Abstract
Background
Research has shown that hepatitis B virus (HBV) genotypes are closely linked to the clinical manifestations, treatment, and prognosis of the disease.
Objective
To study the association between genotype and drug-resistant HBV mutations in 620 Chinese patients with chronic HBV infection.
Methods
HBV DNA levels were determined using real-time quantitative PCR in plasma samples. Microarrays were performed for the simultaneous detection of HBV genotypes (HBV/B, C, and D) and drug-resistance-related hotspot mutations. A portion of the samples analyzed using microarrays was selected randomly and the data were confirmed using direct DNA sequencing.
Results
Most samples were genotype C (471/620; 76.0%), followed by genotype B (149/620; 24.0%). Among the 620 patient samples, 17 (2.7%) had nucleotide analogs (NA) resistance-related mutations. Of these, nine and eight patients carried lamivudine (LAM)-/telbivudine (LdT)-resistance mutations (rtL180M, rtM204I/V) and adefovir (ADV)-resistance mutations (rtA181T/V, rtN236T), respectively. No patients had both lamivudine (LAM)- and either adefovir (ADV) or entecavir (ETV) resistance mutations. Additionally, out of the 620 patient samples, 64.0% (397/620) were also detected with the precore stop-codon mutation (G1896A) by microarray assay.
Conclusion
The results of the current study revealed that the prevalence of nucleotide analogs (NA)-resistance in Chinese hospitalized HBV-positive patients was so low that intensive nucleotide analogs (NA)-resistance testing before nucleotide analog (NA) treatment might not be required. In addition, the present study suggests that chronic HBV patients with genotype C were infected with fitter viruses and had an increased prevalence of nucleotide analogs (NA)-resistance mutations compared to genotype B virus.
Keywords: Chronic HBV, Microarray, Genotyping, Drug-resistance mutations
Introduction
Hepatitis B virus (HBV) infection is a major global health problem that affects more than 240 million people worldwide, causing more than 780,000 deaths annually.1 Therefore, it is becoming increasingly important to take effective measures to control the incidence of HBV in China.
The HBV genotype has formed over a long period, during which point mutations have accumulated due to the asymmetry of HBV reverse transcription and its lack of proofreading enzymes. The HBV mutation rate is high, and the HBV genotype reflects the natural accumulation of mutations as a result of virus evolution. HBV can be divided into several genotypes according to sequence heterogeneity greater than 8%. There are currently eight HBV genotypes (A–H) that have a distinctive geographical distribution and ethnic associations.2 For example, genotypes B and C are highly prevalent in Southeast Asia including China, genotype A is frequent in northwest Europe, and genotype D is common in the Mediterranean region and Central Asia.3 A previous retrospective study revealed that HBV genotype B was associated with earlier HBeAg seroconversion than was genotype C.4 When receiving interferon (IFN) therapy, patients with HBV genotype B have a higher rate of IFN-induced HBeAg clearance compared with those with genotype C.5
In addition to IFN, nucleotide analogs (NAs) are also currently approved for the treatment of HBV infection. NAs inhibit HBV replication and reduce the damage to hepatocytes by competing for incorporation into the viral DNA strands.6 Four NAs have been licensed for clinical use in China: lamivudine (LAM), adefovir (ADV), entecavir (ETV), and telbivudine (LdT). However, long-duration NA treatment increases the prevalence of resistant HBV and leads to treatment failure. For example, LAM, the first widely used antiviral NA, results in a cumulative incidence of resistance as high as 70% after five years of treatment.7 The resistance-related mutations are usually located in the reverse transcriptase region of the HBV polymerase gene.8 The classical LAM-/LdT-resistance mutations are rtM204I and rtM204V.9, 10 Of these, rtL180M is a compensatory mutation that often coexists with rtM204I/V to restore viral replication efficacy.11 In addition, rtN236T and rtA181V are well-recognized ADV-resistance mutations.12 The most prevalent mutation in the precore region (G1896A) creates a TAG stop codon that abolishes the synthesis of HBeAg; thus, the dominance of this mutation could easily account for HBeAg seronegativity.13
The aim of the current study was to determine the HBV genotypes and drug resistance-related mutations in 620 clinical samples simultaneously using microarrays to better understand the correlation between HBV genotypes and drug-resistant mutations.
Materials and methods
Patient samples
A total of 620 patients consisting of 286 males and 334 females with chronic HBV infection were enrolled in the study in Beijing Ditan Hospital. The ages of these patients ranged from one month to 80 years, with a mean of 33.7 ± 12.2 years. Among the 620 patients, 271 received NA-based therapy, 35 received IFN-based therapy, eight received the combination therapy of NA and IFN, and the other 306 were NA/IFN naïve. Informed consent was obtained from adult patients or the guardians of minor patients at the time of whole blood collection for the purpose of HBV testing. The plasma samples obtained from whole blood were used to determine the HBV DNA levels using real-time quantitative PCR. In addition, serum samples were used to determine the HBV genotypes and drug resistance-related mutations.
Quantification of HBV DNA
HBV DNA was quantified using a Cobas AmpliPrep/Cobas Taqman (CAP/CTM) platform (Roche Molecular Systems, Pleasanton, CA, USA). HBV DNA was extracted from 1-mL plasma using a Cobas AmpliPrep automated extractor according to the manufacturer's instructions. The Cobas Taqman 48 Analyzer was used for automated real-time PCR amplification and the detection of PCR products according to the manufacturer's instructions. HBV DNA levels were expressed as IU/mL.
HBV DNA genotyping and drug-resistance-related mutation analysis
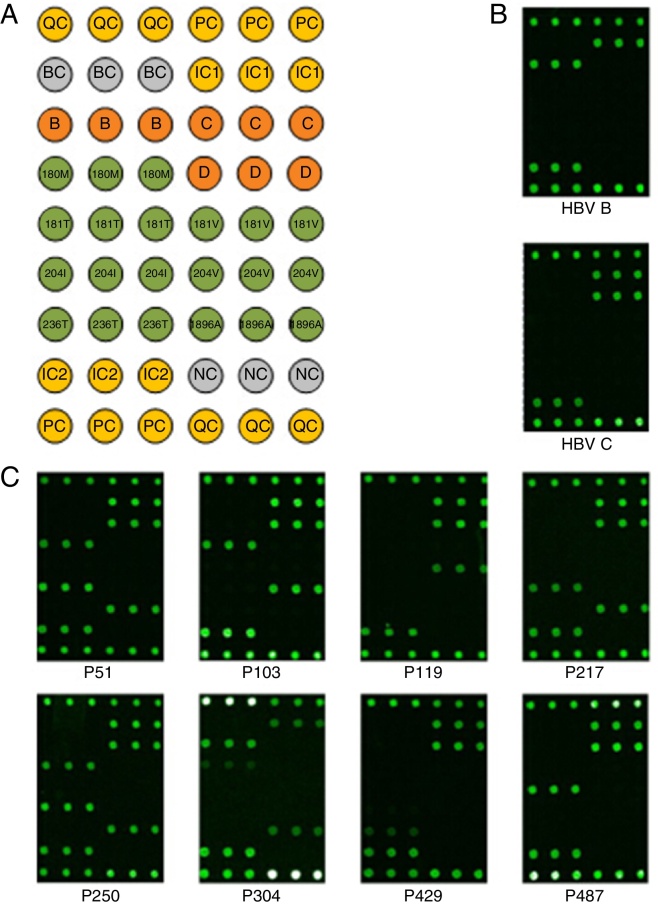
HBV DNA was extracted from 200-μL serum samples using a Magbind CFDNA kit (KWBiotech, Beijing, China). Multiplex PCR, microarray hybridization, and microarray scanning were conducted as recommended by the manufacturer of the microarray kit (CapitalBio Corporation, Beijing, China) for the simultaneous detection of the HBV genotypes (HBV/B, C, and D) and drug-resistance-related hotspot mutations (rtL180M, rtA181T/V, rtM204I/V, rtN236T in the HBV reverse transcriptase region, and G1896A in the HBV precore region). The recommended genotype and mutation-related allele-specific primers were used for multiplex PCR. After amplification, the reaction mixture was hybridized with probes immobilized on the microarray chip at 50 °C for 60 min. Subsequently, the microarray chip was washed in washing buffer (0.1% SDS and 0.3× SSC) at 42 °C for 2 min. The chips were scanned and imaged using a microarray Scanner LuxScan™10K-B (Capitalbio). Assay results were determined based on the fluorescent hybridization signal and distribution of the microarray probes. The microarray probes corresponding to HBV genotypes and drug-resistance-related mutations are shown in Fig. 1A.
Fig. 1.
Microarray-based HBV genotyping and drug resistance-related mutations. (A) Layout of the microarray used in the current study. All probes, with the exception of QC and PC, were spotted in three replicates; QC and PC were spotted in six replicates. QC and BC were the positive and negative controls for array production, respectively. PC and NC were the positive and negative controls for hybridization, and IC1 and IC2 were the internal controls for HBV amplification, respectively. B, C, and D represent HBV genotypes B, C, and D, respectively. 180M, 181T/V, 204I/V, and 236T represent the rtL180M, rtA181T/V, rtM204I/V, and rtN236T, respectively, mutations in the HBV reverse transcriptase region. 1896A denotes the G1896A mutation in the HBV precore region. (B) Hybridization results of the classical HBV samples with genotypes B and C. (C) Hybridization results of eight classical NA-resistant samples.
HBV DNA sequencing
The results of HBV DNA genotyping and drug-resistance-related mutations obtained from the microarrays for a random selection of samples were further confirmed using direct DNA sequencing with the BigDye Terminator v3.1 Cycle Sequencing kit and an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA). The primer sequences and PCR amplification conditions used for direct DNA sequencing have been described elsewhere.14, 15, 16
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software package. All values in the text and figures are expressed as the mean ± standard deviation (SD) of these observations.
Results
HBV DNA quantification
The HBV DNA levels in the 620 enrolled samples ranged from 1.00E5 to 1.50E9 IU/mL, with a mean of 9.89E6 IU/mL (see Supplementary Table S1), as determined by real-time quantitative PCR. The results also confirmed that all studied clinical samples were HBV-positive.
Supplementary Table S1 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bjid.2015.03.012.
Sample information and microarray-based assay results.
HBV DNA genotypes
Two HBV genotypes (B and C) were detected among the 620 HBV DNA positive samples using the Capitalbio microarray assay, as shown in Supplementary Table S1. The microarray hybridization results for the HBV genotypes B and C are shown in Fig. 1B. Of these, 44 samples with genotype B and 151 samples with genotype C were selected randomly and the results were validated using direct DNA sequencing. As shown in Table 1, most samples were identified as genotype C, 471/620 (76.0%), followed by genotype B, 149/620 (24.0%).
Table 1.
HBV genotyping results of the 620 patient samples.
| Gender | HBV/B (%) | HBV/C (%) | Total |
|---|---|---|---|
| Male | 68 (23.8%) | 218 (76.2%) | 286 |
| Female | 81 (24.3%) | 253 (75.7%) | 334 |
| Total | 149 (24.0%) | 471 (76.0%) | 620 |
Drug resistance-related mutations
By microarray assay, NA resistance-related mutational patterns of the 620 patient samples were also directly acquired from hybridization results, as shown in Supplementary Table S1. The mutation patterns of 211 patient samples were then selected and further confirmed using direct DNA sequencing. Among the 620 patient samples, 17 (17/620; 2.7%) carried NA resistance-related mutations, as showed in Table 2 and Fig. 1C. The mean age of these 17 patients was 47.7 ± 12.5 years, which was higher than that of the 620 patients (33.7 ± 12.2). Most of the 17 patient samples belonged to genotype C (14/17, 82.4%), and the remaining to genotype B (3/17, 17.6%). Among the 17 patients, nine carried LAM-/LdT-resistance mutations (rtL180M, rtM204I/V), and eight had ADV-resistance mutations (rtA181T/V, rtN236T). The most common mutations in the LAM-resistant patients were rtM204I and rtM204V, which were found in 35.3% (6/17) of the 17 NA-resistant patients. The compensatory mutation rtL180M was simultaneously detected with rtM204I/V in 17.6% (3/17). The ADV-resistance mutations rtA181V and rtN236T were found in 29.4% (5/17) of these patients. No patient samples with LAM and ADV or ETV-resistance mutations were detected. Among the 17 patients, 4 carried NA-resistance mutations but were NA/IFN-naïve. Ten carried NA-resistance mutations and the NA/IFN-based therapeutic strategy adopted previously was appropriate and effective. The remaining three patients also carried NA-resistance mutations but the previous NA/IFN therapy was not effective and led to treatment failure and was altered. More specifically, patients P119 and P287 both carried rtA181V but received ADV/IFN, P217 carried rtM204I but received LdT. In addition, the microarrays revealed that 397 (64.0%) of the 620 patient samples also carried the precore stop-codon mutation (G1896A), and this was confirmed in 197 samples using direct DNA sequencing.
Table 2.
Assay results of the 17 NA-resistance samples.
| Sample ID | Gender | Age | NA-resistance | Genotype | G1896A | HBV treatment |
|---|---|---|---|---|---|---|
| P51 | Male | 45 | rtL180M, rtM204I | HBV/C | A | Naïve |
| P103 | Male | 52 | rtL180M, rtM204V | HBV/C | G | LAM + ADV |
| P119 | Male | 48 | rtA181V | HBV/C | G | ADV |
| P160 | Male | 43 | rtN236T | HBV/B | A | Naïve |
| P163 | Male | 48 | rtL180M | HBV/C | A | Naïve |
| P208 | Male | 50 | rtM204I | HBV/C | A | ETV |
| P217 | Female | 57 | rtM204I | HBV/C | A | LdT |
| P227 | Female | 52 | rtM204I | HBV/C | A | ETV |
| P250 | Female | 35 | rtL180M, rtM204I | HBV/C | A | LAM + ADV |
| P279 | Female | 33 | rtA181T | HBV/C | G | LdT + ADV |
| P287 | Female | 32 | rtA181V | HBV/C | A | IFN |
| P304 | Male | 74 | rtL180M | HBV/B | A | ETV |
| P393 | Male | 74 | rtL180M | HBV/B | A | ETV |
| P429 | Female | 53 | rtN236T | HBV/C | G | ETV |
| P487 | Female | 37 | rtA181T | HBV/C | G | ETV |
| P564 | Male | 45 | rtA181T | HBV/C | G | ETV + IFN |
| P600 | Male | 34 | rtN236T | HBV/C | A | Naïve |
Discussion
Most of the samples analyzed in the current study belonged to genotype C (471/620; 76.0%) followed by genotype B (149/620; 24.0%). Moreover, there were no marked differences in the distribution of genotypes B and C between male and female patients. These data are consistent with a previous report from northern China, where 78% of the studied HBsAg carriers were infected with genotype C, and 22% carried genotype B.14 However, genotype D was not observed among the patient samples studied. Interestingly, previous reports also identified HBV genotype D in only northwestern China.17, 18
In the current study, 17 patients (17/620; 2.7%) carried NA-resistance-related mutations. These data are consistent with a previous report that preexisting NA-resistance-related mutations were detected in 2.01% of NA-naïve Chinese HBV-infected patients.19 These results suggest that the incidence of NA-resistance in Chinese hospitalized HBV-positive patients is so low that intensive testing for NA-resistance before treatment might not be required, consistent with a previous suggestion.19 In most cases in the current study, NA-resistance mutations were derived from either naturally occurring viral mutations or were transmitted from drug-resistant patients. However, the 17 patients had a mean age of 47.7 ± 12.5 years, which was higher than the mean age of the 620 patients (33.7 ± 12.2). This suggests that the 17 NA-resistance patients had an increased likelihood of being treated using antiviral therapy; therefore, NA resistance testing might be more necessary for HBV-positive elderly patients.
Most of the 17 patients samples belong to genotype C (14/17, 82.4%), and the remaining to genotype B (3/17, 17.6%). The proportion of genotype C in the 17 NA-resistance patients was considerably higher than the above-mentioned data (76.0%) in the 620 HBV-positive patients. These data suggest that HBV patients with genotype C were infected with fitter viruses, which facilitated acquisition of NA-resistance mutations, compared with those with genotype B.
The mutations rtM204I and rtM204V were the most frequent mutations in the LAM-resistant patients, being detected in 35.3% (6/17). The compensatory mutation rtL180M was detected simultaneously with rtM204I/V in 17.6% (3/17) of the patients. The ADV-resistance mutations rtA181V and rtN236T were found in 29.4% (5/17) of the patients. LAM and ADV dual resistance was not detected in any of the 17 patients. Long-term use of NA might have led to multidrug resistance.20 Therefore, NA resistance testing becomes increasingly necessary. For patients P119, P217 and P287 of the 17 patients, the results of NA resistance testing implied in advance that the used NA/IFN therapeutic strategies would lead to treatment failure and had to be altered. It showed that NA resistance testing may be necessary when deciding for the NA/IFN therapeutic strategy.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We are grateful to Guotong Yin and Yunlong Hao (Capitalbio) for their excellent technical assistance. We are also grateful to Dr. Yingying Liu (Capitalbio) for helpful discussions and support.
References
- 1.World Health Organization . 2012. Hepatitis B fact sheets. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/ [updated July 2014] [Google Scholar]
- 2.Bartholomeusz A., Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol. 2004;14:3–16. doi: 10.1002/rmv.400. [DOI] [PubMed] [Google Scholar]
- 3.Miyakawa Y., Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 4.Chu C.J., Hussain M., Lok A.S. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756–1762. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- 5.Wai C.T., Chu C.J., Hussain M., et al. HBV genotype B is associated with better response to interferon therapy in HBeAg (+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425–1430. doi: 10.1053/jhep.2002.37139. [DOI] [PubMed] [Google Scholar]
- 6.Nebbia G., Peppa D., Maini M.K. Hepatitis B infection: current concepts and future challenges. Q J Med. 2012;105:109–113. doi: 10.1093/qjmed/hcr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lok A.S., Zoulim F., Locarnini S., et al. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 8.Wang F., Wang H., Shen H., et al. Evolution of hepatitis B virus polymerase mutations in a patient with HBeAg-positive chronic hepatitis B virus treated with sequential monotherapy and add-on nucleoside/nucleotide analogues. Clin Ther. 2009;31:360–366. doi: 10.1016/j.clinthera.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Lok A.S., McMahon B.J. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 10.Tillmann H.L., McHutchison J.G. Telbivudine versus lamivudine in patients with chronic hepatitis B. N Engl J Med. 2008;358:1517–1518. doi: 10.1056/NEJMc080082. [DOI] [PubMed] [Google Scholar]
- 11.Yuen L.K., Locarnini S.A. Genetic variability of hepatitis B virus and response to antiviral treatments: searching for a bigger picture. J Hepatol. 2009;50:445–448. doi: 10.1016/j.jhep.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Borroto-Esoda K., Miller M.D., Arterburn S. Pooled analysis of amino acid changes in the HBV polymerase in patients from four major adefovir dipivoxil clinical trials. J Hepatol. 2007;47:492–498. doi: 10.1016/j.jhep.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Nie H., Evans A.A., London W.T., et al. Quantitative dynamics of hepatitis B basal core promoter and precore mutants before and after HBeAg seroconversion. J Hepatol. 2012;56:795–802. doi: 10.1016/j.jhep.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J., Zhou B., Tanaka Y., et al. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core ad their clinical implications. J Clin Virol. 2007;39:87–93. doi: 10.1016/j.jcv.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Han Y., Huang L.H., Liu C.M., et al. Characterization of hepatitis B virus reverse transcriptase sequences in Chinese treatment naive patients. J Gastroenterol Hepatol. 2009;24:1417–1423. doi: 10.1111/j.1440-1746.2009.05864.x. [DOI] [PubMed] [Google Scholar]
- 16.Zoulim F., Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Huang Y., Wen S., et al. Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res. 2007;37:S36–S41. doi: 10.1111/j.1872-034X.2007.00102.x. [DOI] [PubMed] [Google Scholar]
- 18.Zeng G., Wang Z., Wen S., et al. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J Viral Hepat. 2005;12:609–617. doi: 10.1111/j.1365-2893.2005.00657.x. [DOI] [PubMed] [Google Scholar]
- 19.Li X., Liu Y., Zhao P., et al. Investigation into drug-resistant mutations of HBV from 845 nucleoside analogue-naïve Chinese patients with chronic HBV infection. Antivir Ther. 2015;20(2):141–147. doi: 10.3851/IMP2813. [DOI] [PubMed] [Google Scholar]
- 20.Song Z.L., Cui Y.J., Zheng W.P., et al. Diagnostic and therapeutic progress of multi-drug resistance with anti-HBV nucleotide analogues. World J Gastroenterol. 2012;18:7149–7157. doi: 10.3748/wjg.v18.i48.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information and microarray-based assay results.