Abstract
The diagnosis of cryptococcosis is usually performed based on cultures of tissue or body fluids and isolation of the fungus, but this method may require several days. Direct microscopic examination, although rapid, is relatively insensitive. Biochemical and immunodiagnostic rapid tests are also used. However, all of these methods have limitations that may hinder final diagnosis. The increasing incidence of fungal infections has focused attention on tools for rapid and accurate diagnosis using molecular biological techniques. Currently, PCR-based methods, particularly nested, multiplex and real-time PCR, provide both high sensitivity and specificity.
In the present study, we evaluated a nested PCR targeting the gene encoding the ITS-1 and ITS-2 regions of rDNA in samples from a cohort of patients diagnosed with cryptococcosis. The results showed that in our hands, this Cryptococcus nested PCR assay has 100% specificity and 100% sensitivity and was able to detect until 2 femtograms of Cryptococcus DNA.
Keywords: Cryptococcus neoformans/Cryptococcus gattii complex, Nested PCR, Cryptococcosis, Molecular diagnosis
Introduction
Cryptococcosis is a fungal infection caused by encapsulated yeasts of the phylum Basidiomycota, genus Cryptococcus. Although over 30 different species of Cryptococcus have been identified to date, just two closely related species – Cryptococcus neoformans and Cryptococcus gattii – cause the majority of human fungal infections.1, 2, 3 Based on specific polysaccharide capsule antigen analysis, subtyping data, and comparisons of the genomic sequences, pathogenic cryptococci have been divided into five capsular serotypes: serotype A (C. neoformans var. grubii), serotypes B and C (both C. gattii), serotype D (C. neoformans var. neoformans), and the hybrid diploid serotype AD.2, 4, 5
C. neoformans is found worldwide, and it causes the majority of cryptococcal infections in people with decreased immunity (primarily AIDS patients, people undergoing immunosuppressive therapies and those with lymphoproliferative disorders), resulting in varying neurological complications.6 In contrast, C. gattii is principally endemic to tropical and subtropical regions, and it causes 70% to 80% of cryptococcal infections in immunocompetent hosts.4, 7, 8
Although reporting fungal infections is not mandatory in Colombia, South America, in 2012 Escandón et al. published the results of a survey on cryptococcosis conducted in Colombia between 2006 and 2010. In this period, 526 reports with at least one case of cryptococcosis were received. These cases originated from 72% of the Colombian political divisions. The most prevalent risk factor reported was HIV infection (83.5%), with cryptococcosis defining AIDS in 23% of the cases. The estimated mean annual incidence rate for cryptococcosis in the general population was one in every 2.4 × 106 habitants, while in AIDS patients this rate rose to one in 3.3 × 103. Neurocryptococcosis was recorded in 81.8% of the cases. Laboratory diagnoses were based on direct examination, culture and latex in 29.3% cases; of 413 Cryptococcus isolates analyzed, 95.6% were identified as C. neoformans var. grubii, 1% C. neoformans var. neoformans, and 3.4% C. gattii.9
Even though the majority of cryptococcosis cases reported correspond to cryptococcal meningitis, the initial infection is generally acquired by the inhalation of airborne fungal propagules from an environmental source.10, 11, 12 Both C. neoformans and C. gattii are capable of causing severe pulmonary and central nervous system (CNS) infections in both immunocompetent and immunosuppressed individuals13, 14, 15; importantly, up to 70% of these individuals will die within three months of infection.16
The diagnosis of cryptococcosis infection is usually based on isolation of the fungus from cultured tissue or body fluids such as sputum, blood and cerebrospinal fluid, but this method may require several days to detect and identify the microorganisms. Although direct microscopic examination is rapid, this method is relatively insensitive. Of rapid biochemical and immunodiagnostic tests, which can be performed on blood and/or cerebrospinal fluid,17, 18 the detection of cryptococcal capsular antigen by latex agglutination is one of the most helpful tests for fungi performed on a routine basis. Its ease of use and sensitivity are better than other conventional immunodiagnostic methods19, 20, 21; however, all of these methods have some limitations that may hinder final diagnosis.15
The diagnostic limitations and increasing incidence of fungal infections have prompted the development of tools for rapid and accurate diagnosis using molecular biological techniques. Currently, molecular methods such as DNA hybridization and PCR-based methods (particularly nested, multiplex and real time PCR) provide both high sensitivity and specificity. Improvements in PCR techniques have allowed the detection of minimal amounts of DNA from the C. neoformans species; in addition, PCR can be used in association with other techniques, making it a valuable tool for molecular epidemiology studies.22, 23, 24
Several target sequences have been utilized to identify the C. neoformans complex, including URA5, CAP59, M13, and ITS (18S, 5.8S, and 28S). The ITS region of rDNA has been the most frequently used region for the detection of fungal sequences because of its high degree of variation compared to that of other ribosomal DNA regions facilitates identification.25, 26, 27
Nested PCR stands out among the most-used PCR-based techniques for detection and identification of C. neoformans and C. gattii. In this technique, the DNA used in the reaction is the product of a previous amplification, and it is very useful when high sensitivity and specificity are desirable.28 The work of Rappelli et al. (1998)26 strongly influenced this area; they developed a nested PCR protocol for the detection of C. neoformans and C. gattii from samples obtained from patients with neurocryptococcosis. The specificity and sensitivity of this technique were tested using DNA from other microorganisms, which were not amplified. Testing different dilutions of fungal DNA samples resulted in the amplification of up to 10 fungal cells/ml.
Recently, Trilles et al. (2014) developed a hyperbranched rolling circle amplification (HRCA) based on the PLB1 locus. Used alone and in combination with a semi-nested PCR, this technique was specific and highly sensitive. This new method has great potential for use in direct diagnosis of cryptococcosis from clinical specimens.29
For the treatment of cryptococcosis, amphotericin B, fluconazole, and itraconazole are recommended as first-line treatments for C. neoformans and C. gattii infections, while voriconazole and posaconazole are used as secondary therapies.30, 31 However, worldwide, approximately 625,000 patients living with HIV/AIDS die from cryptococcal meningitis each year (Centers for Disease Control and Prevention, CDC, Atlanta, USA, http://www.cdc.gov/).
In the present study, we validated the nested PCR described by Rapelli et al. (1998) in e ITS-1 and ITS-2 coding regions of C. neoformans/C. gattii. The gold standard diagnostic technique used in the validation was microorganism culture from clinical specimens.26 We aimed at implementing this molecular assay as an integral component of the diagnostic tests regularly used in our laboratory.
Materials and methods
Clinical samples and isolates
Over the 17-month period from January 2011 to June 2012, 44 human clinical samples from 44 patients with confirmed cryptococcosis were collected. The clinical specimens included: bronchoalveolar lavage (BAL) (n = 10), bronchial lavage (BL) (n = 6), biopsy (n = 4), and cerebrospinal fluid (CSF) (n = 24). All of the specimens were collected at hospitals in Medellín, Colombia, and sent to the Medical and Experimental Mycology Unit of the Corporación para Investigaciones Biológicas (CIB), for mycological diagnosis.
To assess the specificity of the nested PCR, 92 clinical samples collected from patients with other respiratory infections (diagnosed by culture and/or specific stains) were analyzed. These clinical samples included specimens from patients diagnosed with: histoplasmosis (n = 10), paracoccidioidomycosis (n = 21), pneumocystosis (n = 6), candidiasis (n = 16), aspergillosis (n = 19), and tuberculosis (n = 20). As negative controls (n = 51), we used 30 respiratory-negative samples (by culture and/or specific stains for cryptococcosis or other common respiratory infectious pathogens) as well as 21 peripheral blood samples from healthy individuals (Table 1). Additionally, the nested PCR specificity was also evaluated, using purified DNA isolated from cultures of different pathogen microorganisms (n = 35), previously identified by sequencing (Table 2).
Table 1.
Clinical samples used for the Cryptococcus neoformans/Cryptococcus gattii nested PCR validation.
| Kind of patient | Type of sample | Diagnosis | ||
|---|---|---|---|---|
| Clinical samples (n:187) | Negative controls (n:51) | Respiratory symptomatic (n:30) | BAL (n:25) | |
| BL (n:4) | ||||
| Healthy individuals (n:21) | Sputum (n:1) | |||
| Whole blood (n:21) | ||||
| Positive controls (n:44) | Respiratory symptomatic (n:44) | CSF (n:24) | Cryptococcus neoformans (n:44) | |
| BAL (n:10) | ||||
| BL (n:6) | ||||
| Biopsies (n:4) | ||||
| Samples used to evaluate the specificity (n:92) | Respiratory symptomatic (n:92) | Body fluids (n:4) | Aspergillus fumigatus (n:1) | |
| Candida albicans (n:1) | ||||
| Candida tropicalis (n:2) | ||||
| Tracheal aspirates (n:9) | Aspergillus fumigatus (n:1) | |||
| Candida albicans (n:6) | ||||
| Candida tropicalis (n:1) | ||||
| Mycobacterium tuberculosis (n:1) | ||||
| Biopsies (n:25) | Aspergillus fumigatus (n:4) | |||
| Aspergillus flavus (n:2) | ||||
| Candida spp. (n:1) | ||||
| Candida parapsilosis (n:1) | ||||
| Candida guillermondii (n:1) | ||||
| Candida albicans (n:3) | ||||
| Mycobacterium tuberculosis (n:1) | ||||
| Paracoccidioides brasiliensis (n:7) | ||||
| Histoplasma capsulatum (n:5) | ||||
| Sputum (n:20) | Aspergillus fumigatus (n:2) | |||
| Mycobacterium tuberculosis (n:7) | ||||
| Paracoccidioides brasiliensis (n:11) | ||||
| BL (n:10) | Aspergillus fumigatus (n:3) | |||
| Mycobacterium tuberculosis (n:7) | ||||
| BAL (n:24) | Aspergillus fumigatus (n:4) | |||
| Aspergillus flavus (n:1) | ||||
| Aspergillus versicolor (n:1) | ||||
| Mycobacterium tuberculosis (n:4) | ||||
| Paracoccidioides brasiliensis (n:3) | ||||
| Histoplasma capsulatum (n:5) | ||||
| Pneumocystis jirovecii (n:6) |
Table 2.
Purified DNA used to evaluate the Cryptococcus neoformans/Cryptococcus gattii nested PCR specificity.
| Molecular analysis | ||
|---|---|---|
| Microorganisma | Strain source | Nested PCR for the complex C. neoformans/C. gattii |
| Aspergillus flavus | CIBb | − |
| Aspergillus fumigatus | CIB | − |
| Aspergillus terreus | CDCc | − |
| Aspergillus niger | CDC | − |
| Blastomyces dermatitidis | CDC 2008011573 | − |
| Blastomyces dermatitidis | ATCC 26199 | − |
| Candida albicans | CIB | − |
| Candida guillermondii | CIB | − |
| Candida tropicalis | CIB | − |
| Candida parapsilosis | CIB | − |
| Candida dubliniensis | CIB | − |
| Candida glabrata | CIB | − |
| Candida krusei | CIB | − |
| Candida lusitanae | CIB | − |
| Candida bracariensis | CIB | − |
| Candida ohmeri | CIB | − |
| Candida famata | CIB | − |
| Candida ortopsilosis | CIB | − |
| Candida metapsilosis | CIB | − |
| Coccidioides immitis | ATCC 28868 | − |
| Cryptococcus neoformans | CIB | + |
| Cryptococcus gattii | CIB | + |
| Histoplasma capsulatum var. capsulatum | CDC | − |
| Histoplasma capsulatum var. capsulatum | G217B | − |
| Histoplasma capsulatum var. capsulatum | G184B | − |
| Histoplasma capsulatum var. capsulatum | CDC | − |
| Histoplasma capsulatum var. duboisii | CDC/5822 | − |
| Histoplasma capsulatum var. duboisii | CDC/5823 | − |
| Mycobacterium tuberculosis | CIB | − |
| Mycobacterium avium | CIB | − |
| Paracoccidioides brasiliensis | ATCC 60855 | − |
| Paracoccidioides brasiliensis | CIB/Pb339 | − |
| Paracoccidioides brasiliensis | CIB/Pb18 | − |
| Paracoccidioides lutzii | CIB | − |
| Schizophylum commune | CIB | − |
DNA was isolated from cultures of pathogenic microorganisms (n: 35) previously identified by nucleic acid sequencing of genomic targets.
CIB: Corporación para Investigaciones Biológicas, Medellín, Colombia.
CDC: Centers for Disease Control and Prevention, Atlanta, USA. (−) PCR negative result. (+) PCR positive result.
Processing of clinical samples
Both respiratory tract specimens (BAL, BL, sputum) and body fluids (peritoneal, pleural and CSF) were collected in 50 ml sterile Falcon tubes (Becton Dickinson) and centrifuged at 1550 × g for 30 min (Centra MP4R, IEC). Fresh tissues (biopsy) were manually homogenized in 3 ml of sterile saline solution. The pelleted samples and the homogenate obtained were used for culture and stain, and 0.6 ml of each sample was stored at −20° C for subsequent DNA extraction.
Processed specimens were cultured on Sabouraud Dextrose Agar and Mycosel (Becton Dickinson), incubated at room temperature (±18–22° C) for three weeks, and examined weekly for yeast colonies; the identification was performed using microscopic observation, phenotypic type enzyme (urease and phenoloxidase), and carbohydrate assimilation patterns (McTaggart et al., 2011).14 The biovarieties were determined by culturing the isolates on l-canavanine glycine bromothymol blue (CGB) selective medium (Klein et al., 2009).32 These procedures were carried out in a Biosafety Level 3 (BSL3) laboratory.
DNA extraction
Two hundred microliters of each previously processed clinical sample or yeast suspension were used for DNA extraction and purification. The QIAamp® DNA Mini kit (Qiagen, Hildenberg, Germany) was used with some modifications: the initial incubation with lysis buffer was performed at 65° C for one hour, followed by AL buffer incubation at 90 °C for 10 min and an additional incubation with recombinant lyticase (1UI/μl) at 37° C for 45 min. For filamentous fungal isolates, DNA extraction was performed using the phenol-chloroform method or a commercial kit with Genomic G-100 columns (Qiagen Inc., CA) (Sambrook et al., 2001).33 DNA extraction from whole blood was performed using a protocol described by Einsele et al. (1997)34 with some modifications. The relative concentrations of DNA extracted were determined using a NanoDrop ND2000 (Thermo Scientific).
C. neoformans/C. gattii nested-PCR assay
C. neoformans/C. gattii specific primers that target the gene encoding the rDNA internal transcribed regions 1 (ITS-1) and 2 (ITS-2) were used in a nested PCR reaction as described by Rappelli et al. (1998),26 with some modifications. The master mix for the first PCR consisted of 10 μl of purified DNA in a total PCR volume of 50 μl with final concentrations of 2 mM MgCl2 (Invitrogen), 0.2 mM of dNTPs Mix, 0.6 μM of each outer and inner primer (Invitrogen) and 0.02 units of Taq polymerase (Invitrogen). The mixture was incubated at 94 °C for 5 min; 20 cycles of 94 °C for 45 s, 55 °C for 60 s, and 72 °C for 1 min; and a final extension at 72 °C for 5 min. For the second (nested) PCR, the mix was similar to the first, except that 2 μl of the first PCR product was used as template DNA and the reaction mixture was incubated at 94 °C for 5 min; for 30 cycles of 94 °C for 45 s, 70 °C for 60 s, and 72 °C for 1 min, with a final extension at 72 °C for 5 min. The final product of the nested PCR is an 116 bp fragment that indicates the presence of Cryptococcus DNA in the samples analyzed. As a positive control, 10 μl containing 10 ng of purified C. neoformans DNA was used in all PCR assays. To detect any contamination, sterile water was included in the DNA extraction used as a negative control, and additional reaction mixtures without DNA were run during all procedures.
As a control to verify amplifiable DNA or to detect the presence of PCR inhibitors in the clinical samples, a PCR designed to amplify the human gene for β-globin was carried out as described by Bialek et al. (2005).35 All of the PCR reactions were run on a Peltier Thermal Cycler PT100 (MJ Research, USA). The PCR products were visualized by electrophoresis on 2% agarose gels (Sigma Chemical Co., St. Louis, MO, USA), using gel red and a UV transilluminator (Molecular Imager® Gel DocTMXR+ BIORAD). All of the nested PCR products were sequenced to verify that the amplified DNA fragment corresponded to the C. neoformans/C. gattii target.
Detection limit
To establish the detection limit of the nested PCR assay, we extracted and quantified DNA from a C. neoformans yeast suspension and performed serial dilutions (1:2) ranging from 40.4 ng to 1 fg. Each of these dilutions was then used for a specific PCR, to determine the amount of DNA at the assay's detection limit.
Data analysis
The sequences obtained were edited and aligned using Sequencher software (version 4.8), and homology searches of all sequences were carried out using the BLASTn program from the National Center for Biotechnology Information, Washington, DC. The sequences were categorized according to E-values (error probability) as provided by BLASTn, using values lower than 1 × 1040.
Sensitivity and specificity for the C. neoformans/C. gattii nested PCR were calculated using the culture as the gold standard, according to the method of Galen and Gambino (1975).36
Results
Cross-reaction assay
None of the DNA isolated from cultures of related microorganisms previously identified by sequencing tested positive in the Cryptococcus nested PCR assay. By contrast, all purified DNA from the Cryptococcus yeasts, including C. gattii, tested positive in the nested PCR assay (Table 2).
Detection of Cryptococcus DNA in clinical samples
A total of 44 clinical samples from patients with cryptococcosis that was diagnosed by culture were analyzed through nested PCR. All of the BL, BAL, biopsies, and CSF samples were tested positive in the Cryptococcus nested PCR assay. Therefore, the ITS-1 and ITS-2 of rDNA nested PCR for the Cryptococcus neoformans/Cryptococcus gattii complex exhibited a sensitivity of 100% (Table 3A).
Table 3.
Nested PCR results obtained on human clinical samples from patients with cryptococcosis (A) or other respiratory diseases different from cryptococcosis as well as healthy individuals taken as controls (B).
| Specimen type | Proven disease | Number of samples |
|
|---|---|---|---|
| Total | Positive PCR | ||
| (A) | |||
| Bronco alveolar lavage | Cryptococcosis | 10 | 10 |
| Bronchial lavage | Cryptococcosis | 6 | 6 |
| Biopsy | Cryptococcosis | 4 | 4 |
| Cerebrospinal fluid | Cryptococcosis | 24 | 24 |
| 44 | 44 | ||
| (B) | |||
| Bronco alveolar lavage | Paracoccidioidomycosis | 3 | 0 |
| Pneumocystis pneumonia | 6 | 0 | |
| Histoplasmosis | 5 | 0 | |
| Aspergillosis | 6 | 0 | |
| Tuberculosis | 4 | 0 | |
| Bronchial lavage | No disease | 25 | 0 |
| Aspergillosis | 3 | 0 | |
| Tuberculosis | 7 | 0 | |
| No disease | 4 | 0 | |
| Sputum | Aspergillosis | 2 | 0 |
| Paracoccidioidomycosis | 11 | 0 | |
| Tuberculosis | 7 | 0 | |
| No disease | 1 | 0 | |
| Tracheal aspirate | Aspergillosis | 1 | 0 |
| Candidiasis | 7 | 0 | |
| Tuberculosis | 1 | 0 | |
| Biopsy | Candidiasis | 6 | 0 |
| Aspergillosis | 6 | 0 | |
| Paracoccidioidomycosis | 7 | 0 | |
| Tuberculosis | 1 | 0 | |
| Histoplasmosis | 5 | 0 | |
| Pleural fluid | Aspergillosis | 1 | 0 |
| Peritoneal fluid | Candidiasis | 3 | 0 |
| Whole blood | No disease | 21 | 0 |
| 143 | 0 | ||
To assess the specificity of this nested PCR assay, 92 clinical samples collected from patients with other diagnosed respiratory infections by culture and/or specific stains and 51 negative controls (30 respiratory negative samples and 21 peripheral blood samples from healthy individuals) were analyzed. The ITS-1 and ITS-2 of rDNA nested PCR exhibited a specificity of 100% for the negative controls (0 positives/51 samples) as well as, for those samples (0/92) with other diagnosed respiratory infections (Table 3B).
The presence of PCR inhibitors was ruled out because all of the clinical samples with negative results in the Cryptococcus PCR assay allowed amplification of a specific fragment of the human β-globin gene.
Detection limits
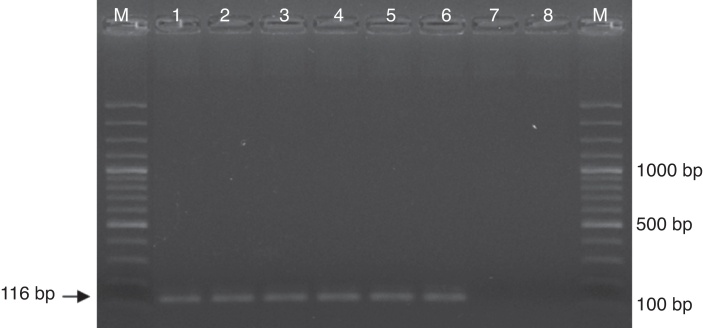
DNA extracted from a C. neoformans yeast suspension was quantified and serial dilutions (1:2) were performed, ranging from 40.4 ng to 1 fg. The optimized nested PCR conditions in our laboratory allowed detection of 2 fg of Cryptococcus DNA (Fig. 1)
Fig. 1.
Electrophoresis in agarose gel was running to make evident the detection limit of the Cryptococcus neoformans/Cryptococcus gattii complex nested PCR. Lanes in gel were loaded with molecular weight marker (M) and amplicons from PCR reactions employing (1) 40 ng, (2) 400 pg, (3) 4 pg, (4) 40 fg, (5) 4 fg, (6) 2 fg, (7) 1 fg, (8) 0 fg of C. neoformans DNA.
Controls
All of the DNA extraction controls tested negative in the nested PCR assays. Therefore, any possible cross-contamination during the extraction procedure was discarded.
Discussion
In our hands, the Cryptococcus nested PCR assay for rDNA ITS-1 and ITS-2 of has 100% sensitivity, as it tested positive for all 44 clinical samples (10 BALs, 6 BLs, 4 biopsies and 24 CSFs) from patients diagnosed with cryptococcosis. The specificity of this assay was also 100% as both the 51 negative controls (25 BALs, 4 BLs, 1 sputum, and 21 whole bloods) and the 92 clinical samples from patients diagnosed with infections other than cryptococcosis (10 histoplasmosis, 21 paracoccidioidomycosis, 6 pneumocystosis, 16 candidiasis, 19 aspergillosis, and 20 tuberculosis) gave negative results. However, in order to obtain negative and positive predictive values, a cohort of patients with syndromes that include C. neoformans/C. gattii as part of the possible causes will be evaluated in the future using both our PCR assays as the gold standard method until a final diagnosis is reached.
Just between the years 2006 and 2010 in our country, 526 reports of at least one case of cryptococcosis were obtained in an epidemiological study conducted by Escandon et al. (2012), even knowing that the reporting of fungal infections was not mandatory in Colombia. In microbiology and mycology laboratories, the diagnosis of fungal infections is based on direct observation of macro/micro morphological characteristics, culture, or biochemical and serological tests that permit identification of the pathogen.37 Nevertheless, the aforementioned diagnostic strategies are not always sufficient for accurate cryptococcosis diagnosis.38 The implementation of molecular diagnostic tests with high sensitivity and specificity would be essential to mycology laboratories in our country. Early diagnosis that leads to adequate and prompt starting of antifungal therapy would be crucial in diminishing the severity of infection.
Accurate diagnosis of infectious fungal diseases is very difficult because nearly 70 thousand fungal species have been described. However, only approximately 270 species have been reported to result in infectious disease in humans, and the majority of them are not well adapted to growth at human body temperature (most of these fungi correspond to dermatophytic species causing superficial infections).39, 40 Among the few species able to produce systemic fungal infections in humans are those included in the genus Cryptococcus, specifically C. neoformans and C. gattii.41
In this context, the PCR technique has much strength compared to conventional methods: it is not laborious, can be used with a small sample, is able to detect a very low fungal load, and is a very rapid technique.22 Although other genetic targets, such as the CAP59 gene (involved in Cryptococcus capsule production) and alternative ribosomal DNA regions (SSU/LSU) have been used to identify different species of the C. neoformans complex, the high degree of variability of the ITS region of the rDNA makes it the most commonly used region for the detection and identification of several fungal sequences. Other studies using different molecular approaches for cryptococcosis diagnosis have also shown high sensitivity and specificity.25, 28, 42, 43, 44, 45
In our study, specific primers targeted to the ITS-1 and ITS-2 regions of rDNA were used according to the description of Rapelli et al. (1998). Additionally, in order to increase the disruption efficiency and liberate cryptococcal DNA from tissue homogenates, the DNA extraction protocol included an incubation period with recombinant lyticase (1 UI/μl) at 37 °C for 45 min.
The current gold standard diagnostic test for cryptococcosis remains culture, despite its lack of sensitivity.38 Antigen tests are also regularly used to detect cryptococcal antigen (CrAg) by both enzyme immunoassay (EA) or latex agglutination (LA). These tests are sensitive and specific but require expertise, special storage, and a central reference laboratory.46, 47 Additionally, cryptococcal antigen titers remain high even five months after effective therapy, which may lead to false positive test results.48 Recently, a lateral flow assay (LFA) with the ability to detect CrAg was developed, but although this rapid test exhibits good sensitivity and specificity, more validation studies are necessary.19, 49, 50
In conclusion, in agreement with the results described by Rapelli et al. (1998),26 we found that Cryptococcus nested PCR is a sensitive, specific, and reproducible method to be used in the analysis of different clinical samples. We confirmed the high sensitivity with the ability to detect amounts down to 2 fg of Cryptococcus DNA. This nested PCR assay may be a useful tool not only for rapid diagnosis of acute cryptococcosis but also for monitoring patients during therapy and confirming clearance of the parasite in follow-up exams.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by: COLCIENCIAS, Bogotá, Colombia (Project No. 2213-519-28916), Research Committee (CODI) of the Universidad de Antioquia through the Sustainability Strategy for Groups A1 and A Program 2013–2014. Medellín, Colombia. Convocatoria Nacional Jóvenes Investigadores e Innovadores of Colciencias, Number 501 and 645 (supporting Marcela Gaviria and Vanessa Rivera). Mycotic Research Branch, Centers for Disease Control and Prevention (CDC), for providing purified DNAs (Table 2).
References
- 1.Srikanta D., Santiago-Tirado F.H., Doering T.L. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast. 2014;31:47–60. doi: 10.1002/yea.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun C.D., Madhani H.D. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzymol. 2010;470:797–831. doi: 10.1016/S0076-6879(10)70033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bovers M., Hagen F., Boekhout T. Diversity of the Cryptococcus neoformans–Cryptococcus gattii species complex. Rev Iberoam Micol. 2008;25:S4–S12. doi: 10.1016/s1130-1406(08)70019-6. [DOI] [PubMed] [Google Scholar]
- 4.Lin X., Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 5.Meyer W., Castañeda A., Jackson S., Huynh M., Castañeda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogliati M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Scientifica (Cairo) 2013:675213. doi: 10.1155/2013/675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escandón P., Sánchez A., Martínez M., Meyer W., Castañeda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6:625–635. doi: 10.1111/j.1567-1364.2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 8.Gullo F.P., Rossi S.a, Sardi J.D.C.O., Teodoro V.L.I., Mendes-Giannini M.J.S., Fusco-Almeida A.M. Cryptococcosis: epidemiology, fungal resistance, and new alternatives for treatment. Eur J Clin Microbiol Infect Dis. 2013;32:1377–1391. doi: 10.1007/s10096-013-1915-8. [DOI] [PubMed] [Google Scholar]
- 9.Escandón P., de Bedout C., Lizarazo J., et al. Cryptococcosis in Colombia: results of the national surveillance program for the years 2006–2010. Biomédica. 2012;32:386–398. doi: 10.1590/S0120-41572012000300009. [DOI] [PubMed] [Google Scholar]
- 10.Botts M.R., Hull C.M. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol. 2010;13:437–442. doi: 10.1016/j.mib.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles S.S., Dagenais T.R.T., Botts M.R., Keller N.P., Hull C.M. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun. 2009;77:3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velagapudi R., Hsueh Y-P., Geunes-Boyer S., Wright J.R., Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid G. Cryptococcus infection in immunocompetent individuals. Int J Infect Dis. 2012;16:e17. [Google Scholar]
- 14.McTaggart L., Richardson S.E., Seah C., Hoang L., Fothergill A., Zhang S.X. Rapid identification of Cryptococcus neoformans var. grubii, C. neoformans var. neoformans, and C. gattii by use of rapid biochemical tests, differential media, and DNA sequencing. J Clin Microbiol. 2011;49:2522–2527. doi: 10.1128/JCM.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huston S.M., Mody C.H. Cryptococcosis: an emerging respiratory mycosis. Clin Chest Med. 2009;30:253–264. doi: 10.1016/j.ccm.2009.02.006. vi. [DOI] [PubMed] [Google Scholar]
- 16.Park B.J., Wannemuehler K.A., Marston B.J., Govender N., Pappas P.G., Chiller T.M. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 17.Qureshi A., Wray D., Rhome R., Barry W., Del Poeta M. Detection of antibody against fungal glucosylceramide in immunocompromised patients: a potential new diagnostic approach for cryptococcosis. Mycopathologia. 2012;173:419–425. doi: 10.1007/s11046-011-9485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeo S.F., Wong B. Current status of nonculture methods for diagnosis of invasive fungal infections. Clin Microbiol Rev. 2002;15:465–484. doi: 10.1128/CMR.15.3.465-484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulware D.R., Rolfes M.A., Rajasingham R., et al. Multisite validation of cryptococcal antigen lateral flow assay and quantification by laser thermal contrast. Emerg Infect Dis. 2014;20:45–53. doi: 10.3201/eid2001.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsley M.D., Mekha N., Baggett H.C., et al. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of cryptococcosis. Clin Infect Dis. 2011;53:321–325. doi: 10.1093/cid/cir379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pongsai P., Atamasirikul K., Sungkanuparph S. The role of serum cryptococcal antigen screening for the early diagnosis of cryptococcosis in HIV-infected patients with different ranges of CD4 cell counts. J Infect. 2010;60:474–477. doi: 10.1016/j.jinf.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Sidrim J.J.C., Costa A.K.F., Cordeiro R.A., et al. Molecular methods for the diagnosis and characterization of Cryptococcus: a review. Can J Microbiol. 2010;56:445–458. doi: 10.1139/w10-030. [DOI] [PubMed] [Google Scholar]
- 23.Leal A.L., Faganello J., Bassanesi M.C., Vainstein M.H. Cryptococcus species identification by multiplex PCR. Med Mycol. 2008;46:377–383. doi: 10.1080/13693780701824429. [DOI] [PubMed] [Google Scholar]
- 24.Magee P.T., Gale C., Berman J., Davis D. Molecular genetic and genomic approaches to the study of medically important fungi. Infect Immun. 2003;71:2299–2309. doi: 10.1128/IAI.71.5.2299-2309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paschoal R.C., Hirata M.H., Hirata R.C., De Souza M., Melhem C. Neurocryptococcosis diagnosis by PCR method. Rev Inst Med Trop SP. 2004;46:203–207. doi: 10.1590/s0036-46652004000400006. [DOI] [PubMed] [Google Scholar]
- 26.Rappelli P., Are R., Casu G., Fiori P.L., Cappuccinelli P., Aceti A. Development of a nested PCR for detection of Cryptococcus neoformans in cerebrospinal fluid. J Clin Microbiol. 1998;36:3438–3440. doi: 10.1128/jcm.36.11.3438-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bovers M., Diaz M.R., Hagen F., et al. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J Clin Microbiol. 2007;45:1874–1883. doi: 10.1128/JCM.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bialek R., Weiss M., Bekure-Nemariam K., et al. Detection of Cryptococcus neoformans DNA in tissue samples by nested and real-time PCR assays. Clin Diagn Lab Immunol. 2002;9:461–469. doi: 10.1128/CDLI.9.2.461-469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trilles L., Wang B., Firacative C., Lazéra M.D.S., Wanke B., Meyer W. Identification of the major molecular types of Cryptococcus neoformans and C. gattii by Hyperbranched rolling circle amplification. PLOS ONE. 2014;9:e94648. doi: 10.1371/journal.pone.0094648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bratton E.W., El Husseini N., Chastain C.A., et al. Approaches to antifungal therapies and their effectiveness among patients with cryptococcosis. Antimicrob Agents Chemother. 2013;57:2485–2495. doi: 10.1128/AAC.01800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perfect J.R., Dismukes W.E., Dromer F., et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein K.R., Hall L., Deml S.M., Rysavy J.M., Wohlfiel S.L., Wengenack N.L. Identification of Cryptococcus gattii by use of l-canavanine glycine bromothymol blue medium and DNA sequencing. J Clin Microbiol. 2009;47:3669–3672. doi: 10.1128/JCM.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J., Russell D.W. CSHL Press; New York: 2001. Commonly used Techniques in Molecular Cloning; p. 2344. [Google Scholar]
- 34.Einsele H., Hebart H., Roller G., et al. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bialek R., Konrad F., Kern J., et al. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J Clin Pathol. 2005;58:1180–1184. doi: 10.1136/jcp.2004.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galen R.S., Gambino S.R. John Wiley & Sons; New York: 1975. Beyond normality the predictive value and efficiency of medical diagnoses; p. 237. [Google Scholar]
- 37.Ramana K.V., Kandi S., Bharatkumar P.V., et al. Invasive fungal infections: a comprehensive review. Am J Infect Dis Microbiol. 2013;1:64–69. [Google Scholar]
- 38.Smith J.A., Kauffman C.A. Pulmonary fungal infections. Respirology. 2012;17:913–926. doi: 10.1111/j.1440-1843.2012.02150.x. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan D., Moran G., Coleman D. In: Fungi biology and applications. second ed. Kavanagh K., editor. John Wiley & Sons, Ltd.; Chichester, UK: 2011. Fungal infections of human; p. 384. [Google Scholar]
- 40.Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson M.D. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother. 2005;56(Suppl 1):i5–i11. doi: 10.1093/jac/dki218. [DOI] [PubMed] [Google Scholar]
- 42.Satoh K., Maeda M., Umeda Y., Miyajima Y., Makimura K. Detection and identification of probable endemic fungal pathogen, Cryptococcus gattii, and worldwide pathogen, Cryptococcus neoformans, by real-time PCR. Microbiol Immunol. 2011;55:454–457. doi: 10.1111/j.1348-0421.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 43.Martins M.L., Ferreira A.S., Sampaio A., Vieira R., Inácio J. Direct and specific identification of Cryptococcus neoformans in biological samples using fluorescently labelled DNA probes. Eur J Clin Microbiol Infect Dis. 2010;29:571–576. doi: 10.1007/s10096-010-0897-z. [DOI] [PubMed] [Google Scholar]
- 44.Veron V., Simon S., Blanchet D., Aznar C. Real-time polymerase chain reaction detection of Cryptococcus neoformans and Cryptococcus gattii in human samples. Diagn Microbiol Infect Dis. 2009;65:69–72. doi: 10.1016/j.diagmicrobio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Enache-Angoulvant A., Chandenier J., Symoens F., et al. Molecular identification of Cryptococcus neoformans serotypes. J Clin Microbiol. 2007;45:1261–1265. doi: 10.1128/JCM.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhana A. Case report diagnosis of cryptococcosis and prevention of cryptococcal meningitis using a novel point-of-care lateral flow assay. Case Rep Med. 2013;2013:640216. doi: 10.1155/2013/640216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binnicker M.J., Jespersen D.J., Bestrom J.E., Rollins L.O. A comparison of four assays for the detection of cryptococcal antigen. Clin Vaccine Immunol. 2012;19:1988–1990. doi: 10.1128/CVI.00446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajasingham R., Meya D.B., Boulware D.R. Integrating cryptococcal antigen screening and pre-emptive treatment into routine HIV care. J Acquir Immune Defic Syndr. 2012;59:85–91. doi: 10.1097/QAI.0b013e31824c837e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen J., Slechta E.S., Gates-Hollingsworth M.A., et al. Large-scale evaluation of the immunomycologics lateral flow and enzyme-linked immunoassays for detection of Cryptococcal antigen in serum and cerebrospinal. Clin Vaccine Immunol. 2013;20:52–55. doi: 10.1128/CVI.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escandón P., Lizarazo J., Agudelo C.I., Chiller T., Castañeda E. Evaluation of a rapid lateral flow immunoassay for the detection of cryptococcal antigen for the early diagnosis of cryptococcosis in HIV patients in Colombia. Med Mycol. 2013;51:765–768. doi: 10.3109/13693786.2013.781692. [DOI] [PubMed] [Google Scholar]