Abstract
Active Fe-superoxide dismutase (SodF) was the third most abundant soluble protein in cells of Nostoc commune CHEN/1986 after prolonged (13 years) storage in the desiccated state. Upon rehydration, Fe-containing superoxide disumutase (Fe-SOD) was released and the activity was distributed between rehydrating cells and the extracellular fluid. The 21-kDa Fe-SOD polypeptide was purified, the N terminus was sequenced, and the data were used to isolate sodF from the clonal isolate N. commune DRH1. sodF encodes an open reading frame of 200 codons and is expressed as a monocistronic transcript (of approximately 750 bases) from a region of the genome which includes genes involved in nucleic acid synthesis and repair, including dipyrimidine photolyase (phr) and cytidylate monophosphate kinase (panC). sodF mRNA was abundant and stable in cells after long-term desiccation. Upon rehydration of desiccated cells, there was a turnover of sodF mRNA within 15 min and then a rise in the mRNA pool to control levels (quantity of sodF mRNA in cells in late logarithmic phase of growth) over approximately 24 h. The extensive extracellular polysaccharide (glycan) of N. commune DRH1 generated superoxide radicals upon exposure to UV-A or -B irradiation, and these were scavenged by SOD. Despite demonstrated roles for the glycan in the desiccation tolerance of N. commune, it may in fact be a significant source of damaging free radicals in vivo. It is proposed that the high levels of SodF in N. commune, and release of the enzyme from dried cells upon rehydration, counter the effects of oxidative stress imposed by multiple cycles of desiccation and rehydration during UV-A or -B irradiation in situ.
The drying of cells leads to crowding of cytoplasmic components, condensation of the nucleoid, and increases in the melting temperature (Tm) of membrane phase transitions, and it imposes stresses upon cell walls (31). Prolonged desiccation may cause damage to proteins, nucleic acids, and membrane components through Maillard reactions and metal-dependent Fenton reactions (17, 31). High photon flux densities and UV irradiation exacerbate the effects of reactive oxygen species in these processes. Mechanisms that protect cells from the effects of desiccation include those which circumvent oxidative damage (5, 7, 16). Of these, the synthesis of superoxide dismutase (SOD) is an important response. SOD mediates the disproportionation of superoxide radicals to hydrogen peroxide and dioxygen; the hydrogen peroxide is then scavenged either by ascorbate peroxidase or catalase (27). In many bacteria, SODs are present in the periplasmic space and their synthesis is repressed under anaerobic conditions (3, 15). Regulation of the expression of genes that encode these periplasmic SODs, as well as genes involved in the synthesis of other antioxidants, may be subject to control by the alternate sigma factor RpoS (13, 15). Other mechanisms that cells use to protect against oxidative damage from desiccation include the synthesis of carotenoids and tocopherols (12, 38, 45) and anthocyanins (in plants [43]).
Cyanobacteria must be especially prone to desiccation damage because they evolve intracellular oxygen through photosynthesis and persist in habitats subject to intense solar irradiation (32). In fact, many terrestrial cyanobacteria withstand the most acute long-term water deficit; how they do so is of considerable practical importance. Cyanobacteria are known to use both Fe- and Mn-containing SODs (Fe- and Mn-SODs) to scavenge superoxide radicals (11), but there is a paucity of information on the roles of these enzymes in response to water deficit. The sod genes of a number of cyanobacteria have been characterized. The genome of Synechocystis sp. strain PCC 6803 contains a single SOD gene (sodB; slr1516 [22]); in contrast, Plectonema boryanum UTEX 485 contains sodB as well as three additional Mn-SOD genes (10). Results from studies with Synechococcus sp. strain PCC 7942, in which sodB was inactivated, suggested that in this cyanobacterium Fe-SOD did not protect photosystem II during oxidative stress but that it did protect photosystem I. The enzyme was also able to protect cells from the effects of chilling in the light (S. K. Herbert and D. J. Thomas, Abstr. VIth Cyanobacterial Workshop, abstr. S15, 1998).
The cyanobacterium Nostoc commune exhibits a marked tolerance of desiccation and was the subject of a study that sought to identify structural, physiological, and molecular mechanisms which contribute to this tolerance (31). It is becoming clear that these mechanisms are both numerous and very diverse. Cells of N. commune CHEN/1986 contain sucrose and trehalose (18), which contribute to a marked depression in the Tm of membranes when these are desiccated and rehydrated (20). In each case, the Tm value is at least 20°C below the ambient temperature of the environment from which the materials were collected. Field colonies of N. commune CHEN/1986 also elaborate a complex extracellular glycan which is secreted in copious amounts by liquid cultures of a derivative strain, N. commune DRH1, and which lends a spherical appearance to colonies grown on solid media (18). The extracellular glycan has unusual rheological properties and can prevent fusion of membrane vesicles at very low relative humidities (−400 MPa) in the presence of a nonreducing sugar (20).
In addition to trehalose, sucrose, and the glycan, significant amounts of at least two major classes of UV-absorbing compounds provide protection to colonies of N. commune (8, 18). The mycosporines (mycosporine-like amino acid derivatives or MAAs) are a series of colorless, low-molecular-weight, water-soluble compounds with a single absorption maximum within the solar UV-R range (40). These MAAs constitute as much as 10% of the total dry weight of desiccated colonies, and they are released and diffuse from colonies upon rehydration (18). The MAAs in N. commune CHEN/1986 form strong interactions with a collection of proteins, the water stress proteins (Wsp [41]), which are also present in substantial amounts in desiccated colonies (approximately 60% of the total soluble protein) and are released from colonies upon rehydration (18).
The regulation of gene expression during desiccation and subsequent rehydration is of considerable interest. One of the most obvious features of gene expression in rehydrating N. commune is an ordered, apparently stringent, recovery of metabolic functions, beginning with respiration and followed by photosynthesis and finally nitrogen fixation (14, 33). The control of these processes is likely to be complex, given what is known about the sensing of other environmental signals in less complex (morphologically) cyanobacteria (42). Studies with field materials of N. commune HUN and N. commune UTEX 584 provided evidence that some, but not all, proteins remain stable despite extended periods of desiccation (18, 41). Although the drying of N. commune UTEX 584 cells led to a rapid cessation of nitrogenase activity (33, 34), no evidence was obtained for hydrolysis of at least one structural component of nitrogenase, Fe protein, which was present in cells of N. commune HUN following 10 years of desiccation (29). The intracellular ATP pool and the protein biosynthetic machinery of desiccated cells remained unperturbed for 30 min and 2 h, respectively, after rapid drying at −99.5 MPa (1, 33, 34). In contrast, short-term drying led to structural changes in the pigment antenna complexes of cyanobacteria, the phycobilisomes. In the light, the phycobiliproteins were degraded, and even in the dark, short-term drying led to subtle changes in the polydispersity of the complexes when they were analyzed in sucrose gradients (30, 41). De novo transcription in rehydrated cells of N. commune UTEX 584 is directed by extant RNA polymerase holoenzyme, which maintains its stability during desiccation, at least long enough for some transcripts to accumulate to control (predrying) levels (46).
During our studies a marked accumulation of active Fe-SOD was found in desiccated field materials of N. commune CHEN/1986 following rehydration. We describe the biochemical characterization of the enzyme and the isolation and structural characterization of sodF from N. commune DRH1 and propose an unusual but otherwise significant role for SodF in the response to desiccation.
MATERIALS AND METHODS
Cyanobacterial strains and growth conditions.
Desiccated colonies of N. commune (Table 1) were collected from field localities (19). All of these belong to one form species as determined through group I intron analysis (data not shown). Colonies were kept desiccated in the dark, for periods between 3 and 130 years, prior to the analyses described here. The relative amounts of each sample were determined, and the samples were used for particular experiments and replicate trials. When necessary, desiccated cells were rehydrated with sterile distilled water at room temperature in the light (see below); rehydrated colonies were dried either passively over a period of 24 h or more rapidly under a forced stream of sterile air.
TABLE 1.
Cyanobacterial strains
| N. commune strain | Collection site | Years dry | Reference |
|---|---|---|---|
| DRH1 | Clonal culture of N. commune CHEN/1986 | NAa | 19 |
| CHEN/1986 | China | 13 | 19 |
| ENG/1996 | Virginia Tech campus, Blacksburg, Va. | 3 | This study |
| WH4/1869 | Heidelberg, Germany | 130 | This study |
| MEL/1968 | Victoria, Australia | 31 | This study |
| ALD8122/1974 | Aldabra Atoll, Indian Ocean | 25 | This study |
| WH2/1939 | Cahul, Romania | 60 | This study |
| WH14/1948 | Indiana | 51 | This study |
NA, not applicable.
Strain N. commune DRH1 is a derivative of N. commune CHEN/1986 and was grown in liquid BG11o (35) in 2-liter airlift fermentors with forced aeration. The photon flux density at the surface of the culture vessel was 166 μmol of photons m−2 s−1. Desiccated colonies of N. commune ENG/1996 were used in transcription and enzyme analyses. Colonies were rehydrated in distilled water in shallow suspensions contained in 14-cm-diameter petri dishes on a rotary shaker operated at 70 rpm at room temperature. Banks of warm-white 15-W fluorescent tubes, soft-white Duluz compact fluorescent 23-W bulbs (Osram Sylvania Ltd., Mississauga, Canada), and a 365-nm UV source (model 36-380 lamp; Spectronics Corporation, Westbury, N.Y.) provided a continuous photon flux density of 2,500 μmol of photons m−2 s−1 at the immediate surface of the cell suspension (measured with a model QSL-100 quantum scalar irradiance meter [Biospherical Instruments Inc., San Diego, Calif.]).
Purification and identification of SodF.
Desiccated colonies of N. commune CHEN/1986 were used to isolate preparative amounts of SodF. To achieve efficient cell lysis, colonies were frozen under liquid nitrogen and ground to a powder with a pestle and mortar. The powder was mixed with sterile distilled water, and cells were allowed to rehydrate for 1 h at room temperature to allow the release of soluble proteins. Phycobiliproteins were removed by batch adsorption with Q-Sepharose (Pharmacia) resin equilibrated in 50 mM Tris · HCl (pH 7.5) buffer. After dialysis, the sample was concentrated with a 10-kDa-cutoff filter and the retentate, containing approximately 14 μg of total protein, was subjected to preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (10%, wt/vol, gels) as described previously (18). Two major classes of soluble protein were resolved: the 27- to 39-kDa Wsp isoforms (41) and a protein with a prominent band at approximately 20 kDa. Proteins were transferred to Immobilon P membrane (Millipore) via Western blotting in CAPS (3-[cyclohexylamino]-1-propoanesulfonic acid) buffer at pH 10.4 in a Hoeffer transfer chamber and visualized with Coomassie brilliant blue as described previously (18). Protein, immobilized on an excised fragment of membrane, was subjected to automated Edman degradation with an Applied Biosystems model 477A protein sequencer.
Enzyme characterization.
Aggregates of desiccated colonies of N. commune ENG/1996 were dispersed gently with a mortar and pestle at room temperature and suspended in sterile distilled water (0.6 g of desiccated cells in 30 ml of sterile distilled water). The suspended cells were incubated quiescently for 5 h in the dark at room temperature and then collected through centrifugation. The supernatant fraction (exudate) was dialyzed at 4°C in 50 mM potassium phosphate–0.1 mM EDTA (pH 7.8) buffer. The exudate at this point was colorless, and only a trace of color (blue; attributed to phycobiliprotein) was observed after subsequent concentration (see below). The cell pellet was resuspended in 6 ml of phosphate buffer, mixed with 0.6 g of alumina (Sigma type A-5), and ground in a mortar for 12 min. The initial slurry, with an additional 6 ml of phosphate buffer, was centrifuged, and the supernatant fraction (extract) was dialyzed in the phosphate buffer. The dialyzed extract and exudate were concentrated approximately fourfold under N2 (Amicon ultra filter, 10-kDa cutoff).
Protein concentration was measured by the bicinchoninic acid method (Pierce Chemical Co.) with bovine serum albumin as a standard. SOD activity was measured by monitoring the inhibition of O2−·-dependent reduction of ferricytochrome c (25). Sodium azide, when present, was added to the assay prior to initiation of the reaction with xanthine oxidase (26). SOD activity was visualized in native polyacrylamide tube gels as described by Salin and McCord (37).
Generation of O2−· from N. commune DRH1 glycan.
The complex extracellular polysaccharide (glycan) of N. commune DRH1 was subjected to exhaustive purification as described previously (20). The glycan was sterilized through autoclaving and used either directly in assays (preparation A) or after solvent extraction to remove monosaccharides released upon autoclaving (preparation B). Glycan (0.9 ml, 10 mg ml−1) was added to a sterile petri dish and mixed with 0.1 ml of 1 mM ferricytochrome c (final concentration, 100 μM). The absorbance at 550 nm (A550) was set to zero, and then the solution was irradiated with a 0.6-A Fisher FBUVLS-150 UV-A and -B radiation source with a spectral intensity maximum at 365 nm. The flux densities at the surfaces of the glycan solutions were measured with digital radiometers (Spectronics Corporation, Westbury, N.Y.) and were 200 μW cm−2 (UV-A) and 20 μW cm−2 (UV-B). The A550 of the solution was measured at time intervals over the course of 70 min. Either 70 or 140 U of bovine erythrocyte Cu-Zn-SOD (1,400 U ml−1) or 1,000 U of bovine liver catalase (14,000 U ml−1) was added to separate aliquots of the glycan-ferricytochrome c solution immediately prior to irradiation. Catalase was assayed by its rate of disproportionation of H2O2, monitored at 245 nm (28). The enzymes were stored in 50 mM potassium phosphate–0.1 mM EDTA (pH 7.8) prior to use.
DNA purification.
Cells of N. commune DRH1 were harvested, lyophilized, and ground to a powder under liquid nitrogen. The powder was rehydrated (approximately 1:1, powder-buffer) in extraction buffer (100 mM Tris · HCl, 1.4 M NaCl, 20 mM EDTA, 1.5% [wt/vol] cetyltrimethylammonium bromide, 1% [vol/vol] β-mercaptoethanol [2]) at 65°C, frozen under liquid nitrogen, and thawed again at 65°C. The cycle was repeated six times. The mixture was extracted with chloroform-isoamyl alcohol (24:1) and centrifuged, and the aqueous phase was mixed with an equal volume of isopropanol. DNA was spooled, dried in air, rehydrated with a minimal volume of Tris-EDTA buffer, and purified further through cesium chloride buoyant-density ultracentrifugation.
Construction of gene libraries.
Genomic DNA of N. commune DRH1 was digested partially with Sau3AI and ligated to Lambda Fix II arms prepared by a partial fill-in reaction with dTTP and dCTP after digestion with XhoI (Stratagene). Ligation reaction mixtures were packaged with Gigapack III XL (Stratagene) packaging extracts to select for inserts approximately 18 to 23 kb in size in recombinant phage grown on Escherichia coli XLI-Blue MRA (P2).
Purification of phage DNA.
E. coli XLI-Blue MRA (P2) was infected with recombinant phage by standard protocols (44), and phage was recovered from plate lysates in SM buffer (50 mM Tris · HCl [pH 7.5], 100 mM NaCl, 8 mM MgSO4, 0.01% [wt/vol] gelatin). Phage DNA was recovered with the Wizards Lambda Preps DNA purification system (Promega Corporation).
Isolation and cloning of sodF.
A 475-bp fragment of sodF was amplified from N. commune DRH1 genomic DNA by a PCR assay. Two 19-mer oligonucleotides were designed based upon a portion of the sequence (PYGMK) at the N-terminal region of N. commune DRH1 SodF (see Results). The two N-terminal oligonucleotides each had a redundancy of 1:64 and differed only in their 3′-terminal purine; the sequence of SODN-2A is 5′GGAA/GCCNTAT/CGGNATGAAA3′ and the sequence of SODN-2G is 5′GGAA/GCCNTAT/CGGNATGAAG3′. Two C-terminal 20-mer oligonucleotides were synthesized based upon the reverse complement of the DNA that encodes the conserved sequence DVWEHAYY found in the carboxy-terminal region of Sod proteins. These two primers had a redundancy of 1:128 and differed only in their 3′-terminal purine; the sequence of SODI-2A is 5′AA/GTANGCA/GTGT/CTCCCANACA3′ and that of SODI-2G is 5′AA/GTANGCA/GTGT/CTCCCANACG3′. All four possible permutations of these primers were tested. Assays were performed with 50-μl reaction mixtures in pH 9.0 buffer (supplied by the manufacturer, Promega Biotec) containing 12.5 nmol (250 μM) of each deoxynucleoside triphosphate, 1 mM MgCl, and 2.5 U of Taq polymerase (Promega Biotec). The annealing (90 s) temperature was at 60°C in the first cycle, dropped 1.5°C per cycle for the first 10 cycles (to 45°C), and was kept constant at 45°C for the remaining 30 cycles. In each cycle, denaturation occurred at 95°C for 1 min and elongation was at 72°C for 90 s. The assay began with a denaturation temperature of 95°C for 2 min and ended with an elongation time of 10 min.
Alkali-labile digoxigenin-11 (DIG)-dUTP (Boehringer Mannheim GmbH) was used to label the 457-bp sodF PCR product for use as a hybridization probe. In this case the concentrations of deoxynucleoside triphosphates were 250 μM each for dATP, dCTP, and dGTP; 130 μM for dTTP; and 70 μM for DIG-dUTP. The product was resolved in an agarose gel, excised, and purified with a Wizard PCR prep kit (Promega Corporation).
The phage library was hybridized with the DIG-dUTP probe by using the buffer and protocols specified in the DIG Easy System manual (Boehringer Mannheim GmbH). Hybridization was overnight at 42°C, with washes first in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M trisodium citrate, pH 7.0)–0.1% (wt/vol) SDS and then at high stringency in 0.1× SSC–0.1% (wt/vol) SDS. Lambda DNA was purified from one phage isolate, and DNA sequencing was performed on an Applied Biosystems 377 Prism DNA sequencer, with the BigDye Terminator system of Perkin-Elmer Applied Biosystems and Taq DNA polymerase.
Distribution of sodF in N. commune.
The distribution of sodF was investigated in desiccated colonies of form species N. commune strains obtained from diverse geographic locations as described in the figure legends (Table 1). Two primers were designed to amplify a 427-bp fragment from within the coding region of N. commune DRH1 sodF (5′GAGTATCACTATGGCAAGCA3′ and 5′CTAAAGTCAATGTAGTAG3′). Conditions of the PCR were as described above.
Transcription analysis.
Approximately 1 g of desiccated colonies of N. commune ENG/1996 or rehydrated colonies which were blotted dry quickly was frozen in liquid nitrogen and ground to a powder. The powder was mixed with 1.5 ml of RNA extraction buffer (0.25 M sucrose, 0.2 M NaCl, 10 mM magnesium acetate, 0.1 M Tris · HCl [pH 9.0]). After the addition of 1.5 ml of buffered phenol, the mixture was sonicated at a setting of 30 (Fisher sonic dismembrator model 300) three times for 15 s each time, with cooling on ice. Glass beads (4-mm diameter) were added (1:1 ratio), and the slurry was vortexed at top speed for 10 min. The aqueous phase was recovered through centrifugation and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) multiple times until no deposit remained at the interface. The aqueous phase was then extracted with chloroform-isoamyl alcohol (24:1), mixed with 2 volumes of ice-cold 100% (vol/vol) ethanol, and left at −20°C for several hours. The precipitate was recovered, dissolved in 1.7 ml of diethyl pyrocarbonate-treated water, mixed with an equivalent volume of 8 M LiCl, and left overnight at 4°C. The pellet was washed with 70% (vol/vol) ice-cold ethanol and stored in 70% (vol/vol) ethanol at −70°C until needed.
RNA was resolved in formaldehyde agarose gels, transferred to a positively charged nylon membrane (Boehringer Mannheim GmbH) through capillary blotting in 50 mM NaOH, and cross-linked to the membrane by UV radiation. Hybridization with DIG-dUTP-labeled PCR fragments of sodF or similarly labeled 23S ribosomal DNA (rDNA) from N. commune DRH1 (data not shown) was at 50°C overnight, with final stringency washes with 0.1× SSC–0.1% (wt/vol) SDS at 60°C (46). DNA-RNA hybrids were visualized with anti-DIG alkaline phosphatase and the chemiluminescent detection system of Boehringer Mannheim GmbH.
Nucleotide sequence accession number.
The DNA sequence data reported here were deposited in GenBank and assigned the accession no. AF177945.
RESULTS
Identification of SodF in desiccated N. commune CHEN/1986.
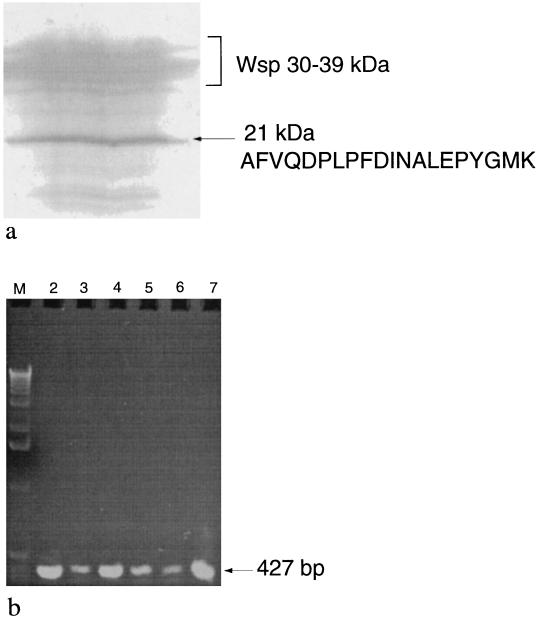
Soluble proteins of N. commune CHEN/1986 were obtained from desiccated colonies after one cycle of freezing and thawing through the addition of water. Resolution of proteins with SDS-polyacrylamide gels identified Wsp isoforms as well as a single prominent band at approximately 21 kDa (Fig. 1a). The 21-kDa protein was recovered, and the unambiguous sequence of the first 20 amino acids of SodF (lacking N-formylmethionine) was determined to be AFVQDPLPFDINALEPYGMK (Fig. 1a). Based on the relative amounts of Wsp proteins, which are the most abundant proteins in desiccated N. commune CHEN/1986 (41), and phycobiliproteins, SodF was judged to be the third most abundant soluble protein in materials of N. commune CHEN/1986 desiccated for 13 years (Fig. 1a).
FIG. 1.
(a) SodF is abundant in desiccated N. commune CHEN/1986. An immunoblot stained with Coomassie blue and used for N-terminal sequence analysis of SodF is shown. Wsp isoforms migrate between 30 and 39 kDa. The N-terminal sequence determined for the 21-kDa peptide (SodF) is indicated. (b) Distribution of sodF in the form species N. commune. An agarose gel (stained with ethidium bromide) of sodF PCR amplification products from samples of N. commune (Table 1) is shown. Lane M, HindIII molecular weight markers (LTI Inc.); lane 2, N. commune ALD8122/1974; lane 3, N. commune MEL/1968; lane 4, N. commune WH2/1939; lane 5, N. commune WH4/1869; lane 6, N. commune WH14/1948; lane 7, N. commune DRH1.
SOD activity in vivo.
After identification of SodF, aliquots of the retentate (see Materials and Methods) stored at −70°C for approximately 1 month, as well as rehydrating cells of N. commune CHEN/1986, were subjected to enzyme assay. In each case azide-inhibitable SOD activity was detected. In one well-controlled experiment, azide-inhibitable SOD activity was distributed almost equally between the rehydrating cells and the extracellular fluid (∼170 U g [dry weight]−1 in each fraction). However, these experiments depleted the supply of desiccated N. commune CHEN/1986. Further experiments were therefore undertaken with other, more abundant, desiccated materials and the readily available clonal strain N. commune DRH1, which was derived from N. commune CHEN/1986.
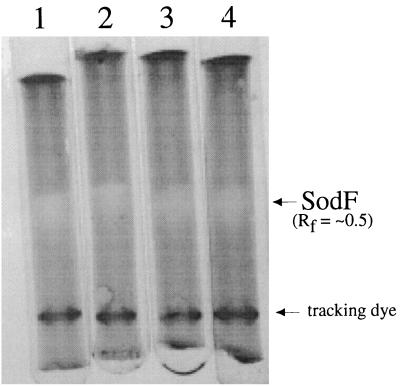
When cells were rehydrated at room temperature, without any physical abrasion or disruption, SOD activity was measured in both the extracellular fluid (exudate) and the cell extracts of rehydrated Nostoc spp. (Table 2). SOD activity was almost equally distributed between the exudates and cell extracts from cultures of strain DRH-1, and SOD-specific activity in the exudate was five times that in cell extracts. SOD was present in the extracellular fluid of rehydrated colonies of N. commune ENG/1996, but approximately 30% of the activity was in the exudate and 70% was in the cell extract. In N. commune ENG/1996, SOD-specific activity was 50% greater in the exudate than in the cell extract. SOD in each fraction was inhibited approximately 50% by 3 mM sodium azide in the assay. Inhibition of this magnitude by azide is consistent with Fe-SOD but not Mn-SOD. Electropherograms of SOD activity in the exudate and cell extract fractions revealed a single band of SOD activity, with approximately the same relative migration distances in each sample (Fig. 2).
TABLE 2.
SOD activity from N. commune cell fractionsa
| N. commune strain | Activity (SOD units g [dry weight]−1) in:
|
Sp act (SOD units mg of soluble protein−1) in:
|
||
|---|---|---|---|---|
| Exudate | Extract | Exudate | Extract | |
| DRH1 | 790 | 890 | 100 | 21 |
| ENG/1996 | 190 | 400 | 65 | 42 |
Values are the means of results of assays performed in triplicate. The data are representative of those obtained in repeat trials.
FIG. 2.
A single Fe-SOD activity is identified in N. commune strains. Shown are SOD activities demonstrated in 7% (wt/vol) native tube gels with 0.7 U of N. commune ENG/1996 cell extract (lane 1), 1.3 U of N. commune ENG/1996 exudate (lane 2), 2.7 U of N. commune DRH1 cell extract (lane 3), and 2.4 U of N. commune DRH1 exudate (lane 4). The relative mobility (Rf value) was calculated by using the migration distance of the tracking dye as a reference.
Cloning of sodF.
Based upon published alignments of cyanobacterial Sod proteins (data not shown), PCR amplification was expected to generate an approximately 483-bp DNA fragment of sodF. A single prominent 475-bp product was obtained with N. commune DRH1 genomic DNA with primers SODN2A and SODI2A; a much less prominent band of equivalent size was produced with primers SODN2G and SODI2A (data not shown). The identity of the former product as a fragment of sodF was confirmed through DNA sequence analysis (data not shown).
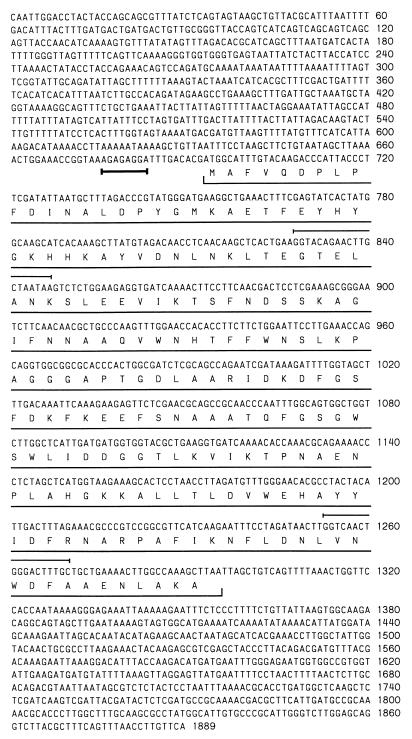
Five positive plaques were isolated from the phage library, and an 18-kbp insert from one of these was sequenced partially. Sequence analysis (with specific primers through the technique of chromosome walking) identified an open reading frame of 200 codons corresponding to sodF (Fig. 3) and located 10 bases downstream of a putative ribosome-binding site (AGAGAGGA). The putative derived amino acid sequence of N. commune DRH1 SodF showed highest sequence similarity to SodF from the filamentous nonheterocystous cyanobacterium P. boryanum UTEX 485 (10). Because detailed alignments of SODs from many sources are published, a comparison of only these two cyanobacterial SODs is presented (Fig. 4). The proximal region upstream of sodF showed highest sequence identity with the region immediately downstream of narB in Anabaena sp. strain PCC 7120 (9). The proximal region downstream of sodF showed high sequence identity to the rbcL-rbcX intergenic region of Nostoc sp. strain NIVA-CYA 124 (36) as well as the region immediately downstream of argF (ornithine carbamoyl transferase) in Nostoc sp. strain PCC 73102 (21).
FIG. 3.
DNA sequence of sodF and flanking regions from N. commune DRH1. Shown is the complete coding region of sodF, with the putative amino acid sequence (underlined) and the putative ribosome-binding site (boldface underlining). Sequences of primers used to amplify the original 475-bp sodF fragment from genomic DNA correspond to positions 768 to 787 (5′ to 3′) and 1192 to 1210 (3′ to 5′) and are underlined; compare the N-terminal amino acid sequence with that of the native protein (Fig. 1a), which shows a single discrepancy (E→D) at position 16 of the open reading frame.
FIG. 4.
SodF from N. commune DRH1 (SODF-NOSDRH1 [this study]) shows greatest sequence similarity to SodF from P. boryanum UTEX 485 (SODF-PLEBO [10]). The C-terminal regions of PanC proteins from Bacillus subtilis (KCY-BACSU), E. coli (KCY-ECOLI), and Haemophilus influenzae (KCY1-HAEIN) correspond to the partial sequence of PanC from N. commune DRH1 (KCY-NOSDRH1 [this study]). A partial alignment of Phr from Synechococcus sp. strain PCC 6301 (PHR-PCC6301) and N. commune DRH1 (PHR-NOSDRH1 [this study]) is shown. Sequences and alignments were manipulated with Lasergene software (DNASTAR, Madison, Wis.) and the software of the Biology Workbench, NCSA, University of Illinois (http://biology.ncsa.uiuc.edu/).
Further sequence analysis of the 18-kbp insert identified regions that may correspond to parts of genes encoding deoxyribodipyrimidine photolyase (phr) and cytidylate monophosphate kinase (panC), upstream and downstream of sodF, respectively. Partial alignments of these proteins with the relevant deduced amino acid sequences from N. commune DRH1 are presented (Fig. 4).
Distribution of sodF in N. commune.
A single amplification product of approximately 427 bp was obtained with sodF-specific primers under stringent conditions of PCR from all strains tested (Fig. 1b), including the three strains (N. commune CHEN/1986, ENG/1996, and DRH1) which were used for the biochemical and enzymatic characterizations of SodF.
Expression of sodF in N. commune ENG/1996.
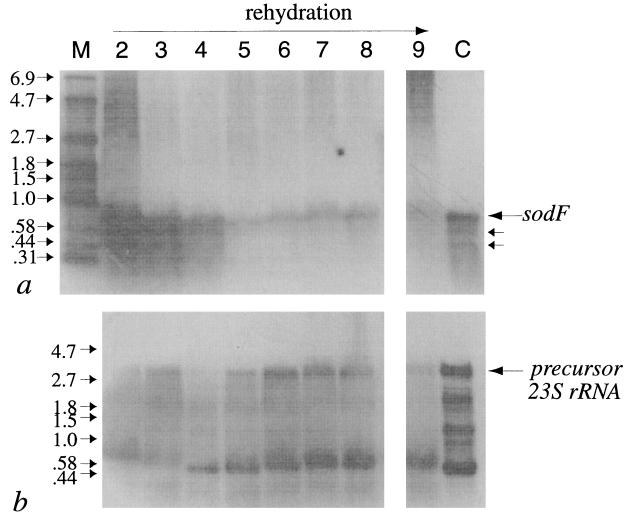
Probing of RNA extracts from desiccated and rehydrating materials of N. commune ENG/1996, as well as liquid cultures of N. commune DRH1 in the late logarithmic phase of growth, identified a major transcript of around 750 bases. Two minor signals between 400 and 600 bases were detected (Fig. 5a, lanes 2, 3, 4 and C), but their resolution was varied in repeat trials. sodF mRNA was abundant in cells after 3 years of desiccation (Fig. 5a, lane 2) and remained so up to 15 min after the onset of rehydration (Fig. 5a, lanes 3 and 4). After this time the intensity of the signals diminished and then gradually increased over the next 24 h (Fig. 5a, lanes 5 through 9). The decrease in sodF mRNA after 15 min of rehydration accompanied an increase in the intensity of signals attributed to 23S rRNA that appeared to reach steady state after approx 3 h of rehydration (Fig. 5b). Multiple bands detected when we used the 23S rDNA probe may represent precursor and processed forms of one or more copies of 23S rRNA (1). The fluctuation in the amounts of sodF mRNA in cells, apparently according to their degree of hydration, raised the question of the variability in mRNA content within different colonies. However, the pattern was consistent in repeat trials with different samples of cells of N. commune ENG/1996; most notably, the depletion in sodf mRNA after 15 min of rehydration and the subsequent increase in the pool of sodF mRNA to controls levels after approximately 24 h were reproducible (data not shown). The patterns in the de novo synthesis of 23S rRNA in response to rehydration (Fig. 5c) were consistent when different samples of the desiccated colonies of N. commune ENG/1996 were used (data not shown).
FIG. 5.
(a) sodF expression in N. commune ENG/1996 by Northern blotting. Lane M contains RNA molecular weight markers (in kilobases). Other lanes contain mRNAs from colonies desiccated 3 years (lane 2) or rehydrated for 5 min (lane 3), 15 min (lane 4), 1 h (lane 5), 3 h (lane 6), 6 h (lane 7), 12 h (lane 8), or 24 h (lane 9). A liquid culture of N. commune DRH1 in late logarithmic phase of growth is shown in lane C. (b) 23S rRNA in N. commune ENG/1996 by Northern blotting. The lanes contain identical amounts of the same samples applied in panel a.
Function of SodF in vivo.
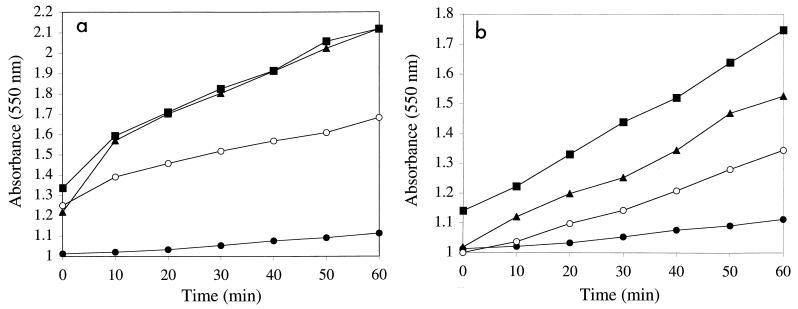
In view of the release of significant amounts of SodF upon cell rehydration, we questioned the physiological relevance of this extracellular activity and the possible role of the glycan, because there is some evidence that superoxide radicals can lead to depolymerization of polysaccharides (10). Ferricytochrome c was reduced in a time-dependent fashion when it was irradiated with UV in the presence of the glycan (Fig. 6). The rates of ferricytochrome c reduction in the presence of glycan preparation A (Fig. 6a) or glycan preparation B (Fig. 6b) were approximately 150 × 10−3 min−1 and 80 × 10−3 min−1, respectively, in comparison to the autoreduction rate in these experiments (20 × 10−3 min−1). Reduction of ferricytochrome c was inhibited approximately 50% in the presence of SOD, and the degrees of inhibition were equivalent when two different concentrations of SOD were used (see Materials and Methods). Catalase did not prevent the reduction of ferricytochrome c in this system (Fig. 6).
FIG. 6.
Glycan of N. commune DRH1 generates O2−· when it is exposed to UV irradiation. (a) Glycan preparation A; (b) glycan preparation B. Shown is the reduction of ferricytochrome c (measured as increases in A550) in the presence of glycan (▴), glycan plus SOD (○), glycan plus catalase (■), and the control (no glycan present in assay) (●). Values are determinations from single experiments; experiments were repeated four times, and the data presented are representative of those obtained in each trial. Variations in the A550 at zero time are due to differences in the buffer solutions of the different components. The slopes of relevant curves were used to estimate the rates described in the text.
DISCUSSION
Genetic mechanisms that contribute to desiccation tolerance remain poorly understood. It was proposed, in view of the destructive effects of oxygen, that genes involved in oxygen-scavenging mechanisms are likely to be important in the responses of prokaryotes to air drying (31). The quantities of active SodF in N. commune strains CHEN/1986 and ENG/1996, despite prolonged desiccation of cells, suggest that SOD is an important component of the repair process of this cyanobacterium. The fact that SodF remains active after such long-term storage may suggest that the enzyme has structural features which permit it to remain in the native state despite removal of most of the water from the cells or that it may be sequestered in a particular cell microenvironment. The amount of water in such desiccated (anhydrobiotic) cells can be as small as 0.02 g g (dry weight) of cells−1 in which proteins lack a monolayer coverage of water (31, 32). However, not all proteins remain stable under these conditions (30, 41).
SOD activity is located in extracellular fluids, such as synovial fluid (24), or the periplasmic space of E. coli (6) and other bacteria (3, 13, 15), and Nocardia asteroides synthesizes and secretes a SOD which adheres to the outer cell membrane (4). In each of these instances, SOD intercepts extracellular superoxide radicals and diminishes damage to the cell. In none of these examples, however, does the quantity of SOD released from the cells approach the magnitude of that released by rehydrating N. commune. This finding is all the more significant considering the time the cells were stored in a dormant state.
SodF does not have a recognizable N-terminal signal sequence, and microsequencing of the protein through Edman degradation confirmed that there was no processing at the N-terminal region in vivo. Three observations suggest that SodF is secreted in the exudate specifically, rather than via large-scale breakage of cells. First, the specific activity of extracellular Fe-SOD was significantly greater (fivefold in N. commune DRH-1) in the exudate than in the cell extract. Second, there was only a limited number of proteins observed upon electrophoretic examination of the exudate compared to the number present in the crude cell extract. Third, the exudate was colorless and carried only a faint tinge of color following concentration; i.e., there was an inverse correlation between the presence of phycobiliprotein (a sensitive indicator of cell lysis) and SodF activity distributed between the exudate and cell extract. The mechanism of release of SodF from N. commune is unknown, but it represents the third secretion event we have identified that may be of physiological relevance to rehydrating N. commune. Both the predominant acidic water stress proteins (Wsp) and the water-soluble UV-absorbing pigments (MAAs) are released in conspicuous amounts upon rehydration of desiccated cells in the absence of cell lysis (18, 19). A recent characterization of the wsp gene suggests that Wsp, like SodF, lacks an N-terminal signal sequence for secretion (data not shown).
It is reasonable to suspect that crowding and elevated salt concentrations within desiccated cells, as well as intense solar irradiation, contribute to the production of both intra- and extracellular superoxide radicals during rehydration as cells become metabolically active in an environment laced with organic compounds. Metabolic activity exacerbates the damage acquired during desiccation, because cells recover the capacity for photosynthesis and thus oxygen evolution (39).
Cells are metabolically inactive following dehydration and during subsequent storage in the dry state. Transcription analysis indicates that there is turnover of sodF mRNA immediately upon rewetting and that then sodF mRNA synthesis resumes. Although not directly measured, sodF mRNA synthesis must continue to be made and/or be stable as water is removed from cells (to account for the large amounts in all desiccated samples analyzed). The cells after 24 h of rehydration (Fig. 5a, lane 9) are in recovery phase, which may be equated with exponential growth. However, the system is complex. For example, in previous studies we demonstrated a sequential recovery of metabolic functions upon rehydration, beginning with respiration and followed by photosynthesis, etc. (for references, see reference 31). Nitrogen fixation is recovered after 24 h of rehydration. The fact that sodF mRNA is abundant after 24 h of rehydration emphasizes the possible importance of SodF during the recovery phase.
The extracellular glycan may be the primary potential source of damage for rehydrating N. commune in view of its abundance, its capacity to generate significant quantities of superoxide radicals upon UV irradiation, and the fact that it completely envelops the cells and makes intimate contact with the outer membrane (18, 19; in particular, see Fig. 1 in reference 20). Autoclaving the glycan leads to release of ribose from the structure (data not shown) and thus an increase in the concentration of free reducing ends. This finding is consistent with the higher rate of ferricytochrome c reduction obtained with glycan preparation A (Fig. 6a) and mediated through increased O2−· generation. Again, this may be of physiological relevance in situ, because the ribose linkage is labile, and changes in the rheological properties of the glycan during growth of the cells may involve release of ribose through enzyme hydrolysis (data not shown). In situ, the UV-absorbing MAAs (especially in the presence of reducing sugars) may also be a source of superoxide; they were largely removed during purification of the glycan for the experiments described here. We therefore interpret the release of SodF to be a beneficial biological response to rehydration of the desiccated cells. During this upshift, cells must deal with damage accumulated during desiccation and in part in response to UV radiation. There is clear evidence that SODs can mediate such repair. For example, Deinococcus radiodurans R1 is extremely resistant to oxidative stress and sodA mutants of this organism are much more sensitive to ionizing radiation than the wild type (23). The flux densities encountered by N. commune ENG/1996 in situ on a clear sunny day were measured as 2,150 and 300 μW cm−2 for UV-A and UV-B, respectively. These values are an order of magnitude greater than those used in the experiments with the purified glycan. The contribution of the glycan to superoxide generation in situ may therefore be significant.
Although sodF is transcribed as a monocistronic transcript, its proximity to at least two genes involved in DNA repair and synthesis may suggest that this region of the genome of N. commune DRH1 functions as part of a global response to environmental stress.
ACKNOWLEDGMENTS
This work was supported by NSF grant IBN9513157 (M.P.) and DARPA grant N00173-98-1-G005 (M.P. and R.F.H.).
REFERENCES
- 1.Angeloni S V, Potts M. Polysome turnover in immobilized cells of Nostoc commune (Cyanobacteria) exposed to water stress. J Bacteriol. 1986;168:1036–1039. doi: 10.1128/jb.168.2.1036-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley and Sons; 1992. p. I-index 25. [Google Scholar]
- 3.Barnes A C, Balebona M C, Horne M T, Ellis A E. Superoxide dismutase and catalase in Photobacterium damselae subsp. piscicida and their roles in resistance to reactive oxygen species. Microbiology. 1999;145:483–494. doi: 10.1099/13500872-145-2-483. [DOI] [PubMed] [Google Scholar]
- 4.Beaman B L, Scates S M, Moring S E, Deem R, Misra H P. Purification and properties of a unique superoxide dismutase from Nocardia asteroides. J Biol Chem. 1983;258:91–96. [PubMed] [Google Scholar]
- 5.Beckman K B, Ames B N. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 6.Benov L, Chang L Y, Day B, Fridovich I. Copper, zinc superoxide dismutase in Escherichia coli: periplasmic location. Arch Biochem Biophys. 1995;319:508–511. doi: 10.1006/abbi.1995.1324. [DOI] [PubMed] [Google Scholar]
- 7.Berlett B S, Stadtman E R. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 8.Böhme G A, Pfleiderer W, Böger P, Scherer S. Structure of a novel oligosaccharide-mycosporine amino acid ultraviolet-A/B sunscreen pigment from the terrestrial cyanobacterium Nostoc commune. J Biol Chem. 1995;270:8536–8539. doi: 10.1074/jbc.270.15.8536. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell W S, Laudenbach D E. Characterization of four superoxide dismutase genes from a filamentous cyanobacterium. J Bacteriol. 1995;177:964–972. doi: 10.1128/jb.177.4.964-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canini A, Civitareale P, Marini S, Grilli Caiola M, Rotilio G. Purification of iron superoxide dismutase from the cyanobacterium Anabeana cylindrica Lemm. and localization of the enzyme in heterocysts by immunogold labeling. Planta. 1992;187:438–444. doi: 10.1007/BF00199961. [DOI] [PubMed] [Google Scholar]
- 12.Ehling-Schulz M, Bilger W, Scherer S. UV-B-induced synthesis of photoprotective pigments and extracellular polysaccharides in the terrestrial cyanobacterium Nostoc commune. J Bacteriol. 1997;179:1940–1945. doi: 10.1128/jb.179.6.1940-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang F C, DeGroote M A, Foster J W, Baumler A J, Ochsner U, Testerman T, Bearson S, Giard J C, Xu Y S, Campbell G, Laessig T. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao K, Qiu B, Xia J, Yu A. Light dependency of the photosynthetic recovery of Nostoc flagelliforme. J Appl Phycol. 1998;10:51–53. [Google Scholar]
- 15.Gort A S, Ferber D M, Imlay J A. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol Microbiol. 1999;32:179–191. doi: 10.1046/j.1365-2958.1999.01343.x. [DOI] [PubMed] [Google Scholar]
- 16.Haslekas C, Stacy R A P, Nygaard V, Culiáñez-Macia F A, Aalen R B. The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol Biol. 1998;36:833–845. doi: 10.1023/a:1005900832440. [DOI] [PubMed] [Google Scholar]
- 17.Henle E S, Linn S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 18.Hill D R, Hladun S L, Scherer S, Potts M. Water stress proteins of Nostoc commune (Cyanobacteria) are secreted with UV-A/B-absorbing pigments and associate with 1,4-β-d-xylanxylanohydrolase activity. J Biol Chem. 1994;269:7726–7734. [PubMed] [Google Scholar]
- 19.Hill D R, Peat A, Potts M. Biochemistry and structure of the glycan secreted by desiccation-tolerant Nostoc commune (Cyanobacteria) Protoplama. 1994;182:126–148. [Google Scholar]
- 20.Hill D R, Keenan T W, Helm R F, Potts M, Crowe L M, Crowe J H. Extracellular polysaccharide of Nostoc commune (Cyanobacteria) inhibits fusion of membrane vesicles during desiccation and freeze-drying. J Appl Phycol. 1997;9:237–248. [Google Scholar]
- 21.Jansson E, Lindblad P. Cloning and molecular characterization of a presumptive argF, a structural gene encoding ornithine carbamoyl transferase (OCT), in the cyanobacterium Nostoc PCC 73102. Physiol Plant. 1998;103:347–353. [Google Scholar]
- 22.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 23.Markillie L M, Varnum S M, Hradecky P, Wong K K. Targeted mutagenesis by duplication insertion in the radioresistant bacterium Deinococcus radiodurans: radiation sensitivities of catalase (katA) and superoxide dimutase (sodA) mutants. J Bacteriol. 1999;181:666–669. doi: 10.1128/jb.181.2.666-669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marklund S L. Extracellular superoxide dismutase in human tissue and human cell lines. J Clin Investig. 1984;74:1398–1403. doi: 10.1172/JCI111550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCord J M, Fridovich I. Superoxide dismutase: an enzymatic function for erythrocuprein. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 26.Misra H P, Fridovich I. Inhibition of superoxide dismutase by azide. Arch Biochem Biophys. 1978;189:317–322. doi: 10.1016/0003-9861(78)90218-7. [DOI] [PubMed] [Google Scholar]
- 27.Miyake C, Michihata F, Asada K. Scavenging of hydrogen peroxide in prokaryotic and eukaryotic algae: acquisition of ascorbate peroxidase during evolution of cyanobacteria. Plant Cell Physiol. 1991;32:33–43. [Google Scholar]
- 28.Nelson D P, Kiesow L A. Enthalpy of decomposition of hydrogen peroxide by catalase at 25°C (with molar extinction coefficients of H2O2 in the uv) Anal Biochem. 1972;49:474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- 29.Peat A, Powell N, Potts M. Ultrastructural analysis of the rehydration of desiccated Nostoc commune HUN (Cyanobacteria) with particular reference to the immunolabelling of NifH. Protoplasma. 1988;146:72–80. [Google Scholar]
- 30.Potts M. Protein synthesis and proteolysis in immobilized cells of Nostoc commune UTEX 584 (Cyanobacteria) subjected to water stress. J Bacteriol. 1985;164:1025–1031. doi: 10.1128/jb.164.3.1025-1031.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potts, M. Mechanisms for desiccation tolerance. Eur. J. Phycol., in press.
- 33.Potts M, Bowman M A. Sensitivity of Nostoc commune UTEX 584 (Cyanobacteria) to water stress. Arch Microbiol. 1985;141:51–56. [Google Scholar]
- 34.Potts M, Bowman M A, Morrison N S. Control of matric water potential (ψm) in immobilized cultures of cyanobacteria. FEMS Microbiol Lett. 1984;24:193–196. [Google Scholar]
- 35.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 36.Rudi K, Skulberg O M, Jakobsen K S. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol. 1998;180:3453–3461. doi: 10.1128/jb.180.13.3453-3461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salin M L, McCord J M. Superoxide dismutase in polymorphonuclear leucocytes. J Clin Investig. 1974;54:1005–1009. doi: 10.1172/JCI107816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandmann G, Kuhn S, Böger P. Evaluation of structurally different carotenoids in Escherichia coli transformants as protectants against UV-B radiation. Appl Environ Microbiol. 1998;64:1972–1974. doi: 10.1128/aem.64.5.1972-1974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherer S, Ernst A, Chen T-W, Böger P. Rewetting of drought-resistant blue-green algae: time course of water uptake and reappearance of respiration, photosynthesis, and nitrogen fixation. Oecologia (Berlin) 1984;62:418–423. doi: 10.1007/BF00384277. [DOI] [PubMed] [Google Scholar]
- 40.Scherer S, Chen T-W, Böger P. A new UVA/B protecting pigment in the terrestrial cyanobacterium Nostoc commune. Plant Physiol. 1988;88:1055–1057. doi: 10.1104/pp.88.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scherer S, Potts M. Novel water stress protein from a desiccation-tolerant cyanobacterium: purification and partial characterization. J Biol Chem. 1989;264:12546–12553. [PubMed] [Google Scholar]
- 42.Schwarz R, Grossman A R. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under adverse extreme conditions. Proc Natl Acad Sci USA. 1998;95:11008–11013. doi: 10.1073/pnas.95.18.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwin H W, Farrant J M. Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regul. 1998;24:203–210. [Google Scholar]
- 44.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 45.Stahl W, Sies H. Physical quenching of singlet oxygen and cis-trans isomerization of carotenoids. Ann N Y Acad Sci. 1993;691:10–19. doi: 10.1111/j.1749-6632.1993.tb26153.x. [DOI] [PubMed] [Google Scholar]
- 46.Xie W-Q, Tice D, Potts M. Cell water deficit regulates expression of rpoC1C2 (RNA polymerase) at the level of mRNA in desiccation-tolerant Nostoc commune UTEX 584 (Cyanobacteria) FEMS Microbiol Lett. 1995;126:159–164. [Google Scholar]