicians in selecting the right drug at the first trial.
This cohort study tests a deep learning model using readily available clinical information to predict treatment success with the first antiseizure medication for individual patients.
Introduction
Epilepsy affects 50 million people worldwide.1 The treatment goal for newly diagnosed epilepsy is seizure freedom, usually defined as no seizures for 12 months or more,2 achieved as soon as possible with minimal or no treatment-related adverse effects. This goal is achieved using antiseizure medications (ASMs), which suppress the occurrence of seizures without modifying the underlying pathology.3 Choosing the first ASM is of paramount importance because an inadequate response to the first prescribed drug is a strong predictor of poor subsequent long-term outcome.4,5,6 A proportion of patients whose first ASM treatment fails will respond to subsequent treatments,6 which implies that they might have become seizure free sooner if they were given the “right drug” at the outset.
Presently, for a patient with newly diagnosed epilepsy, an ASM is selected mainly based on broad seizure types (focal vs generalized onset) classified according to clinical history and investigation findings. However, for each type of seizure, many drugs have a similar effectiveness when analyzed on a group basis in head-to-head randomized clinical trials.7,8,9,10 It is not possible to predict which particular drug will be most effective for a given patient, and typically various drugs are sequentially trialed (prioritized on availability, tolerability, and safety considerations) if seizures persist.11 Under this trial-and-error approach, patients may have to endure multiple trials of ineffective treatments before the right drug is found.6 The drug selection algorithms recommended to date are based on expert opinion with untested clinical utility.12,13
A more reliable way to predict response to different ASMs is needed so that the most effective drug can be selected for an individual patient at the time of treatment initiation. We hypothesized that this objective might be achieved by finding patterns linking treatment outcomes to patients’ health data using machine learning techniques.14 Machine learning is a subset of artificial intelligence that is able to improve automatically through experience. A previous attempt to develop an algorithm for appropriate ASM selection for first monotherapy applied traditional machine learning techniques, such as linear regression and a support vector machine using a drug-dispensing data set.15 However, diagnostic coding may not be reliable in these data sets, which do not capture detailed information on treatment response or the individual or disease characteristics that are potentially important predictors of seizure control.
In this cohort study, we applied deep learning, a type of machine learning, to train and test a model to predict response to the first prescribed ASM monotherapy using longitudinal clinical data sets of patients with newly diagnosed epilepsy, focusing on the first year of commencing treatment. The model incorporated a broad range of demographic and epilepsy-related factors. The performance and robustness of the model were validated both internally and externally. We also compared our model against other machine learning models to assess whether the performance of current machine learning algorithms are near ready for recommending ASMs.
Methods
Study Settings and Cohorts
We included 5 independent cohorts of patients with newly diagnosed and treated epilepsy for model development and validation. In each cohort, only patients who were aged 18 years or older at the time of treatment initiation and were followed up for at least 1 year or until the failure of the first ASM treatment and had no missing data were included. The largest cohort was of patients seen at the Epilepsy Unit of the Western Infirmary in Glasgow, Scotland. The setting of this cohort has been previously described.4,6,16,17,18 In this analysis, we included patients seen between July 1, 1982, and October 31, 2012, who were followed up until April 30, 2016, or death. The other cohorts included patients treated at the University of Malaya Medical Centre in Kuala Lumpur, Malaysia (from January 1, 2002, to December 31, 2020, and followed up until October 1, 2021)19; the WA Adult Epilepsy Service in Perth, Australia (from May 1, 1999, to May 31, 2016, and followed up until April 1, 2018)18,20; and the First Affiliated Hospital of Chongqing Medical University in Chongqing (from September 1, 2003, to July 31, 2019, and followed up until December 14, 2021) and The First Affiliated Hospital, Sun Yat-Sen University in Guangzhou (from June 1, 2004, to June 30, 2020, and followed up until July 25, 2021), both in China.
Because the Glasgow cohort was the largest cohort in this study, we included only those ASMs prescribed as initial monotherapy for at least 10 patients in the Glasgow registry to train the model. Patients using gabapentin as their first monotherapy were excluded (n = 24) because this ASM was not used in other cohorts for initial monotherapy. Patients were included from the other cohorts if they were treated with the same ASMs as the first monotherapy.
The study was approved by the research ethics committees of the participating hospitals. The requirement of informed consent was waived by the research ethics committees of the participating hospitals because the data were deidentified prior to analysis. The study was also registered at the Monash University Human Research Ethics Committee.
Treatment Approach and Follow-up
Monotherapy was the preferred mode of therapy in all cohorts. After commencement of treatment, patients were instructed to keep a record of any breakthrough seizures for review during follow-up visits. Details of treatment approach and follow-up protocols have been described previously for the Glasgow cohort,4,6,21 the Kuala Lumpur cohort,19 and the Perth cohort.22,23
Definitions
Epilepsy was diagnosed and classified according to the guidelines of the International League Against Epilepsy.24,25 Epilepsy was diagnosed after the occurrence of 2 or more unprovoked seizures more than 24 hours apart or a single unprovoked seizure with a high likelihood of seizure recurrence based on neuroimaging findings, electroencephalographic abnormalities, or remote symptomatic etiology.24 Epilepsy type was classified as generalized, focal, combined generalized and focal, or unclassified.25 For each patient, seizure control was assessed at 1 year after commencement of the first ASM regimen. Treatment was deemed successful if the patient was seizure free while still taking the first ASM during the entire first year of treatment. This definition is akin to per-protocol analysis in randomized clinical trials.7,8,9 The proportion of patients who had successful treatment at 1 year was calculated by dividing the number of patients who had successful treatment by the total number of patients who commenced treatment. Treatment was deemed unsuccessful if the patient had recurrent seizures of any type, if the first ASM was replaced with a different drug or another ASM was added owing to persistent seizures, or if the first ASM was withdrawn owing to adverse effects.
Data Collection
Similar information was collected and aligned from both development and validation cohorts. Information on demographic characteristics, medical history, family history, epilepsy risk factors, number of pretreatment seizures, and investigation results were collected at baseline. The presence or absence of neuroimaging lesions considered to be epileptogenic26 and epileptiform abnormalities detected on electroencephalograms (EEGs)27 were documented. Data on ASM regimens and treatment response were collected during subsequent follow-up visits. Patients with persistent poor treatment adherence unrelated to the effectiveness or tolerability of the drug were excluded from inclusion in the study cohorts.6 Information in all cohorts was collected in a manner that maintained the time series nature of the longitudinal registries (ie, treatment response was traceable for each ASM taken with the start and end dates).
Model Architecture
We developed an attention-based deep learning model called the transformer model28 to predict the probability of treatment success with the first prescribed ASM. This model comprises an encoder and decoder, both of which have a multihead attention mechanism. The multihead attention mechanism in the transformer model splits the patient information into separate parts, allowing the transformer model to use the information of specific variables for different drug responses. A further description of the model and an overview of a single encoder-decoder pair of the transformer network and the attention map can be found in the eAppendix and eFigure 1 and eFigure 2 in the Supplement.
Input Variables
Input variables for the model were clinical factors known or hypothesized to predict seizure freedom. Table 1 shows the final list of input variables and how they were categorized. The variables were binarized for each category. Age at treatment initiation was trichotomized based on tertiles of the complete Glasgow cohort (n = 1504) to reduce the complexity for model training. Similar to previous reports,29,30 number of pretreatment seizures was dichotomized as 5 or fewer or more than 5. For patients who had undergone both magnetic resonance imaging and computed tomography brain scans, only the magnetic resonance imaging findings were used for model training. Patients who had both epileptiform and nonspecific abnormalities detected on EEGs and brain imaging, respectively, were considered as having only epileptiform abnormalities. The drug prescribed for each patient was also included as a variable, but the drug dosage was not included.
Table 1. Input Variables for the Machine Learning Models.
| Input variable | Categorization |
|---|---|
| Sex | Male or female |
| Age at treatment initiation | Age groups (tertiles), ya |
| History | |
| Febrile convulsions | Yes or no |
| Central nervous system infection in childhood | Yes or no |
| Significant head trauma | Yes or no |
| Cerebral hypoxic injury | Yes or no |
| Substance abuse | Yes or no |
| Alcohol abuse | Yes or no |
| Epilepsy in first-degree relatives | Yes or no |
| Presence of | |
| Cerebrovascular disease | Yes or no |
| Intellectual disability | Yes or no |
| Psychiatric disorder | Yes or no |
| No. of pretreatment seizures | ≤5 or >5 |
| Type of epilepsy | Focal, generalized, or unclassified |
| Electroencephalography findings | Normal, abnormal epileptiform, or abnormal nonepileptiform |
| Brain imaging findingsb | Normal, abnormal epileptogenic, or abnormal nonepileptogenic |
| Drug used | Carbamazepine, lamotrigine, levetiracetam, oxcarbazepine, phenytoin, topiramate, or valproate |
Tertiles are 18 to 29 years, older than 29 to 46 years, and older than 46 years.
Computed tomography or magnetic resonance imaging.
Model Performance Metrics
The ability of the model to predict treatment success with the prescribed ASM was evaluated with a series of metrics, including sensitivity, specificity, area under the receiver operating characteristic curve (AUROC), and accuracy.31 Sensitivity (the true-positive rate) and specificity (the true-negative rate) reflected the ability of the model to make true-positive and true-negative predictions, respectively. The false-positive rate (1 − specificity) represented the rate of misclassification of positive outcomes (ie, treatment success). To identify the optimal probability cutoff to classify successful treatment outcomes, thresholds at intervals of 0.01 were used to calculate these metrics. The receiver operating characteristic (ROC) curve was then constructed by plotting the true-positive rate (y-axis) against the false-positive rate (x-axis) to derive the AUROC, which measured the model’s ability to distinguish the dichotomized treatment outcomes (ie, treatment success or not), whereby an AUROC of greater than 0.5 indicated better performance than a random classifier. Weighted balanced accuracy was used to account for the class imbalance.32
Model Development and Validation
We conducted 2 sets of experiments. In the first experiment, all 5 cohorts were pooled together to form a comprehensive cohort distribution for modeling. We randomly selected 80% of the patients in the pooled cohort as the training set and the remaining 20% as the test set. The trained model was evaluated on the test set. Subgroup analysis was performed for focal and generalized epilepsy types.
In the second experiment, the model was trained using the entire Glasgow data set as the development set, and performance was externally validated, in turn, on each of the other 4 cohorts without further fine-tuning. The same input variables as those used for model development were extracted for the patients in the validation cohorts.
The performance of the transformer model was compared with the performance of the multilayered perceptron model,33 which is another deep learning model commonly studied for classification problems34,35 and treatment response prediction36,37,38 in diverse fields. Further, we compared performance with traditional machine learning models, such as the extreme gradient boosting,39 support vector machine,40 random forest,41 and logistic regression models.42 We also recorded the computational times for the various machine learning models.
Cohort Comparability and Feature Importance
Cohort comparability was visually assessed using t-distributed stochastic neighbor embedding analysis43 for projection of clusters of similar patients as viewed by last encoder layer of the transformer network. To ascertain which input variables play an important role in model prediction, we performed deep analysis using Shapley additive explanations (ie, SHAP values).44
Statistical Analysis
All input variables were categorical and summarized using frequency and percentage; a 5-fold cross-validation was performed to calculate mean (SD) values. Point estimates of performance with 95% CIs were reported for each metric. The mean (SD) SHAP value of each input variable is provided. We examined the pattern of missing data in each cohort and stratified by the seizure outcome and performed the Little χ2 test for missing completely at random (MCAR). Statistical significance was set at 2-sided P < .05. All model building was performed by using Python, version 3.8, and the Little χ2 test for MCAR was performed by using the Stata, version 16 (StataCorp LLC) user-written program “mcartest.”45
The study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline. The software implementing the transformer model can be downloaded from Github.46
Results
Patients
The 5 registries included a total of 2404 eligible adult patients, of whom 606 (25.2%) were excluded from the final cohort because of missing information in 1 or more variables (eTable 1 in the Supplement). A total of 1798 patients were included in the pooled cohort (54.5% female; median age, 34 years [IQR, 24-50 years]). This pooled cohort comprised 1065 patients from Glasgow, 242 from Kuala Lumpur, 191 from Chongqing, 189 from Perth, and 111 from Guangzhou. The clinical characteristics of the patients and the data on the first ASM monotherapy used in each of the 5 cohorts are shown in Table 2.26 The sex distribution was similar across the cohorts. The 2 Chinese cohorts included younger patients (101 of 191 patients [52.9%] in the Chongqing cohort and 68 of 111 patients [61.3%] in the Guangzhou chort who were aged 18 to ≤29 years) compared with the Glasgow cohort (378 of 1065 [35.5%] aged 18 to ≤29 years), the Kuala Lumpur cohort (95 of 242 [39.3%] aged 18 to ≤29 years), and the Perth cohort (61 of 189 [32.3%] aged 18 to ≤29 years). The rate of substance abuse was higher in the Glasgow and Perth cohorts compared with the other cohorts (101 of 1065 patients [10.3%] in the Glasgow cohort, 1 of 242 patients [0.4%] in the Kuala Lumpur cohort, 0 patients in the Chongqing cohort, 25 of 189 patients [13.2%] in the Perth cohort, and 0 patients in the Guangzhou cohort); the rate of alcohol abuse was higher in the Glasgow cohort compared with the other cohorts (220 of 1065 patients [20.7%] in the Glasgow cohort, 3 of 242 patients [1.2%] in the Kuala Lumpur cohort, 2 of 191 patients [1.0%] in the Chongqing cohort, 9 of 189 patients [4.8%] in the Perth cohort, and 1 of 111 patients [0.9%] in the Guangzhou cohort); and the rate of the history of cerebrovascular disease was higher in the Glasgow and Perth cohorts compared with the other cohorts (115 of 1065 patients [10.8%] in the Glasgow cohort, 18 of 242 patients [7.4%] in the Kuala Lumpur cohort, 6 of 191 patients [3.1%] in the Chongqing cohort, 26 of 189 patients [13.8%] in the Perth cohort, and 4 of 111 patients [3.7%] in the Guangzhou cohort). Epileptiform abnormalities detected on EEGs were twice as frequent in the Kuala Lumpur and Guangzhou cohorts compared with the other cohorts (328 of 1065 patients [30.8%] in the Glasgow cohort, 147 of 242 patients [60.7%] in the Kuala Lumpur cohort, 45 of 191 patients [23.6%] in the Chongqing cohort, 41 of 189 patients [21.7%] in the Perth cohort, and 73 of 111 patients [65.8%] in the Guangzhou cohort). Epileptogenic abnormalities detected on neuroimaging were relatively uncommon in the Glasgow cohort compared with the other cohorts (123 of 1065 patients [11.5%] in the Glasgow cohort, 97 of 242 patients [40.1%] in the Kuala Lumpur cohort, 68 of 191 patients [35.7%] in the Chongqing cohort, 78 of 189 patients [41.2%] in the Perth cohort, and 39 of 111 patients [35.1%] in the Guangzhou cohort), likely reflecting its recruitment in an era prior to modern neuroimaging (from 1982). Focal epilepsy was the predominant type of epilepsy in all of the cohorts except the Chongqing cohort (898 of 1065 patients [84.3%] in the Glasgow cohort, 201 of 242 patients [83.1%] in the Kuala Lumpur cohort, 76 of 191 patients [39.8%] in the Chongqing cohort, 114 of 189 patients [60.3%] in the Perth cohort, and 103 of 111 patients [92.8%] in the Guangzhou cohort). Each patient was treated with 1 of 7 commonly used ASMs as the first monotherapy (carbamazepine, lamotrigine, levetiracetam, oxcarbazepine, phenytoin, topiramate, or valproate). Valproate was either the most common (3 cohorts) or among the 3 most commonly used drugs. Lamotrigine was the most frequently used drug in the Glasgow cohort but less so in the other cohorts (320 of 1065 patients [30.0%] in the Glasgow cohort, 16 of 242 patients [6.6%] in the Kuala Lumpur cohort, 18 of 191 patients [9.4%] in the Chongqing cohort, 17 of 189 patients [9.0%] in the Perth cohort, and 15 of 111 patients [13.5%] in the Guangzhou cohort). Carbamazepine and phenytoin were rarely used in the the Chinese cohorts (3 of 191 patients [1.6%] in the Chongqing cohort and 5 of 111 patients [4.5%] in the Guangzhou cohort). Seizure outcomes and the reasons for drug changes within the first 12 months of commencing treatment in each cohort are provided in eTable 2 in the Supplement.
Table 2. Clinical Characteristics of Patients and Antiseizure Medications Used in Each Cohort (N = 1798).
| Characteristic | Cohort, No. (%) of patients | ||||
|---|---|---|---|---|---|
| Glasgow | Kuala Lumpur | Chongqing | Perth | Guangzhou | |
| Total No. | 1065 | 242 | 191 | 189 | 111 |
| Sex | |||||
| Male | 498 (46.8) | 113 (46.7) | 91 (47.6) | 70 (37.0) | 46 (41.4) |
| Female | 567 (53.2) | 129 (53.3) | 100 (52.4) | 119 (63.0) | 65 (58.6) |
| Age groups at treatment initiation, y | |||||
| 18 to ≤29 | 378 (35.5) | 95 (39.3) | 101 (52.9) | 61 (32.3) | 68 (61.3) |
| >29 to ≤46 | 364 (34.2) | 65 (26.9) | 53 (27.8) | 61 (32.3) | 26 (23.4) |
| >46 | 323 (30.3) | 82 (33.9) | 37 (19.4) | 67 (35.4) | 17 (15.3) |
| History | |||||
| Febrile convulsions | |||||
| Yes | 13 (1.2) | 10 (4.1) | 18 (9.4) | 6 (3.2) | 7 (6.3) |
| No | 1052 (98.8) | 232 (95.9) | 173 (90.6) | 183 (96.8) | 104 (93.7) |
| Central nervous system infection in childhood | |||||
| Yes | 11 (1.0) | 16 (6.6) | 10 (5.2) | 3 (1.6) | 4 (3.6) |
| No | 1052 (98.8) | 226 (93.4) | 181 (94.8) | 186 (98.4) | 107 (96.4) |
| Significant head trauma | |||||
| Yes | 11 (1.0) | 0 | 3 (1.6) | 4 (2.1) | 1 (0.9) |
| No | 1054 (99.0) | 242 (100) | 188 (98.4) | 185 (97.9) | 110 (99.1) |
| Cerebral hypoxic injury | |||||
| Yes | 157 (14.7) | 26 (10.7) | 21 (11.0) | 23 (12.2) | 7 (6.3) |
| No | 908 (85.3) | 216 (89.3) | 170 (89.0) | 166 (87.8) | 104 (93.7) |
| Substance abuse | |||||
| Yes | 110 (10.3) | 1 (0.4) | 0 | 25 (13.2) | 0 |
| No | 955 (89.7) | 241 (99.6) | 191 (100) | 164 (86.8) | 111 (100) |
| Alcohol abuse | |||||
| Yes | 220 (20.7) | 3 (1.2) | 2 (1.0) | 9 (4.8) | 1 (0.9) |
| No | 845 (79.3) | 239 (98.8) | 189 (99.0) | 180 (95.2) | 110 (99.1) |
| Epilepsy in first-degree relative | |||||
| Yes | 166 (15.6) | 40 (16.5) | 19 (10.0) | 25 (13.2) | 7 (6.3) |
| No | 899 (84.4) | 202 (83.5) | 172 (90.0) | 164 (86.8) | 104 (93.7) |
| Cerebrovascular disease | |||||
| Yes | 115 (10.8) | 18 (7.4) | 6 (3.1) | 26 (13.8) | 4 (3.6) |
| No | 950 (89.2) | 224 (92.6) | 185 (96.9) | 163 (86.2) | 107 (96.4) |
| Intellectual disability | |||||
| Yes | 29 (2.7) | 9 (3.7) | 2 (1.0) | 11 (5.8) | 3 (2.8) |
| No | 1036 (97.3) | 233 (96.3) | 189 (99.0) | 178 (94.2) | 108 (97.2) |
| Psychiatric disorder | |||||
| Yes | 298 (28.0) | 25 (10.3) | 1 (0.5) | 51 (27.0) | 3 (2.7) |
| No | 767 (72.0) | 217 (89.7) | 190 (99.5) | 138 (73.0) | 108 (97.3) |
| No. of pretreatment seizures | |||||
| >5 | 515 (48.4) | 27 (11.2) | 49 (25.7) | 18 (9.5) | 38 (34.2) |
| ≤5 | 550 (51.6) | 215 (88.8) | 142 (74.3) | 171 (90.5) | 73 (65.8) |
| Type of epilepsy | |||||
| Focal | 898 (84.3) | 201 (83.1) | 76 (39.8) | 114 (60.3) | 103 (92.8) |
| Generalized or unclassified | 167 (15.7) | 41 (16.9) | 115 (60.2) | 75 (39.7) | 8 (7.2) |
| Electroencephalography findings | |||||
| Epileptiform abnormality | 328 (30.8) | 147 (60.7) | 45 (23.6) | 41 (21.7) | 73 (65.8) |
| Nonepileptiform abnormality | 314 (29.5) | 34 (14.0) | 24 (12.6) | 68 (36.0) | 23 (20.7) |
| Normal | 423 (39.7) | 61 (25.2) | 122 (63.9) | 80 (42.3) | 15 (13.5) |
| Brain imaging findingsa | |||||
| Epileptogenic abnormality | 123 (11.5) | 97 (40.1) | 68 (35.7) | 78 (41.2) | 39 (35.1) |
| Nonepileptogenic abnormalityb | 317 (29.8) | 80 (33.1) | 16 (8.3) | 15 (8.0) | 26 (23.4) |
| Normal | 625 (58.7) | 65 (26.9) | 107 (56.0) | 96 (50.8) | 46 (41.4) |
| Name of the first prescribed antiseizure medication | |||||
| Lamotrigine | 320 (30.0) | 16 (6.6) | 18 (9.4) | 17 (9.0) | 15 (13.5) |
| Valproate | 267 (25.1) | 102 (42.2) | 68 (35.6) | 88 (46.6) | 27 (24.3) |
| Carbamazepine | 227 (21.3) | 41 (16.9) | 3 (1.6) | 34 (18.0) | 5 (4.5) |
| Levetiracetam | 145 (13.6) | 49 (20.2) | 56 (29.3) | 16 (8.5) | 30 (27.0) |
| Oxcarbazepine | 51 (4.8) | 1 (0.4) | 34 (17.8) | 0 | 30 (27.0) |
| Topiramate | 44 (4.1) | 4 (1.7) | 12 (6.3) | 4 (2.1) | 3 (2.7) |
| Phenytoin | 11 (1.0) | 29 (12.0) | 0 | 30 (15.9) | 1 (0.9) |
Computed tomography or magnetic resonance imaging.
Nonepileptogenic abnormalities include small vessel ischemic changes, unspecified lesion, white matter hyperintensity, other cystic lesions, nonspecific T2 signal, cerebral atrophy, hippocampal structures asymmetry, developmental venous anomaly, Chiari I malformation, demyelination, calcification, lipoma, cerebral aneurysm, brachycephaly, cerebellar atrophy, congenital asymmetry of ventricles, cortical thickening, thinning of frontal lobes, hydrocephalus, hypoplastic right vertebral artery, pontine myelinolysis, cerebrospinal fluid space prominence, basal ganglia perivascular prominence, inferior tonsillar herniation, ventriculomegaly, cerebellar tonsillar herniation, and tortuous basilar artery.26
Experiment 1: Pooled Cohort for Model Training and Testing
eTables 3 and 4 in the Supplement show the tuned hyperparameters and computational times, respectively, for the machine learning models. In the first experiment, the transformer model was trained on the development set (80%) of the pooled cohort combining longitudinal patient data from the 5 cohorts (n = 1438), and internal validation was performed on the test set (20%) of this pooled cohort (n = 360). The AUROC of the transformer model on the training set was 0.72 (95% CI, 0.70-0.74), with a weighted balanced accuracy of 0.65 (95% CI, 0.63-0.67) (Figure, A). The AUROC shows that a probability threshold of 0.50 is the optimal cutoff for classifying treatment success (Figure, B). This threshold also resulted in the highest weighted balanced accuracy. The AUROC of the transformer model in the test set was 0.65 (Figure, C). With the use of this threshold, the model had a sensitivity of 0.69 and a specificity of 0.55 in predicting success of the first ASM monotherapy in the test set (Table 3).
Figure. Receiver Operating Characteristic (ROC) Curves and Weighted Balanced Accuracy Curve for the Transformer Model Developed Using a Pooled Cohort.
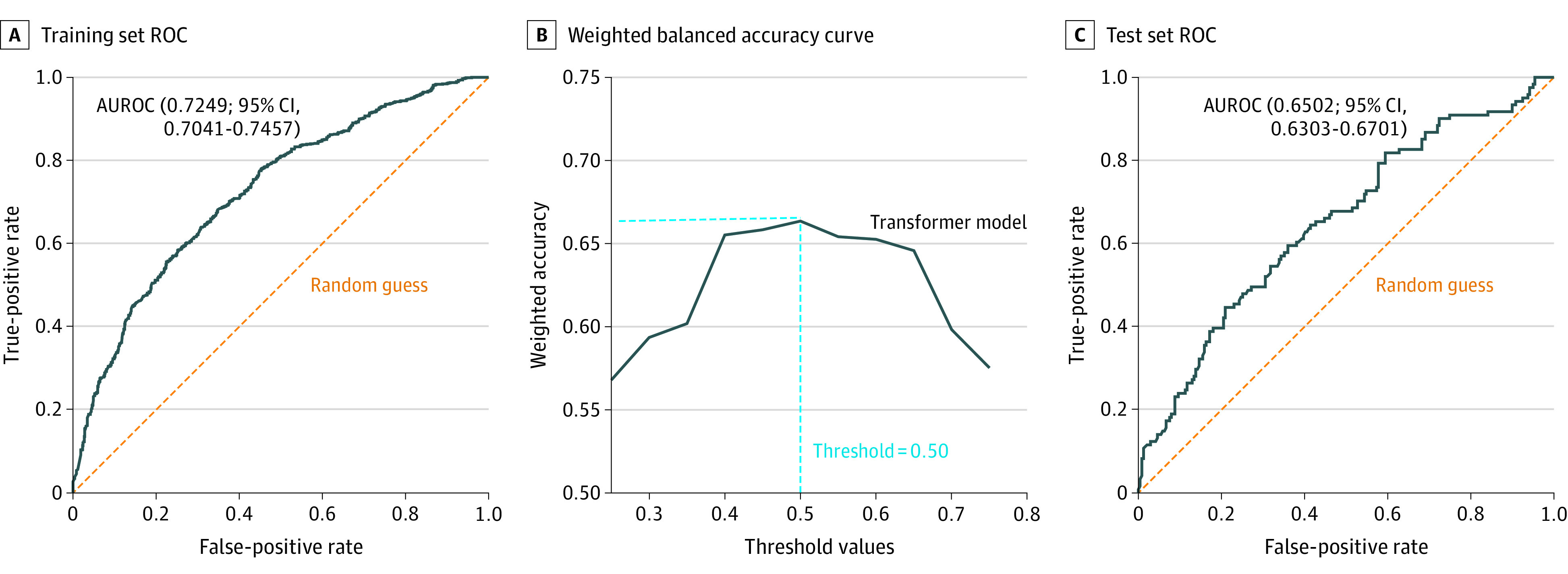
A, ROC curve on the training set. B, Weighted balanced accuracy curve at different threshold values of probability. The highest weighted balanced accuracy was obtained at a threshold of 0.5. The optimal threshold value is indicated by the intersection of dashed blue lines. C, ROC curve on the test set. AUROC indicates area under the receiver operating characteristic curve.
Table 3. Comparison of Model Performance on the Test Set of the Pooled Cohort.
| Model parameter | Transformer | Multilayered perceptron | Logistic regression | Support vector machine | XGBoost | Random forest |
|---|---|---|---|---|---|---|
| Mean AUROC (95% CI) | 0.65 (0.63-0.67) | 0.63 (0.60-0.66) | 0.61 (0.58-0.64) | 0.61 (0.59-0.63) | 0.60 (0.58-0.62) | 0.58 (0.56-0.60) |
| Weighted balanced accuracy (95% CI) | 0.62 (0.60-0.64) | 0.59 (0.57-0.61) | 0.60 (0.58-0.62) | 0.57 (0.55-0.59) | 0.59 (0.57-0.61) | 0.59 (0.57-0.61) |
| Sensitivity (95% CI) | 0.69 (0.66-0.72) | 0.59 (0.55-0.63) | 0.54 (0.52-0.56) | 0.65 (0.62-0.68) | 0.54 (0.52-0.56) | 0.47 (0.44-0.50) |
| Specificity (95% CI) | 0.55 (0.52-0.58) | 0.60 (0.57-0.63) | 0.63 (0.60-0.66) | 0.52 (0.49-0.55) | 0.61 (0.58-0.64) | 0.62 (0.59-0.65) |
Abbreviation: AUROC, area under the receiver operating characteristic curve.
The transformer model outperformed the extreme gradient boosting (AUROC, 0.60 [95% CI, 0.58-0.62]) and random forest (AUROC, 0.58 [95% CI, 0.56-0.60]) models with nonoverlapping 95% CIs. Although the point estimate of the mean AUROC of the transformer model was higher than the other 3 models (support vector machine, logistic regression, and multilayered perceptron), the 95% CIs overlapped. Similarly, the transformer model demonstrated the highest weighted balanced accuracy, but the lower limit of its 95% CI overlapped with other models except for the support vector machine model (Table 3).
Performance of the transformer model for subgroups of patients with focal epilepsy and patients with generalized or unclassified epilepsy revealed an AUROC of 0.64 (95% CI, 0.62-0.66) and 0.58 (95% CI, 0.56-0.60), respectively, and a weighted balanced accuracy of 0.62 (95% CI, 0.60-0.64) and 0.60 (95% CI, 0.58-0.62), respectively. eTable 5 in the Supplement shows the comparison of model performance in these subgroups.
Experiment 2: Cross-Cohort Validation
In the second experiment, the model was developed using the entire Glasgow cohort and externally validated on each of the other 4 cohorts separately. The AUROC of our transformer model on the Glasgow cohort (n = 1065) was 0.73 (95% CI, 0.71-0.75) (eFigure 3A in the Supplement). The AUROC shows that a probability threshold of 0.50 is the optimal cutoff for classifying treatment success (eFigure 3B in the Supplement). The AUROCs of the transformer model on the 4 external validation sets ranged from 0.52 to 0.60 (eFigure 3C-F in the Supplement), and weighted balanced accuracy ranged from 0.51 to 0.62. Using 0.50 as a probability threshold, the sensitivity of the transformer ranged from 0.41 to 0.61, and specificity ranged from 0.55 to 0.66 in predicting success of the first ASM monotherapy in the external validation cohorts (Table 4). As shown in Table 4, the transformer model generally demonstrated similar or better point estimates of the mean AUROC and weighted balanced accuracy than the other machine learning models in the external validation cohorts, although their 95% CIs overlapped.
Table 4. Model Performance After Training Exclusively on the Glasgow Cohort (N = 1065).
| Model parameter | Type of modela | |||||
|---|---|---|---|---|---|---|
| Transformer | Multilayered perceptron | Logistic regression | Support vector machine | XGBoost | Random forest | |
| Kuala Lumpur cohort (n = 242) | ||||||
| Mean AUROC | 0.58 (0.57-0.59) | 0.55 (0.53-0.57) | 0.57 (0.55-0.59) | 0.57 (0.55-0.59) | 0.57 (0.55-0.59) | 0.46 (0.44-0.48) |
| Weighted balanced accuracy | 0.58 (0.56-0.60) | 0.52 (0.50-0.54) | 0.56 (0.54-0.58) | 0.54 (0.52-0.56) | 0.55 (0.53-0.57) | 0.49 (0.47-0.51) |
| Sensitivity | 0.46 (0.44-0.48) | 0.55 (0.51-0.59) | 0.56 (0.52-0.60) | 0.59 (0.55-0.63) | 0.50 (0.47-0.53) | 0.38 (0.35-0.41) |
| Specificity | 0.65 (0.61-0.69) | 0.50 (0.46-0.54) | 0.56 (0.53-0.59) | 0.50 (0.47-0.53) | 0.59 (0.56-0.62) | 0.55 (0.53-0.57) |
| Chongqing cohort (n = 191) | ||||||
| Mean AUROC | 0.60 (0.58-0.62) | 0.57 (0.55-0.59) | 0.58 (0.55-0.61) | 0.59 (0.57-0.61) | 0.57 (0.55-0.59) | 0.56 (0.54-0.58) |
| Weighted balanced accuracy | 0.62 (0.60-0.64) | 0.56 (0.54-0.58) | 0.55 (0.53-0.57) | 0.56 (0.55-0.57) | 0.57 (0.55-0.59) | 0.53 (0.51-0.55) |
| Sensitivity | 0.61 (0.58-0.64) | 0.63 (0.59-0.67) | 0.65 (0.61-0.69) | 0.65 (0.61-0.69) | 0.65 (0.61-0.69) | 0.52 (0.50-0.54) |
| Specificity | 0.62 (0.59-0.65) | 0.49 (0.46-0.52) | 0.48 (0.45-0.51) | 0.49 (0.47-0.51) | 0.51 (0.48-0.54) | 0.57 (0.54-0.60) |
| Perth cohort (n = 189) | ||||||
| Mean AUROC | 0.53 (0.51-0.55) | 0.51 (0.49-0.53) | 0.52 (0.51-0.53) | 0.53 (0.51-0.55) | 0.54 (0.52-0.56) | 0.53 (0.51-0.55) |
| Weighted balanced accuracy | 0.57 (0.55-0.59) | 0.51 (0.49-0.53) | 0.55 (0.53-0.57) | 0.54 (0.52-0.56) | 0.53 (0.50-0.56) | 0.56 (0.54-0.58) |
| Sensitivity | 0.41 (0.39-0.43) | 0.50 (0.47-0.53) | 0.50 (0.47-0.53) | 0.50 (0.47-0.53) | 0.59 (0.56-0.62) | 0.42 (0.39-0.45) |
| Specificity | 0.66 (0.63-0.69) | 0.52 (0.50-0.54) | 0.57 (0.54-0.60) | 0.56 (0.53-0.59) | 0.46 (0.42-0.50) | 0.64 (0.61-0.67) |
| Guangzhou cohort (n = 111) | ||||||
| Mean AUROC | 0.52 (0.50-0.54) | 0.49 (0.47-0.51) | 0.51 (0.49-0.53) | 0.49 (0.47-0.51) | 0.51 (0.49-0.53) | 0.45 (0.43-0.47) |
| Weighted balanced accuracy | 0.51 (0.49-0.53) | 0.48 (0.46-0.50) | 0.52 (0.50-0.54) | 0.49 (0.47-0.51) | 0.49 (0.47-0.52) | 0.45 (0.43-0.47) |
| Sensitivity | 0.47 (0.44-0.50) | 0.47 (0.44-0.50) | 0.53 (0.50-0.56) | 0.49 (0.46-0.52) | 0.49 (0.46-0.52) | 0.44 (0.40-0.48) |
| Specificity | 0.55 (0.52-0.58) | 0.50 (0.46-0.54) | 0.50 (0.47-0.53) | 0.47 (0.44-0.50) | 0.51 (0.49-0.53) | 0.46 (0.43-0.49) |
Abbreviations: AUROC, area under the receiver operating characteristic curve; XGBoost, extreme gradient boosting.
The numbers in parentheses are 95% CIs.
Model Generalizability and Feature Importance
The t-distributed stochastic neighbor embedding analysis demonstrated the overall generalizability of the model developed using the Glasgow cohort to the other 4 cohorts, although there are distinct clusters not captured (eFigure 4 in the Supplement). These distinct clusters are likely due to the differences in the distribution of baseline characteristics between the cohorts.
The results of the SHAP analysis for both experiments are provided in eFigure 5 in the Supplement. More than 5 pretreatment seizures, the presence of psychiatric disorders, and EEG and imaging findings were the most important determinants of model prediction in both experiments.
Discussion
Using a large, pooled data set of 5 longitudinal cohorts of patients with newly diagnosed epilepsy across 4 countries, we have developed an attention-based deep learning model to potentially predict response to specific ASMs as the first monotherapy at an individual level. The model uses routinely collected data on clinical history and investigation results without additional data (such as genomics, raw EEG signals, or magnetic resonance imaging scans). The model performance was internally tested as well as externally validated in 4 cohorts of patients separately.
The final adapted transformer model, developed using a pooled data set of 5 cohorts (experiment 1), had an AUROC of 0.65 and a weighted balanced accuracy of 0.62. During subgroup analysis, the transformer model performed better for patients with focal epilepsy than for patients with generalized or unclassified epilepsy. This outcome may be associated with class imbalance because the majority of patients in the pooled cohort had focal epilepsy (n = 1392 [77.4%]). During external validation (experiment 2), by training our model on the largest (Glasgow) cohort and separately assessing its performance in the other 4 cohorts, we observed a reduction in model performance. This reduction might be due to the smaller sample size in these cohorts,47 or it might be due to differences in the distribution of input variables as highlighted in the t-distributed stochastic neighbor embedding analysis. Differences in clinical characteristics and ASM choices are expected across patients managed in different health care settings. It is imperative that a clinically useful prediction model is able to account for these differences and does not overfit associations learned during the model training phase. The comparison of individual input features and the pattern of drug use across the validation cohorts did not reveal any obvious trends explaining variability in model performance during external validation. Nonetheless, the lowest limit of the 95% CI of the AUROC was above 0.50 across 3 of 4 external validation cohorts, indicating model stability and better performance than random chance. An evaluation in more health care settings is needed to further determine the generalizability of the model.
Machine learning methods, particularly deep learning algorithms, have demonstrated a superior ability to mine hidden correlations between input variables and model nonlinearities (features having an unclear association with the outcome) in the data set,48 and they can uncover relevant associations that are masked from the human eye and standard statistical methods.49 We used a relatively new attention-based deep learning model called transformer that uses an attention mechanism to simultaneously focus on different parts of an input sequence of data.28 The transformer model was used in this setting owing to its ability to capture hidden, latent dependencies between individual patient’s ASM trial and the ability of the attention mechanism to focus on different ASM treatments based on individual patient data (eFigure 2 in the Supplement). The transformer, developed using the pooled data set, outperformed 2 of the other 5 models tested in terms of AUROC. Although a single time point per patient was applied in the present study, use of the transformer model will facilitate its future adaptation for predicting treatment response to sequential antiseizure medications over multiple time points.
Few studies have used machine learning approaches to predict treatment response in patients with epilepsy. A previous study used a traditional machine learning approach (random forest algorithm) to develop a predictive algorithm of ASM change based on records in the US medical claims database.15 The model showed good predictive power, achieving an AUROC of 0.72. The study was limited by the lack of clinical details to allow verification of the reasons for change of treatment. Further, potentially important clinical predictors (for instance, seizure type, etiology, and EEG and magnetic resonance imaging results) were not included. In a smaller study with retrospectively identified cases, a support vector machine classifier was able to predict seizure freedom for patients receiving levetiracetam as monotherapy (accuracy, 72.2%; AUROC, 0.96).50 More recently, a study developed both traditional models (ie, linear models and decision tree models) and a deep learning neural network model using clinical and genomic information from regulatory placebo-controlled trials to predict response to adjunctive brivaracetam in patients with drug-resistant epilepsy.51 A gradient-boosted trees classifier model achieved the highest AUROC in development (0.76 [95% CI, 0.74-0.77]) and validation (0.75 [95% CI, 0.60-0.90]) cohorts. Of note, the study demonstrated that integrating genomic variables implicated in disease pathogenesis and drug response enhanced the model performance compared with models trained on individual data modalities. This outcome underscores that, ultimately, the treatment response to ASMs is likely governed by a complex interplay of various clinical and genetic variables.
The SHAP analysis enhances model transparency by quantifying the contribution of individual input features to the outcome predicted by the model.44 Number of pretreatment seizures (>5), presence of psychiatric comorbidity, and EEG and brain imaging findings contributed strongly toward predicted outcomes. A comparable trend was reported for the model for predicting response to adjunctive brivaracetam.51 In this model, the top 3 factors with the highest SHAP value were number of seizure days, temporal lobe localization, and anxiety at baseline. In studies using traditional statistical analysis, higher pretreatment seizure density52 and history of psychiatric comorbidity53 also have been shown to predict poor treatment outcome. Such consistency with prior knowledge supports the “internal validity” of our machine learning model. It should be noted that the results of the SHAP analysis do not imply that ASM treatment per se has less of a contribution to predicting seizure control compared with other input variables given that only patients treated with ASMs were included in developing the model and there were no untreated patients. The relative importance of the clinical variables in our model varied between the 2 experiments, possibly reflecting their different distribution between the cohorts used for model development.
Limitations
Although our model benefited from the use of well-characterized longitudinal cohorts of newly diagnosed epilepsy,4,6,17,18,19,20 it has limitations. The model was developed for adults and may not be generalizable to children who have different age-related epilepsy syndromes with a different prognosis. Subgroup analysis was limited to broad epilepsy types. We were unable to perform subgroup analysis on other determinants of treatment outcome, such as intellectual disability, because of the marked class imbalance or on socioeconomic status because the information was not available. We applied seizure freedom from all seizure types as the outcome, which is justified by previous studies demonstrating that only seizure freedom is consistently associated with improved quality of life in patients with epilepsy.54 Nonetheless, we acknowledge that there may be clinical scenarios in which the physician and patient decide to accept ongoing, nondisabling seizures as an outcome. Understanding stakeholder acceptance of medical artificial intelligence in clinical care should be the foremost goal of future prospective studies.55,56 The model was developed to predict response to 7 first- and second-generation ASMs as initial monotherapy.10 The model may be adapted to incorporate more ASMs as relevant data sets become available in the future through transfer learning57 or incremental learning.58 Given the limited sensitivity and specificity, it is acknowledged that our model needs further improvement before it can be applied in practice. Model performance may potentially be enhanced by incorporating unstructured clinical information using natural language processing59 and genetic determinants of ASM responsiveness,60 as well as quantitative EEG61 and imaging data62,63 as input features.
Conclusions
We have developed and validated a machine learning model for potential personalized prediction of treatment response to commonly used ASMs in adults with newly diagnosed epilepsy. Although its current performance is modest, our model serves as a foundation to further improve clinical applicability to and comparison with expert consensus decision-making algorithms. Because our model may predict an individual’s response to a range of ASMs, it can potentially be used as a clinical decision support tool to inform drug selection. To do so, its performance would require enhancement (eg, by adding other data modalities). This enhancement would assist clinicians by presenting a personalized management strategy to start treatment with the right drug, replacing the century-old trial-and-error approach. It is hoped that this personalized approach to medicine will enable more patients to attain seizure freedom faster, thereby improving their quality of life and reducing the societal cost of epilepsy.
eAppendix. Description of the Transformer Model
eReferences.
eFigure 1. Overview of a Single Encoder-Decoder Pair of The Transformer Network
eFigure 2. Attention Map of Transformer Network
eFigure 3. Performance of Transformer Model Developed Using Glasgow Cohort Only
eFigure 4. t-SNE Analysis
eFigure 5. SHAP Analysis
eTable 1. Summary of Missing Data in Each Cohort
eTable 2. Comparison of Seizure Outcomes and Treatment Status at 12 Months in Individual Cohort
eTable 3. Tuned Hyperparameters for the Machine Learning Models
eTable 4. Computational Time (Inference Time) for the Machine Learning Models
eTable 5. Comparison of Model Performance in Subgroups of Focal and Generalized/unclassified Epilepsy
References
- 1.Fiest KM, Sauro KM, Wiebe S, et al. Prevalence and incidence of epilepsy: a systematic review and meta-analysis of international studies. Neurology. 2017;88(3):296-303. doi: 10.1212/WNL.0000000000003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwan P, Arzimanoglou A, Berg AT, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51(6):1069-1077. doi: 10.1111/j.1528-1167.2009.02397.x [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Brodie MJ, Kwan P. What has been the impact of new drug treatments on epilepsy? Curr Opin Neurol. 2020;33(2):185-190. doi: 10.1097/WCO.0000000000000803 [DOI] [PubMed] [Google Scholar]
- 4.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342(5):314-319. doi: 10.1056/NEJM200002033420503 [DOI] [PubMed] [Google Scholar]
- 5.Dhamija R, Moseley BD, Cascino GD, Wirrell EC. A population-based study of long-term outcome of epilepsy in childhood with a focal or hemispheric lesion on neuroimaging. Epilepsia. 2011;52(8):1522-1526. doi: 10.1111/j.1528-1167.2011.03192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Brodie MJ, Liew D, Kwan P. Treatment outcomes in patients with newly diagnosed epilepsy treated with established and new antiepileptic drugs: a 30-year longitudinal cohort study. JAMA Neurol. 2018;75(3):279-286. doi: 10.1001/jamaneurol.2017.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marson AG, Al-Kharusi AM, Alwaidh M, et al. ; SANAD Study group . The SANAD study of effectiveness of carbamazepine, gabapentin, lamotrigine, oxcarbazepine, or topiramate for treatment of partial epilepsy: an unblinded randomised controlled trial. Lancet. 2007;369(9566):1000-1015. doi: 10.1016/S0140-6736(07)60460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marson AG, Al-Kharusi AM, Alwaidh M, et al. ; SANAD Study group . The SANAD study of effectiveness of valproate, lamotrigine, or topiramate for generalised and unclassifiable epilepsy: an unblinded randomised controlled trial. Lancet. 2007;369(9566):1016-1026. doi: 10.1016/S0140-6736(07)60461-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marson A, Burnside G, Appleton R, et al. ; SANAD II collaborators . The SANAD II study of the effectiveness and cost-effectiveness of valproate versus levetiracetam for newly diagnosed generalised and unclassifiable epilepsy: an open-label, non-inferiority, multicentre, phase 4, randomised controlled trial. Lancet. 2021;397(10282):1375-1386. doi: 10.1016/S0140-6736(21)00246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perucca E, Brodie MJ, Kwan P, Tomson T. 30 Years of second-generation antiseizure medications: impact and future perspectives. Lancet Neurol. 2020;19(6):544-556. doi: 10.1016/S1474-4422(20)30035-1 [DOI] [PubMed] [Google Scholar]
- 11.St Louis EK. Truly “rational” polytherapy: maximizing efficacy and minimizing drug interactions, drug load, and adverse effects. Curr Neuropharmacol. 2009;7(2):96-105. doi: 10.2174/157015909788848929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legros B, Boon P, Ceulemans B, et al. Development of an electronic decision tool to support appropriate treatment choice in adult patients with epilepsy—Epi-Scope®. Seizure. 2012;21(1):32-39. doi: 10.1016/j.seizure.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 13.Asadi-Pooya AA, Beniczky S, Rubboli G, Sperling MR, Rampp S, Perucca E. A pragmatic algorithm to select appropriate antiseizure medications in patients with epilepsy. Epilepsia. 2020;61(8):1668-1677. doi: 10.1111/epi.16610 [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Rollo B, Antonic-Baker A, et al. New era of personalised epilepsy management. BMJ. 2020;371:m3658. doi: 10.1136/bmj.m3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devinsky O, Dilley C, Ozery-Flato M, et al. Changing the approach to treatment choice in epilepsy using big data. Epilepsy Behav. 2016;56:32-37. doi: 10.1016/j.yebeh.2015.12.039 [DOI] [PubMed] [Google Scholar]
- 16.Mohanraj R, Brodie MJ. Diagnosing refractory epilepsy: response to sequential treatment schedules. Eur J Neurol. 2006;13(3):277-282. doi: 10.1111/j.1468-1331.2006.01215.x [DOI] [PubMed] [Google Scholar]
- 17.Brodie MJ, Barry SJ, Bamagous GA, Norrie JD, Kwan P. Patterns of treatment response in newly diagnosed epilepsy. Neurology. 2012;78(20):1548-1554. doi: 10.1212/WNL.0b013e3182563b19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson HD, Foster E, Ademi Z, et al. Markov modelling of treatment response in a 30-year cohort study of newly diagnosed epilepsy. Brain. 2021;145(4):1326-1337. doi: 10.1093/brain/awab401 [DOI] [PubMed] [Google Scholar]
- 19.Fong SL, Lim KS, Mon KY, Bazir SA, Tan CT. How many more seizure remission can we achieve with epilepsy surgeries in a general epilepsy population? Neurol Asia. 2020;25(4):467-472. [Google Scholar]
- 20.Sharma S, Chen Z, Rychkova M, et al. Short- and long-term outcomes of immediate and delayed treatment in epilepsy diagnosed after one or multiple seizures. Epilepsy Behav. 2021;117:107880. doi: 10.1016/j.yebeh.2021.107880 [DOI] [PubMed] [Google Scholar]
- 21.Brodie MJ, Kwan P. Staged approach to epilepsy management. Neurology. 2002;58(8)(suppl 5):S2-S8. doi: 10.1212/WNL.58.8_suppl_5.S2 [DOI] [PubMed] [Google Scholar]
- 22.Kho LK, Lawn ND, Dunne JW, Linto J. First seizure presentation: do multiple seizures within 24 hours predict recurrence? Neurology. 2006;67(6):1047-1049. doi: 10.1212/01.wnl.0000237555.12146.66 [DOI] [PubMed] [Google Scholar]
- 23.Lawn N, Chan J, Lee J, Dunne J. Is the first seizure epilepsy—and when? Epilepsia. 2015;56(9):1425-1431. doi: 10.1111/epi.13093 [DOI] [PubMed] [Google Scholar]
- 24.Fisher RS, Acevedo C, Arzimanoglou A, et al. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55(4):475-482. doi: 10.1111/epi.12550 [DOI] [PubMed] [Google Scholar]
- 25.Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512-521. doi: 10.1111/epi.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakami T, McIntosh A, Todaro M, et al. MRI-identified pathology in adults with new-onset seizures. Neurology. 2013;81(10):920-927. doi: 10.1212/WNL.0b013e3182a35193 [DOI] [PubMed] [Google Scholar]
- 27.Tatum WO, Olga S, Ochoa JG, et al. American Clinical Neurophysiology Society guideline 7: guidelines for EEG reporting. J Clin Neurophysiol. 2016;33(4):328-332. doi: 10.1097/WNP.0000000000000319 [DOI] [PubMed] [Google Scholar]
- 28.Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. arXIV. Preprint posted online June 12, 2017. doi: 10.48550/arXiv.1706.03762 [DOI]
- 29.Alsfouk BAA, Hakeem H, Chen Z, Walters M, Brodie MJ, Kwan P. Characteristics and treatment outcomes of newly diagnosed epilepsy in older people: a 30-year longitudinal cohort study. Epilepsia. 2020;61(12):2720-2728. doi: 10.1111/epi.16721 [DOI] [PubMed] [Google Scholar]
- 30.Alsfouk BAA, Brodie MJ, Walters M, Kwan P, Chen Z. Tolerability of antiseizure medications in individuals with newly diagnosed epilepsy. JAMA Neurol. 2020;77(5):574-581. doi: 10.1001/jamaneurol.2020.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861-874. doi: 10.1016/j.patrec.2005.10.010 [DOI] [Google Scholar]
- 32.Gupta A, Tatbul N, Marcus R, Zhou S, Lee I, Gottschlich J. Class-weighted evaluation metrics for imbalanced data classification. arXiv. Preprint posted online October 12, 2020. doi: 10.48550/arXiv.2010.05995 [DOI]
- 33.Naraei P, Abhari A, Sadeghian A. Application of multilayer perceptron neural networks and support vector machines in classification of healthcare data. In: Proceedings of the 2016 Future Technologies Conference (FTC). IEEE; 2017:848-852. [Google Scholar]
- 34.Bottaci L, Drew PJ, Hartley JE, et al. Artificial neural networks applied to outcome prediction for colorectal cancer patients in separate institutions. Lancet. 1997;350(9076):469-472. doi: 10.1016/S0140-6736(96)11196-X [DOI] [PubMed] [Google Scholar]
- 35.Lin CH, Hsu KC, Johnson KR, et al. ; Taiwan Stroke Registry Investigators . Evaluation of machine learning methods to stroke outcome prediction using a nationwide disease registry. Comput Methods Programs Biomed. 2020;190:105381. doi: 10.1016/j.cmpb.2020.105381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giuseppe C, Antonino S, Giuliana F, et al. A multilayer perceptron neural network-based approach for the identification of responsiveness to interferon therapy in multiple sclerosis patients. Inf Sci. 2010;180(21):4153-4163. doi: 10.1016/j.ins.2010.07.004 [DOI] [Google Scholar]
- 37.Lin CC, Wang YC, Chen JY, et al. Artificial neural network prediction of clozapine response with combined pharmacogenetic and clinical data. Comput Methods Programs Biomed. 2008;91(2):91-99. doi: 10.1016/j.cmpb.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 38.Hassan MR, Al-Insaif S, Hossain MI, Kamruzzaman J. A machine learning approach for prediction of pregnancy outcome following IVF treatment. Neural Comput Appl. 2020;32:2283-2297. doi: 10.1007/s00521-018-3693-9 [DOI] [Google Scholar]
- 39.Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge, Discovery and Data Mining. Association for Computing Machinery; 2016:785-794. [Google Scholar]
- 40.Hearst MA, Dumais ST, Osuna E, Platt J, Scholkopf B. Support vector machines. IEEE Intell Syst Their Appl. 1998;13(4):18-28. doi: 10.1109/5254.708428 [DOI] [Google Scholar]
- 41.Breiman L. Random forests. Machine Learning. 2001;45(1):5-32. [Google Scholar]
- 42.DeMaris A. A tutorial in logistic regression. J Marriage Fam. 1995;57(4):956-968. doi: 10.2307/353415 [DOI] [Google Scholar]
- 43.Van Der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(11):2579-2605. [Google Scholar]
- 44.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Proceedings of the 31st International Conference on Neural Information Processing Systems. Curran Associates Inc; 2017:4765-4774. [Google Scholar]
- 45.Li C. Little’s test of missing completely at random. Stata J. 2013;13(4):795-809. doi: 10.1177/1536867X1301300407 [DOI] [Google Scholar]
- 46.fengweie/transformer_ep. GitHub, Inc. Accessed July 19, 2022. https://github.com/fengweie/transformer_ep
- 47.Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236-1246. doi: 10.1093/bib/bbx044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigsby J, Kramer RE, Schneiders JL, Gates JR, Brewster Smith W. Predicting outcome of anterior temporal lobectomy using simulated neural networks. Epilepsia. 1998;39(1):61-66. doi: 10.1111/j.1528-1157.1998.tb01275.x [DOI] [PubMed] [Google Scholar]
- 49.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349(6245):255-260. doi: 10.1126/science.aaa8415 [DOI] [PubMed] [Google Scholar]
- 50.Zhang JH, Han X, Zhao HW, et al. Personalized prediction model for seizure-free epilepsy with levetiracetam therapy: a retrospective data analysis using support vector machine. Br J Clin Pharmacol. 2018;84(11):2615-2624. doi: 10.1111/bcp.13720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Jong J, Cutcutache I, Page M, et al. Towards realizing the vision of precision medicine: AI based prediction of clinical drug response. Brain. 2021;144(6):1738-1750. doi: 10.1093/brain/awab108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shorvon SD, Goodridge DM. Longitudinal cohort studies of the prognosis of epilepsy: contribution of the National General Practice Study of Epilepsy and other studies. Brain. 2013;136(pt 11):3497-3510. doi: 10.1093/brain/awt223 [DOI] [PubMed] [Google Scholar]
- 53.Kanner AM. Do psychiatric comorbidities have a negative impact on the course and treatment of seizure disorders? Curr Opin Neurol. 2013;26(2):208-213. doi: 10.1097/WCO.0b013e32835ee579 [DOI] [PubMed] [Google Scholar]
- 54.Birbeck GL, Hays RD, Cui X, Vickrey BG. Seizure reduction and quality of life improvements in people with epilepsy. Epilepsia. 2002;43(5):535-538. doi: 10.1046/j.1528-1157.2002.32201.x [DOI] [PubMed] [Google Scholar]
- 55.Budd S, Robinson EC, Kainz B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med Image Anal. 2021;71:102062. doi: 10.1016/j.media.2021.102062 [DOI] [PubMed] [Google Scholar]
- 56.Kovarik CL. Patient perspectives on the use of artificial intelligence. JAMA Dermatol. 2020;156(5):493-494. doi: 10.1001/jamadermatol.2019.5013 [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Gao X. Transfer adaptation learning: a decade survey. medRxiv. Preprint posted online March 12, 2019. doi: 10.48550/arXiv.1903.04687 [DOI]
- 58.Luo Y, Yin L, Bai W, Mao K. An appraisal of incremental learning methods. Entropy (Basel). 2020;22(11):E1190. doi: 10.3390/e22111190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234-1240. doi: 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balestrini S, Sisodiya SM. Pharmacogenomics in epilepsy. Neurosci Lett. 2018;667:27-39. doi: 10.1016/j.neulet.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Croce P, Ricci L, Pulitano P, et al. Machine learning for predicting levetiracetam treatment response in temporal lobe epilepsy. Clin Neurophysiol. 2021;132(12):3035-3042. doi: 10.1016/j.clinph.2021.08.024 [DOI] [PubMed] [Google Scholar]
- 62.Kim HC, Kim SE, Lee BI, Park KM. Can we predict drug response by volumes of the corpus callosum in newly diagnosed focal epilepsy? Brain Behav. 2017;7(8):e00751. doi: 10.1002/brb3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao F, Koepp MJ, Zhou D. Pharmaco-fMRI: a tool to predict the response to antiepileptic drugs in epilepsy. Front Neurol. 2019;10:1203. doi: 10.3389/fneur.2019.01203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Description of the Transformer Model
eReferences.
eFigure 1. Overview of a Single Encoder-Decoder Pair of The Transformer Network
eFigure 2. Attention Map of Transformer Network
eFigure 3. Performance of Transformer Model Developed Using Glasgow Cohort Only
eFigure 4. t-SNE Analysis
eFigure 5. SHAP Analysis
eTable 1. Summary of Missing Data in Each Cohort
eTable 2. Comparison of Seizure Outcomes and Treatment Status at 12 Months in Individual Cohort
eTable 3. Tuned Hyperparameters for the Machine Learning Models
eTable 4. Computational Time (Inference Time) for the Machine Learning Models
eTable 5. Comparison of Model Performance in Subgroups of Focal and Generalized/unclassified Epilepsy


