Abstract
We report a simple, postsynthetic strategy for synthesis of oligonucleotides containing 2,6-diaminopurine nucleotides and 2-aminoadenine conjugates using 2-fluoro-6-amino-adenosine. The strategy allows introduction of 2,6-diaminopurine and other 2-amino group-containing ligands. The strongly electronegative 2-fluoro deactivates 6-NH2 obviating the need for any protecting group on adenine, and simple aromatic nucleophilic substitution of fluorine makes reaction with aqueous NH3 or R-NH2 feasible at the 2-position.
The 2,6-diaminopurine (DAP) nucleobase was discovered in S-L2 cyanophage DNA in 1977.1 DAP can form three hydrogen bonds with thymidine in DNA (Figure 1) and uridine in RNA,2−4 enhancing duplex stability by approximately 1–2 °C per modification relative to adenine.5,6 NMR and circular dichroism studies showed that a duplex between DNA modified with DAP and complementary RNA adopts the A-form conformation7,8 and that the duplex formed between DNA modified with DAP and complementary DNA adopts the B-form, which transitions to the A-form in high salt.9,10 Thus, the DAP modification does not significantly alter the conformation of nucleic acid duplexes.
Figure 1.
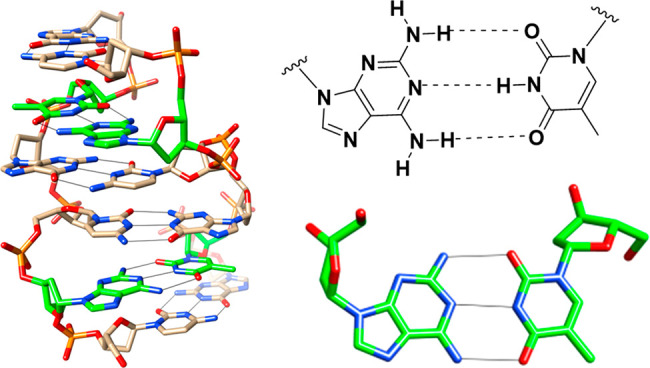
Three hydrogen bonds between DAP (D) and thymine as seen in the crystal structure of [d(CDCGTG)]2 (PDB ID 1VTY).2
The DAP modification has been studied in the context of numerous nucleic acid sugar modifications, including 2′-O-methyl (2′-OMe),11−13 2′-O-allyl,14 and 2′-O-propargyl,15 as well as in combination with anhydrohexitol,16,17 threose nucleic acid,18 locked nucleic acid (LNA),13,19,20 unlocked nucleic acid,21 double headed dimers,22 2′-5′ linked dimers,23−25 N3′-P5′ linked dimers,26,27 peptide nucleic acid,28 and serinol nucleic acid.29,30 Generally, DAP building blocks have been obtained from the guanosine analogue, but yields are low.3,4,31−34 Synthetic routes have been developed using halogenated purines such as the 6-chloro-2-aminopurine,11,13,14,16,17,20,35 2,6-dicholoropurine,36−38 and the commercially available 2,6-diaminopurine.29,39 However, these strategies involve multiple, tedious synthetic steps. Because of the different reactivities of the two amines, the protection and deprotection strategies are difficult, and purification can be a challenge.
Starting from the free 2,6-diaminopurine containing nucleosides, various protected DAP phosphoramidites have been obtained using homoprotecting group approaches with benzoyl,5,19 acetyl,13 isobutyl,13 dimethylformamidine,40 phenoxyacetyl,11 or Fmoc.20,41,40 Again, because of the different reactivities of the two amines, protection and deprotection are difficult. Thus, the yields of protected DAP phosphoramidites are generally low, and deprotection is inefficient.42 The heteroprotecting group strategy has also been employed, but synthesis becomes more complicated.15,39
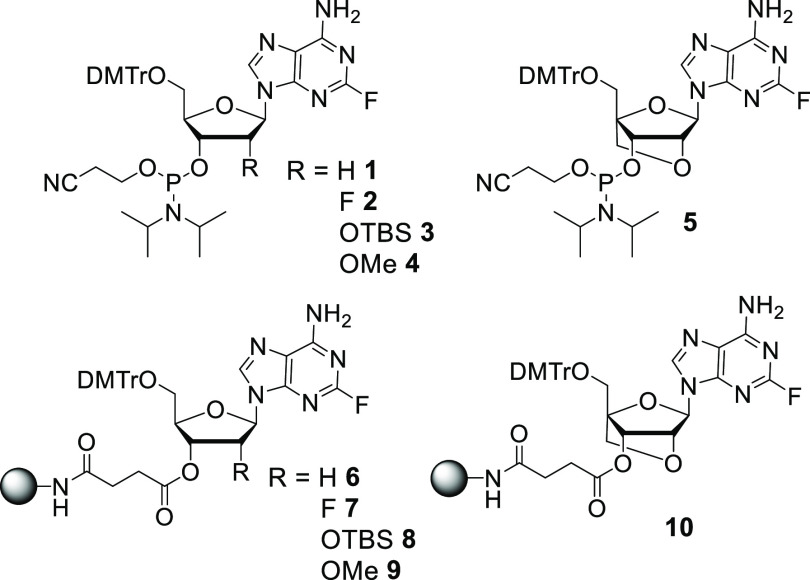
We previously reported the use of 2-fluoro-6-amino purines as novel precursors for DAP,43,44 but these patent protocols have not been embraced. This prompted us to revisit this area, and herein we describe a synthetic route for simple and efficient incorporation of DAP into oligonucleotides using a postsynthetic strategy. To avoid oligonucleotide deprotection issues, we used the 2-fluoro-6-amino-adenosine monomer as the oligonucleotide building block. The combination of the fluorine inductive effect (−I) and the resonance effect (+R) of the N6 amino meant that no protecting group was required during phosphoramidite or solid-support synthesis. We describe synthesis of 2′-deoxy, 2′-OMe, 2′-fluoro (2′-F), ribo, and LNA phosphoramidites and solid supports containing 2-fluoro-6-aminopurine (Figure 2). A postsynthetic treatment with ammonia yielded the DAP-modified oligonucleotide or, if an amine functionality was used, a 2-position conjugate.
Figure 2.
2-fluoro-6-aminopurine phosphoramidites and solid supports synthesized.
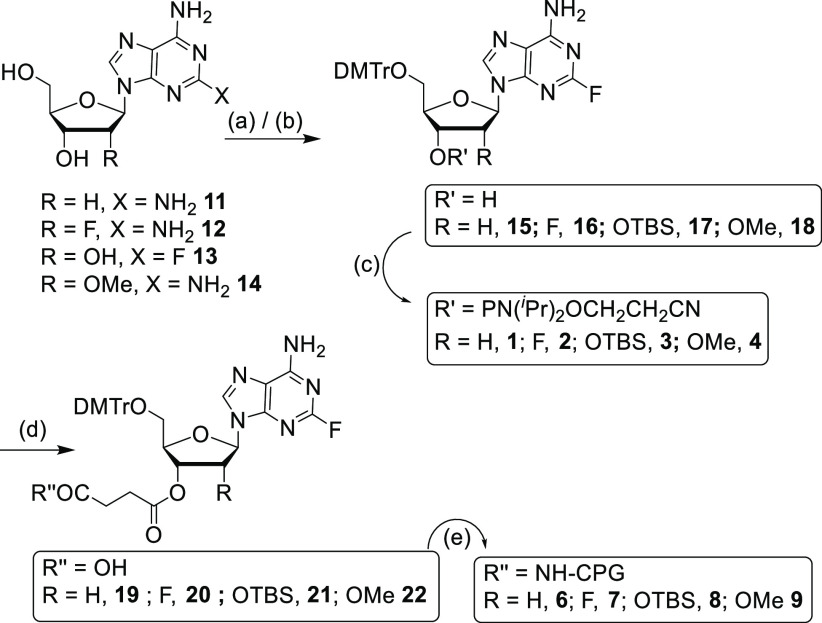
Two different synthetic approaches were undertaken to afford five different 2-fluoro-6-aminopurine nucleosides depending on the available starting materials. For 2′-deoxy, 2′-F, and 2′-OMe analogues, 2,6-diaminopurine nucleosides were used as the starting material. Following the literature procedure,45 2-fluoro-6-aminopurine nucleosides were obtained from 2,6-diaminopurine (Schemes 1 and 2). Diazotization reactions of the diaminopurines 11, 12, and 14(11) in the presence of 70% HF-pyridine and tert-butyl nitrite afforded the 2-fluoro analogues. Diazotization is highly selective for the 2-NH2 vs 6-NH2 as demonstrated earlier.45,46
Scheme 1. Synthesis of Phosphoramidites 1–4 and CPGs 6–9.
Reaction conditions: (a) (i) 70% HF-pyridine, tert-butyl nitrite, (ii) DMTrCl, pyridine for 15 (27%), 16 (66%), and 18 (60%); (b) (i) DMTrCl, pyridine; (ii) AgNO3, pyridine, TBDMSCl, separation of 3′-isomer for 17 (17%); (c) PClN(iPr)2OCH2CH2CN, DIPEA, NMI, DCM, room temperature, 1 h for 1 (71%), 2 (84%), 3 (89%), and 4 (85%); (d) succinic anhydride, DMAP, DCM, room temperature, 3 h for 19 (71%), 20 (77%), 21 (93%), and 22 (78%); (e) NH2–CPG (171 μmol/g), HBTU, DIPEA, acetonitrile for 6 (96 μmol/g), 7 (107 μmol/g), 8 (89 μmol/g), and 9 (124 μmol/g). Yields are over two steps for 15–18.
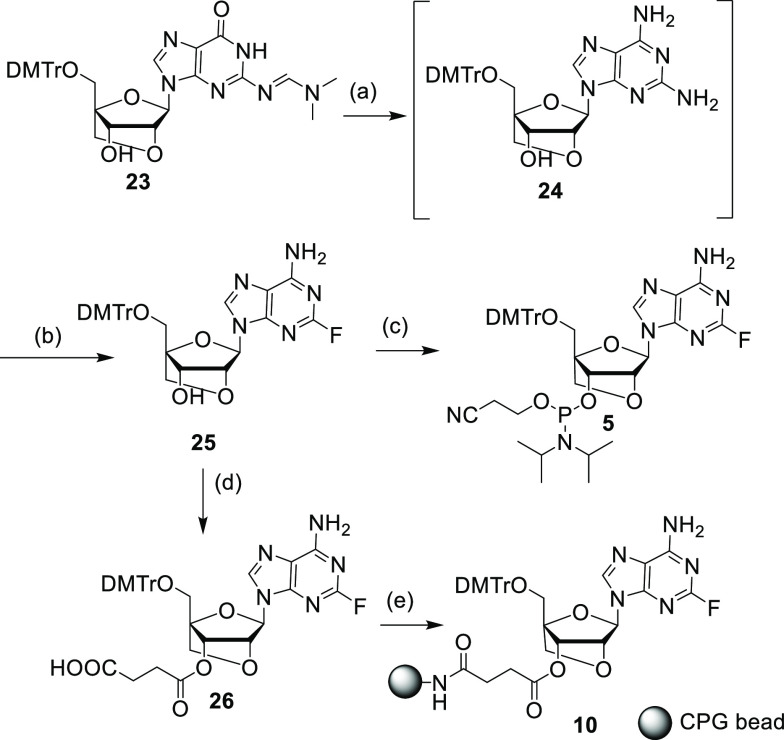
Scheme 2. Synthesis of Phosphoramidite 5 and CPG 10.
Reaction conditions: (a) (i) NaOH, MeOH, 1 M HCl, (ii) trifluoroacetic anhydride, liq. NH3, −60 °C; (b) 70% HF/30% pyridine, tert-butyl nitrite for 25 (34% over two steps); (c) PClN(iPr)2OCH2CH2CN, DIPEA, NMI, DCM, room temperature, 1 h for 5 (85%); (d) succinic anhydride, DMAP, DCM, room temperature, 3 h for 26 (95%); (e) NH2–CPG (171 μmol/g), HBTU, DIPEA, acetonitrile for 10 (104 μmol/g).
The 5′-OH groups were then protected with 4,4′-dimethoxytriphenylmethyl chloride (DMTrCl) to afford the 5′-O-DMTr-2-fluoro-6-aminopurines 15, 16, and 18, respectively. Subsequent phosphitylation reactions of 15, 16, and 18 with 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite under basic conditions afforded the 2′-deoxy, 2′-F, and 2′-OMe analogues of 2-fluoro-6-aminopurine phosphoramidites 1, 2, and 4, respectively, in good yields. The 2′-ribo analogue 13 was synthesized from the 2,6-diaminopurine ribonucleoside following the literature procedure.47 The 5′-OH group of 13 was protected first with DMTr, and then the 2′-OH group was protected with TBS to afford compound 17. The 3′-O-TBS-protected minor isomer was separated from the desired product 17 by column chromatography, and 17 was converted to the corresponding phosphoramidite 3 in good yield. To obtain solid supports on controlled pore glass (CPG),4815–18 were reacted with succinic anhydride in the presence of N,N-dimethylaminopyridine (DMAP) to afford compounds 19–22, respectively. The carboxylic acid groups of these succinates were coupled with the free amine groups of the CPG beads under standard coupling conditions.49,50 Unreacted sites on CPG were then capped with acetic anhydride, and loading values were calculated for the resulting nucleoside-modified CPGs 6–9 (Scheme 1).
For the LNA analogue, commercially available guanosine nucleoside 23 was converted to corresponding 2,6-diaminopurine compound 24 and used for the next step without further purification. Compound 24 was converted to the corresponding 2-fluoro-6-aminopurine compound 25 via diazotization, followed by fluorination, with moderate yield over the two steps. Compound 25 was then converted to the corresponding phosphoramidite 5 and CPG 10 in good yields (Scheme 2).
To evaluate the efficacy of the postsynthetic conversion of 2-fluoro-6-aminopurine to DAP, we synthesized several oligonucleotides containing single or multiple incorporations using standard solid-phase synthesis methods. As models, we used previously described small interfering RNA (siRNA) targeting mouse Ttr,51 single stranded antisense oligonucleotides called REVERSIRs,52 and an antagomir targeting miR-122.53 For oligonucleotide sequences, see Supporting Information (SI) Tables S1–S4. The standard ammonia deprotection step at 60 °C for 5 h caused displacement of 2-fluoro by ammonia and deprotection of the oligonucleotides and simultaneous cleavage from solid support, yielding the DAP-modified strands for all oligonucleotides with the exception of the oligonucleotides modified with the ribo monomer. For the oligonucleotides modified with the ribo 2-fluoro-6-aminopurine, ammonia treatment at room temperature followed by Et3N.3HF treatment resulted in 2′-silyl deprotection without 2-fluoro substitution.
To overcome this problem, a second ammonia treatment for 5 h at 60 °C was attempted; however, the second ammonia treatment led to degradation of RNA due to base hydrolysis. Therefore, to obtain the pure ribo-DAP-modified oligonucleotides, oligonucleotides were first treated with ammonia at 65 °C for 5 h followed by triethylamine trihydrofluoride treatment to yield the desired oligonucleotides (see SI Table S3).
After purification, the strands were analyzed and characterized by ion exchange chromatography and LCMS respectively. The LCMS analyses of representative strands modified with deoxy-2,6-diaminopurine (ON5), 2′-F-2,6-diaminopurine (ON10), LNA-2,6-diaminopurine (ON15), 2′-OMe-2,6-diaminopurine (ON21), and mixed 2′-F- and 2′-OMe-2,6-diaminopurine (ON22) demonstrated that 2,6-diaminopurine was present and that there were no unexpected modifications to other positions (Figure 3).
Figure 3.
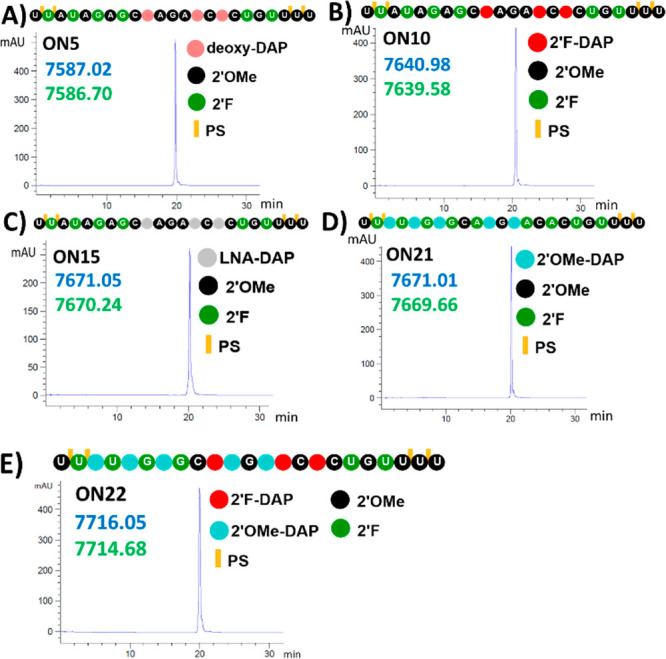
LCMS analysis of purified oligonucleotides (A) ON5, (B) ON10, (C) ON15, (D) ON21, and (E) ON22 demonstrating incorporation of 2,6-diaminopurines with calculated (blue) and observed (green) masses. In bead diagrams of strands, black and green indicate 2′-OMe and 2′-F sugar modifications, respectively. Pink, red, gray, and light blue beads represent deoxy-DAP, 2′-fluoro-DAP, LNA-DAP, and 2′-OMe-DAP, respectively. Phosphorothioate linkages are indicated by a yellow vertical line.
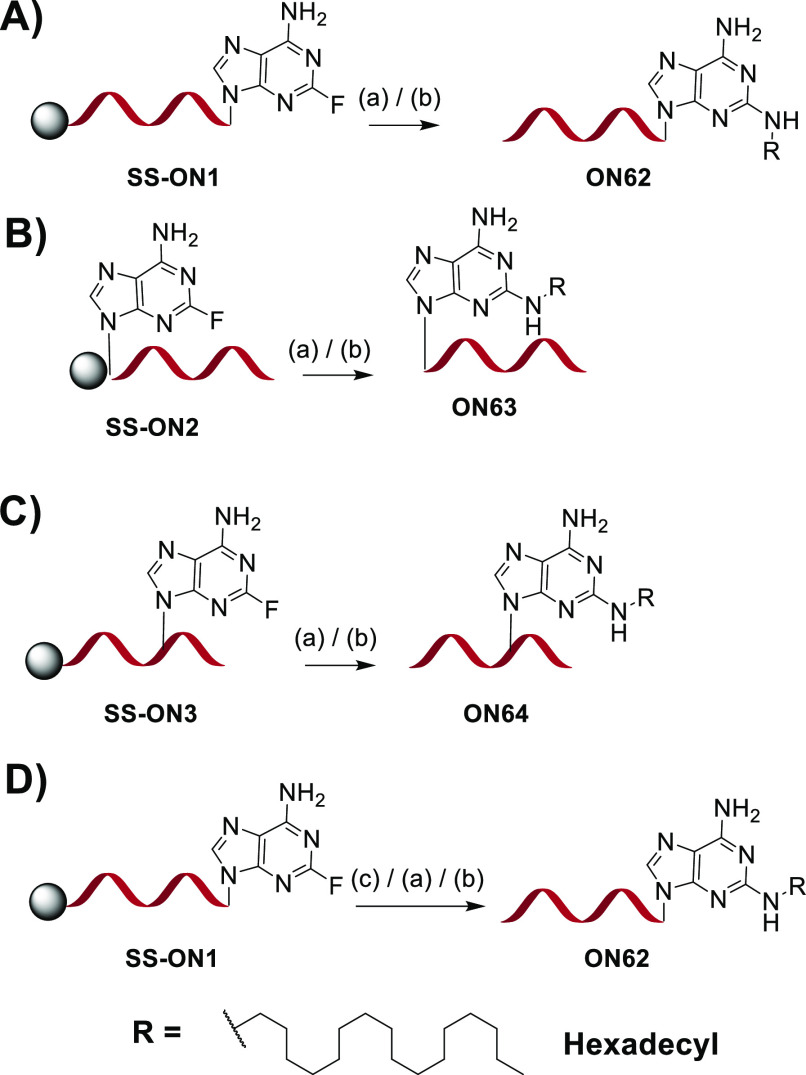
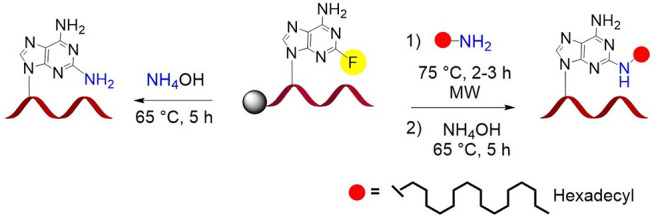
Delivery of RNA molecules to the appropriate tissue or cell has been a challenge. However, liver hepatocyte-specific delivery of three clinically approved siRNAs, givosiran, lumasiran, and inclisiran is made possible by conjugation to N-acetylgalactosamine (GalNAc), the ligand for the asialoglycoprotein receptor.51 In earlier examples, conjugation to lipophilic molecules like cholesterol and fatty acids resulted in broad tissue distribution and cellular uptake in liver and central nervous system.54−57 We have recently demonstrated that siRNAs can be targeted to extrahepatic tissues using a 2′-O-lipophile-functionalized nucleoside conjugate.58 As fluorine is a good leaving group for the nucleophilic aromatic substitution reaction, we reasoned that 2-fluoro-6-aminopurine could serve as a site for postsynthetic conjugation of lipid amines at the 2-position of adenine of an oligonucleotide on solid support or in solution. To test this, the 2-fluoro-6-aminopurine monomer was incorporated at the 5′ end or the 3′ end of the Ttr siRNA sense strand or at position 5 of the Ttr siRNA antisense strand (see SI Figure S1–S2 and Tables S5 and S6). Hexadecylamine was used as the ligand (Figure 4A–C). The CPGs were first heated by microwave for 2 to 3 h. Due to high loading and the narrow space (500 Å) of the CPG pore size, 3′-end conjugation needed to be performed at 90 °C. In contrast, 5′ conjugation was completed in 2 h at 75 °C. Conjugation at an internal position was also performed at 90 °C.
Figure 4.
(A–C) Schematic of conjugation of hexadecylamine at position 2 of adenosine at (A) the 5′ end, (B) the 3′ end, and (C) an internal position of the oligonucleotide on solid support. (D) Schematic of solution-phase conjugation of hexadecylamine at position 2 of adenosine at the 5′ end of an oligonucleotide. Reaction conditions: (a) 0.1 M hexadecylamine in DMSO/EtOH/H2O (v/v/v, 1:2:1), DIPEA, 90 °C, MW, 2–4 h; (b) 28–30% NH4OH, 60 °C, 5 h; (c) 28–30% NH4OH, room temperature, 1 h.
Inspired by synthesis of the 2-fluoro derivative at the N2 position of deoxyguanosine,59 we developed a solution route to the synthesis of the 5′-end conjugate ON62 (Figure 4D). The oligonucleotide was cleaved from solid support by treatment with concentrated NH4OH solution for 1 h at room temperature. Under these conditions, the 2-fluoro substituent remained intact. Ammonia was evaporated from the solution, and conjugation with hexadecylamine was performed. More than 80% conjugation was achieved after 4 h at 75 °C assisted by microwave (Figure 5, see SI Figure S2 and Table S7).
Figure 5.
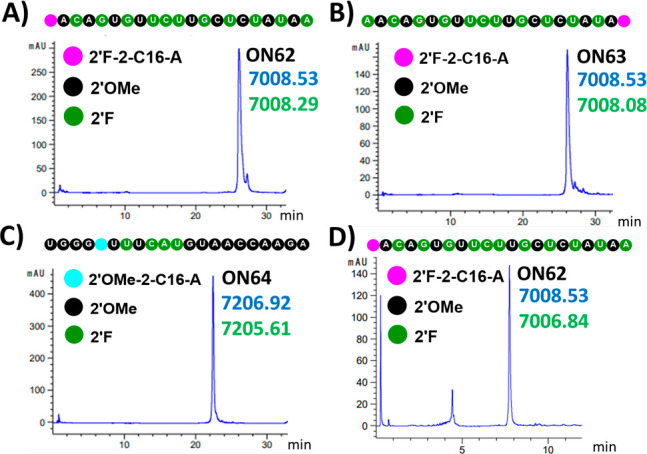
(A–C) LCMS analysis of purified (A) 5′-end conjugate ON62, (B) 3′-end conjugate ON63, and (C) internal conjugate ON64 obtained after on solid support conjugation. (D) LCMS analysis of crude reaction of 5′-end conjugate ON62 obtained by solution-phase conjugation. Calculated masses are in blue, and observed masses are in green. In the bead diagrams, black, green, light blue, and pink beads indicate 2′-OMe, 2′-F, 2-(-NHC16H33)-2′-O-methyl-adenosine, and 2-(-NHC16H33)-2′-fluoro-adenosine, respectively.
In summary, we have described the synthesis of 2-fluoro-6-aminopurine nucleoside phosphoramidites and CPG building blocks with five different sugar modifications (2′-H, 2′-OH, 2′-OMe, 2′-F, and LNA) and their use in oligonucleotide synthesis. No protecting groups were required on the 2-fluoro-6-amino-adenosine. Oligonucleotides containing these monomers were also conjugated to an amine-containing lipophilic ligand to yield oligonucleotide conjugates at the position 2 of the adenine by the displacement of fluorine. Conjugation was carried out both on solid support and in solution. Work to use this conjugation strategy to attach bulky ligands, such as trivalent GalNAc,51 in the minor groove is in progress. We also anticipate that strategic placement of conjugate within both sense strand and antisense strands of siRNAs apart from terminal ends will be valuable.60,61 The effect of the DAP modification on RNAi activity is also under investigation. Based on our previous work, chemical modifications are tolerated in most of the positions by the enzymes of the RNAi machinery,61 and the stabilizing effect of DAP on duplex formation may enhance siRNA activity in strategic positions. Overall, our methodologies will enable synthesis of oligonucleotides containing DAP, various N2-position minor groove conjugates, and other modifications for therapeutic and diagnostic applications and also for structural studies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.2c01848.
Experimental, and compound characterization (PDF)
The authors declare the following competing financial interest(s): All authors, except Professor Egli and Dr. Ross, are employees of Alnylam Pharmaceuticals.
Dedication
We dedicate this work to Dr. P. Dan Cook for his contributions to nucleoside, nucleotide, and oligonucleotide based therapeutics.
Supplementary Material
References
- Kirnos M. D.; Khudyakov I. Y.; Alexandrushkina N. I.; Vanyushin B. F. 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 1977, 270, 369–370. 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- Coll M.; Wang A. H.; van der Marel G. A.; van Boom J. H.; Rich A. Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs. J. Biomol. Struct. Dyn. 1986, 4, 157–72. 10.1080/07391102.1986.10506337. [DOI] [PubMed] [Google Scholar]
- Gaffney B. L.; Marky L. A.; Jones R. A. The influence of the purine 2-amino group on DNA conformation and stability-II: Synthesis and physical characterization of d[CGT(2-NH2)ACG], d[CGU(2-NH2)ACG], and d[CGT(2-NH2)AT(2-NH2)ACG]. Tetrahedron 1984, 40, 3–13. 10.1016/0040-4020(84)85098-X. [DOI] [Google Scholar]
- Gaffney B. L.; Marky L. A.; Jones R. A. The influence of the purine 2-amino group on DNA conformation and stability. Synthesis and conformational analysis of d[T(2-aminoA)]3. Nucleic Acids Res. 1982, 10, 4351–4361. 10.1093/nar/10.14.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A.; Cech T. R.; Usman N.; Beigelman L. The 2,6-Diaminopurine Riboside.cntdot.5-Methylisocytidine Wobble Base Pair: An Isoenergetic Substitution for the Study of G·U Pairs in RNA. Biochemistry 1994, 33, 13824–13835. 10.1021/bi00250a037. [DOI] [PubMed] [Google Scholar]
- Gryaznov S.; Schultz R. G. Stabilization of DNA:DNA and DNA:RNA duplexes by substitution of 2′-deoxyadenosine with 2′-deoxy-2-aminoadenosine. Tetrahedron Lett. 1994, 35, 2489–2492. 10.1016/S0040-4039(00)77151-6. [DOI] [Google Scholar]
- Howard F. B.; Miles H. T. 2NH2 A·T helixes in the ribo- and deoxypolynucleotide series. Structural and energetic consequences of 2NH2A substitution. Biochemistry 1984, 23, 6723–6732. 10.1021/bi00321a068. [DOI] [PubMed] [Google Scholar]
- Kikuchi K.; Taniyama Y.; Marumoto R. Evaluation of the 2 NH2A—T Pair in Hybridization, I Synthesis of the DNA/RNA Hybrid Oligomers Containing 2-Aminoadenosines. Zeitschrift für Naturforschung B 1988, 43, 623–630. 10.1515/znb-1988-0523. [DOI] [Google Scholar]
- Borah B.; Cohen J. S.; Howard F. B.; Miles H. T. Poly(d2NH2A-dT): Two-Dimensional NMR Shows a B to A Conversion in High Salt. Biochemistry 1985, 24, 7456–7462. 10.1021/bi00346a064. [DOI] [PubMed] [Google Scholar]
- Borah B.; Howard F. B.; Miles H. T.; Cohen J. S. Conversions of poly(2-aminodeoxyadenylate-5-halodeoxyuridylate) from B to A forms in high salt. An NMR and circular dichroism study. Biochemistry 1986, 25, 7464–7470. 10.1021/bi00371a031. [DOI] [PubMed] [Google Scholar]
- Sproat B. S.; Beijer B.; Iribarren A. New synthetic routes to protected purine 2′-O-methylriboside-3′-O-phosphoramidites using a novel alkylation procedure. Nucleic Acids Res. 1990, 18, 41–49. 10.1093/nar/18.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K.; Gozu H.; Hosaka H.; Sakamoto K.; Yokoyama S.; Takai K.; Takaku H. Cleavage effect of oligoribonucleotides substituted at the cleavage sites with modified pyrimidine- and purine-nucleosides. Biochim. Biophys. Acta 1997, 1354, 211–218. 10.1016/S0167-4781(97)00099-7. [DOI] [PubMed] [Google Scholar]
- Pasternak A.; Kierzek E.; Pasternak K.; Turner D. H.; Kierzek R. A chemical synthesis of LNA-2,6-diaminopurine riboside, and the influence of 2′-O-methyl-2,6-diaminopurine and LNA-2,6-diaminopurine ribosides on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2007, 35, 4055–4063. 10.1093/nar/gkm421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproat B. S.; Iribarre A. M.; Garcia R. G.; Beijer B. New synthetic routes to synthons suitable for 2′-O-allyloligoribonucleotide assembly. Nucleic Acids Res. 1991, 19, 733–738. 10.1093/nar/19.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujari S. S.; Leonard P.; Seela F. Oligonucleotides with “Clickable” Sugar Residues: Synthesis, Duplex Stability, and Terminal versus Central Interstrand Cross-Linking of 2′-O-Propargylated 2-Aminoadenosine with a Bifunctional Azide. J. Org. Chem. 2014, 79, 4423–4437. 10.1021/jo500392j. [DOI] [PubMed] [Google Scholar]
- Boudou V.; Kerremans L.; De Bouvere B.; Lescrinier E.; Schepers G.; Busson R.; Van Aerschot A.; Herdewijn P. Base pairing of anhydrohexitol nucleosides with 2,6-diaminopurine, 5-methylcytosine and uracil as base moiety. Nucleic Acids Res. 1999, 27, 1450–1456. 10.1093/nar/27.6.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudou V.; Rothenbacher K.; Aerschot A. V.; Herdewijn P. Oligonucleotides with 2,6-Diaminopurine Base Replacing for Adenine: Synthesis and Properties. Nucleosides and Nucleotides 1999, 18, 1429–1431. 10.1080/07328319908044743. [DOI] [Google Scholar]
- Wu X.; Delgado G.; Krishnamurthy R.; Eschenmoser A. 2,6-Diaminopurine in TNA: Effect on Duplex Stabilities and on the Efficiency of Template-Controlled Ligations. Org. Lett. 2002, 4, 1283–1286. 10.1021/ol020016p. [DOI] [PubMed] [Google Scholar]
- Koshkin A. A. Syntheses and Base-Pairing Properties of Locked Nucleic Acid Nucleotides Containing Hypoxanthine, 2,6-Diaminopurine, and 2-Aminopurine Nucleobases. J. Org. Chem. 2004, 69, 3711–3718. 10.1021/jo0303923. [DOI] [PubMed] [Google Scholar]
- Rosenbohm C.; Pedersen D. S.; Frieden M.; Jensen F. R.; Arent S.; Larsen S.; Koch T. LNA guanine and 2,6-diaminopurine. Synthesis, characterization and hybridization properties of LNA 2,6-diaminopurine containing oligonucleotides. Bioorg. Med. Chem. 2004, 12, 2385–2396. 10.1016/j.bmc.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Langkjær N.; Wengel J.; Pasternak A. Watson–Crick hydrogen bonding of unlocked nucleic acids. Bioorg. Med. Chem. Lett. 2015, 25, 5064–5066. 10.1016/j.bmcl.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Hornum M.; Stendevad J.; Sharma P. K.; Kumar P.; Nielsen R. B.; Petersen M.; Nielsen P. Base-Pairing Properties of Double-Headed Nucleotides. Chem.—Eur. J. 2019, 25, 7387–7395. 10.1002/chem.201901077. [DOI] [PubMed] [Google Scholar]
- Muraoka M.; Iida A.; Takahashi S.; Ebata T.; Uesugi S. Synthesis and Properties of 2′4′- and 3′-5′-Linked Ribodinucleoside Monophosphates Containing 2-Aminoadenosine and Uridine. Nucleosides and Nucleotides 1991, 10, 1317–1332. 10.1080/07328319108047065. [DOI] [Google Scholar]
- Muraoka M.; Takahashi S.; Uesugi S. Interaction of (2′−5′) and (3′−5′) Linked 2-Aminoadenylyl-3-aminoadenosines with Polyuridylic Acid. Nucleosides and Nucleotides 1995, 14, 1503–1518. 10.1080/15257779508009488. [DOI] [Google Scholar]
- Muraoka M.; Takahashi S.; Yamaguchi N.; Oda Y.; Uesugi S. Synthesis and Properties of Dinucleoside Monophosphates Containing 2-Aminoadenine 8,2′-S- and Uracil 6,2′-O-Cyclonucleosides. Nucleosides and Nucleotides 1990, 9, 205–221. 10.1080/07328319008045133. [DOI] [Google Scholar]
- Matray T.; Gamsey S.; Pongracz K.; Gryaznov S. A Remarkable Stabilization of Complexes Formed by 2,6-Diaminopurine Oligonucleotide N3′→P5′ Phosphoramidates. Nucleosides, Nucleotides & Nucleic Acids 2000, 19, 1553–1567. 10.1080/15257770008045446. [DOI] [PubMed] [Google Scholar]
- Matray T. J.; Gryaznov S. M. Synthesis and properties of RNA analogs—oligoribonucleotide N3′→P5′ phosphoramidates. Nucleic Acids Res. 1999, 27, 3976–3985. 10.1093/nar/27.20.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaiima G.; Hansen H. F.; Christensen L.; Dahl O.; Nielsen P. E. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 1997, 25, 4639–4643. 10.1093/nar/25.22.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y.; Donoshita Y.; Kamimoto H.; Murayama K.; Ariyoshi J.; Asanuma H. Introduction of 2,6-Diaminopurines into Serinol Nucleic Acid Improves Anti-miRNA Performance. ChemBioChem. 2017, 18, 1917–1922. 10.1002/cbic.201700272. [DOI] [PubMed] [Google Scholar]
- Kamiya Y.; Sato F.; Murayama K.; Kodama A.; Uchiyama S.; Asanuma H. Incorporation of Pseudo-complementary Bases 2,6-Diaminopurine and 2-Thiouracil into Serinol Nucleic Acid (SNA) to Promote SNA/RNA Hybridization. Chem.—Asian J. 2020, 15, 1266–1271. 10.1002/asia.201901728. [DOI] [PubMed] [Google Scholar]
- Reza F.; Bhaswati G.; Pei-Pei K.; Gaffney B. L.; Jones R. A. Synthesis of 6-substituted 2′-deoxyguanosine derivatives using trifluoroacetic anhydride in pyridine. Tetrahedron Lett. 1990, 31, 319–321. 10.1016/S0040-4039(00)94543-X. [DOI] [Google Scholar]
- Gao H.; Fathi R.; Gaffney B. L.; Goswami B.; Kung P. P.; Rhee Y.; Jin R.; Jones R. A. 6-O-(Pentafluorophenyl)-2’-deoxyguanosine: a versatile synthon for nucleoside and oligonucleotide synthesis. J. Org. Chem. 1992, 57, 6954–6959. 10.1021/jo00051a051. [DOI] [Google Scholar]
- Porcher S.; Pitsch S. Synthesis of 2′-O-[(Triisopropylsilyl)oxy]methyl (= tom)-Protected Ribonucleoside Phosphoramidites Containing Various Nucleobase Analogues. Helv. Chim. Acta 2005, 88, 2683–2704. 10.1002/hlca.200590209. [DOI] [Google Scholar]
- Ross B.; Springer R.; Ravikumar V. An Efficient and Scalable Synthesis of 2,6-Diaminopurine Riboside. Nucleosides, Nucleotides & Nucleic Acids 2008, 27, 67–69. 10.1080/15257770701571990. [DOI] [PubMed] [Google Scholar]
- Liu C.; Dumbre S. G.; Pannecouque C.; Korba B.; De Jonghe S.; Herdewijn P. Synthesis and antiviral evaluation of base-modified deoxythreosyl nucleoside phosphonates. Org. Biomol. Chem. 2017, 15, 5513–5528. 10.1039/C7OB01265A. [DOI] [PubMed] [Google Scholar]
- Vorličková M.; Sági J.; Szabolcs A.; Szemzö A.; Ötvös L.; Kypr J. Conformation of the synthetic DNA poly(amino 2 dA-dT) duplex in high-salt and aqueous alcohol solutions. Nucleic Acids Res. 1988, 16, 279–289. 10.1093/nar/16.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen L. F.; Broom A. D.; Robins M. J.; Bloch A. Synthesis and biological activity of selected 2,6-disubstituted(2-deoxy-.alpha.- and -.beta.-D-erythro-pentofuranosyl)purines. J. Med. Chem. 1972, 15, 735–739. 10.1021/jm00277a010. [DOI] [PubMed] [Google Scholar]
- Wright G. E.; Hildebrand C.; Freese S.; Dudycz L. W.; Kazimierczuk Z. Convenient synthesis of 2-halo-2’-deoxyadenosines. J. Org. Chem. 1987, 52, 4617–4618. 10.1021/jo00229a037. [DOI] [Google Scholar]
- Arico J. W.; Calhoun A. K.; McLaughlin L. W. Preparation of the 2′-Deoxynucleosides of 2,6-Diaminopurine and Isoguanine by Direct Glycosylation. J. Org. Chem. 2010, 75, 1360–1365. 10.1021/jo902616s. [DOI] [PubMed] [Google Scholar]
- Luyten I.; Van Aerschot A.; Rozenski J.; Busson R.; Herdewijn P. Protection of 2,6-Diaminopurine 2′-Deoxyriboside. Nucleosides and Nucleotides 1997, 16, 1649–1652. 10.1080/07328319708006247. [DOI] [Google Scholar]
- Srivastava S. C.; Srivastava N. P.. N-fmoc nucleosides and phosphoramidites useful in the synthesis of oligonucleotides. WO2010062404A2, 2010.
- Kutyavin I. V.; Rhinehart R. L.; Lukhtanov E. A.; Gorn V. V.; Meyer R. B.; Gamper H. B. Oligonucleotides Containing 2-Aminoadenine and 2-Thiothymine Act as Selectively Binding Complementary Agents. Biochemistry 1996, 35, 11170–11176. 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- Ross B. S.; Springer R. H.; Bharadwaj R.; Symons A. M.; Manoharan M. A New, Efficient Exocyclic Amine Protection Scheme for 2-Aminoadenosine and Derivatives for Incorporation into Oligonucleotides. Nucleosides and Nucleotides 1999, 18, 1203–1204. 10.1080/07328319908044662. [DOI] [Google Scholar]
- Ross B. S.; Manoharan M.. Improved process for the synthesis of oligonucleotides incorporating 2-aminoadenosine. WO2000012563A1, 2000.
- Krolikiewicz K.; Vorbrüggen H. The Synthesis of 2-Fluoropurine Nucleosides. Nucleosides and Nucleotides 1994, 13, 673–678. 10.1080/15257779408013271. [DOI] [Google Scholar]
- Robins M. J.; Uznański B. Nucleic acid related compounds. 34. Non-aqueous diazotization with tert-butyl nitrite. Introduction of fluorine, chlorine, and bromine at C-2 of purine nucleosides. Can. J. Chem. 1981, 59, 2608–2611. 10.1139/v81-375. [DOI] [Google Scholar]
- Montgomery J. A.; Hewson K. Convenient method for the synthesis of 2-fluoro-adenosine. J. Org. Chem. 1968, 33, 432–4. 10.1021/jo01265a094. [DOI] [PubMed] [Google Scholar]
- Mikami A.; Erande N.; Matsuda S.; Kel’in A.; Woods L. B.; Chickering T.; Pallan P. S.; Schlegel M. K.; Zlatev I.; Egli M.; Manoharan M. Synthesis, chirality-dependent conformational and biological properties of siRNAs containing 5′-(R)- and 5′-(S)-C-methyl-guanosine. Nucleic Acids Res. 2020, 48, 10101–10124. 10.1093/nar/gkaa750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeur E.; Bradley M. Amide bond formation: beyond the myth of coupling reagents. Chem. Soc. Rev. 2009, 38, 606–631. 10.1039/B701677H. [DOI] [PubMed] [Google Scholar]
- El-Faham A.; Albericio F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011, 111, 6557–6602. 10.1021/cr100048w. [DOI] [PubMed] [Google Scholar]
- Nair J. K.; Willoughby J. L. S.; Chan A.; Charisse K.; Alam M. R.; Wang Q.; Hoekstra M.; Kandasamy P.; Kel’in A. V.; et al. Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- Zlatev I.; Castoreno A.; Brown C. R.; Qin J.; Waldron S.; Schlegel M. K.; Degaonkar R.; Shulga-Morskaya S.; Xu H.; et al. Reversal of siRNA-mediated gene silencing in vivo. Nat. Biotechnol. 2018, 36, 509–511. 10.1038/nbt.4136. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J.; Rajewsky N.; Braich R.; Rajeev K. G.; Tuschl T.; Manoharan M.; Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs. Nature 2005, 438, 685–689. 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Soutschek J.; Akinc A.; Bramlage B.; Charisse K.; Constien R.; Donoghue M.; Elbashir S.; Geick A.; Hadwiger P.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Prakash T. P.; Mullick A. E.; Lee R. G.; Yu J.; Yeh S. T.; Low A.; Chappell A. E.; Østergaard M. E.; Murray S.; Gaus H. J.; Swayze E. E.; Seth P. P. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019, 47, 6029–6044. 10.1093/nar/gkz354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M.; Sena-Esteves M.; Chase K.; Sapp E.; Pfister E.; Sass M.; Yoder J.; Reeves P.; Pandey R. K.; et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Nat. Acad. Sci. 2007, 104, 17204. 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. F.; Khvorova A. Improving siRNA Delivery In Vivo Through Lipid Conjugation. Nucleic Acid Ther 2018, 28, 128–136. 10.1089/nat.2018.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. M.; Nair J. K.; Janas M. M.; Anglero-Rodriguez Y. I.; Dang L. T. H.; Peng H.; Theile C. S.; Castellanos-Rizaldos E.; Brown C.; Foster D.; et al. Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nat. Biotechnol. 2022, 10.1038/s41587-022-01334-x. [DOI] [PubMed] [Google Scholar]
- DeCorte B. L.; Tsarouhtsis D.; Kuchimanchi S.; Cooper M. D.; Horton P.; Harris C. M.; Harris T. M. Improved Strategies for Postoligomerization Synthesis of Oligodeoxynucleotides Bearing Structurally Defined Adducts at the N2 Position of Deoxyguanosine. Chem. Res. Toxicol. 1996, 9, 630–637. 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- Schlegel M. K.; Janas M. M.; Jiang Y.; Barry J. D.; Davis W.; Agarwal S.; Berman D.; Brown C. R.; Castoreno A.; LeBlanc S.; et al. From bench to bedside: Improving the clinical safety of GalNAc–siRNA conjugates using seed-pairing destabilization. Nucleic Acids Res. 2022, gkac539. 10.1093/nar/gkac539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M.; Manoharan M. Re-Engineering RNA Molecules into Therapeutic Agents. Acc. Chem. Res. 2019, 52, 1036–1047. 10.1021/acs.accounts.8b00650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.