Abstract
Leprosy, whose etiological agent is Mycobacterium leprae, is a chronic infectious disease that mainly affects the skin and peripheral nervous system. The diagnosis of leprosy is based on clinical evaluation, whereas histopathological analysis and bacilloscopy are complementary diagnostic tools. Quantitative PCR (qPCR), a current useful tool for diagnosis of infectious diseases, has been used to detect several pathogens including Mycobacterium leprae. The validation of this technique in a robust set of samples comprising the different clinical forms of leprosy is still necessary. Thus, in this study samples from 126 skin biopsies (collected from patients on all clinical forms and reactional states of leprosy) and 25 slit skin smear of leprosy patients were comparatively analyzed by qPCR (performed with primers for the RLEP region of M. leprae DNA) and routine bacilloscopy performed in histological sections or in slit skin smear. Considering clinical diagnostic as the gold standard, 84.9% of the leprosy patients were qPCR positive in skin biopsies, resulting in 84.92% sensitivity, with 84.92 and 61.22% positive (PPV) and negative (NPV) predictive values, respectively. Concerning bacilloscopy of histological sections (BI/H), the sensitivity was 80.15% and the PPV and NPV were 80.15 and 44.44%, respectively. The concordance between qPCR and BI/H was 87.30%. Regarding the slit skin smear, 84% of the samples tested positive in the qPCR. Additionally, qPCR showed 100% specificity, since all samples from different mycobacteria, from healthy individuals, and from other granulomatous diseases presented negative results. In conclusion, the qPCR technique for detection of M. leprae using RLEP primers proved to be specific and sensitive, and qPCR can be used as a complementary test to diagnose leprosy irrespective of the clinical form of disease.
Keywords: Mycobacterium leprae, qPCR, Leprosy, Bacilloscopy
Introduction
Leprosy, whose etiologic agent is Mycobacterium leprae (M. leprae), is a chronic infectious disease that affects primarily the skin and the peripheral nervous system. The clinical spectrum proposed by Ridley and Jopling (R&J) consists of two poles: tuberculoid (TT) and lepromatous (LL), and three intermediate forms: borderline-tuberculoid (BT), borderline-borderline (BB), and borderline-lepromatous (BL).1 In addition, there are clinical forms associated with two reactional forms: type 1 reaction (or reversal reaction – RR) and type 2 reaction (or erythema nodosum leprosum – ENL).2
So far, M. leprae cannot grow in vitro, and this is a limiting factor to the study of the disease. The diagnosis of leprosy is based on clinical evaluation of patients, whereas histopathological analysis and bacilloscopy are complementary diagnostic exams. In this context, the bacilloscopy of histological sections (BI/H) and bacilloscopy of slit skin smear (BI/S) have an important role for patient's diagnosis and follow up, and for choosing the adequate chemotherapeutic regimen. In spite of that, these techniques are time consuming and have limited sensitivity. Additionally, in tuberculoid patients, in which bacilli are rare or nonexistent, there is always a limitation in using bacilloscopy as an auxiliary exam, once negative results do not rule out the disease. Then, the implementation of new, more sensitive techniques to detect M. leprae are necessary to reduce the time for a definitive diagnosis.
Currently, quantitative PCR (qPCR) has been considered a rapid, sensitive and specific method for detection of pathogens, including M. leprae.3, 4 However, validation of this technique in a large number of samples collected from different clinical forms of the leprosy spectrum and reactional states (RR and ENL) is still necessary.
In order to improve the odds of the molecular detection of M. leprae DNA, the gene sequence selected for the design of primers in this study was based on the overlapping of Specific Repetitive Element (RLEP) regions located along the M. leprae genome. The use of RLEP has the advantage of being more sensitive than targets located in other gene regions because it provides multiple copies.5, 6, 7, 8 Assays using the RLEP target proved to be more sensitive in the detection of M. leprae when compared to other single copy targets like Ag85B, sodA and 16S rRNA.5 Furthermore, PCR assays based on the use of RLEP have shown rapid and objective results as a tool for molecular detection and quantification of mycobacteria in clinical samples.9 Additionally, this sequence was chosen because it has no homology to other mycobacteria or bacterial species providing, therefore, the specificity required for a test designed to improve the diagnosis of leprosy via M. leprae detection.
Thus, the objective of this study was to validate a RLEP based qPCR for the detection of M. leprae in skin lesions biopsies collected from patients on all clinical forms of leprosy and reactional states, and also in slit skin smears. The importance of this work is that a large number of samples were evaluated by qPCR, and skin biopsies were representative of all clinical forms of leprosy, including reactional manifestations of the disease. In addition, in order to compare the ability to detect M. leprae in other types of samples, qPCR was also tested in DNA extracted from slit skin smear slides.
Material and methods
Patients
Biopsies of skin lesions (single 5 or 6 mm3 punchs) of leprosy patients were collected (in RNAlater) during one year at the Lauro de Souza Lima Institute and Health Center Jardim Guanabara. A total of 126 skin biopsies used to assess BI/H from patients classified according to R&J criteria1 were included. Of those, there were 38 tuberculoid (TT), 21 borderline tuberculoid (BT), 18 borderline borderline (BB), 12 borderline lepromatous (BL), and 13 lepromatous (LL). Biopsies of reactional patients were collected either at diagnosis, during or after the course of multidrug therapy (MDT); however, none of the patients had received any corticosteroids or thalidomide to treat the reactional event. Reactional patients were divided into two subgroups: 12 reversal reaction (RR) and 12 erythema nodosum leprosum (ENL). Additionally, for analysis the samples (n = 126) were also divided into paucibacillary (PB), with BI/H 0 to 1+, and multibacillary (MB) with BI/H equal or higher than 2+.
For baciloscopic index of slit skin smear (BI/S), 25 samples were collected randomly in the period of one month from the Microbiology Laboratory of the Lauro de Souza Lima Institute. These samples had positive BI/S ranging from 1+ to 5+, and were collected from index points: earlobes, elbows, knees, and lesion.10 Of note, these samples were multibacillary and did not match skin lesions biopsies of patients submitted to skin biopsy.
Furthermore, for both BI/H and BI/S, negative bacilloscopy results show baciloscopic indexes (BI) equal 0+, and positive results show BI between 1+ and 6+. BI was defined according to the literature, so that 0+ means absence of bacilli in 100 fields evaluated; 1+ presence of 1–10 bacilli in 100 fields evaluated; 2+ presence of 1–10 bacilli in 10 fields evaluated; 3+ presence of 1–10 bacilli per field on average; 4+ presence of 10–100 bacilli per field on average; 5+ presence of 100–1000 bacilli per field on average; 6+ presence more than 1000 bacilli per field on average.
Additionally, 10 skin biopsies from patients with other granulomatous skin diseases [pemphigus foliaceus (2), pemphigus vulgaris (1), nummular eczema (1), lichenified eczema (1), porphyria (2), and erythematous lupus (3)] and 10 skin biopsies from healthy individuals (undergoing cosmetic surgery without previous diagnosis of leprosy or any other skin disease) were used as negative controls. This study was approved by the Ethical Committee of Lauro de Souza Lima Institute (protocol n° 046/2009) and informed consent was obtained from all participants.
Mycobacteria samples
Purified DNA from M. leprae11 was used as positive control and DNA from different mycobacterial species (M. tuberculosis, M. fortuitum, M. avium, M. smegmatis, M. intracellulare, M. chimera, M. nonchromogenicum, M. colombiense and M. abscessus subspboletti) were used as negative controls.
Extraction of DNA from skin biopsies
Extraction of total DNA from the biopsies was performed using approximately 20 mg of material using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's specifications, including a step of overnight Proteinase K pre-digestion under agitation (1400 rpm) at 56 °C. After extraction, the concentration and purity of the samples were assessed using the Nanodrop2000 equipment (Thermo Fisher Scientific, Waltham, MA, USA). To perform the qPCR technique, the DNA concentration used was 10 ng of total DNA, and depending on DNA concentration, variable volume (uL) was used to perform the qPCR.
DNA extraction from slit skin smear slides
For DNA extraction from slit skin smear slides, excess immersion oil used for counting bacilli, was removed by pressure with absorbent paper. Then, 90 μl ultrapure water was added on top of the smear which was scrapped off with an unused scalpel blade, and the content was pipetted into the tubes where the suspensions were centrifuged at 10,000 rpm for 20 min. After the centrifugation step, the supernatant was discarded and the pellet subjected to DNA extraction using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. After extraction, the samples were concentrated by vacuum method in the miVAC DNA Concentrator (Genevac, Ipswich, Suffolk, UK) for 2 h at room temperature, obtaining a final volume of approximately 50 μl. The concentration and purity of the samples were assessed using the Nanodrop 2000 equipment (Thermo Fisher Scientific, Waltham, MA, USA). To perform the qPCR technique, the DNA concentration used was 10 ng of total DNA, and depending on DNA concentration, variable volume (uL) was used to perform the qPCR.
Quantitative PCR (real-time PCR assays)
The gene sequence selected for construction of the primers was based on overlapping of RLEPs 1, 2, 3 and 4. The pair of primers was designed using the Primer Express 2.0 (Thermo Fisher Scientific, Waltham, MA, USA), followed by confirmation of homology through the software Basic Local Alignment Search Tool (NCBI/BLAST). These primers differ from others previously described.5, 6, 7, 9, 12, 13
Assays for detection of M. leprae were performed in duplicates using 10 ng of total DNA (for samples from skin lesions and slit skin smear) in the StepOnePlus equipment (Thermo Fisher Scientific, Waltham, MA, USA), using SYBRGreen chemistry and the pair of primers (sense 5′ATTTCTGCCGCTGGTATCGGT 3′, antisense 5′ TGCGCTAGAAGGTTGCCGTAT 3′) (ThermoFisher Scientific, Waltham, MA, USA).
The reaction consisted of two minutes at 50 °C, two minutes at 95 °C, and 40 cycles of 30 s at 95 °C, 30 s 62.5 °C (annealing temperature) and one minute at 72 °C, and a final cycle of 20 min with increasing temperature from 60 to 95 °C to obtain the dissociation of the reaction (melting curve). The melting curve was used for the analysis of specificity of the amplification products curve. The results were obtained in accordance with the first fluorescent signal detection Cycle Threshold (CT) and the sample was considered positive when it showed CT smaller than 40 (cutoff). The quality of the DNA sample was verified using primers specific for the constitutive Beta-actin gene (sense 5′ GGTGTCACCAAAGCAAAGGC 3′, antisense 5′ TAAGGGAGCTTTCGGAGCTGT 3′).
Standard curve and limit of detection
To check the detection limit, a standard curve was prepared from the purified DNA of M. leprae extracted after processing a footpad of nude mice (six months after inoculation). The footpad was removed and the suspension (1 × 109) was processed in 500 μl sterile saline solution using a tissue homogenizer (Turrax/IKA, Guangzhou, China) running five pulses 15′′/speed 4.10 The DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's specifications, including a step of overnight Proteinase K pre-digestion under agitation (1400 rpm) at 56 °C. The standard curve consisted of six points, and it was performed by serial dilution (1:10) with the initial and final points 0.3 ng and 3 fg, respectively. This curve was performed in three different days to confirm reproducibility.
Statistical analysis
The correlations between CT values and bacilloscopy (BI/H and BI/S), and CT and DNA concentration of M. leprae were calculated using the Spearman's test. For detection of M. leprae in the paucibacillary and multibacillary samples, according to the CT values, the Mann Whitney test was used. For detection of M. leprae in clinical forms of the disease and in slit skin smears, according to the CT values, the Kruskal–Wallis test was used. All analyses were performed using the GraphPad Prism 5.0 (GraphPad Software, California, USA). Sensitivity, specificity, concordance, positive predictive value, and negative predictive value were determined using the GraphPad Quick Calcs (GraphPad Software, California, USA). Additionally, these parameters were also calculated to each group, after selecting equal numbers of samples by random selection (random.org) to avoid under or overrepresentation of any group in the overall analysis. This procedure was performed four times to ensure homogeneous distribution and the values were expressed as average value.
Results
qPCR in skin lesions
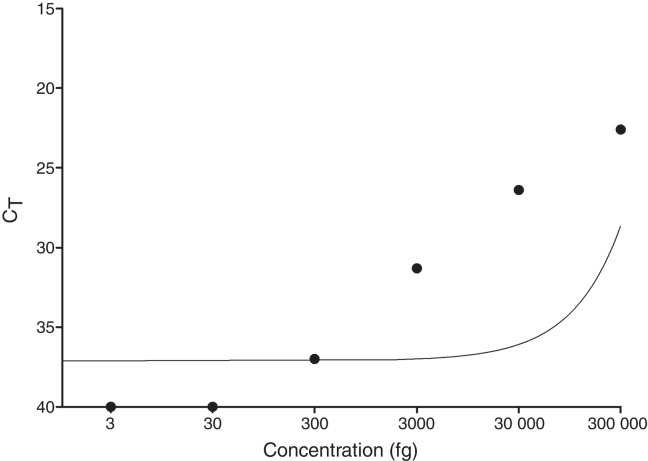
As shown in Fig. 1, the limit of detection for qPCR was equal to 300 fg, since the technique did not detect concentrations below 300 fg (30 fg and 3 fg). Additionally, the standard curve showed reproducibility and there was a statistically significant negative correlation between CT and DNA concentration of M. leprae (p = 0.0028).
Fig. 1.
Quantitative PCR standard curve. A standard curve was prepared from the purified DNA of M. leprae. It was performed by serial dilution (1:10) with the initial and final points 0.3 ng and 3 fg, respectively, and the limit of detection was equal to 300 fg.
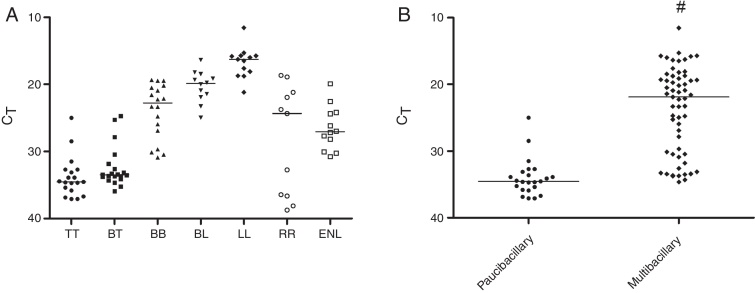
When evaluating the positivity of the samples selected for this study by qPCR, 84.9% were M. leprae positive, being 55.2% TT, 95.2% BT, 91.7% RR, and 100% BB, BL, LL, and ENL groups (Table 1). Regarding the PB and MB group, 61.7% (29/47) and 98.7 (78/79) were M. leprae positive, respectively. As shown in Fig. 2A, considering only the qPCR positive results, the CT averages observed in different clinical groups were 33.8 (24.9–37.1) for TT, 32.4 (24.7–35.9) for BT, 23.9 (19.3–30.9) for BB, 20.3 (16.3–24.9) for BL, and 16.7 (11.5–21.2) for LL. In the reactional patients, the RR group had a higher CT (CT average 28.3) than the ENL group in whom the average value was 26.5. When all groups were evaluated, there was a statistically significant negative correlation between CT and BI/H (p < 0.0001). A significant difference was observed between CT values of multibacillary (CT average 23.7) and paucibacillary patients (CT average 34) (Fig. 2B).
Table 1.
Number of samples (skin lesions) detected by quantitative PCR.
| Clinical form | Positive qPCR/total number (%) | qPCR positive/total number (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| BI/H 0+ | BI/H 1+ | BI/H 2+ | BI/H 3+ | BI/H 4+ | BI/H 5+ | BI/H 6+ | ||
| TT | 21/38 (55.2) | 9/22 (40.9) | 12/16 (75) | – | – | – | – | – |
| BT | 20/21 (95.2) | – | 4/4 (100) | 12/13 (92.3) | 4/4 (100) | – | – | – |
| BB | 18/18 (100) | – | – | 2/2 (100) | 2/2 (100) | 6/6 (100) | 8/8 (100) | |
| BL | 12/12 (100) | – | – | – | – | – | 6/6 (100) | 6/6 (100) |
| LL | 13/13 (100) | – | – | – | – | – | – | 13/13 (100) |
| RR | 11/12 (91.7) | 2/3 (66.6) | 2/2 (100) | 1/1(100) | 1/1 (100) | 2/2 (100) | 3/3 (100) | – |
| ENL | 12/12 (100) | – | – | 4/4 (100) | 6/6 (100) | – | – | 2/2 (100) |
| Total | 107/126 (84.9) | 11/25 (44) | 18/22 (81.8) | 19/20 (95) | 13/13 (100) | 8/8 (100) | 17/17 (100) | 21/21 (100) |
qPCR, quantitative PCR; TT, tuberculoid; BT, borderline-tuberculoid; BB, borderline-borderline; BL, borderline-lepromatous; LL, lepromatous; RR, reversal reaction; ENL, erythema nodosum leprosum.
Distribution according to the clinical form and bacilloscopic index of histological sections (BI/H).
Fig. 2.
Detection of M. leprae in biopsy samples by qPCR technique. Data reported in scatter dot plot as medians, excluding negative results. (A) Patients divided in seven groups (n = 107). (B) Patients grouped as paucibacillary (BI/H ≤ 1+) and multibacillary (BI/H ≥ 2+) forms, excluding negative results (n = 107). BI/H: bacilloscopy of histological sections, TT: tuberculoid, BT: borderline-tuberculoid, BB: borderline-borderline, BL: borderline-lepromatous, LL: lepromatous, RR: reversal reaction, ENL: erythema nodosum leprosum. #p < 0.05.
Additionally, when considering only TT patients with negative BI/H (0+) the qPCR was capable of detecting 9 (40.9%) out of 22 samples. In TT patients with BI/H1+, the qPCR was able to detect 12 (75%) out of 16 samples (Table 1). When considering only the 101 BI/H positive samples (BI/H 1+ to 6+), 95% (96/101) of the samples were detected by the qPCR and there was a statistically significant negative correlation between CT and BI/H (p < 0.0001).
Regarding specificity, qPCR showed to be 100% specific, as M. leprae was not detected in any negative controls including different mycobacteria, healthy individuals, and other granulomatous diseases, with CT value higher than 40 (Table 2).
Table 2.
Specificity of quantitative PCR.
| Species (n = 10) | CT value |
|---|---|
| Mycobacterium leprae | 12.5 ± 0.5 |
| Mycobacterium tuberculosis | >40 |
| Mycobacterium avium | >40 |
| Mycobacterium fortuitum | >40 |
| Mycobacterium smegmatis | >40 |
| Mycobacterium intracellulare | >40 |
| Mycobacterium chimera | >40 |
| Mycobacterium nonchromogenicum | >40 |
| Mycobacterium colombiense | >40 |
| Mycobacterium abscessus subspboletti | >40 |
| Other dermatological diseases (n = 10) | |
| Pemphigus foliaceus (n = 2) | >40 |
| Pemphigus vulgaris (n = 1) | >40 |
| Nummular eczema (n = 1) | >40 |
| Lichenified eczema (n = 1) | >40 |
| Porphyria (n = 2) | >40 |
| Lupus erythematosus (n = 3) | >40 |
| Healthy individuals (n = 10) | |
| Normal skin | >40 |
qPCR, quantitative PCR; CT, cycle threshold.
Specificity of the primers relative to other species of mycobacteria, dermatological diseases (non leprosy) and healthy individuals.
When qPCR and bacilloscopy (BI/H) were compared in the group of 126 leprosy patients, 96 patients were positive and 14 patients were negative for both technique, with a concordance of 87.30% (110/126) between qPCR and BI/H. The discordance was 12.6% (16/126) with five patients BI/H positive and qPCR negative, and 11 patients BI/H negative and qPCR positive (Table 3). For all the patients (126) qPCR sensitivity was 84.9%, and the positive (PPV) and negative (NPV) predictive values were 84.9 and 61.2%, respectively (Table 4). Concerning bacilloscopy (BI/H), the sensitivity was 80.1%, and the positive (PPV) and negative (NPV) predictive values were 80.1 and 44.4%, respectively (Table 4). Additionally, when equal number of samples was randomly selected (n = 10), sensitivity and PPV for qPCR (both 52.5%) and BI/H (both 37.5%) decreased for TT group, while NPV values increased (86.3 for qPCR and 61.5% for BI/H). For the RR group, sensitivity and PPV for qPCR increased (both 95%) and decreased (both 75%) for BI/H, and for both NPV values increased (98.3 for qPCR and 80% for BI/H). Regarding other groups all the parameters were 100% (Table 4).
Table 3.
Comparison between qPCR and BI/H techniques.
| Contrasts | qPCR positive (+) | qPCR negative (−) | Total |
|---|---|---|---|
| BI/H positive (+) | 96 | 5 | 101 |
| BI/H negative (−) | 11 | 14 | 25 |
| Total | 107 | 19 | 126 |
The concordance between qPCR and BI/H was 87.30%. qPCR, real time PCR; BI/H, bacilloscopy of histological sections.
Table 4.
Sensitivity, PPV and NPV values for the qPCR and BI/H (skin lesions).
| BI/H |
qPCR |
|||||
|---|---|---|---|---|---|---|
| Patients | Sensitivity (%) | PPV (%) | NPV (%) | Sensitivity (%) | PPV (%) | NPV (%) |
| Total (n = 126) | 80.1 | 80.1 | 44.4 | 84.9 | 84.9 | 61.2 |
| TT (n = 10) * | 37.5 | 37.5 | 61.5 | 52.5 | 52.5 | 86.3 |
| BT (n = 10) * | 100 | 100 | 100 | 100 | 100 | 100 |
| BB (n = 10) * | 100 | 100 | 100 | 100 | 100 | 100 |
| BL (n = 10) * | 100 | 100 | 100 | 100 | 100 | 100 |
| LL (n = 10) * | 100 | 100 | 100 | 100 | 100 | 100 |
| RR (n = 10) * | 75 | 75 | 80 | 95 | 95 | 98.3 |
| ENH (n = 10) * | 100 | 100 | 100 | 100 | 100 | 100 |
qPCR, real time PCR; BI/H, bacilloscopy of histological sections; PPV, positive predictive value; NPV, negative predictive value.
To avoid overrepresentation of the groups with small sample size, the sensitivity, PPV and NPV values for the individual groups (leprosy clinical forms and reactional forms) were calculated after a random selection of the samples to comprise groups with the equal n = 10.
qPCR in slit skin smear
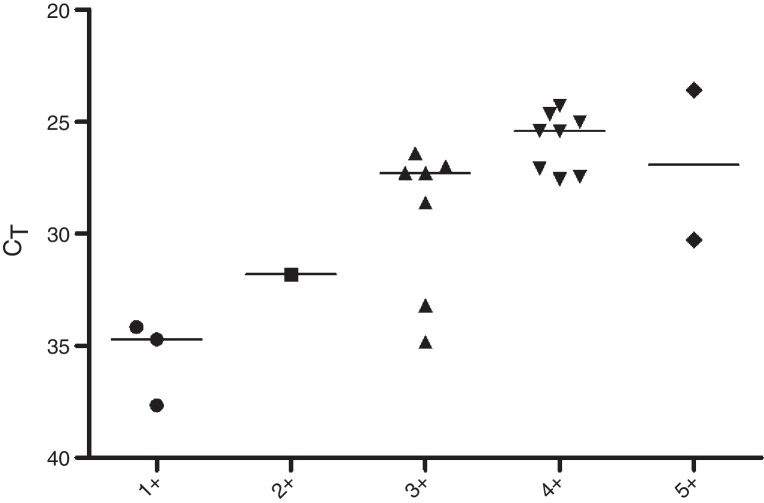
Regarding the CT values obtained from the DNA of slit skin smears, good results were observed with detection of 84% of the samples tested, 60% in BI/S1+, 50% in BI/S2+, 88.8% in BI/S3+, 100% in BI/S4+, and BI/S5+ (Table 5). As shown in Fig. 3, considering only the qPCR positive results, the CT averages observed in different BI/S were 35.5 (34.1–37.6) to BI/S1+, 31.8 (34.7–37.7) to BI/S2+, 29.2 (26.4–34.8) to BI/S3+, 25.8 (24.3–27.5) to BI/S4+, and 26.9 (26.3–30.2) to BI/S5+. Additionally, there was a statistically significant negative correlation between CT and BI/S (p < 0.0001).
Table 5.
Number of samples (slit skin smear) detected by quantitative PCR.
| Positive qPCR/total number (%) | qPCR positive/total number (%) |
||||
|---|---|---|---|---|---|
| BI/S1+ | BI/S 2+ | BI/S 3+ | BI/S 4+ | BI/S 5+ | |
| 21/25 (84) | 3/5 (60) | 1/2 (50) | 8/9 (88.8) | 7/7 (100) | 2/2 (100) |
qPCR, quantitative PCR; BI/S, bacilloscopy of slit skin smear.
Fig. 3.
Detection of M. leprae in slit skin smear samples by qPCR technique. Data reported in scatter dot plot as medians, excluding negative results (n = 21). Samples grouped according to bacilloscopy of slit skin smears (BI/S).
Discussion
In this study, we describe the use of the qPCR technique for detection of M. leprae in biopsy samples and slit skin smear of from leprosy patients with the different clinical forms of the disease. Our results demonstrate that this is a simple and reproducible technique using detection of M. leprae genomic DNA. Additionally, in contrast to other studies,3, 4, 5, 14, 15 this work included a large number of patient samples (n = 126) from different forms of the leprosy spectrum, including reactional states (RR and ENL), besides the operational classification as paucibacillary (PB) and multibacillary (MB). This strategy allowed the evaluation of a robust set of samples.
In this work, for skin lesions, 84.92% of the leprosy patients were qPCR positive, showing high sensitivity. Moreover, qPCR showed significant correlation and good concordance (87.30%) with direct bacillary counting (BI/H). Regarding lower positivity displayed by the TT group (n = 38, 55.2%), compared to the other clinical forms evaluated, this was probably due to the high number of BI/H 0+ patients (n = 22). In addition, qPCR showed positivity in 9/22 samples (40.9%), demonstrating an improved sensitivity of this method compared to BI/H. Opposed to that, other study obtained higher percentage of TT positive samples5; however, it included only two TT patients and one pure neural leprosy case. Note that after random selection, by analyzing each group separately, the value of the qPCR sensitivity decreased for the TT patients when compared to the sensitivity for all samples (n = 126). On the other hand, for the TT patients, BI/H sensitivity was lower than qPCR, showing again an improved sensitivity of this method compared with BI/H.
Some of the BI/H 1+ and 2+ were qPCR negative (n = 5), affecting the sensitivity of this method. One of the reasons for this result was the collection of two skin biopsies from the same patient's lesion, for different purposes (qPCR and bacilloscopy), so that the number of bacilli might have varied between them. It is important to consider that positive qPCR in lesions with BI/H 0+, as discussed above, may also have resulted positive because different biopsies were collected from the same lesion. Additionally, the negative bacilloscopy sometimes obtained in a sample in the tuberculoid side of leprosy, does not indicate that the method is not capable of detecting bacilli; instead, it shows that the patient may have minimum bacillary load. Also, a negative bacilloscopy in leprosy may be found in immunocompetent patients (polar TT patients, capable of spontaneous cure) after clearance.
Similar to the TT group, in the present study two RR samples were qPCR positive, despite being BI/H 0+, showing again the good sensitivity of the method. Interestingly, the result of the RR group, shown in Fig. 2A, reveals the variability within the group, which corroborates the clinical findings in the RR forms.16 In one hand, qPCR positive and low CT values samples were related to regressive disease. On the other hand, in the same RR group, samples with high CT values were related to progressive disease.
Based on the present results, it was observed that using the qPCR methodology, it was not possible to establish a cutoff CT for leprosy clinical forms, reactional forms or for operational classification of patients as PB and MB, since there was large variation in the samples CT when assessed individually (Fig. 2). Similar to that, other study using pPCR for the 85B gene showed significant difference between MB and PB patients, but again it was not possible to establish a cutoff CT for leprosy.4 On the other hand, this technique showed to be useful for detection of paucibacillary samples, especially from TT patients. Therefore, the qPCR technique used for the detection of M. leprae may contribute for the detection of bacilli in PB patients (BI/H 0+ and 1+).
In respect to specificity, the qPCR resulted negative (with CT > 40) in all non-leprae mycobacteria used in the assay. Additionally, samples from other dermatological diseases and from healthy individuals were all negative, demonstrating 100% specificity. Note that bacilloscopy is also considered a 100% specific method, even without comparison with negative controls, since there are not other tests for detection of acid-fast bacilli in BI/H or BI/S as there is for leprosy, therefore no false-positive results occur.
Other studies also used qPCR technique based on RLEP region,5, 12 but there are some differences between them and the present study. For example, they used TaqMan qPCR and this study used Sybr Green qPCR. Although different chemistry was used, the number of positive results was similar to different forms of the leprosy spectrum, especially for the BB, BL and LL groups (100% positive). For the TT group, as mentioned above, Martinez et al. (2011)5 showed higher percentage of TT positive samples (66.7%); however, they included only two TT patients and one pure neural leprosy case, while this study included 38 TT patients. In another study,12 the percentage of positive results in the TT group was similar (50% TT positive). On the other hand, among the PB group, a higher rate of positive results were reported (74.5%) when compared with this study (61.7%). Additionally, the present study showed 100% specificity compared to 73%5 of that study, and although our detection limit (300 fg) was bigger than other studies,5, 12 this result did not affect significantly the method sensitivity.
In this work, RLEP gene was chosen because this region repeats 37 times along the bacillus genome.8 Therefore, the advantage of using qPCR based on RLEP is that it would amplify the power of detection when compared to assays based on the use of a single copy gene.6, 13, 16 Indeed the study using primers to sodA, 16S, and Ag85B showed lower positivity in BT patients, with positive results ranging between 18% and 36.4%, and no TT patients were positive to these target genes.5 On the other hand, other studies using primers to 16S14 and Proline-rich antigen15 showed similar results to PB patients (with BI 0+), with positive results ranging between 44%15 and 53%.14 However, another study showed higher number of positive PB patients (80%),4 but in this case, differently from this study, it was necessary to decrease the CT value to avoid detection of negative controls. Yet, most of the aforementioned studies showed similar results for MB patients, except one, that showed lower positive results (46%).15
Concerning the study design, it is important to emphasize that only one study classified patients based on BI of histological sections,12 while in other studies the classification of patients was based on BI of the slit skin smear. However, in this case the PCR performed on skin biopsies could have different bacilloscopic indexes. In the present study, both the classification of the patients and qPCR were based on skin biopsies, thus the results cannot be fully compared to previous studies.4, 5, 14, 15
In respect to DNA amplification in slit skin smear slides, satisfactory results were observed (84% of positive samples) by qPCR; however, all samples were BI/S positive. Similar results were previously obtained by other authors, with 83% of samples showing positive amplification using simple PCR assay.13 On the other hand, in another study using qPCR for 85A-C only 38.5% of the MB patients showed positive results.17 For further confirmation of our results, it would be necessary to include samples with all baciloscopic indexes, including BI zero (PB patients). It is important to point out that the DNA used for qPCR was extracted from slit skin smear slides, therefore previous fixing and staining may have impaired the quality of the DNA. Indeed, Ruiz-Fuents et al. (2015)18 showed that different extraction methods to obtain M. leprae DNA can affect its amplification. Thus, the present study shows that the extraction of DNA from slit skin smear slides, after microscopic analysis, can be used as an alternative to detect the M. leprae bacillus, even though this is not the ideal type of sample for this purpose. Therefore, it may be used when a biopsy is unavailable. Furthermore, DNA extracted from slit skin smear can be used in other molecular techniques, such as verification of mutations associated with drug resistance.
In conclusion, qPCR technique for detection of M. leprae proved to be specific and sensitive, and has an additional advantage as a large number of samples can be processed simultaneously within a period of three hours, whereas for the bacilloscopy the time required to stain and count bacilli is considerably higher, particularly for PB cases. On the other hand, the qPCR technique has some limitations as it requires equipment, techniques and reagents of high cost unlike microscopy. In some cases qPCR may become limited to a few Reference Centers making it difficult to be performed in field. However, it is perfectly applicable to research purposes and experimental quantification of M. leprae DNA. Additionally, qPCR cannot detect all PB patients and, according to what was mentioned above, it was not possible to establish a cutoff CT for leprosy clinical forms, reactional forms or for operational classification of patients (PB and MB), since there was large variation in the samples CT when assessed individually. However, outside of Reference Centers often there is no expertise in evaluating samples of leprosy patients, thus the qPCR technique, when well executed, can decrease the chance of false-negative results when used as a complementary diagnostic tool.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by grants and scholarships from FAPESP (2009/06122-5 and 2011/09218-3).
References
- 1.Ridley D.S., Jopling W.H. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34:255–273. [PubMed] [Google Scholar]
- 2.Scollard D.M., Adams L.B., Gillis T.P., Krahenbuhl J.L., Truman R.W., Williams D.L. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19:338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phetsuksiri B., Rudeeaneksin J., Supapkul P., Wachapong S., Mahotarn K., Brennan P.J. A simplified reverse transcriptase PCR for rapid detection of Mycobacterium leprae in skin specimens. FEMS Immunol Med Microbiol. 2006;48:319–328. doi: 10.1111/j.1574-695X.2006.00152.x. [DOI] [PubMed] [Google Scholar]
- 4.Martinez A.N., Britto C.F., Nery J.A., et al. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J Clin Microbiol. 2006;44:3154–3159. doi: 10.1128/JCM.02250-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez A., Ribeiro-Alves M., Sarno E. Evaluation of qPCR-based assays for leprosy diagnosis directly in clinical specimens. PLoS Negl Trop Dis. 2011;5:e1354. doi: 10.1371/journal.pntd.0001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truman R.W., Andrews P.K., Robbins N.Y., Adams L.B., Krahenbuhl J.L., Gillis T. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2:e328. doi: 10.1371/journal.pntd.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang T.J., Kim S.K., Lee S.B., Chae G.T., Kim J.P. Comparison of two different PCR amplification product (the 18 kDa protein gene vs. RLEP repetitive sequence) in the diagnosis of Mycobacterium leprae. Clin Exp Dermatol. 2003;28:420–424. doi: 10.1046/j.1365-2230.2003.01300.x. [DOI] [PubMed] [Google Scholar]
- 8.Cole S., Supply P., Honoré N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr Rev. 2001;72:449–461. [PubMed] [Google Scholar]
- 9.Martinez A.N., Talhari C., Moraes M.O., Talhari S. PCR-based techniques for leprosy diagnosis: from the laboratory to the clinic. PLoS Negl Trop Dis. 2014;8:e2655. doi: 10.1371/journal.pntd.0002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brasil, Ministério da Saúde Secretaria de Vigilância em Saúde. Departamento de Vigilância Epidemiológica. Guia de Procedimentos Técnicos. Baciloscopia em Hanseníase. Brasília (DF); 2010.
- 11.Trombone A.P.F., Pedrini S.C.B., Diório S.M., Belone A.F.F., Fachin L.R.V., Nascimento D.C. Optimized protocols for Mycobacterium leprae strain management: frozen stock preservation and maintenance in athymic nude mice. J Vis Exp. 2014;23 doi: 10.3791/50620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan W., Xing Y., Yuan L.C., et al. Application of RLEP real-time PCR for detection of M. leprae DNA in paraffin-embedded skin biopsy specimens for diagnosis of paucibacillary leprosy. Am J Trop Med Hyg. 2014;90:524–529. doi: 10.4269/ajtmh.13-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turankar R.P., Pandey S., Lavania M., et al. Comparative evaluation of PCR amplification of RLEP: 16S rRNA, rpoT and SodA gene targets for detection of M. leprae DNA from clinical and environmental samples. Int J Mycobacteriol. 2015;4:54–59. doi: 10.1016/j.ijmyco.2014.11.062. [DOI] [PubMed] [Google Scholar]
- 14.Rudeeaneksin J., Srisungngam S., Sawanpanyalert P., et al. LightCycler TM real-time PCR for rapid detectionand quantitation of Mycobacterium leprae in skin specimens. Immunol Med Microbiol. 2008;54:263–270. doi: 10.1111/j.1574-695X.2008.00472.x. [DOI] [PubMed] [Google Scholar]
- 15.Kramme S., Bretzel G., Panning M., Kawuma J., Drosten C. Detection and quantification of Mycobacterium leprae in tissue samples by real-time PCR. Med Microbiol Immunol. 2004 Nov;193:189–193. doi: 10.1007/s00430-003-0188-8. [DOI] [PubMed] [Google Scholar]
- 16.Ridley D.S., Radia K.B. The histological course of reactions in borderline leprosy and their outcome. Int J Lepr Other Mycobact Dis. 1981;49:383–392. [PubMed] [Google Scholar]
- 17.Rosa F.B., Souza V.C., Almeida T.A., et al. Detection of Mycobacterium leprae in saliva and the evaluation of oral sensitivity in patients with leprosy. Mem Inst Oswaldo Cruz. 2013;108:572–577. doi: 10.1590/0074-0276108052013006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Fuentes J.L., Díaz A., Entenza A.E., et al. Comparison of four DNA extraction methods for the detection of Mycobacterium leprae from Ziehl-Neelsen-stained microscopic slides. Int J Mycobacteriol. 2015;4:284–289. doi: 10.1016/j.ijmyco.2015.06.005. [DOI] [PubMed] [Google Scholar]