Abstract
Enterobacteria-producing extended-spectrum β-lactamases (ESBL) play an important role in healthcare infections, increasing hospitalization time, morbidity and mortality rates. Among several ESBLs that emerge from these pathogens, CTX-M-type enzymes had the most successful global spread in different epidemiological settings. Latin America presents high prevalence of CTX-M-2 in ESBL-producing enterobacterial infections with local emergence of the CTX-M-1 group. However, this high prevalence of the CTX-M-1 group has not yet been reported in Chile. The aim of this study was to identify ESBLs among enterobacteria isolated from clinical samples of critically ill patients from southern Chile. One-hundred thirty seven ESBL-producing bacteria were isolated from outpatients from all critical patient units from Hernán Henríquez Aravena Hospital. Phenotype characterization was performed by antibiogram, screening of ESBL, and determination of minimum inhibitory concentration (MIC). PCR was used for genetic confirmation of resistance. Molecular typing was performed by ERIC-PCR. ESBL-producing isolates were identified as Klebsiella pneumoniae (n = 115), Escherichia coli (n = 18), Proteus mirabilis (n = 3), and Enterobacter cloacae (n = 1), presenting multidrug resistance profiles. PCR amplification showed that the strains were positive for blaSHV (n = 111/81%), blaCTX-M-1 (n = 116/84.7%), blaTEM (n = 100/73%), blaCTX-M-2 (n = 28/20.4%), blaCTX-M-9 (0.7%), blaPER-1 (0.7%), and blaGES-10 (0.7%). The multiple production of ESBL was observed in 93% of isolates, suggesting high genetic mobility independent of the clonal relationship. The high frequency of the CTX-M-1 group and a high rate of ESBL co-production are changing the epidemiology of the ESBL profile in Chilean intensive care units. This epidemiology is a constant and increasing challenge, not only in Chile, but worldwide.
Keywords: ESBL-producing enterobacteria, Intensive care units, CTX-M-1 group
Introduction
The massive use of expanded-spectrum cephalosporins in the treatment of infections caused by enterobacteriaceae, generated a selective pressure, followed by the rapid emergence of new β-lactamases that are able to degrade and confer resistance to these compounds, named extended-spectrum β-lactamases (ESBLs).1 Infections by these enterobacteria resistant to third-generation cephalosporins are a high concern in intensive care units.2, 3, 4, 5 Regional and multicenter studies carried out by epidemiological surveillance programs have highlighted the participation of ESBL-producing enterobacteria as a clinical and epidemiological emergency. This is due to an increasing prevalence of these resistant isolates of clinical origin in recent years in different parts of the world.6, 7 In Latin America, the prevalence of ESBL-producing bacteria is 53% among Klebsiella pneumoniae and 25% in Escherichia coli.7, 8 However, in Chile, the incidence of K. pneumoniae strains resistant to third generation cephalosporins mediated by ESBLs is around 60%.9, 10
ESBLs like TEM, SHV, and CTX-M are the main β-lactamases, especially CTX-M with an emergence prevalence reaching rates of over 85% in some regions of the world.11, 12 The CTX-M-1 group is one of the most prevalent CTX-M groups in Europe and North America, spreading worldwide.13, 14 Although the prevalence of this group has increased in the last years in Latin America,15, 16 the CTX-M-2 group remains the main enzyme group reported in the region.9, 11, 17 Relatively little is known about the spread of these enzymes in healthcare facilities in Chile,18, 19 especially in critically ill patient units, because there are no nationwide surveillance programs for this bacterial resistance mechanism.
In intensive care units, knowledge of the epidemiology, spectrum, and nature of infections, along with the susceptibilities of the causative organism, are extremely valuable for efficient and opportune treatment of several infections. There is a crucial need for surveillance and an early warning system that can detect signs of emerging and/or increasing antimicrobial resistance rates at local, regional, and national levels in order to minimize the impact of resistance and dissemination.4, 5 In this context, the aim of this study was to characterize the ESBL-carrying enterobacteria identified in critically ill patients of intensive care units in one of the main high-complexity hospital centers in southern Chile.
Methods
Setting and bacterial samples
Hernan Henriquez Aravena Hospital is a high-complexity public medical center in La Araucania Region, and a main referral center for cardiology in southern Chile. The total capacity of the critical patient units is 129 beds, divided between intensive care (ICU) and intensive therapy units (ITU) of the adult, pediatric, cardiology and neonatology departments.
One hundred and thirty seven ESBL-producing enterobacteria were isolated from patients of all critical units between December 2014 and October 2015 in the laboratory of the study hospital, and then transported to the Applied Molecular Biology Laboratory of Universidad de La Frontera. Specimens were isolated from infection sites such as blood, bronchoalveolar lavage, eye secretion, urethral secretion, and infected catheters. All bacteria samples were inoculated in 2 mL of Muller Hinton broth and incubated at 37 °C for 24 h in a rotating shaker at 150 rpm. Subsequently the samples were inoculated in MacConckey agar and once again incubated at 37 °C for 18–24 h. This last culture was used for phenotype tests and genetic material extraction, and stored at −80 °C with 20% glycerol.
Phenotype resistance profile
The susceptibility profiles of bacterial isolates were determined by antibiogram using the Kirby-Bauer method on Muller Hinton agar (Becton Dickinson, New Jersey, USA), following the standard procedures of the Clinical and Laboratory Standards Institute.20 For this purpose, the following antibiotics were used: amoxicillin/clavulanic acid (10 μg/4 μg), ampicillin-sulbactam (10 μg/10 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), cefoxitin (30 μg), piperacillin-tazobactam (100/10 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), sulphamethoxazole/trimethoprim (25 μg), tetracycline (30 μg), ertapenem (10 μg), cefuroxime (30 μg), chloramphenicol (30 μg), and doripenem (10 μg) (OXOID, Hampshire, United Kingdom). Minimum inhibitory concentration (MIC) against selected β-lactams of ceftazidime, cefotaxime, aztreonam, cefepime, imipenem, meropenem, and ertapenem were performed in micro-dilution broth in concentrations of 0.03–32 μg/mL with and without β-lactamase inhibitors such clavulanic acid (4 μg/mL) and phenyl boronic acid (400 μg/mL) (Sigma Aldrich, Stenheim, Germany). E. coli ATCC 25922 was used as the control organism for antibiotic susceptibility. Procedures and interpretations of antibiotic susceptibility results were carried out according to the CLSI 2016 guidelines.20, 21
Phenotype detection of ESBL, MBL, AmpC and KPC β-lactamases
The β-lactamases were screened for ESBL, AmpC, and carbapenemases by double disc diffusion test with discs containing clavulanic acid (4 μg/mL), boronic acid (400 μg/mL) and EDTA (320 μg/mL).20, 21, 22 The appearance of either an enhanced or a phantom zone between the antimicrobial agents and the inhibitor disc was considered a positive result and indicative of ESBL, AmpC and MBL production, respectively. Finally, the phenotype test for KPC detection was performed by the Modified Hodge Test following the CLSI guidelines.20
Bacterial DNA extraction and PCR amplification
Total DNA extraction was carried out using conventional DNA extraction by the boiling method.23 Detection of gene encoding for TEM, SHV, VEB, GES, PER, and CTX-M- type was performed by PCR assay. The PCR master mix was as follows: 2.5 μL of PCR buffer 10×, 2.5 μL of 2 mM dNTPs, 0.125 μL of Taq polymerase (New England Biolabs, Ipswich, MA, USA), 16.875 μL double distilled sterile water and 1 μL each of the forward and reverse primers. Aliquots of 24 μL of the master mix were taken in individual PCR reaction tubes, and then 1 μL of the extracted DNA was added to each tube for a total volume of 25 μL. The reaction mixture was initially denatured for 5 min at 94 °C, subjected to 30 denaturation cycles at 94 °C for 1 min, annealed at optimal melting temperature of each primer for 1 min, extended at 72 °C for 1 min, extended at 72 °C for 10 min and finally soaked at 4 °C. The primer sequences and the respective melting temperatures are listed in Table 1. The amplified PCR products were analyzed using 1% agarose gel electrophoresis. Gels were stained with GelRed (Biotium Inc, Fremont, CA, USA) and visualized by UV trans-illuminator.
Table 1.
Primer sequences and melting temperatures for identification of β-lactamases.
| Primers | Gene | Sequence 5′–3′ | Tm (°C) |
|---|---|---|---|
| SHV_Fw | blaSHVlike | AGCCGCTTGAGCAAATTAAA | 60 |
| SHV_Rv | TCCCGCAGATAAATCACCAC | ||
| TEM_Fw | blaTEMlike | GCCTTCCTGTTTTTGCTCAC | 60 |
| TEM_Rv | GATACGGGAGGGCTTACCAT | ||
| CTXM1_Fw | blaCTX-M-1group | TTAGGAARTGTGCCGCTGT | 59 |
| CTXM1_Rv | TACAAACCGTTGGTGACGA | ||
| CTXM2_Fw | blaCTX-M-2group | GACGCTACCCCTGCTATTTA | 58 |
| CTXM2_Rv | AGAAACCGTGGGTTACGATT | ||
| CTXM8_Fw | blaCTX-M-8group | TCGCGTTAAGCGGATGATGCT | 64 |
| CTXM8_Rv | TCGGTGACGATTTTCGCGGCA | ||
| CTXM9_Fw | blaCTX-M-9group | TGGTGACAAAGAGARTGCAA | 58 |
| CTXM9_Rv | GATTCTCGCCGCTGAAG | ||
| CTXM25_Fw | blaCTX-M-25group | CGACAGCCTGTGTTTCGCTGC | 64 |
| CTXM25_Rv | TCGGTGACWATTCTGGCGGCA | ||
| VEB_Fw | blaVEBlike | TTCAAATGCTCAARCTGACAA | 58 |
| VEB_Rv | TCCACGTTATTTTTGCAATG | ||
| GES1_Fw | blaGES-1like | CGCTTCATTCACGCACTATT | 60 |
| GES1_Rv | CGTGCTCAGGATGAGTTGTG | ||
| GES10_Fw | blaGES-10like | GAGAAGCTAGAGCGCGAAAA | 60 |
| GES10_Rv | ACTTGACCGACAGAGGCAAC | ||
| PER1_Fw | blaPER-1like | ACTGTAGGCGTTGCAGTGTG | 60 |
| PER1_Rv | TAATTTGGGCTTAGGGCAGA | ||
| PER2_Fw | blaPER-2like | GTTCTGCATCAGGTCGATCA | 60 |
| PER2_Rv | CCATCAGGCAACATAATGACG |
Genotyping analysis
All ESBL-producing isolates were subjected to Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR) with ERIC2 primer (5′-AAGTAAGTGACTGGGGTGAGCG-3′). The PCR conditions were previously described by Aydin et al.,24 and interpretation was by alignment of similarity using the DICE coefficient with a similarity of 90% in the GelJ free software.25
Results
Distribution of ESBL-infection in intensive care units
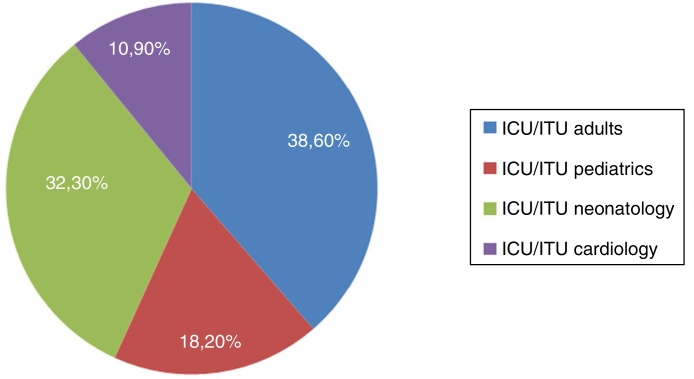
During the study period, 137 patients from all critical patient units were associated with infections by ESBL-producing enterobacteria. Among critically ill patients, the highest frequency of specimens by unit was at the adult unit with 38.6% (n = 53) of all isolates (28.4% from ITU and 10.2% from ICU), followed by neonatology unit with 32.3% (n = 44) of all isolates (21% ICU and 11.3% ITU), pediatric unit with 18.2% (n = 25) (10.9% ICU, 7.3% ITU) and cardiology unit with 10.9% (n = 15) (8% ICU and 2.9% ITU) (Fig. 1). Bacterial isolates were recovered from urinary tract infections (n = 61; 44.6%), blood infections (n = 27; 19.7%), soft tissue infections (n = 27; 19.7%) and respiratory tract infections (n = 22; 16%). After clinical bacteriology analysis, the 137 ESBL-producing isolates were identified as belonging to four species of enterobacteria: K. pneumoniae (n = 115; 84%), E. coli (n = 18; 13.1%), Proteus mirabilis (n = 3; 2.2%), and Enterobacter cloacae (n = 1; 0.7%). The distribution of species by critical patient unit is described in Table 2.
Fig. 1.
Distribution of ESBL-producing enterobacteria infections by critical patient units. ICU, intensive care unit; ITU, intensive therapy unit.
Table 2.
Distribution of enterobacteria species by critical patient units.
| Specie | ICU/ITU-A | ICU/ITU-C | ICU/ITU-N | ICU/ITU-P | No. isolates |
|---|---|---|---|---|---|
| K. pneumoniae | 40 | 13 | 40 | 22 | 115 |
| E. coli | 10 | 1 | 4 | 3 | 18 |
| P. mirabilis | 2 | 1 | 0 | O | 3 |
| E. cloacae | 1 | 0 | 0 | 0 | 1 |
| Total | 53 | 15 | 44 | 25 | 137 |
Legends: ICU, intensive care unit; ITU, intensive therapy unit; A, adults; C, cardiology; N, neonatology; P, pediatric.
Resistance profile
All ESBL-producing isolates exhibited multidrug resistance (MDR) profiles with resistance to three or more antibiotic classes. Among non-β-lactam drugs, high resistance rates were observed against ciprofloxacin (n = 115; 84%) sulfamethoxazole/trimethoprim (n = 120; 87%), and gentamicin (n = 104; 76%). However, amikacin and tetracycline were much more efficient with susceptibility rates of 80% (n = 110) and 84% (n = 115), respectively. Another antibiotic with greater then >50% activity was chloramphenicol.
The most active β-lactam among the ESBL-producing enterobacteria was imipenem with susceptibility rate of 90.2% (n = 123), followed by meropenem (n = 110; 80%), doripenem (n = 100; 73%), and ertapenem (n = 93; 67%). High resistance rates were observed with all types of cephalosporins (>88%) and β-lactams with inhibitor showed a resistance rate between 61% and 93%, with piperacillin/tazobactam being the most active. Cephamycin cefoxitin, highly similar to cepahlosporins, presented better activity with a susceptibility rate of 56% (n = 77). All susceptibility rates are shown in Table 3.
Table 3.
Antimicrobial susceptibility profile of ESBL-producing enterobacteria isolated from critical patient units.
| Specie | No. isolates | Susceptibility rates % (no. isolates) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | STX | TET | CIP | CHL | CTX | CXM | AMC | TZP | SAM | FOX | CAZ | FEP | ATM | ETP | MER | IPM | DOR | ||
| K. pneumoniae | 115 | 83 (95) | 20 (23) | 9 (10) | 90 (104) | 9 (10) | 57 (66) | 0 | 0.9 (1) | 10 (12) | 33 (38) | 4 (5) | 52 (60) | 7 (8) | 5 (6) | 3 (3) | 64 (74) | 76 (88) | 90 (104) | 71 (82) |
| E. coli | 18 | 83 (15) | 50 (9) | 39 (7) | 56 (10) | 67 (12) | 67 (12) | 0 | 0 | 33 (6) | 78 (14) | 17 (3) | 89 (16) | 44 (8) | 11 (2) | 28 (5) | 94 (17) | 100 (18) | 94 (17) | 89 (16) |
| P. mirabilis | 3 | 0 | 33 (1) | 0 | 0 | 0 | 33 (1) | 33.3 (1) | 0 | 67 (2) | 67 (2) | 33 (1) | 33 (1) | 33 (1) | 33 (1) | 67 (2) | 67 (2) | 100 (3) | 33 (1) | 33 (1) |
| E. cloacae | 1 | 0 | 0 | 0 | 100 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 (1) | 100 (1) | 100 (1) |
| Total | 137 | 80 (110) | 24 (33) | 12 (17) | 84 (115) | 16 (22) | 58 (79) | 2.6 (4) | 0.7 (1) | 15 (20) | 39 (54) | 7 (9) | 56 (77) | 12 (17) | 7 (9) | 7 (10) | 67 (93) | 80 (110) | 90 (123) | 73 (100) |
AMK, amikacin; GEN, gentamicin; STX, sulfamethoxazole/trimethoprim; TET, tetracycline; CIP, ciprofloxacin; CHL, chloramphenicol; CTX, cefotaxime; CXM, cefuroxime; AMC, amoxicillin/clavulanic acid; TZP, piperacillin/tazobactam; SAM, ampicillin/sulbactam; FOX, cefoxitin; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; ETP, ertapenem; MEM, meropenem; IPM, impenem; DOR, doripenem.
The MIC50 values for β-lactams were >32 μg/mL for ceftazidime, cefotaxime, cefepime, and aztreonam, 8 μg/mL for cefoxitin, 1 μg/mL for imipenem, 0.25 μg/mL and 0.125 μg/mL for ertapenem and meropenem, respectively. The use of β-lactamase inhibitors like clavulanic acid decreased the MIC by 76% of cephalosporin-resistant isolates (n = 104); the MIC50 decreased to 1 and 0.25 for ceftazidime and cefotaxime, respectively. Phenyl boronic acid did not reduce the MIC of any isolate except for one K. pneumoniae (Kp123).
Molecular identification of ESBL
PCR amplification showed that ESBL-producing isolates harbored a large amount of ESBL genes. Most of the strains were positive for blaSHV-like (n = 111/81%), blaCTX-M-1 group (n = 116/84.7%), and blaTEM-like (n = 100/73%) (Table 4). For the other CTX-M groups, the blaCTX-M-2 group was the second most prevalent in ESBL-producing enterobacteria (n = 28/20.4%) and just one isolate was positive for the blaCTX-M-9 group (0.7%); blaPER-1-like and blaGES-10-like were detected in only one isolate. The most prevalent group of ESBL-producing K. pneumoniae were CTX-M-1 group and SHV-like, carried in 85% and 89% of the isolates, respectively; E. coli carried CTX-M group 1 and TEM in 83% and 67% of the isolates, respectively. These were the most prevalent enzymes in this species. Fourteen ESBL profiles were observed in the 137 isolates, and the production rate of multiple ESBL in the isolates studied was very high with 93% of isolates producing more than one ESBL enzyme; the most prevalent co-production was three ESBL enzymes (n = 74) followed by two enzymes (n = 44). However, one K. pneumoniae (kp121) presented co-production of five ESBL enzymes, carrying SHV-like, CTX-M-1 group, CTX-M-2 group, PER-1-like, and TEM-like (Table 5). The most frequent profile observed in the isolates was the simultaneous production of the CTX-M-1 group, SHV-like, and TEM-like (n = 57) followed by two profiles of two enzymes CTX-M-1 group and SHV-like (n = 25), and CTX-M-1 group and TEM-like (n = 13).
Table 4.
Prevalence and distribution of ESBL by enterobacteria species.
| Specie (no. isolates) | ESBL produced by enterobacteria % (no. isolates) |
||||||
|---|---|---|---|---|---|---|---|
| CTX-M1 group | CTX-M2 group | CTX-M9 group | TEM-like | SHV-like | GES-10 | PER-1 | |
| K. pneumoniae (115) | 85.2 (98) | 21.7 (25) | 0.9 (1) | 71.3 (82) | 89.6 (103) | 0.9 (1) | 0.9 (1) |
| E. coli (18) | 83.3 (15) | 11.1 (2) | 0 | 66.7 (12) | 33.3 (6) | 0 | 0 |
| P. mirabilis (3) | 100 (3) | 0 | 0 | 100 (3) | 66 (2) | 0 | 0 |
| E. cloacae (1) | 0 | 100 (1) | 0 | 100 (1) | 0 | 0 | 0 |
| Total (137) | 84.7 (116) | 20.4 (28) | 0.7 (1) | 71.5 (98) | 81 (111) | 0.7 (1) | 0.7 (1) |
Table 5.
Prevalence of multiple ESBL production by enterobacteria species.
| Specie (no. isolates) | Rate of multiple ESBLproduction % (no. isolates) |
||||
|---|---|---|---|---|---|
| 1 enzyme | 2 enzymes | 3 enzymes | 4 enzymes | 5 enzymes | |
| K. pneumoniae (115) | 3.5 (4) | 28.7 (33) | 59.1 (68) | 7.8 (9) | 0.9 (1) |
| E. coli (18) | 58 (5) | 50 (9) | 22 (4) | – | – |
| P. mirabilis (3) | – | 33.3 (1) | 67 (2) | – | – |
| E. cloacae (1) | – | 100 (1) | – | – | – |
| Total (137) | 6.6 (9) | 32.1 (44) | 54 (74) | 6.7 (9) | 0.7 (1) |
Genotyping analysis
Thirty-eight genotypically unrelated strains of K. pneumoniae and 19 similarity patterns (>85%) were identified, showing four major clusters with six to 10 strains by cluster. Two of these large clusters were restricted to neonatology and adult services. Twelve out of 19 clusters of K. pneumoniae were constituted by strains isolated from several critical patient units, showing vertical dissemination between these services; however, four of them were exclusive of pediatric services. In the E. coli strains, one pattern of similarity (CE1) and nine other genotypically unrelated strains were identified. The three P. mirabilis showed no relatedness between isolates (data not shown).
Discussion
Antibiotic resistance is one of the greatest concerns in public health, since infections by multi-resistant microorganisms can cause great economic losses as well as deterioration in the patient's quality of life. Deteriorated health conditions and the selective pressure of antibiotics in the hospital environment are important factors for the occurrence of these infections, which is why intensive care units are a cause for concern in this emerging issue that requires urgent surveillance and control.26 The aim of our study was to identify and evaluate the genetic profile of resistance associated with ESBL enzymes in infections in critical patient units of the most complex hospital in southern Chile.
In this study, 137 infections caused by enterobacteria with an ESBL profile were identified, with a similar distribution between adult and pediatric services. The high percentage of infections in the pediatric service was associated with a high isolation rate in neonatology (32%). Recent studies have indicated that this population is at high risk from this type of infection,27 leading to high mortality and morbidity in this type unit.28 A meta-analysis conducted by Li et al. identified 13 risk factors for infection and colonization of ESBL in neonatal patients; among them are low birth-weight, parenteral nutrition and gestational age.29 In addition, transversal risk factors of intensive care units, such as length of hospital stay, previous antibiotic treatment, mechanical ventilation, catheter use, and previous colonization of ESBL-producing enterobacteria, make the neonatology service an area of great concern for these infections.29, 30
Fecal carriage of ESBL-producing enterobacteria was another risk factor strongly related to infection by ESBL-producing bacteria in the present study. In a descriptive hospital study from India, up to 65% of patients hospitalized in critically ill patient units had ESBL-producing enterobacteriaceae, with 26.6% of colonized patients becoming infected.31 Even though in Europe the colonization rate by ESBL-producing enterobacteria is lower than 25.9%, infection rates remain between 12.5% and 23%.26, 32, 33 All these risk factors, such as fecal carriage, length of hospital stay, previous antibiotic treatment, mechanical ventilation, and catheter use are very prevalent conditions in critical patient units and could be strongly associated with the high isolation of ESBL-producing enterobacteria observed in our study.
Despite this high isolation rate, we observed a low diversity of ESBL-producing enterobacteria: only four species were isolated from ESBL infections showing a high prevalence of K. pneumoniae (83.9%) followed by E. coli (13.1%). This predominance has been reported in several epidemiological studies in Latin America, but with lower margins between species.7, 34, 35 In a multicenter study of 13 European ICUs carried out between 2008 and 2011, Gurieva et al. observed that K. pneumoniae isolates in intensive care units had 3.7 times greater transmission capacity than E. coli.26 Curiously, a high percentage of the epidemiological reports from Europe and some places in Africa and in the Middle East show a greater predominance of isolates of E. coli, although with less significant differences in distribution compared to K. pneumoniae.5, 33, 36
Our susceptibility results showed a high rate of resistance to multiple drugs; this MDR profile is frequent in ESBL-positive infections, where antibiotic activity is restricted to carbapenems, such as meropenem and imipenem, and to other classes like tigecycline and amikacin.33, 35 This multidrug resistance has been associated mainly with membrane impermeability, caused by the selective pressure of hospital environments and plasmid resistance.37, 38 In Chile, Elgorriaga et al. (2012) reported plasmid resistance to quinolones and aminoglycosides associated with ESBL-producing enterobacteria from hospital infections.39 However, in our plasmid analyses, we observed transfer of resistance only to β-lactams like cephalosporin, with a higher level of resistance to cefotaxime, suggesting the plasmid transfer of the CTX-M type enzyme but no association with multidrug resistance (data not shown).
Since their discovery in Germany, the number of CTX-M variants has increased substantially and they predominate in Europe and South America.1, 17 Currently, more than 165 variants of CTX-M have been identified worldwide (www.lahey.org/Studies). The CTX-M-1 group was one of the most frequently identified ESBLs in strains isolated from critical care services in our samples, with an over 80% prevalence. Inside the CTX-M-1 group, CTX-M-15 is one of the prevailing enzymes in Europe and North America, with spreading worldwide predominance.13, 14 In Latin America, although the prevalence of this group has increased in recent years,15, 16 the CTX-M-2 group remains the main enzyme reported in the region.9, 11, 17, 18, 19 However, data from our study differ significantly from those previously reported, since the prevalence of the CTX-M-2 group was secondary in our study. It was identified in only 20.4% of the isolates studied, a significantly lower frequency than that observed for the CTX-M-1 group. This suggests the establishment of a new molecular profile of resistance, which implies changes in both dissemination mechanisms and preventive actions. The genetic environment of each enzyme group is different; CTX-M-2 is mostly associated with a complex class 1 integron, while CTX-M-1 is associated with insertion sequences.19 This implies that dissemination of CTX-M-1 is usually associated with an individual determinant of resistance, unlike the multidrug resistance of a class 1 integron carrying the CTX-M-2 group.
In the 137 strains studied, 14 different molecular profiles were found for ESBL-producing bacteria although only six different types of ESBL were observed (CTX-M-1 group, CTX-M-2 group, CTX-M-9 group, TEM-like, SHV-like, PER-1-like, and GES-10-like). The high prevalence of CTXM-1 (86%), SHV (81%) and TEM (73%) in several profiles suggests great versatility in the dissemination mechanisms exhibited by these enzymes. The different profiles suggest an individual mobility of each resistance determinant, mainly observed in K. pneumoniae. The versatility of profiles observed in this species reveals the plasticity of its genome, and its ability to adapt and carry out gene uptake from hospital environments.
Due to the high prevalence of certain enzymes, co-production of ESBL enzymes was observed in a large proportion of our isolates (93%). Jened et al. reported a similar prevalence of co-production in isolates from a tertiary hospital.36 However, in their study the presence of three ESBL enzymes was observed in only 17.5% of the isolates, significantly lower than that observed in our study, where 60.8% of the isolates presented three or more ESBL enzymes, again suggesting a high degree of gene mobility, independent of the clonal relationship. In fact, the diversity observed in the genotyping of our isolated species showed that, despite observing clonal dissemination in some of our isolates, reporting the presence of vertical transmission in the same or among different units of critical care, a high percentage of strains (38/115 K. pneumoniae, 9/18 of E. coli and all three P. mirabilis) were characterized as clonally unrelated.
Critical care units remain the epicenter of the antibiotic resistance crisis in hospitalized patients because they are places where multiple risk factors converge. Moreover, the diminished immunity of patients leads to the need for prolonged therapy and, consequently, to permanent selective antibiotic pressure.2, 33 Our study confirms that the high prevalence of ESBL in enterobacteria obtained from critical patients is a constant and increasing challenge in Chile as well, an issue of concern not only for our country but worldwide.26, 30, 33, 40
Conclusions
High prevalence of CTX-M type enzymes is an emerging endemic problem in Latin America. A high frequency of isolates carrying the enzyme of the CTX-M-1 group observed in this study, suggests a change in the epidemiology of ESBL profile in Chile. Epidemiological and molecular data obtained on the prevalence of ESBL genes provide effective tools for understanding the dissemination mechanisms involved and the trends in antibiotic resistance in critical patient services in southern Chile, allowing for better surveillance of these bacterial pathogens and their resistance profiles.
Ethical approval
The protocol of the present study was approved by the Ethics Committee of Universidad de La Frontera, decision number 032/2015.
Funding
The present study was supported by DIUFRO at Universidad de La Frontera, Chile (Grant #DI15-0070). Claudia Troncoso is recipient of fellowships from CONICYT-Chile (Doctoral Grant: CONICYT–PFCHA/Doctorado Nacional/2017–21171513).
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.D’Andrea M.M., Arena F., Pallecchi L., Rossolini G.M. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Martin S.J., Yost R.J. Infectious diseases in the critically ill patients. J Pharm Pract. 2011;24:35–43. doi: 10.1177/0897190010388906. [DOI] [PubMed] [Google Scholar]
- 3.Vincent J.L., Rello J., Marshall J., et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 4.Sader H.S., Farrell D.J., Flamm R.K., Jones R.N. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in US and European hospitals: results from the SENTRY Antimicrobial Surveillance Program, 2009–2012. Int J Antimicrob Agents. 2014;43:328–334. doi: 10.1016/j.ijantimicag.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Parajuli N.P., Acharya S.P., Mishra S.K., Parajuli K., Rijal B.P., Pokhrel B.M. High burden of antimicrobial resistance among gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob Resist Infect Control. 2017;6:67. doi: 10.1186/s13756-017-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner P.J. MYSTIC Europe 2007: activity of meropenem and other broad-spectrum agents against nosocomial isolates. Diagn Microbiol Infect Dis. 2009;63:217–222. doi: 10.1016/j.diagmicrobio.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Jones R.N., Guzman-Blanco M., Gales A.C., et al. Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011) Braz J Infect Dis. 2013;17:672–681. doi: 10.1016/j.bjid.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gales A.C., Castanheira M., Jones R.N., Sader H.S. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010) Diagn Microbiol Infect Dis. 2012;73:354–360. doi: 10.1016/j.diagmicrobio.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Guzmán-Blanco M., Labarca J.A., Villegas M.V., Gotuzzo E., LAWGoB Resistance Extended spectrum β-lactamase producers among nosocomial Enterobacteriaceae in Latin America. Braz J Infect Dis. 2014;18:421–433. doi: 10.1016/j.bjid.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chile I. Boletin Instituto de Salud Publica de Chile [Internet]; 2015. Vigilancia de resistencia a antimicrobianos en bacterias que pueden producir infecciones asociadas a atencion en salud; p. 5. [Google Scholar]
- 11.Bello H., Trabal N., Ibáñez D., et al. [beta-Lactamases other than TEM and SHV among strains of Klebsiella pneumoniae subsp pneumoniae isolated from Chilean hospitals] Rev Med Chil. 2005;133:737–739. doi: 10.4067/s0034-98872005000600018. [DOI] [PubMed] [Google Scholar]
- 12.Cantón R., González-Alba J.M., Galán J.C. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiral E., Pons M.J., Vubil D., et al. Epidemiology and molecular characterization of multidrug-resistant. Infect Drug Resist. 2018;11:927–936. doi: 10.2147/IDR.S153601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merino I., Hernández-García M., Turrientes M.C., et al. Emergence of ESBL-producing Escherichia coli ST131-C1-M27 clade colonizing patients in Europe. J Antimicrob Chemother. 2018;73:2973–2980. doi: 10.1093/jac/dky296. [DOI] [PubMed] [Google Scholar]
- 15.Seki L.M., Pereira P.S., de Souza Conceição M., et al. Molecular epidemiology of CTX-M producing Enterobacteriaceae isolated from bloodstream infections in Rio de Janeiro, Brazil: emergence of CTX-M-15. Braz J Infect Dis. 2013;17:640–646. doi: 10.1016/j.bjid.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sennati S., Santella G., Di Conza J., et al. Changing epidemiology of extended-spectrum β-lactamases in Argentina: emergence of CTX-M-15. Antimicrob Agents Chemother. 2012;56:6003–6005. doi: 10.1128/AAC.00745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocha F.R., Pinto V.P., Barbosa F.C. The spread of CTX-M-type extended-spectrum β-lactamases in Brazil: a systematic review. Microb Drug Resist. 2016;22:301–311. doi: 10.1089/mdr.2015.0180. [DOI] [PubMed] [Google Scholar]
- 18.Garcia P., Rubilar C., Vicentini D., et al. Caracterizacion clínica y molecular de bacteremias causadas por enterobacterias productoras de B-lactamasas de espectro extendido. 2004-2007. Rev Chil Infectol. 2011;28:563–571. [PubMed] [Google Scholar]
- 19.Carrasco-Anabalón S., Vera-Leiva A., Quezada-Aguiluz M., et al. Genetic platforms of blaCTX-M in carbapenemase-producing strains of K. pneumoniae isolated in Chile. Front Microbiol. 2018;9:324. doi: 10.3389/fmicb.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI . 26th ed. Clinical and Laboratory Standard Institute; Wayne, PA: 2016. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 21.Coudron P.E. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J Clin Microbiol. 2005;43:4163–4167. doi: 10.1128/JCM.43.8.4163-4167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picão R.C., Andrade S.S., Nicoletti A.G., et al. Metallo-beta-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46:2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lincopan N., McCulloch J.A., Reinert C., Cassettari V.C., Gales A.C., Mamizuka E.M. First isolation of metallo-beta-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J Clin Microbiol. 2005;43:516–519. doi: 10.1128/JCM.43.1.516-519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aydin F., Gümüşsoy K.S., Atabay H.I., Iça T., Abay S. Prevalence and distribution of Arcobacter species in various sources in Turkey and molecular analysis of isolated strains by ERIC-PCR. J Appl Microbiol. 2007;103:27–35. doi: 10.1111/j.1365-2672.2006.03240.x. [DOI] [PubMed] [Google Scholar]
- 25.Heras J., Dominguez C., Mata E., Pascual V. GelJ-A tool for analizyng DNA fingerprint gel images. BMC bioinformat. 2005:270. doi: 10.1186/s12859-015-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurieva T., Dautzenberg M.J.D., Gniadkowski M., Derde L.P.G., Bonten M.J.M., Bootsma M.C.J. The transmissibility of antibiotic-resistant Enterobacteriaceae in intensive care units. Clin Infect Dis. 2018;66:489–493. doi: 10.1093/cid/cix825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zingg W., Hopkins S., Gayet-Ageron A., et al. Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey. Lancet Infect Dis. 2017;17:381–389. doi: 10.1016/S1473-3099(16)30517-5. [DOI] [PubMed] [Google Scholar]
- 28.Stone P.W., Gupta A., Loughrey M., et al. Attributable costs and length of stay of an extended-spectrum beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003;24:601–606. doi: 10.1086/502253. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Xu X., Yang X., et al. Risk factors for infection and/or colonisation with extended-spectrum β-lactamase-producing bacteria in the neonatal intensive care unit: a meta-analysis. Int J Antimicrob Agents. 2017;50:622–628. doi: 10.1016/j.ijantimicag.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 30.Repessé X., Artiguenave M., Paktoris-Papine S., et al. Epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae in an intensive care unit with no single rooms. Ann Intensive Care. 2017;7:73. doi: 10.1186/s13613-017-0295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulki S.S., Ramamurthy K., Bhat S. Fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in intensive care unit patients. Indian J Crit Care Med. 2017;21:525–527. doi: 10.4103/ijccm.IJCCM_112_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kluytmans-van den Bergh M.F., Verhulst C., Willemsen L.E., Verkade E., Bonten M.J., Kluytmans J.A. Rectal carriage of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in hospitalized patients: selective preenrichment increases yield of screening. J Clin Microbiol. 2015;53:2709–2712. doi: 10.1128/JCM.01251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sader H.S., Farrell D.J., Flamm R.K., Jones R.N. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011) Diagn Microbiol Infect Dis. 2014;78:443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Sader H.S., Castanheira M., Farrell D.J., Flamm R.K., Mendes R.E., Jones R.N. Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011–2014) Int J Antimicrob Agents. 2016;48:144–150. doi: 10.1016/j.ijantimicag.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Vega S., Dowzicky M.J. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann Clin Microbiol Antimicrob. 2017;16:50. doi: 10.1186/s12941-017-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jena J., Debata N.K., Sahoo R.K., Gaur M., Subudhi E. Molecular characterization of extended spectrum β-lactamase-producing Enterobacteriaceae strains isolated from a tertiary care hospital. Microb Pathog. 2018;115:112–116. doi: 10.1016/j.micpath.2017.12.056. [DOI] [PubMed] [Google Scholar]
- 37.Pavez M., Vieira C., de Araujo M.R., et al. Molecular mechanisms of membrane impermeability in clinical isolates of Enterobacteriaceae exposed to imipenem selective pressure. Int J Antimicrob Agents. 2016;48:78–85. doi: 10.1016/j.ijantimicag.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Elgorriaga-Islas E., Guggiana-Nilo P., Domínguez-Yévenes M., et al. [Prevalence of plasmid-mediated quinolone resistance determinant aac(6′)-Ib-cr among ESBL producing enterobacteria isolates from Chilean hospitals] Enferm Infecc Microbiol Clin. 2012;30:466–468. doi: 10.1016/j.eimc.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Hanberger H., Arman D., Gill H., et al. Surveillance of microbial resistance in European Intensive Care Units: a first report from the Care-ICU programme for improved infection control. Intensive Care Med. 2009;35:91–100. doi: 10.1007/s00134-008-1237-y. [DOI] [PubMed] [Google Scholar]