Abstract
The mechanisms contributing to persistence of coagulase-negative staphylococci are diverse; to better understanding of their dynamics, the characterization of nosocomial isolates is needed. Our aim was to characterize phenotypic and molecular characteristics of Staphylococcus epidermidis and Staphylococcus haemolyticus human blood isolates from two tertiary care hospitals in Mexico, the Hospital Universitario in Monterrey and the Hospital Civil in Guadalajara.
Antimicrobial susceptibility was determined. Biofilm formation was assessed by crystal violet staining. Detection of the ica operon and Staphylococcal Cassette Chromosome mec typing were performed by PCR. Clonal relatedness was determined by Pulsed-fiel gel electrophoresis and Multi locus sequence typing.
Methicillin-resistance was 85.5% and 93.2% for S. epidermidis and S. haemolyticus, respectively. Both species showed resistance >70% to norfloxacin, clindamycin, levofloxacin, trimethoprim/sulfamethoxazole, and erythromycin. Three S. epidermidis and two S. haemolyticus isolates were linezolid-resistant (one isolate of each species was cfr+). Most isolates of both species were strong biofilm producers (92.8% of S. epidermidis and 72.9% of S. haemolyticus). The ica operon was amplified in 36 (43.4%) S. epidermidis isolates. SCCmec type IV was found in 47.2% of the S. epidermidis isolates and SCCmec type V in 14.5% of S. haemolyticus isolates. No clonal relatedness was found in either species. Resistance to clindamycin, levofloxacin, erythromycin, oxacillin, and cefoxitin was associated with biofilm production for both species (p < 0.05). A G2576T mutation in 23S rRNA gene was detected in an S. haemolyticus linezolid-resistant isolate. All linezolid-resistant S. epidermidis isolates belonged to ST23; isolate with SCCmec type IV belonged to ST7, and isolate with SCCmec type III belonged to ST2. This is the first report of ST7 in Mexico.
There was a high genetic diversity in both species, though both species shared characteristics that may contibute to virulence.
Keywords: Linezolid resistance, Staphylococcal Cassette Chromosome mec, Coagulase-negative staphylococci, Multilocus Sequence Typing
Introduction
Coagulase-negative staphylococci (CoNS) are among the main causative agents of bacteremia.1 Staphylococcus epidermidis and Staphylococcus haemolyticus are the CoNS species most frequently isolated from blood.2 These species are often associated with infections in immunocompromised patients who have medical device implants.3 These species persist on medical devices because they can form biofilms, bacterial clusters that attach to materials such as plastics. Biofilm formation has been associated with the presence of the ica operon that encodes for the production of a polysaccharide intercellular adhesin (PIA). This operon contains the icaA, icaB, icaC, icaD, and icaR genes; expression of these genes have been found to be involved in biofilm formation.4 Furthermore, biofilm production has been associated with an increased resistance to antibiotics.5 CoNS strains may present a high proportion of resistance to antibiotics,6, 7 particularly methicillin resistance, thus complicating the management of these infections.
Methicillin resistance in Staphylococcus aureus was first reported in 1961.8 Methicillin-resistant S. aureus strains produce a penicillin-binding protein, known as PBP2a or PBP2′, that has low binding affinity for β-lactams. PBP2a is encoded by mecA, which is contained within the Staphylococcal Cassette Chromosome mec (SCCmec).9 Also, methicillin-resistance has been found in CoNS species more often than in S. aureus species isolated from clinical samples.10 To date, 11 types of SCCmec have been described in S. aureus (http://www.sccmec.org/), and evidence suggests that SCCmec structures may be more diverse in CoNS. These various structures may contain combinations of mec and ccr complexes not described for S. aureus or may contain multiple ccr complexes.11 Since methicillin-resistance is more frequent in CoNS, methicillin-resistant CoNS (MR-CoNS) may serve as a large reservoir of SCCmec and may contribute to the formation of methicillin-resistant S. aureus (MRSA) strains.10
Since CoNS are components of the human skin microbiota, often endogenous strains are capable of causing infections in immunocompromised individuals. However, there are reports of persisting strains in hospital wards.12, 13 The genetic relatedness between these strains has been determined by Pulsed-Field Gel Electrophoresis (PFGE); a technique that has been widely used for molecular typing of nosocomial pathogens. Nevertheless, Multilocus Sequence Typing (MLST), based on sequencing of conserved housekeeping genes, is proving to be the most appropriate tool for the study of the global epidemiology, allowing the comparison of isolates from different countries and the naming of international clones. To date, a widely acceptable MLST scheme and database have been developed for S. epidermidis only but not for S. haemolyticus.14
The persistence of strains of these species may be due to their increasing high resistance to antimicrobials, which may enhance their fitness to hospital environments, including linezolid-resistance. At the same time, the production of biofilm has been associated with antibiotic resistance and the persistence of the strains in medical devices. Furthermore, the presence of SCCmec worsens the scenario, since horizontal transference of these elements may contribute to antibiotic resistance. Finally, the information of genetic relatedness would allow us to know the dynamics of transmission of these infections. We hypothesized that the presence of strains with traits such as biofilm formation, high resistance to antibiotics and diverse SCCmec elements may contribute to the persistence of these strains. Thus, we aimed to characterize phenotypic and molecular characteristics of S. epidermidis and S. haemolyticus blood isolates from two Mexican tertiary-care hospitals, the Hospital Universitario in Monterrey and the Hospital Civil in Guadalajara, in order to determine distribution of SCCmec elements, antibiotic resistance, genetic relatedness, and biofilm formation, as well as to examine its relationship to drug resistance. Both settings are teaching hospitals that offer a variety of services, educational programs, a space for clinical research and health-related community services working as a reference facility for two of the major metropolitan areas in Mexico.
Materials and methods
Hospital setting and identification of clinical isolates
This study was performed at the Hospital Civil de Guadalajara “Fray Antonio Alcalde”, a 1000-bed tertiary care teaching hospital, with approximately 30,000 admissions annually, located in Guadalajara, Jalisco, and at the Hospital Universitario “Dr. José Eleuterio González”, a 450-bed tertiary-care teaching hospital with an average hospital admittance rate of 26,500 patients yearly, located in Monterrey, Nuevo León.
Isolates of S. epidermidis (n = 83) and S. haemolyticus (n = 59) were collected from the two tertiary care hospitals in Mexico from 2006 to 2013. Isolates were obtained from blood cultures. Only one isolate per patient was included. Regarding the unit ward of precedence, 93/142 (65.5%) of isolates proceeded from intensive care units and 90/142 (63.4%) of patients were male.
Blood cultures were cultivated using Versa TREK REDOX bottles and processed using the Versa TREK blood culture system (Thermo Scientific, Oakwood Village, OH, USA). Cultures were incubated at 37 °C and the bottles were monitored for seven days before being discarded as negative. When positive, the bottle was subcultured in blood agar and incubated at 37 °C up to 72 h. Isolates were identified by biochemical and molecular methods. For the biochemical method, Sensititre Panels (TREK Diagnostic Systems Inc., Cleveland, OH, USA) were used according to the manufacturer's instructions. For the molecular method, DNA was obtained by phenol-chloroform extraction and species were identified by multiplex PCR of the nuc gene fragment as described previously.15 S. epidermidis ATCC 14990 and S. haemolyticus ATCC 29970 were used as the reference strains. All isolates were stored in Brucella broth containing 15% glycerol at −70 °C.
Methicillin-resistance and antimicrobial susceptibility testing
Methicillin-resistance was evaluated using the cefoxitin disk test described in the M02-A11 document of the Clinical and Laboratory Standards Institute (CLSI). Minimum inhibitory concentration (MIC) ranges, MIC50 and MIC90 were determined using the broth microdilution method. Panels from Sensititre (TREK Diagnostic Systems Inc.) were used according to the manufacturer's instructions. The antimicrobial agents tested were oxacillin, vancomycin, erythromycin, tetracycline, levofloxacin, clindamycin, trimethoprim-sulfamethoxazole, and linezolid. Methicillin-resistance and antimicrobial susceptibility test results were interpreted according to the M100-S24 document of the CLSI. S. aureus ATCC 29213 and S. aureus BAA-44 were used as quality controls.
Sequencing of 23S RNA gene and detection of cfr
Domain V of 23S rRNA gene was amplified to identify possible mutations in linezolid-resistant isolates, using the primers previously described.16 PCR products were purified by using a precipitation protocol with 3 M sodium acetate and absolute ethanol, at −20 °C. The precipitate was recovered by centrifugation and washed with 70% ethanol. The precipitate was dissolved in sterile nuclease-free water. The products were then sequenced (Macrogen, Korea) and aligned with the corresponding nucleotide sequence from reference strains of S. aureus and Escherichia coli (GenBank accession numbers X68425 and AF053966, respectively). Presence or absence of the cfr gene in the linezolid-resistant isolates was determined as described previously.17 Extraction of plasmid DNA was performed using an alkaline lysis protocol.
The biofilm formation assay and amplification of the ica operon
Biofilm formation was evaluated by crystal violet staining. Overnight cultures grown in tryptic soy broth were diluted 1:100 in tryptic soy broth supplemented with 1% glucose or tryptic soy broth supplemented with 3% NaCl. Each dilution was dispensed into four wells of flat-bottom polystyrene plates (Falcon, Franklin Lakes, NJ). After incubation for 24 h at 37 °C, the absorbance of planktonic cells was determined at 595 nm. The broth was eliminated and the wells were washed twice with sterile PBS (pH 7.3). Biofilms were stained with Hucker's crystal violet for 15 min; the stain was removed, and the wells were washed with sterile deionized water. The stained biofilms were dissolved in 200 μL of 30% acetic acid, and optical densities (OD) were measured at 595 nm. Biofilm formation was evaluated using the cut-off value suggested by Christensen et al.18 If the OD were less than or equal to 0.120, we classified the strain as non-biofilm producer. If the OD was above 0.240, the strain was classified as strong biofilm producer. Strains with OD greater than 0.120 but less than or equal to 0.240 were classified as weak biofilm producers. The biofilm index (biofilm absorbance/planktonic cell absorbance) was also calculated, which normalizes for differences in growth rates.
Genes in the ica operon were detected by multiplex PCR as described by Arciola et al.19 All five ica genes, icaA, icaD, icaB, icaC, and icaR, were tested, and PCR products were visualized on a 2.5% agarose gel stained with 1 mg/mL of ethidium bromide.
Detection of mecA and SCCmec typing
Amplification of mecA and SCCmec typing were performed by multiplex PCR as described by Zhang et al.20 and Kondo et al.21 The multiplex PCR assay by Kondo et al. contained a modification to the primers used to amplify mec complex class C and ccr complex type 4 and type 5 as described by Ruppé et al.11
Clonal relatedness
Clonal relatedness of isolates was determined by PFGE as described previously,10 but with modifications for CoNS in running conditions: the pulse times were 1–35 s for 24 h at 200 V. DNA was digested with the restriction enzyme SmaI, on a CHEF-DRIII instrument (Bio-Rad Laboratories, Hercules, CA) electrophoresis was performed. Gels were stained with ethidium bromide and images were obtained using the Labworks 4.5 software package. Banding patterns were analyzed visually, a database was created, and statistical analysis was performed using the statistical software package SPSS 20.0 (IBM Corporation, Somers, NY).
Multi locus sequence typing of S. epidermidis isolates
Primers and conditions were obtained from a previously described scheme.14 Determination of alleles and sequence type (ST) were performed using the S. epidermidis MLST database (http://sepidermidis.mlst.net/).22
Statistical analysis
The correlation between drug resistance and biofilm production was analyzed using a chi-square test and OpenEpi software version 3.03 (Rollins School of Public Health, Emory University). A p-value <0.05 was considered significant.
Results
Methicillin-resistance and antimicrobial susceptibility profiles
Of the S. epidermidis isolates tested, 85.5% (71/83) were methicillin-resistant, whereas 93.2% (55/59) of the S. haemolyticus isolates were methicillin-resistant, both by the cefoxitin disk test. In S. epidermidis, all methicillin-resistant isolates were also resistant to oxacillin and contained mecA; however, a methicillin-susceptible isolate was resistant to oxacillin and contained mecA. In S. haemolyticus, all methicillin-resistant isolates were also resistant to oxacillin and contained mecA.
Resistance to oxacillin, erythromycin, levofloxacin, clindamycin, and trimethoprim/sulfamethoxazole was >70% for both species (Table 1). Twelve percent of the S. epidermidis isolates were tetracycline-resistant, whereas 22% of the S. haemolyticus isolates were tetracycline-resistant. Three (3.6%) of the S. epidermidis and two (3.4%) of the S. haemolyticus isolates were resistant to linezolid.
Table 1.
Antimicrobial susceptibility of S. epidermidis and S. haemolyticus isolates.
| Antimicrobial |
S. epidermidis |
S. haemolyticus |
||||||
|---|---|---|---|---|---|---|---|---|
| MIC50a (μg/mL) | MIC90b (μg/mL) | Resistant n (%) |
Susceptible n (%) |
MIC50a (μg/mL) | MIC90b (μg/mL) | Resistant n (%) |
Susceptible n (%) |
|
| Oxacillin | >0.5 | >0.5 | 72 (86.7) | 11 (13.3) | >0.5 | >0.5 | 55 (93.2) | 4 (6.8) |
| Vancomycin | 2 | 4 | 0 (0) | 83 (100) | 2 | 2 | 0 (0) | 59 (100) |
| Erythromycin | >4 | >4 | 66 (79.5) | 16 (19.3) | >4 | >4 | 53 (89.8) | 6 (10.2) |
| Tetracycline | ≤2 | >8 | 10 (12) | 71 (85.6) | ≤2 | >8 | 13 (22.0) | 44 (74.6) |
| Levofloxacin | >2 | >2 | 62 (74.7) | 18 (21.7) | >2 | >2 | 54 (91.5) | 4 (6.8) |
| Norfloxacin | >8 | >8 | 64 (77.1) | 18 (21.7) | >8 | >8 | 53 (89.8) | 6 (10.2) |
| Clindamycin | >2 | >2 | 68 (81.9) | 12 (14.5) | >2 | >2 | 53 (89.8) | 5 (8.5) |
| Trimethoprim/sulfamethoxazole | >2/38 | >2/38 | 60 (72.3) | 23 (27.7) | >2/38 | >2/38 | 45 (76.3) | 14 (23.7) |
| Linezolid | ≤2 | 4 | 3 (3.6) | 80 (96.4) | ≤2 | ≤2 | 2 (3.4) | 57 (96.6) |
Minimum inhibitory concentration (MIC) ranges, MIC50 and MIC90 were determined using the broth microdilution method. Panels from Sensititre (TEK Diagnostic Systems Inc.) were used according to the manufacturer's instructions.
MIC50, minimal inhibitory concentration for 50% of the isolates.
MIC90, minimum inhibitory concentration for 90% of the isolates.
Sequencing of 23S RNA gene and detection of cfr
Analysis of domain V of 23S rRNA in linezolid-resistant isolates showed a G to T mutation at position 2576 (E. coli numbering) in a S. haemolyticus isolate (isolate 2975). Remaining isolates were negative for mutations in domain V. The cfr gene was detected in one linezolid-resistant S. epidermidis isolate (isolate 14565) and one linezolid-resistant S. haemolyticus isolate (isolate 9976) (Table 2). Also, plasmid DNA extraction was performed in cfr-negative isolates in order to determine the presence of the cfr gene in plasmids. The cfr gene was not amplified in this isolates.
Table 2.
SCCmec typing, susceptibility profile and biofilm production of linezolid-resistant isolates.
| Isolate | Species | SCCmec typing |
Susceptibility profilea |
Linezolid resistanceb |
Biofilm production |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mecA | mec class | ccr type | FOX | OX | TET | MIC (μg/mL) | cfr | Glucose | NaCl | ica operon | ||
| 14583 | S. epidermidis | + | A | 3 + 5 | R | R | S | 8 | Negative | Strong | Strong | Negative |
| 14565 | S. epidermidis | + | A | 3 + 5 | R | R | I | >32 | Positive | Strong | Strong | Positive |
| 12701 | S. epidermidis | + | A | 3 | R | R | S | 8 | Negative | Strong | Strong | Positive |
| 9976 | S. haemolyticus | − | NA | NA | S | S | S | 8 | Positive | NP | NP | Negative |
| 2975 | S. haemolyticus | + | NT | NT | R | R | R | 32 | Negative | Strong | NP | Negative |
NA, not applicable; NT, not typable; FOX, cefoxitin; OX, oxacillin; TET, tetracycline; MIC, minimum inhibitory concentration; NP, non-producer.
Methicillin-resistance was evaluated using the cefoxitin disk test, minimum inhibitory concentration was determined using the broth microdilution method. Biofilm formation was assessed by crystal violet staining. Amplification of the ica operon, mecA and SCCmec typing were performed by multiplex PCR.
All isolates were resistant to norfloxacin, clindamycin, levofloxacin, trimethoprim/sulfamethoxazole, and erythromycin. All isolates were susceptible to vancomycin.
Mutation G2576T was found in isolate 2975.
All linezolid-resistant isolates, except one S. haemolyticus isolate, were also resistant to methicillin. None of the isolates was resistant to vancomycin.
Biofilm production and presence of the ica operon
Of the S. epidermidis isolates, 77 (92.8%) were strong biofilm producers, 5 (6%) were weak biofilm producers, and 1 (1.2%) did not form biofilms in glucose-supplemented broth. In NaCl-supplemented broth, 72 (86.7%) of the isolates were strong biofilm producers, 7 (8.4%) were weak biofilm producers, and 4 (4.8%) did not form biofilms.
Similar results were found with the S. haemolyticus isolates; 43 (72.9%) were strong biofilm producers, 12 (20.3%) were weak biofilm producers, and 4 (6.8%) did not form biofilms in glucose-supplemented broth. In NaCl-supplemented broth, 30 (50.8%) were strong biofilm producers, 7 (11.9%) were weak biofilm producers, and 22 (37.3%) did not form biofilms.
Genes within the ica operon were present in 36 (43.4%) of the S. epidermidis isolates but were not found in any of the S. haemolyticus isolates. All the ica-positive isolates contained icaA, icaD, icaC, icaB, and icaR. All but one of the ica-positive isolates produced strong biofilms in the glucose-supplemented broth. Thirty-two of the 36 ica-positive isolates produced strong biofilms in the NaCl-supplemented broth. For both S. epidermidis and S. haemolyticus, an association was found between strong biofilm production and resistance to norfloxacin, clindamycin, levofloxacin, erythromycin, oxacillin, and cefoxitin (p < 0.05) when testing biofilm production in the glucose-supplemented broth.
Detection of mecA and SCCmec typing
The mecA gene was found in 72/83 (86.7%) of S. epidermidis isolates and 55/59 (93.2%) of S. haemolyticus isolates. Among the S. epidermidis isolates containing mecA, 5 (6.9%) were classified as SCCmec type III (ccr type 3, class A mec) and 34 (47.2%) were classified as type IV (ccr type 2, class B mec). Also, 22 (64.7%) of the SCCmec type IV isolates were subtyped as SCCmec IVa and one (2.9%) was subtyped as SCCmec IVb. The remaining isolates could not be subtyped to SCCmec IVc or IVd. S. epidermidis with SCCmec type IV were more likely to be resistant to norfloxacin and erythromycin. The five SCCmec type III isolates were subtyped as subtype III.1. Among the remaining isolates, more diverse SCCmec combinations were found; four isolates (5.6%) had ccr and mec combinations not included in the classification scheme for S. aureus, and 23 (31.9%) isolates amplified for more than one ccr complex. Four (5.6%) of the isolates could not be typed since mec or ccr complexes were not amplified in these strains (Table 3).
Table 3.
SCCmec typing, resistance profile, and biofilm production of S. epidermidis isolates.
| No. of isolates | SCCmec typing |
Resistance profilea n (%) |
Biofilm productionb n (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mec class | ccr type | SCCmec | NOR | CLI | TET | LEV | LZD | SXT | ERY | Glucose | NaCl | |
| 34 | B | 2 | IV | 30 (88) | 30 (88) | 3 (8.8) | 30 (88) | 0 (0) | 25 (74) | 30 (88) | 33 (97) | 30 (88) |
| 5 | A | 3 | III | 5 (100) | 4 (80) | 0 (0) | 5 (100) | 1 (20) | 4 (80) | 4 (80) | 5 (100) | 4 (80) |
| 3 | A | 1 | New | 3 (100) | 3 (100) | 2 (67) | 3 (100) | 0 (0) | 2 (67) | 3 (100) | 3 (100) | 2 (67) |
| 1 | A | 5 | New | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| 1 | A | 1 + 2 | Variant | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) |
| 5 | B | 1 + 2 | Variant | 5 (100) | 5 (100) | 1 (20) | 5 (100) | 0 (0) | 4 (80) | 5 (100) | 5 (100) | 4 (80) |
| 1 | A | 2 + 3 | Variant | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| 7 | A | 3 + 5 | Variant | 7 (100) | 7 (100) | 1 (14) | 7 (100) | 2 (29) | 7 (100) | 7 (100) | 7 (100) | 7 (100) |
| 5 | B | 2 + 5 | Variant | 3 (60) | 5 (100) | 1 (20) | 2 (40) | 0 (0) | 3 (60) | 4 (80) | 5 (100) | 5 (100) |
| 2 | A | 2 + 4 | Variant | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| 1 | A | 3 + 4 | Variant | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 1 (100) |
| 1 | B | 2 + 4 | Variant | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| 2 | A | 1 + 4 + 5 | Variant | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 1 (50) |
| 1 | NT | 2 + 5 | NT | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| 1 | NT | 2 | NT | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| 2 | A | NT | NT | 2 (100) | 2 (100) | 1 (50) | 1 (50) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
NT, not typeable; CLI, clindamycin; TET, tetracycline; LEV, levofloxacin; LZD, linezolid; SXT, trimethoprim/sulfamethoxazole; ERY, erythromycin.
Methicillin-resistance was evaluated using the cefoxitin disk test, minimum inhibitory concentration was determined using the broth microdilution method. Biofilm formation was assessed by crystal violet staining. Amplification of the ica operon and SCCmec typing were performed by multiplex PCR.
All isolates were resistant to cefoxitin and oxacillin. All isolates were susceptible to vancomycin.
Strong producers.
Of the S. haemolyticus isolates containing mecA, only 8 (14.5%) isolates could be typed and they were classified as SCCmec type V (ccr complex type 5, mec complex class C2). SCCmec subtype V.1 was not detected. Four (7.3%) isolates contained more than one ccr complex. 43 (78.1%) of the isolates did not contain mec complex, but 13 (26.6%) of them contained ccr. More than half of the isolates (30/55, 54.5%) did not contain ccr or mec complexes (Table 4).
Table 4.
SCCmec typing, resistance profile, and biofilm production of S. haemolyticus isolates.
| No. of isolates | SCCmec typing |
Resistance profilea |
Biofilm productionb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| mec class | ccr type | SCCmec | CLI | TET | LZD | ERY | Glucose | NaCl | |
| 8 | C2 | 5 | V | 7 (88) | 3 (38) | 0 (0) | 7 (88) | 7 (88) | 4 (50) |
| 1 | C2 | 1 + 2 + 5 | Variant | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| 3 | C2 | 1 + 4 + 5 | Variant | 3 (100) | 3 (100) | 0 (0) | 3 (100) | 3 (100) | 2 (67) |
| 12 | NT | 4 | NT | 10 (83) | 3 (25) | 0 (0) | 11 (92) | 8 (67) | 5 (42) |
| 1 | NT | 5 | NT | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) |
| 30 | NT | NT | NT | 30 (100) | 3 (10) | 1 (3) | 30 (100) | 23 (77) | 18 (60) |
NT, not typeable; CLI, clindamycin; TET, tetracycline; LZD, linezolid; ERY, erythromycin.
Methicillin-resistance was evaluated using the cefoxitin disk test, minimum inhibitory concentration was determined using the broth microdilution method. Biofilm formation was evaluated by crystal violet staining. Amplification of the ica operon and SCCmec typing were performed by multiplex PCR.
All isolates were resistant to cefoxitin, oxacillin, norfloxacin, levofloxacin and trimethoprim/sulfamethoxazole. All isolates were susceptible to vancomycin.
Strong producers.
Clonal diversity
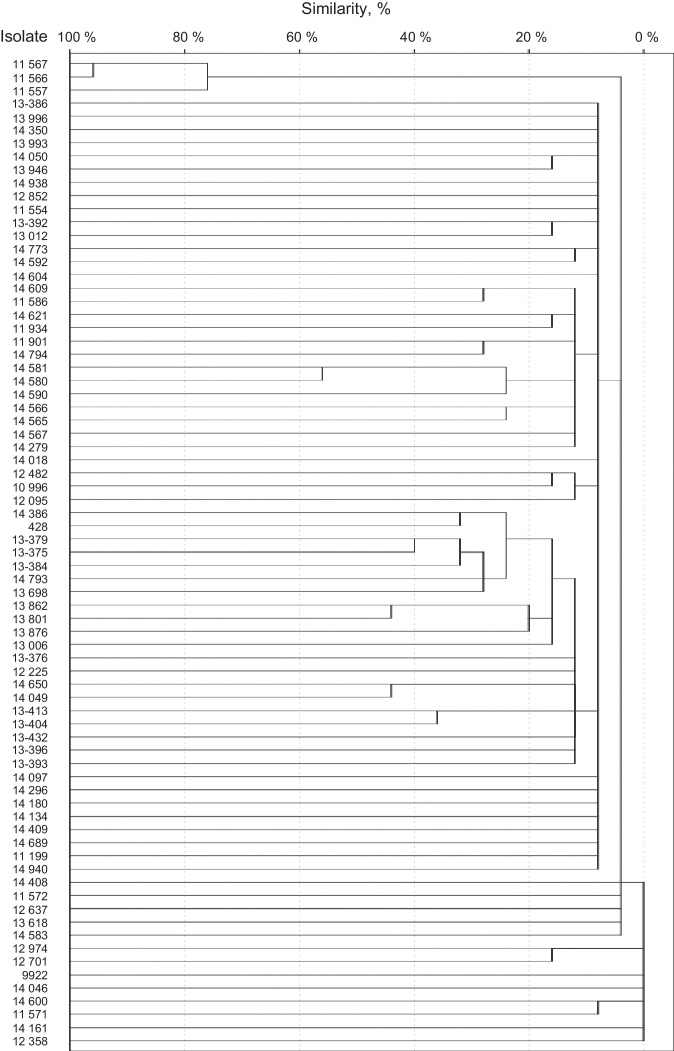
SmaI restriction digestion of 76 S. epidermidis isolates generated 7–17 fragments. For seven of the isolates, three or fewer fragments were generated. Therefore, these isolates were not considered for further analysis. A total of 75 different restriction patterns with similarities ranging from 0 to 95% were obtained. Only two of the isolates (isolates 11566 and 11567) were 95% homologous and were considered as a clone (Fig. 1). Both strains were classified as SCCmec type III by the methodology described by Zhang et al., but were classified as variants by Kondo et al. methodology, since they also amplified ccrC. Both were resistant to norfloxacin, clindamycin, levofloxacin, trimethoprim/sulfamethoxazole, erythromycin, oxacillin, and cefoxitin, but susceptible to linezolid and vancomycin. Also, both strains were classified as strong biofilm producers by both supplemented broths and proceed from intensive care units.
Fig. 1.
Dendrogram of S. epidermidis isolates.
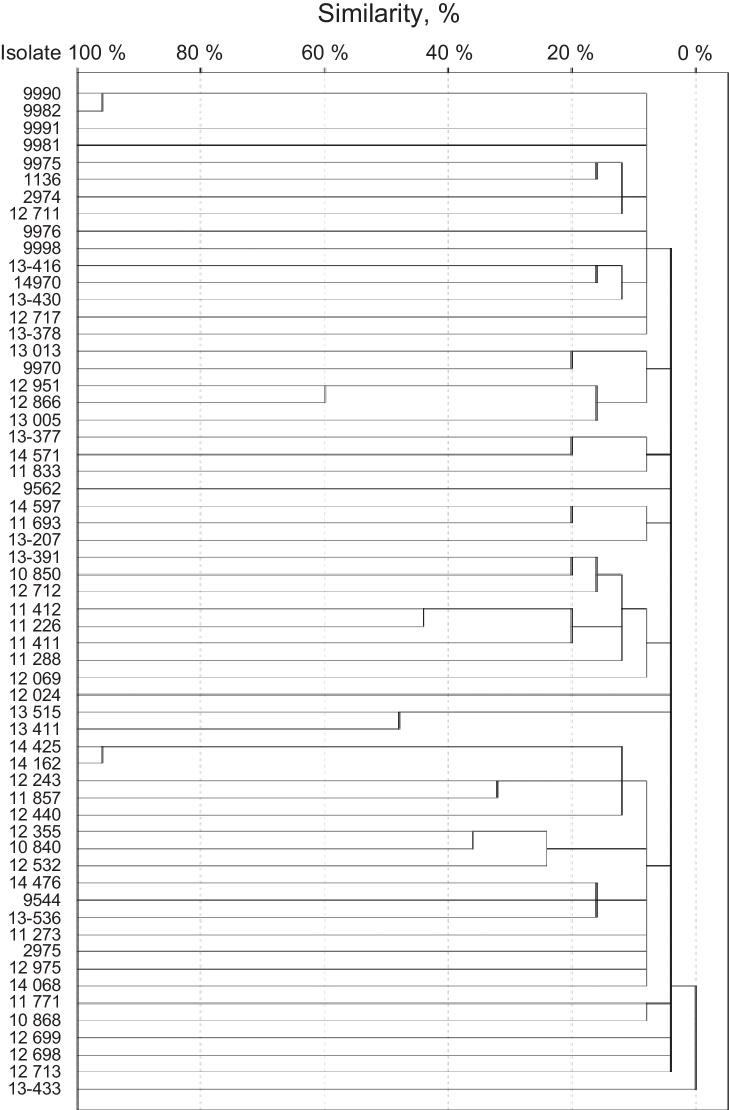
SmaI restriction digestion of the S. haemolyticus isolates generated 7–15 fragments, and 57 different patterns were produced. Isolates 9990 and 9982 were considered clone A, and isolates 14425 and 14162 were considered clone B (Fig. 2). Both clones had 95% of similarity. The remaining isolates had similarities of 60% or less.
Fig. 2.
Dendrogram of S. haemolyticus isolates.
Isolates 9990 and 9982 were classified as non-typable SCCmec. Both were resistant to clindamycin, levofloxacin, trimethoprim/sulfamethoxazole, erythromycin, oxacillin, and cefoxitin. Also, both strains were classified as strong biofilm producers by NaCl-supplemented broths and proceed from intensive care units. Isolates 14425 and 14162 were classified as SCCmec type V. Both were resistant to norfloxacin, clindamycin, levofloxacin, trimethoprim/sulfamethoxazole, erythromycin, oxacillin, and cefoxitin. Also, both strains were classified as strong biofilm producers by both supplemented broths and proceed from intensive care units.
MLST typing
Five S. epidermidis isolates were chosen to be analyzed by MLST: the linezolid-resistant isolates, an isolate representative of the group harboring SCCmec type IV, and an isolate representative of the group harboring SCCmec type III. MLST analysis showed that all the linezolid-resistant S. epidermidis isolates (n = 3) belonged to ST23 while the isolate with SCCmec type IV and the isolate with SCCmec type III belonged to ST7 and ST2, respectively.
Discussion
CoNS infections are associated with high antimicrobial resistance, which makes them difficult to treat. In this study, we analyzed S. epidermidis and S. haemolyticus human blood isolates for biofilm formation, antimicrobial resistance, SCCmec type, and clonal diversity.
We identified three linezolid-resistant S. epidermidis and two linezolid-resistant S. haemolyticus isolates. Four of these were also methicillin-resistant. Linezolid is a synthetic oxazolidinone that inhibits protein synthesis by binding to ribosomal peptidyl transferase, and it is recommended to treat multidrug-resistant Gram-positive infections. Although linezolid resistance is rare (<1% in S. aureus and <2% in CoNS), the emergence of linezolid-resistant strains is a serious healthcare concern.23 In the Hospital Civil in Guadalajara, linezolid is the most commonly used antibiotic for nosocomial pneumonia, surgical wound infections, and is also used for bloodstream infections not associated with catheter. In order to reduce the spreading of linezolid-resistant strains a stewardship antibiotic consumption program limiting linezolid use has been implemented.
In two of the isolates, linezolid-resistance was associated with the presence of the cfr gene, which encodes for an adenine methyltransferase that modifies the adenosine at position 2503 in 23S rRNA. cfr also confers resistance to phenicol compounds, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A.24 cfr has primarily been found on plasmids in S. epidermidis and S. haemolyticus isolates,25, 26 but can also be found on the chromosome as a result of the activities of transposons and insertion sequences.27 The presence of cfr on mobile genetic elements, which may carry additional resistance genes, probably facilitates cfr dissemination. The two cfr-containing isolates had different MIC values for linezolid; isolate 9976 had an MIC of 8 μg/mL, whereas isolate 14565 had an MIC > 32 μg/mL. These differences suggested the presence of additional resistance mutations in isolate 14565, so we sought for mutations in domain V of 23S rRNA in all linezolid-resistant isolates. However, only isolate 2975 harbored the G2576T mutation, which is the most frequently reported.28 This isolate had an MIC of 32 μg/mL and no cfr gene. Linezolid-resistance can also be conferred by mutations in ribosomal proteins L3 and/or L4.29 Our linezolid-resistant isolates should be analyzed further to identify additional mutations explaining the differences in MIC.
In Mexico, S. epidermidis STs 2, 23, 46, 61, 71, and 82 have been reported.30 The three linezolid-resistant S. epidermidis isolates analyzed belonged to ST23. Linezolid-resistance has been reported in the ST23 clone. In a study from Italy, clinical isolates from blood and cerebrospinal fluid were analyzed. The most frequently ST found in linezolid-resistant strains were ST23; also, almost the half of the ST23 strains were cfr positive (22/50), which suggested an association between the genetic background with the cfr gene.31 ST23 has also been reported in Argentina, Germany, Greece, Hungary, Iceland, Poland, Portugal, United States and Uruguay (http://sepidermidis.mlst.net).
One of our isolates was ST2. Linezolid-resistant strains belonging to ST2 have been described in a Brazilian tertiary-care hospital in isolates from skull, blood, and catheter cultures.32 ST2 has also been described in Germany, Argentina, Italy, Poland, Spain, Mexico, Cape Verde, Denmark, Greece, Hungary, Uruguay; Bulgaria and Colombia, Japan, Netherlands, and United States. (http://sepidermidis.mlst.net). In Mexico, Juarez-Verdayes et al. analyzed isolates from healthy skin, healthy conjunctiva, and ocular infections; ST2 lineage was the most frequent among the isolates (50% for healthy skin, 25% for healthy conjunctiva and 46.5% for ocular infections).33 Similarly, Flores-Paez et al. reported ST2 and ST23 from isolates from ocular infections.34 None of ST23 or ST2 has been previously reported from sterile sites in Mexico. To the best of our knowledge S. epidermidis ST7 has not been reported in Mexico. ST7 has been reported in isolates from catheter-related bloodstream infections35 and prosthetic valve endocarditis.14
Both S. epidermidis and S. haemolyticus have been shown to be strong biofilm producers, and the ica operon has been associated with biofilm production in S. epidermidis.36 We found the ica operon to be present in 36 of the S. epidermidis isolates, and 35 of these isolates were strong biofilm producers. On the other hand, the ica operon was not found in any of the S. haemolyticus isolates; this is consistent with a report by Fredheim et al. in which nearly all of their S. haemolyticus isolates were ica-negative.37 Although S. haemolyticus has been associated with ica-independent biofilm formation, a report by Pereira et al. showed that 58% of their isolates harbored icaA.38 Genes such as aap, bap, bhp have been shown to be involved in ica-independent biofilm formation.4 Moreover, studies on biofilm detachment have shown that PIA is not a major component of S. haemolyticus biofilms.37
All three linezolid-resistant S. epidermidis isolates were strong biofilm producers, and two were positive for the ica operon. Biofilm formation has long been associated with antibiotic resistance, and increased MICs have been linked to the presence of biofilms.39 One S. haemolyticus linezolid-resistant isolate was a strong biofilm producer, but neither S. haemolyticus linezolid-resistant isolate contained the ica operon. Resistance to norfloxacin, clindamycin, levofloxacin, erythromycin, cefoxitin, and oxacillin were associated with strong biofilm production (p < 0.05) for both the S. epidermidis and S. haemolyticus isolates.
As previously described, SCCmec type IV, especially SCCmec type IVa, was frequently found (47.2%) among the S. epidermidis isolates. This frequency has been found among isolates from both inpatients and carriers. In a study that analyzed 44 blood isolates, SCCmec type IV was found at a frequency of 36%,40 whereas SCCmec type IVa was found at a frequency of 65% among healthy subjects.41 Wisplinghoff et al. suggested that genetic information was transmitted between S. epidermidis and S. aureus since SCCmec IV sequences from both species were >98% homologous.40 Transmission of SCCmec between these species may lead to an increase in beta-lactam antibiotic resistance in S. aureus; currently, methicillin-resistance in S. aureus is not as high as in CoNS.
In this study, we found several combinations of mec and ccr complexes in S. epidermidis. Only two SCCmec types (IV and III) were identified. The other S. epidermidis isolates had mec and ccr combinations distinct from the 11 SCCmec types reported to date or could not be typed. A problem with amplification of more than one ccr complex is that it is unknown whether the amplified ccr complexes are located within the same SCCmec.11, 42
In S. haemolyticus isolates the only SCCmec found was SCCmec type V, and it has been found at frequencies as high as 55%.6, 11 Similar to the transfer of SCCmec type IV from S. epidermidis to S. aureus, SCCmec type V may be transferred from S. haemolyticus to S. aureus. Notably, we found that most of the S. haemolyticus isolates could not be typed. Likewise, Barros et al.6 reported that 43% (24 isolates) of their S. haemolyticus isolates could not be typed. There appears to be great SCCmec diversity among S. haemolyticus isolates.
Despite being commensals, both S. epidermidis, and S. haemolyticus have been found to be clonal, particularly S. haemolyticus.7 However, only a clone composed of two S. epidermidis isolates was found. In addition, two clones with two strains or S. haemolyticus each one were detected in this study. Thus, the hypothesis of clonal transmission at the two source hospitals is discarded. Since both species are components of the normal microbiota, it is likely that the infections were endogenous.
The actual knowledge of the molecular epidemiology of both species includes an extremely high genetic diversity and recombination. PFGE is mostly useful for an outbreak or short-term epidemiological investigations and may not detect clonal relatedness in isolates recovered through the years. On the contrary, MLST is more useful, particularly for the S. epidermidis isolates since it allows to determine their genetic backgrounds and to compare to those previously described from other countries. Due to the relevance of the linezolid-resistant isolates, MLST analysis was considered for these isolates.
In conclusion, this study is the first performed in Mexico that characterizes biofilm production, antimicrobial susceptibility, SCCmec and clonal relatedness of S. epidermidis and S. haemolyticus blood isolates. Both species were found to be highly resistant to antibiotics. We also detected linezolid-resistance, which is a concern for infection control practices.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by granting CB-2011-01-167802 from CONACyT (Mexican Council for Science and Technology). The authors thank Lucy Acevedo for her assistance in the laboratory.
References
- 1.Favre B., Hugonnet S., Correa L., Sax H., Rohner P., Pittet D. Nosocomial bacteremia: clinical significance of a single blood culture positive for coagulase-negative staphylococci. Infect Control Hosp Epidemiol. 2005;26:697–702. doi: 10.1086/502605. [DOI] [PubMed] [Google Scholar]
- 2.Koksal F., Yasar H., Samasti M. Antibiotic resistance patterns of coagulase-negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiol Res. 2009;164:404–410. doi: 10.1016/j.micres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Longauerova A. Coagulase negative staphylococci and their participation in pathogenesis of human infections. Bratisl Lek Listy. 2006;107:448–452. [PubMed] [Google Scholar]
- 4.Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 5.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barros E.M., Ceotto H., Bastos M.C., Dos Santos K.R., Giambiagi-Demarval M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J Clin Microbiol. 2012;50:166–168. doi: 10.1128/JCM.05563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristof K., Kocsis E., Szabo D., et al. Significance of methicillin-teicoplanin resistant Staphylococcus haemolyticus in bloodstream infections in patients of the Semmelweis University hospitals in Hungary. Eur J Clin Microbiol Infect Dis. 2011;30:691–699. doi: 10.1007/s10096-010-1142-5. [DOI] [PubMed] [Google Scholar]
- 8.Barber M. Methicillin-resistant staphylococci. J Clin Pathol. 1961;14:385–393. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.IWG-SCC Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanssen A.M., Kjeldsen G., Sollid J.U. Local variants of Staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48:285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruppe E., Barbier F., Mesli Y., et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother. 2009;53:442–449. doi: 10.1128/AAC.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnie J.P., Naderi-Nasab M., Loudon K.W., Matthews R.C. An epidemiological study of blood culture isolates of coagulase-negative staphylococci demonstrating hospital-acquired infection. J Clin Microbiol. 1997;35:1746–1750. doi: 10.1128/jcm.35.7.1746-1750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingenberg C., Ronnestad A., Anderson A.S., et al. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin Microbiol Infect. 2007;13:1100–1111. doi: 10.1111/j.1469-0691.2007.01818.x. [DOI] [PubMed] [Google Scholar]
- 14.Thomas J.C., Vargas M.R., Miragaia M., Peacock S.J., Archer G.L., Enright M.C. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol. 2007;45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotaki S., Sasaki T., Kuwahara-Arai K., Hiramatsu K. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J Clin Microbiol. 2011;49:3627–3631. doi: 10.1128/JCM.00488-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong T., Li X., Wang J., Sloan C., Cicogna C. Sequential linezolid-resistant Staphylococcus epidermidis isolates with G2576T mutation. J Clin Microbiol. 2007;45:3277–3280. doi: 10.1128/JCM.02048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehrenberg C., Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother. 2006;50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen G.D., Simpson W.A., Younger J.J., et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arciola C.R., Gamberini S., Campoccia D., et al. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J Biomed Mater Res A. 2005;75:408–413. doi: 10.1002/jbm.a.30445. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K., McClure J.A., Elsayed S., Louie T., Conly J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo Y., Ito T., Ma X.X., et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aanensen D.M., Spratt B.G. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 2005;33(Web Server issue):W728–W733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamm R.K., Mendes R.E., Ross J.E., Sader H.S., Jones R.N. An international activity and spectrum analysis of linezolid: ZAAPS Program results for 2011. Diagn Microbiol Infect Dis. 2013;76:206–213. doi: 10.1016/j.diagmicrobio.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Long K.S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tewhey R., Gu B., Kelesidis T., et al. Mechanisms of linezolid resistance among coagulase-negative staphylococci determined by whole-genome sequencing. MBio. 2014;5 doi: 10.1128/mBio.00894-14. e00894–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L., Wang Y., Li Y., et al. Cfr-mediated linezolid-resistance among methicillin-resistant coagulase-negative staphylococci from infections of humans. PLOS ONE. 2013;8:e57096. doi: 10.1371/journal.pone.0057096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehrenberg C., Aarestrup F.M., Schwarz S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother. 2007;51:483–487. doi: 10.1128/AAC.01340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiles-Melero I., Gomez-Gil R., Romero-Gomez M.P., et al. Mechanisms of linezolid resistance among Staphylococci in a tertiary hospital. J Clin Microbiol. 2013;51:998–1001. doi: 10.1128/JCM.01598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendes R.E., Deshpande L.M., Jones R.N. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat. 2014;17:1–12. doi: 10.1016/j.drup.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Miragaia M., Thomas J.C., Couto I., Enright M.C., de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanile F., Mongelli G., Bongiorno D., et al. Worrisome trend of new multiple mechanisms of linezolid resistance in staphylococcal clones diffused in Italy. J Clin Microbiol. 2013;51:1256–1259. doi: 10.1128/JCM.00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Almeida L.M., Lincopan N., de Araujo M.R., Mamizuka E.M. Dissemination of the linezolid-resistant Staphylococcus epidermidis clone ST2 exhibiting the G2576T mutation in the 23S rRNA gene in a tertiary-care hospital, Brazil. J Antimicrob Chemother. 2012;67:768–769. doi: 10.1093/jac/dkr538. [DOI] [PubMed] [Google Scholar]
- 33.Juarez-Verdayes M.A., Ramon-Perez M.L., Flores-Paez L.A., et al. Staphylococcus epidermidis with the icaA(−)/icaD(−)/IS256(−) genotype and protein or protein/extracellular-DNA biofilm is frequent in ocular infections. J Med Microbiol. 2013;62(Pt 10):1579–1587. doi: 10.1099/jmm.0.055210-0. [DOI] [PubMed] [Google Scholar]
- 34.Flores-Paez L.A., Zenteno J.C., Alcantar-Curiel M.D., et al. Molecular and phenotypic characterization of Staphylococcus epidermidis isolates from healthy conjunctiva and a comparative analysis with isolates from ocular infection. PLOS ONE. 2015;10:e0135964. doi: 10.1371/journal.pone.0135964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherifi S., Byl B., Deplano A., Nonhoff C., Denis O., Hallin M. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol. 2013;51:1541–1547. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziebuhr W., Heilmann C., Gotz F., et al. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fredheim E.G., Klingenberg C., Rohde H., et al. Biofilm formation by Staphylococcus haemolyticus. J Clin Microbiol. 2009;47:1172–1180. doi: 10.1128/JCM.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira P.M., Binatti V.B., Sued B.P., et al. Staphylococcus haemolyticus disseminated among neonates with bacteremia in a neonatal intensive care unit in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2014;78:85–92. doi: 10.1016/j.diagmicrobio.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Dunne W.M., Jr. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wisplinghoff H., Rosato A.E., Enright M.C., Noto M., Craig W., Archer G.L. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother. 2003;47:3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamaluddin T.Z., Kuwahara-Arai K., Hisata K., et al. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol. 2008;46:3778–3783. doi: 10.1128/JCM.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanssen A.M., Sollid J.U. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob Agents Chemother. 2007;51:1671–1677. doi: 10.1128/AAC.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]