Abstract
Neisseria gonorrhoeae is naturally competent for DNA transformation. Under most conditions encountered in vivo, gonococci express one or more opacity (Opa) proteins on their surfaces. Recently, it was shown that DNA preferentially binds to the surfaces of Opa-expressing organisms compared to those of isogenic Opa-negative strains, presumably due to the numerous cationic residues in the predicted surface-exposed loops of the Opa protein. This study examined whether Opa-DNA interactions actually influence DNA transformation of the gonococcus. The data show that Opa-expressing gonococci are more efficient recipients of DNA for transformation and are more susceptible to exogenous DNase I treatment at early stages during the DNA transformation process than non-Opa expressors. Furthermore, inhibition of the transformation process was demonstrable for Opa+ populations when either nonspecific DNA or the polyanion heparin was used. Overall, the data suggest that Opa expression, with its presumptive positive surface charge contribution, promotes DNA transformation by causing a more prolonged sequestration of donor DNA at the cell surface, which translates into more efficient transformation over time.
The members of the genus Neisseria are naturally competent for DNA transformation, and there is considerable evidence indicating that horizontal exchange of chromosomal and plasmid DNAs occurs in these bacteria in vivo (23). For most naturally transformable bacteria, an organism (e.g., Bacillus subtilis or Haemophilus influenzae) becomes competent in vitro through starvation and/or other manipulations of the culture conditions (14, 29). However, the neisseriae are a little different in that competence is constitutively expressed in vitro (5). Nonetheless, some limitations are imposed on the gonococcal transformation process, as only genus-specific DNA can be efficiently taken up into a DNase I-resistant state due to a strong preference for a specific neisserial DNA uptake sequence within the donor DNA (10, 15). Consequently, gonococci can be transformed in the presence of excess amounts of nonspecific DNA, whereas only small amounts of neisserial DNA are required to competitively inhibit the process (8, 16).
Studies of the mechanism of gonococcal transformation have relied heavily on the isolation of organisms with specific mutations that block some step in the transformation process. Using this approach, several gonococcal gene products that assist in the movement of donor DNA across the cell envelope have been identified. Gonococci exhibit (i) a general requirement for the presence of pili on the cell surface and Tpc protein for genetic competence (5, 13), (ii) involvement of PilT and PilC proteins in sequestering donor DNA into a DNase I-resistant state (4, 26, 38), (iii) involvement of ComL in the movement of DNA across the peptidoglycan layer (12), and (iv) involvement of ComA in transporting donor DNA across the cytoplasmic membrane (11). Furthermore, it has been shown that once the donor DNA has translocated into the cytoplasm, the RecA protein facilitates its incorporation into the host chromosome (21). Recently, the RecBCD recombination pathway was also shown to be involved in the transformation process (24). However, unlike all other organisms, gonococcal recD mutants are not hyperrecombinogenic for DNA transformation (6).
For most DNA transformation systems, uptake of donor DNA into a DNase I-resistant state is generally considered to be the first step in the process. However, prior to this uptake is the actual recruitment of donor DNA onto the bacterial cell surface, and very little is known about the bacterial surface component(s) that participates in this early step. However, when considered simplistically, it would seem that any surface component that could contribute a positive surface charge should be a candidate for an electrostatic attractant of donor DNA (a complex polyanion) to the bacterial cell surface.
Gonococci may provide an ideal model system to test this hypothesis (as well as explore the more general hypothesis that changes in cell surface charge can influence the biological properties of an organism) because, under most conditions encountered in vivo, gonococci express a unique group of proteins on their cell surfaces (the opacity [Opa] proteins) (9, 19, 20, 34) that theoretically should endow the organism with a positive cell surface charge. Colonies of cells expressing Opa protein display increased opacity when viewed under a phase-contrast microscope (30, 31). Moreover, the Opa proteins belong to a complex multigene family (3) and are one of three gonococcal surface constituents that vary in vivo (20, 34, 35). However, the unique feature of Opa expression that is pertinent to this study is that secondary structural algorithms of their predicted primary polypeptide sequences indicate that their surface-exposed segments should contain an excess of positively charged amino acids (3, 32). Consequently, if Opa expression does endow the organism with a positive cell surface charge, then Opa protein may preferentially attract complex polyanions (such as DNA) to the cell surface. This apparently is the case, as immobilized Opa proteins are able to bind radiolabelled DNA in a blot format, as well as promote DNA accretion in situ on intact bacteria (32). Furthermore, the endowment of a positive cell surface charge through Opa expression is further indicated by the results of physical studies that examined the relative electrophoretic mobility of Opa-expressing gonococci (32).
In this study, I examined whether Opa-DNA interactions actually translate into an enhanced transformation phenotype for the gonococcus. The data show that Opa expression invariably correlates with an increased transformation efficiency. Moreover, by growing gonococci under conditions in which binding of nonspecific polyanions to the cell surface is limited, then the DNA transformability of the organism is significantly enhanced, with transformation now being inhibitable by nonspecific DNA and/or the complex polyanion heparin. Overall, the data suggest that the elaboration of a specific cell surface charge may indeed influence a basic biological function of the gonococcus.
MATERIALS AND METHODS
Strains.
Three isogenic series of Neisseria gonorrhoeae MS11 that expressed various Opa proteins in different pilus antigenic backgrounds were used (the different pilus backgrounds reflect pili of differing antigenic composition due to the expression of different pilin polypeptides from the following N. gonorrhoeae MS11 pilE alleles: pilE7302 [2], pilE3 [18], and pilE6 [18]). All presented a LosB phenotype as defined by their reactivity to a LosB-specific monoclonal antibody (7). The majority of the experiments reported herein utilized bacteria expressing OpaA in a pilE7302 background due to the distinctive morphologic features of the colonies of such organisms, which allowed easier monitoring, by phase-contrast microscopy, for maintenance of the appropriate phenotype. The various isogenic Opa variants were derived by plating a single colony on solid medium and identifying opaque variants by phase-contrast microscopy. Each Opa variant was passaged once prior to typing against known standards by immunoblotting with an Opa-specific monoclonal antibody (4B12) (data not shown) (33, 34).
Growth conditions.
N. gonorrhoeae strains were propagated on one of the following solid media: (i) PO42−-buffered (pH 8) agar-based clear gonococcal typing medium (GTM) (0.375% Trypticase peptone [BBL], 0.75% Thiotone E [VWR Scientific], 23 mM K2HPO4, 7 mM KH2PO4, 0.5% NaCl, 0.1% soluble starch [Baker], 1% Bacto Agar, and 1% IsoVitaleX [BBL]) (31), (ii) a PO42−-buffered (pH 8) agarose-based medium (with the same composition as GTM except for the addition of 0.8% agarose [Seakem; Gold lot no. 132893] instead of agar [37]); or (iii) a HEPES-buffered (pH 8) agarose-based medium (with the same composition as GTM except that the phosphate salts were replaced by 0.2% HEPES, Na+ salt [Calbiochem, La Jolla, Calif.] and 0.5% HEPES acid [Calbiochem]). Gonococci were passaged daily on solid medium and incubated at 37°C in a 5% CO2 atmosphere. Liquid cultures utilized the same media as described above except for the omission of agar or agarose. When gonococci were grown in HEPES-buffered liquid medium, IsoVitaleX was also omitted.
DNA transformation.
High-cell-density transformations were performed essentially as described by Seifert et al. (27). Briefly, piliated gonococci were swabbed from plates and resuspended to an approximate initial density of 108 cells per ml in liquid GTM medium. DNA was added to the cell suspension at a concentration of 1 μg/ml, and the suspension was incubated for 20 min at 37°C prior to being diluted 1:10 with prewarmed medium; the cells were then incubated for a further 5 h prior to being plated on selective medium. Low-cell-density DNA transformations were performed as follows. Single colonies from an 18-h culture were lifted from their respective plates with sterile pieces of Whatman no. 3 filter paper, and the cells were resuspended in 1-ml volumes of liquid medium containing 10 mM MgCl2 (average density, approximately 1 × 106 to 5 × 106 cells/ml). Unless otherwise indicated, cells were then incubated for 5 h at 37°C with MS11 chromosomal DNA (1 μg/ml) carrying either a pilE::cat drug resistance marker or a point mutation that conferred resistance to rifampin prior to being plated on selective medium (agar-based GTM containing chloramphenicol [10 μg/ml] or rifampin [10 μg/ml]). Conducting the experiments under low-cell-density conditions allowed (i) the number of colony-forming units to increase over time to various degrees depending on which Opa protein was being expressed and (ii) maintenance of Opa expression throughout the entire experiment. Therefore, when comparisons are made between Opa+ and Opa− culture transformation efficiencies, only the results of experiments performed with equivalent cell densities are compared. For mixing experiments, both cell types were mixed prior to the addition of the donor DNA.
For transformations performed in the presence of competing DNA or heparin, herring sperm DNA or a sodium salt of heparin (Sigma, St. Louis, Mo.) was present in the transforming medium at all times. For transformations performed in the presence of DNase I (molecular biology grade; United States Biochemicals), 1 U of enzyme was added at various times after the addition of the donor DNA. Under these conditions, the presence of DNase I in the culture did not impede growth (data not shown).
The data are presented as means of values obtained from experiments performed on the same day. There were day-to-day variations in the absolute values obtained; however, the trends as reported were consistent over many reiterations of the experiment. Total colony counts were performed at the beginning as well as at the end of each experiment. The initial, final, and transformant (i.e., those organisms that grew on selective plates) populations were also scored microscopically for the percentage of Opa+ colonies in the culture.
Because gonococcal populations do not exhibit 100% competence (5), the degrees of competence exhibited by the Opa+ and Opa− cultures were determined by using the method of Goodgal and Herriot (14). Briefly, two unlinked markers (pilE::CAT and rif) were used, and the actual frequency of double transformation to chloramphenicol and rifampin resistance was compared to the expected frequency (i.e., the product of each individual frequency) by using the formula f = N1 × N2/ND × V, where f is the fraction of bacteria that are competent; N1, N2, and ND are the number of transformants transformed to chloramphenicol resistance, rifampin resistance, and both, respectively; and V is the number of bacteria.
The percent error factor presented in Table 1, which assesses whether clumping of bacteria following plating on selective and nonselective media had a significant impact on the procurement of the data, was calculated as follows. In any given population of bacteria, the overall transformation frequency is the sum of the individual subpopulations' transformation frequencies (for our purposes, the transformation frequencies of the Opa+ and Opa− populations). To obtain each subpopulation's transformation frequency, the final and transformant populations were scored microscopically to determine the percentages of Opa+ and Opa− colonies (on the basis of opacity differences) that were present, which then allows each subpopulation's transformation frequency to be calculated. Therefore, comparison of the overall transformation frequency (which is obtained without regard to phenotype) to the value obtained by summing the individual subpopulations' frequencies (which requires two microscopic evaluations; if clumping is a factor, then this value should be noticeably different from the overall frequency determined without regard to phenotype) gives an indication of the degree of error between the two computations. Strong concordance between the two values would indicate little effect of clumping with respect to plating in these experiments.
TABLE 1.
Transformation of N. gonorrhoeae expressing different Opa proteins
| Opa phenotype | Oa | Transformation frequency ([no. of transformants/μg of DNA/CFU] × 103)
|
% Errord | ||||
|---|---|---|---|---|---|---|---|
| Overallb | Ma | Subpopulationc
|
Pa | ||||
| Opa+ | Opa− | ||||||
| Opa− | NAe | 0.77 | NA | NA | NA | NA | NA |
| OpaA | 0.88 | 2.83 | 0.18 | 7.45 | 1.88 | 0.47 | 2 |
| OpaF | 0.93 | 3.35 | 0.16 | 7.25 | 2.61 | 0.35 | <1 |
| OpaH | 0.54 | 1.86 | 0.08 | 2.73 | 1.73 | 0.13 | 3 |
| OpaI | 0.79 | 2.26 | 0.16 | 3.65 | 1.88 | 0.27 | <1 |
O, M, and P values refer to the fraction of Opa+ organisms in the initial, final, and transformant populations, respectively.
Overall frequency represents the mean value (n = 5) determined without phenotypic evaluation of the culture.
Subpopulation frequencies were obtained for each Opa+ and Opa− subpopulation within a culture as described in Materials and Methods.
% Error was calculated as indicated in Materials and Methods.
NA, not applicable.
Statistical analyses utilized Student's t test.
Preparation of donor DNA.
Donor DNA was prepared as described by Ausubel et al. (1), using 10% CTAB (cetyltrimethylammonium bromide; United States Biochemical Corporation). Precipitated DNA was washed in 70% ethanol, dried, and resuspended in Tris-EDTA buffer (pH 8).
RESULTS
Opa protein expression leads to elevated DNA transformation rates.
Swanson's data (32) strongly suggest that Opa expression promotes the binding of exogenous DNA to the cell surface. Therefore, I examined whether these presumptive Opa-DNA interactions actually influenced the gonococcal transformation process. An arbitrary isogenic series of Opa+ and Opa− bacteria was established in an MS11 pilE7302 genetic background, and the transformation rates of variants that either expressed no Opa or expressed OpaA, OpaF, OpaH, or OpaI were compared. High-cell-density transformations (27) were performed in PO42−-buffered medium (pH 8), and, irrespective of which Opa protein was being expressed, transformation frequencies for the Opa+ cultures were consistently elevated compared to those of the isogenic Opa− control (Table 1). When genetic competence of the population was measured (by the method outlined by Goodgal and Herriot [14]), Opa expression also correlated with increased competence (Opa-negative cultures exhibited 10% competence, OpaA cultures demonstrated 42% competence, OpaH cultures showed 20% competence, and OpaI cultures demonstrated 30% competence).
During each transformation experiment, each population (i.e., the initial, final, and transformant populations) was examined by phase-contrast microscopy to assess the phenotypic quality of the bacteria (i.e., whether Opa expression was maintained throughout the entire experiment). The microscopic analysis revealed considerable Opa heterogeneity in all populations (Table 1). Consequently, it was concluded that the overall transformation frequency was actually the sum of two individual subpopulation frequencies (i.e., one subpopulation being Opa+ and the other being Opa−). When transformation rates for each subpopulation were independently assessed (by microscopically determining the percentage of Opa-negative bacteria within the culture on the basis of opacity), it became apparent that the subpopulations exhibited different transformation rates (Table 1), with the Opa+ subpopulations consistently demonstrating higher relative transformation frequencies than their corresponding Opa− subpopulations.
In addition to the observed Opa heterogeneity, it also became apparent that considerable cell lethality was associated with transformations performed at high cell densities. This was particularly pronounced with Opa+ cultures, for which cell lethality ranged from 10 to 95% depending on the initial Opa phenotype (data not shown). However, cell lethality was not specifically associated with Opa expression, as Opa-negative cultures also showed comparable declines in viability, thus confirming a previous report that described similar culture lethality (5). Moreover, cell lethality and Opa heterogeneity occurred irrespective of (i) which Opa was being expressed, (ii) the presence or absence of pili on the cells, (iii) medium composition, (iv) pH, and (v) the presence or absence of different cations, and they occurred with cells carrying either the recA or dud-1 mutation (data not shown).
Culture conditions influence transformation efficiency.
The considerable Opa protein heterogeneity and the precipitous declines in cell viability which were observed when traditional transformation protocols were employed necessitated the development of new regimens in which lethality and clumping of bacteria were kept to a minimum. Studies of colloidal suspensions have long dealt with problems associated with clumping of particles (17). Therefore, by equating bacteria in solution to simple particles in solution, the basic principles of colloidal chemistry were applied in an effort to overcome the problems associated with bacterial cell clumping (17). In a similar vein, it was also reasoned that some of the observed effects regarding Opa-expressing cultures might reflect the presence or absence of various polyanions in the solid medium (e.g., polysulfated anions, which are a component of agar) which, when bound at the cell surface, masked or modified some cell surface component(s) being expressed. Consequently, culture conditions were optimized with respect to (i) the cell density at which the transformation reactions were performed (this modification eliminated density-dependent cell lethality over time [cultures now grew] and tempered the effects of clumping [when starting cultures were initially assessed by Gram staining] [data not shown]) and (ii) the composition of the medium used for the propagation of bacteria prior to and during the transformation assay. Scanning electron microscopy (data not shown) indicated that the cellular integrity of OpaA-expressing bacteria was best maintained when bacteria were propagated on a HEPES-buffered, agarose-based medium prior to undergoing transformation in HEPES-buffered liquid medium in the absence of any supplement; optimum cell densities were in the range of ≤106 CFU per ml (which necessitated the performance of single-colony transformations). Overall, these newly defined conditions allowed (i) OpaA-expressing bacteria to remain viable (immediate [i.e., <5 min] density-dependent cell lysis was reduced to a minimum [<5%]), (ii) an increase in CFU over the course of the experiment, (iii) direct comparison of isogenic Opa+ and Opa− colonies lifted from a single streak, and (iv) maintenance of the Opa and pilus phenotypes in all relevant populations (i.e., initial, final, and transformant).
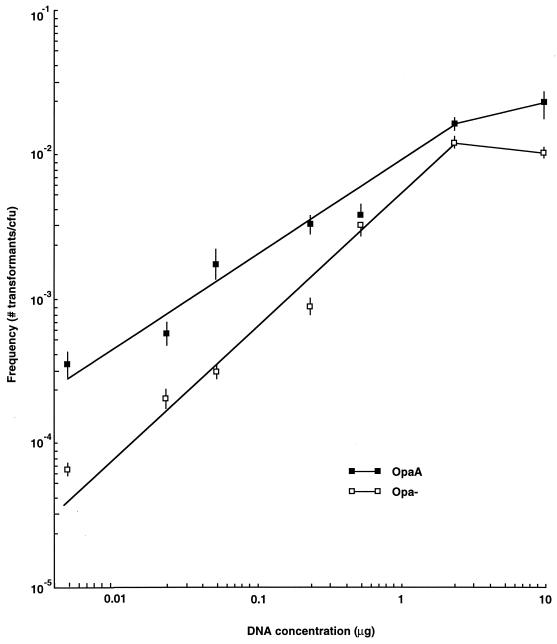
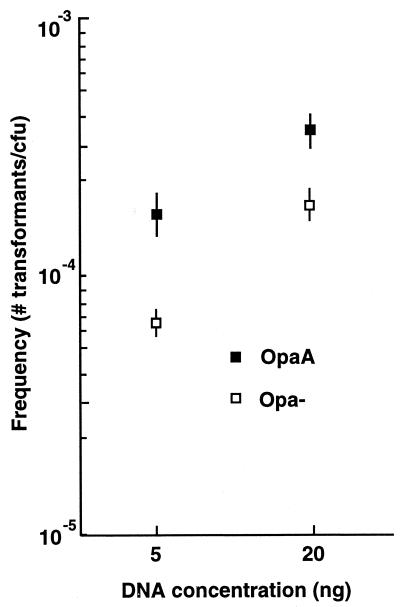
Using these newly defined conditions, the transformation efficiencies of OpaA and Opa− isogenic pairs were then compared over a range of donor DNA concentrations, and it was found that OpaA-expressing bacteria were more efficient at transformation than their Opa− counterparts at all donor DNA concentrations tested (Fig. 1), with saturation of the transformation process for OpaA-expressing bacteria occurring when the donor DNA concentration exceeded 5 μg/ml. Moreover, the data indicated that OpaA's effect on transformation was more pronounced at low donor DNA concentrations (between 5 and 100 ng of donor DNA), which was likewise manifest during mixing experiments using low donor DNA concentrations, when OpaA and Opa− bacteria had to actively compete for uptake of the genetically marked donor DNA (at 5 ng of donor DNA, P < 0.05; at 20 ng of donor DNA, P < 0.02 [n = 6]) (Fig. 2).
FIG. 1.
Transformation efficiency versus donor DNA concentration. OpaA and Opa-negative bacteria were transformed with various amounts of donor DNA. Each data point represents the mean transformation efficiency value at that DNA concentration ± standard error (n = 6). Frequencies are presented as the number of drug-resistant transformants per CFU.
FIG. 2.
Transformation of mixed OpaA and Opa-negative populations at low DNA concentrations. OpaA and Opa− bacteria were mixed prior to the addition of the donor DNA, with the percentage of Opa+ and Opa− bacteria in each population being scored on the basis of colony opacity following microscopic evaluation. The data presented are mean transformation efficiencies ± standard errors (n = 6).
Opa expression increases the susceptibility of in situ-bound DNA to exogenous DNase I treatment.
If donor DNA does preferentially bind to the cell surface of Opa-expressing bacteria, then such in situ-bound DNA should be susceptible to DNase I hydrolysis during a transformation reaction and Opa-expressing bacteria should exhibit a higher level of sensitivity than Opa− bacteria. To test this possibility, single-colony transformations were performed with isogenic, piliated, PO42−-buffered-agar-grown OpaA and Opa− bacteria, with 1 U of DNase I being added 20 min following the addition of the genetically marked donor DNA (DNase I was present throughout the remainder of the experiment and was not detrimental to bacterial growth). The results (Table 2) showed that OpaA-expressing bacteria underwent transformation at significantly higher rates than isogenic Opa-negative cultures (in the absence of DNase; P < 0.001, n = 15) and that OpaA-expressing gonococci were more susceptible to the in vitro effects of DNase I hydrolysis than the Opa-negative control during transformation (Table 2). Therefore, these data genetically confirm previous physical observations (e.g., the binding of nonspecific DNA to Opa proteins immobilized on nitrocellulose and differences in electrophoretic migration of whole cells in an applied electric field [32]) that Opa proteins expressed in situ can indeed bind exogenous DNA.
TABLE 2.
Effect of medium composition and DNase I treatment on transformation
| Opa phenotype | DNase I treatmenta | Values for organisms grown on:
|
|||
|---|---|---|---|---|---|
| Agar-based medium
|
Agarose-based medium
|
||||
| Transformation frequencyb | Ratioc | Transformation frequencyb | Ratioc | ||
| Opa− | − | 5.19 ± 0.82 | 0.063 | 9.52 ± 1.97 | 0.047 |
| + | 0.33 ± 0.09 | 0.45 ± 0.08 | |||
| OpaA | − | 54.9 ± 1.54 | 0.018 | 161.0 ± 2.87 | 0.045 |
| + | 0.99 ± 0.28 | 7.28 ± 1.28 | |||
± 1 U of DNase I.
Calculated with the following formula: (no. of transformants/μg of DNA/CFU) × 103. Values are means ± standard errors for data from 15 experiments.
Mean frequency in the presence of DNase I/mean frequency in the absence of DNase I.
However, the growth medium's effect on transformation was noticeable in this aspect of the study, because when bacteria were grown on a PO42−-buffered, agarose-based medium prior to transformation in broth medium, both Opa-negative and Opa-expressing bacteria showed equal susceptibilities to DNase I hydrolysis (Table 2). Furthermore, accompanying this change in DNase I susceptibility was an increase in transformation efficiency for both Opa-negative and OpaA cultures (again, in the absence of DNase, OpaA-expressing bacteria underwent transformation at significantly higher rates than Opa-negative cultures; P < 0.001, n = 15). Yet, despite this common upward trend in transformation efficiency for both types of bacteria, the effect of growth on agarose-based medium on transformation still remained more pronounced for OpaA-expressing organisms than for the corresponding Opa-negative culture (i.e., a 2.93-fold relative increase compared to a 1.8-fold relative increase in transformation efficiency, respectively, in the absence of DNase I). Therefore, these observations indicate that culture conditions can have a profound effect on the transformability of the gonococcus.
Pilus antigenic composition has little effect on the Opa transformation effect.
Gonococci expressing differing pilin polypeptides (the different pilins tested were expressed from the following genes in an MS11 background: pilE7302, pilE3, and pilE6) were used to determine whether the elevated-transformation phenotype associated with Opa expression occurred independently of the antigenic composition of the pilus organelle. The data presented in Table 3 show that in most cases, transformation rates of OpaA- and OpaI-expressing bacteria were higher than those of isogenic Opa-negative control strains derived from a common bacterial streak, irrespective of which pilin was being expressed. However, from the data it is apparent that certain pilus-Opa configurations occasionally influence transformation of the gonococcus, perhaps by presenting different molecular arrays at the cell surface. For example, when gonococci expressed the pilE3 allele, coexpression of OpaA did not significantly elevate the transformation efficiency compared to that of its isogenic Opa-negative control strain (Table 3). Analysis of the predicted primary amino acid sequence of the pilE3 pilin indicates that it contains numerous cationic residues (18). Consequently, the pilE3 Opa-negative strain may have an artificially high transformation frequency that masks OpaA's effect. Consistent with this viewpoint is the finding that pilE3 Opa-negative cultures are more efficient at transformation than are bacteria expressing the pilE7302 allele (mean transformation frequencies ± standard errors, 1.34 × 10−2 ± 0.05 and 7.74 × 10−3 ± 0.27, respectively; P < 0.05, n = 20).
TABLE 3.
Effect of different pilins on transformation under low-density conditions
| Expt | Opa phenotype | Values for organism with pilin encoded by:
|
|||||
|---|---|---|---|---|---|---|---|
|
pilE7302
|
pilE6
|
pilE3
|
|||||
| Transformation frequencya | Ratio (P)b | Frequencya | Ratio (P)b | Frequencya | Ratio (P)b | ||
| 1 | Opa− | 0.99 ± 0.13 | 18.9 (<0.02) | 0.71 ± 0.13 | 4.94 (<0.01) | 1.75 ± 0.63 | 1.7 (NS) |
| OpaA | 18.8 ± 5.25 | 3.55 ± 0.67 | 2.98 ± 0.86 | ||||
| 2 | Opa− | 0.92 ± 0.23 | 4.68 (<0.05) | 0.67 ± 0.14 | 7.17 (NS) | 1.22 ± 0.17 | 4.03 (<0.02) |
| OpaI | 4.33 ± 1.06 | 4.84 ± 2.01 | 4.92 ± 0.90 | ||||
Calculated as follows: (no. of transformants/μg of DNA/CFU) × 103. Values are means ± standard errors for data from five experiments.
Mean frequency for Opa+ organisms/mean frequency for Opa− organisms. P values indicate the degree of significance between the reported Opa+ and Opa− transformation frequencies. NS, not significant at a P value of 0.05.
Preferential inhibition of transformation of OpaA-expressing gonococci with nonspecific DNA and heparin sulfate.
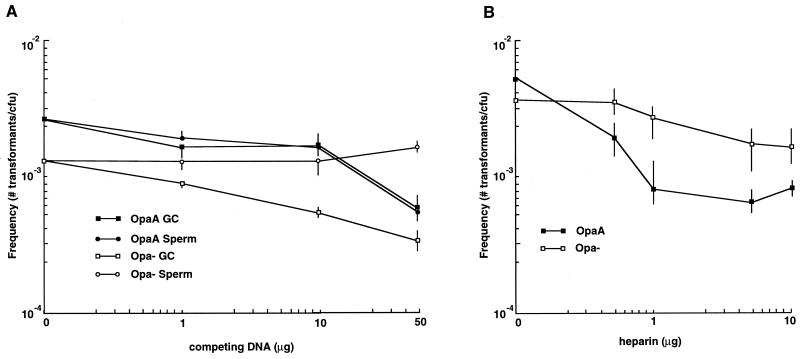
Since nonspecific DNA binds to immobilized Opa proteins in a blot format (presumably due to the excessive number of positively charged amino acids found within their primary polypeptide sequences [3, 32]), Opa-mediated effects on transformation should also be inhibitable with nonspecific polyanions. This was tested by performing transformations in the presence of increasing amounts of either nonspecific herring sperm DNA (Fig. 3A) or the polyanion heparin (Fig. 3B). In both sets of experiments, transformation of OpaA-expressing bacteria was inhibited by the competing polyanion whereas the nonspecific polyanions had less of an impact on the transformability of the Opa-negative culture. Therefore, these observations further indicate that OpaA-expressing bacteria can preferentially bind polyanions at the cell surface. However, the effects of the two polyanions were noticeably different; the presence of the nonspecific DNA appeared to simply competitively inhibit the Opa-dependent increase in transformation, whereas the presence of heparin may well have blocked some aspect of DNA uptake in Opa-expressing cells by causing the transformation efficiency of heparin-treated Opa-expressing bacteria to noticeably lag behind that of comparably treated Opa-negative cultures (compare the effects of the supplemented polyanions on the Opa transformation efficiency of OpaA-expressing bacteria relative to those of the Opa-negative culture [Fig. 3]). The basis for the latter observation is currently being investigated.
FIG. 3.
DNA transformation in the presence of competing polyanions. Bacteria (either Opa+ or Opa−) were picked from HEPES-buffered (pH 8)-agarose-based plates and were transformed with 1-μg quantities of donor DNA in the presence of various amounts of either unmarked gonococcal chromosomal (GC) or herring sperm (Sperm) DNA (A) or sodium heparin (B). The data presented are mean transformation frequencies ± standard errors (n = 6 [A] or 12 [B]).
Expressed Opa increases the transformation efficiency by sequestering donor DNA at the cell surface for extended periods of time.
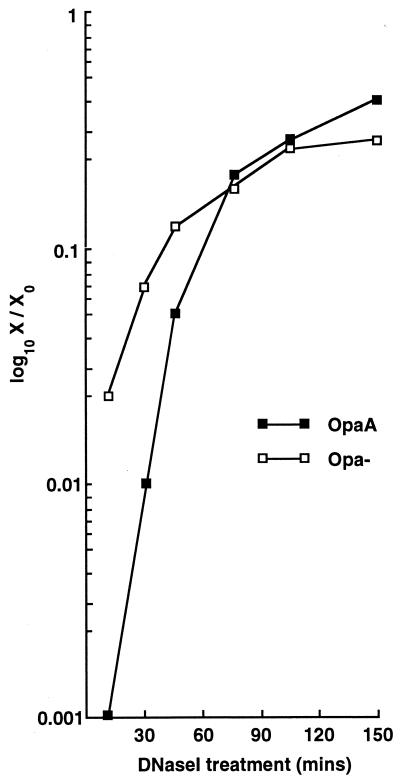
The results presented in Table 2 indicate that under low-cell-density conditions many transformants actually arise from donor DNA that makes the transition into a DNase I-resistant state only following a considerable period of incubation (>20 min) with the bacteria. To test the relative rates of donor DNA uptake into a DNase I-resistant state, a kinetic analysis was performed with HEPES-buffered (pH 8)-agarose-grown bacteria, in which 1 U of DNase I was added at various time points following addition of the donor DNA. The data presented in Fig. 4 compare the transformation frequencies that were obtained at the various time points following DNase I treatment against the frequency obtained for the non-DNase I-treated control, and they show that (i) at short incubation periods, Opa− cultures are more efficient at sequestering donor DNA in a DNase I-resistant state than the corresponding OpaA-expressing cultures; (ii) over extended incubation periods, OpaA-expressing organisms overcome the barrier of uptake into a DNase I-resistant state; and (iii) a larger proportion of OpaA transformants are derived from donor DNA that is sequestered in a DNase I-resistant state at later time points during the reaction than are derived from donor DNA that is taken up at earlier time points. Therefore, these data indicate that the basis for the effect of Opa proteins on transformation is a prolonged sequestration, at the cell surface, of donor DNA, which over time eventually makes the transition into a DNase I-resistant state.
FIG. 4.
DNase I susceptibility over time. Bacterial colonies (either Opa+ or Opa−) were picked from HEPES-buffered (pH 8)-agarose-based plates and were transformed with 1-μg quantities of donor DNA. Incubations were allowed to proceed for various periods of time prior to being terminated through the addition of 1 U of DNase I. The data were obtained by dividing the transformation frequency obtained following the addition of DNase I (X) by the transformation frequency obtained for a non-DNase I-treated sample (X0). Transformation frequencies were the means of values obtained for groups of six transformed on the same day.
DISCUSSION
The data presented here demonstrate that when gonococci express Opa, they are more efficient at DNA transformation than they are in the Opa− state provided that the (presumptive) surface-exposed cationic groups on the Opa proteins are not masked through the accretion of nonspecific polyanions. The fact that this Opa-mediated effect was inhibitable by both nonspecific DNA and heparin indicates that the basis for the elevated transformation rates lies to some degree with the higher positive cell surface charge afforded by Opa expression. However, the noticeable difference in the inhibitory effects of nonspecific DNA and heparin indicates that the effect of Opa protein expression may not be simply electrostatic but may also result in movement of DNA across the outer membrane.
Aqueous suspensions of bacteria have long been known to exhibit strong induced dipole moments when placed within an electric field. From initial attempts at studying the electrostatic properties of the bacterial cell surface, it was concluded that cell surface charge is moderated through the adsorption of counterions (25). However, the application of electrophoretic analysis to the study of physical effects at the bacterial cell surface was limited, to some degree, by a lack of knowledge of the actual molecular components that were present at the cell surface. Generally, assumptions were made concerning the three-dimensional array of specific surface components, with few studies providing direct physical support for the various contentions by using intact organisms. Recently, electric light scattering, in conjunction with changes in electrophoretic mobility, has proven to be a potent tool for studying the electrostatic properties of the gonococcal cell surface in intact organisms (32). Moreover, since the gonococcus changes several surface antigens (e.g., pilin polypeptide, Opa proteins, and lipopolysaccharide components [35]), comparisons of surface charges of the different defined variants can now be readily made, allowing more precise determinations of cell surface charge differences. Consequently, by using a combination of physical techniques on a series of defined mutants, contributions of the porin polypeptide and lipooligosaccharide to the physical cell surface properties of the gonococcus were monitored with a high degree of precision at the whole-organism level (32, 36). Moreover, these types of studies indicated that the expression of Opa proteins by gonococci also impacted the charge distribution at the cell surface, apparently causing the organisms to preferentially bind exogenous DNA (32).
This study investigated whether changing the presumptive electrostatic composition of the gonococcal cell surface through the expression of Opa proteins actually influenced a basic biological property of the organism, namely DNA transformation. From the data presented, it is evident that Opa expression clearly correlates with enhanced transformation efficiency. However, culture conditions had a noticeable impact on the analysis, requiring the development of transformation regimens that did not overly impinge on the innate physical properties of the gonococcal cell surface. Early on it became apparent that when transformations were performed under high-cell-density conditions, two distinct subpopulations arose (to various degrees) within an Opa+ culture (i.e., Opa+ and Opa− bacteria). Consequently, it was realized that each transformation frequency actually represented a composite value that was determined by the relative contribution of each subpopulation to the overall transformation frequency (Table 1). In addition, high-cell-density Opa+ and Opa− cultures were especially prone to undergo cell lysis (which ranged from 10 to 95% depending on the starting cell density), which had the effect of masking the charge contributions of the Opa proteins (presumably due to chromosomal DNA, released from lysed sibling cells, sticking to the cell surface). Furthermore, under these conditions, one could not confidently eliminate the impact of bacterial clumping on the observed elevation in transformation frequency. Nonetheless, these early studies, performed under nonoptimal conditions, consistently showed that Opa expression somehow contributed to an enhanced-transformation phenotype. When different transformation regimens were implemented (especially growth of bacteria on agarose-based medium prior to transformation), many of the difficulties associated with the early experiments (e.g., culture lethality, Opa protein heterogeneity, and cell clumping) were successfully overcome, with the new conditions allowing both Opa+ and Opa− cultures to grow during the course of the experiment. Accordingly, transformation rates increased, with Opa-expressing cultures showing significantly higher rates (P values of <0.001) (Table 2) than Opa-negative cultures. Furthermore, under these newly defined conditions, transformation of OpaA-expressing bacteria was now inhibitable by nonspecific DNA present within the transformation menstruum.
Overall, the data support a two-step model for conversion of donor DNA into a DNase I-resistant state with its subsequent translocation across the cell envelope. In this model, the elaboration of a positive cell surface charge through Opa expression causes an initial tight binding of donor DNA at the cell surface (step 1), which impedes the rapid transition of the bound DNA into a DNase I-resistant state (step 2). Therefore, by tightly binding the donor DNA at the cell surface, Opa protein expressed there effectively increases the relative donor DNA concentration at the cell surface over time by significantly decreasing the off rate (i.e., for the on/off rate constants, k1 ≫ k−1). In contrast, binding of donor DNA to the cell surface in Opa-negative cultures is probably in equilibrium, with a significant off rate (i.e., k1 ≈ k−1). Therefore, for Opa-expressing organisms, by effectively increasing the substrate concentration over time, this property overcomes the slower kinetics of transition into a DNase I-resistant state, which eventually translates into more donor DNA being translocated into the cytoplasm, resulting in more efficient transformation.
The fact that nonspecific polyanions in the transformation reaction mixture could impact the transformation efficiency of Opa-expressing bacteria may have important ramifications with respect to the horizontal exchange of DNA at the mucosal surface. In vivo, gonococci are predominantly Opa+ (9, 20, 34) and are likely bathed with mucosal secretions which will include heparin (22). Furthermore, with the onset of neutrophil infiltration, gonococci will also encounter a potent DNase (S. A. Hill and W. Shafer, unpublished observation). Therefore, the data suggest that Opa expression may actually hinder horizontal transmission at the mucosal surface because it results in the maintenance of gonococcal transforming DNA in a more DNase I-susceptible state for longer periods of time, as well as allowing competitive inhibition by components within mucosal secretions and nonspecific human chromosomal DNA that is released from damaged epithelial cells and/or polymorphonuclear leukocytes. Therefore, a more protective physical role can be envisaged for Opa-DNA interactions in vivo, which may help gonococci survive within the harsh chemical environment that is likely to be encountered on an inflamed mucosa. Such a survival strategy may not be unique to the gonococcus. Much of the pathology associated with the disease cystic fibrosis is caused by the persistent colonization of Pseudomonas aeruginosa within viscous purulent lung secretions, of which DNA is a major component (28). Therefore, perhaps binding of DNA to the cell surface of mucosal pathogens represents a common survival strategy for this unique group of pathogens.
ACKNOWLEDGMENT
I thank John Swanson for insights and encouragement during this project.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, Suppl. 6. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. p. 2.4.2. [Google Scholar]
- 2.Bergstrom S, Robbins K, Koomey J M, Swanson J. Piliation control mechanisms in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1986;83:3890–3894. doi: 10.1073/pnas.83.11.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jahnig F, Stern A, Kupsch E-M, Meyer T F, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 4.Biswas G D, Lacks S A, Sparling P F. Transformation-deficient mutants of piliated Neisseria gonorrhoeae. J Bacteriol. 1989;171:657–664. doi: 10.1128/jb.171.2.657-664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas G D, Sox T, Blackman E, Sparling P F. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee M S, Wilson J, Hill S A. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology. 1999;145:389–400. doi: 10.1099/13500872-145-2-389. [DOI] [PubMed] [Google Scholar]
- 7.Chen T, Swanson J, Wilson J, Belland R J. Heparin protects Opa+Neisseria gonorrhoeae from the bactericidal action of normal human serum. Infect Immun. 1995;63:1790–1795. doi: 10.1128/iai.63.5.1790-1795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty T J, Asmus A, Tomasz A. Specificity of DNA uptake in genetic transformation of gonococci. Biochem Biophys Res Commun. 1979;86:97–104. doi: 10.1016/0006-291x(79)90386-3. [DOI] [PubMed] [Google Scholar]
- 9.Draper D L, James J F, Brooks G F, Sweet R L. Comparison of virulence markers of peritoneal and fallopian tube isolates with endocervical Neisseria gonorrhoeae isolates from women with acute salpingitis. Infect Immun. 1980;27:882–888. doi: 10.1128/iai.27.3.882-888.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Facius D, Meyer T F. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol Microbiol. 1993;10:699–712. doi: 10.1111/j.1365-2958.1993.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 12.Fussenegger M, Facius D, Meier J, Meyer T F. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;19:1095–1105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 13.Fussenegger M, Kahrs A F, Facius D, Meyer T F. Tetrapac (tpc), a novel genotype of Neisseria gonorrhoeae affecting epithelial cell invasion, natural transformation competence and cell separation. Mol Microbiol. 1996;19:1357–1372. doi: 10.1111/j.1365-2958.1996.tb02479.x. [DOI] [PubMed] [Google Scholar]
- 14.Goodgal S H, Herriott R M. Studies on transformation of Hemophilus influenzae. I. Competence. J Gen Physiol. 1961;44:1201–1227. doi: 10.1085/jgp.44.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graves J F, Biswas G D, Sparling P F. Sequence-specific DNA uptake in transformation of Neisseria gonorrhoeae. J Bacteriol. 1982;152:1071–1077. doi: 10.1128/jb.152.3.1071-1077.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris R H, Mitchell R. The role of polymers in microbial aggregation. Annu Rev Microbiol. 1973;27:27–50. doi: 10.1146/annurev.mi.27.100173.000331. [DOI] [PubMed] [Google Scholar]
- 18.Hill S A, Morrison S G, Swanson J. The role of direct oligonucleotide repeats in gonococcal pilin gene variation. Mol Microbiol. 1990;4:1341–1352. doi: 10.1111/j.1365-2958.1990.tb00713.x. [DOI] [PubMed] [Google Scholar]
- 19.James J F, Swanson J. Color/opacity colony variants of Neisseria gonorrhoeae and their relationship to the menstrual cycle. In: Gotschlich E C, Holmes K K, Sawyer W D, Young F E, editors. Immunobiology of Neisseria gonorrhoeae. Washington, D.C.: American Society for Microbiology; 1978. pp. 338–343. [Google Scholar]
- 20.Jerse A E, Cohen M S, Drown P M, Whicker L G, Isbey S F, Seifert H S, Cannon J G. Multiple gonococcal opacity proteins are expressed during experimental urethral infection in the male. J Exp Med. 1994;179:911–920. doi: 10.1084/jem.179.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koomey J M, Falkow S. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol. 1987;169:790–795. doi: 10.1128/jb.169.2.790-795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D A. Heparin binding and neutralizing proteins. In: Lane D A, Lindahl U, editors. Heparin. Boca Raton, Fla: CRC Press; 1989. pp. 363–391. [Google Scholar]
- 23.Maynard-Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4389. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehr I J, Seifert H S. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 25.Morris V J, Jennings B R. The effect of neomycin and streptomycin on the electrical polarisability of aqueous suspensions of Escherichia coli. Biochim Biophys Acta. 1975;392:328–334. doi: 10.1016/0304-4165(75)90014-8. [DOI] [PubMed] [Google Scholar]
- 26.Rudel T, Facius D, Barten R, Scheuerpflug I, Nonnenmacher E, Meyer T F. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1995;92:7986–7990. doi: 10.1073/pnas.92.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifert H S, Ajioka R S, Marchal C, Sparling P F, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- 28.Sferra T J, Collins F S. The molecular biology of cystic fibrosis. Annu Rev Med. 1993;44:133–144. doi: 10.1146/annurev.me.44.020193.001025. [DOI] [PubMed] [Google Scholar]
- 29.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson J. Studies on gonococcus infection. XII. Colony color and opacity variants of gonococci. Infect Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson J. Colony opacity and protein II compositions of gonococci. Infect Immun. 1982;37:359–368. doi: 10.1128/iai.37.1.359-368.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson J. Effects of Opa proteins and lipooligosaccharides on surface charge and biological behaviour of gonococci. In: Kado C I, Crosa J H, editors. Molecular mechanisms of bacterial virulence. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 109–125. [Google Scholar]
- 33.Swanson J, Barrera O. Immunological characteristics of gonococcal outer membrane protein II assessed by immunoprecipitation, immunoblotting, and coagglutination. J Exp Med. 1983;157:1405–1420. doi: 10.1084/jem.157.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson J, Barrera O, Sola J, Boslego J. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J Exp Med. 1988;168:2121–2129. doi: 10.1084/jem.168.6.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson J, Belland R J, Hill S A. Neisserial surface variation: how and why? Curr Opin Genet Dev. 1992;2:805–811. doi: 10.1016/s0959-437x(05)80143-1. [DOI] [PubMed] [Google Scholar]
- 36.Swanson J, Dorward D, Lubke L, Kao D. Porin polypeptide contributes to surface charge of gonococci. J Bacteriol. 1997;179:3541–3548. doi: 10.1128/jb.179.11.3541-3548.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson J, Hill S A, Fischer S H. Growth on different solid media markedly affects the properties and behaviors of Opa+ gonococci. In: Conde-Glez C J, Morse S, Rice P, Sparling F, Calderon E, editors. Proceedings of the Eighth International Pathogenic Neisseria Conference. Cuernavaca, Mexico: Instituto Nacional de Salud Publica; 1994. pp. 771–776. [Google Scholar]
- 38.Wolfgang M, Park H S, Hayes S F, van Putten J P, Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]