Abstract
Recently, SARS-CoV-2 Omicron variant (B.1.1.529) was first identified in Botswana in November 2021. In a short period of time, this highly mutated variant replaced the previous dominant Delta variant, causing an exponential increase in the number of COVID-19 cases, resulting in a new wave of pandemic. This current research article aims to analyze and summarize information about the genetic characteristics, amino acid mutations and epidemiological data providing scientific findings to enrich the SARS-CoV-2 knowledge. More importantly, we describe here, for the first time, the identification of a new Omicron variant of concern: Omicron-L452R in Brazil.
Keywords: SARS-CoV-2, S:L452R, Mutational scanning, VOC
1. Introduction
In November 2021, researchers in Botswana and South Africa identified a new SARS-CoV-2 variant through whole-genome sequencing (WGS), posteriorly announced by WHO as a Variant of Concern (on November 26, 2021) and named as Omicron (B.1.1.529) (WHO, 2021). Omicron has spread rapidly, increasing COVID-19 cases throughout the world (Mohapatra et al., 2022). On December 25, 2021, over 108 countries reported confirmed cases of Omicron variant isolate. In Brazil, the first description was reported a few weeks after the first case in South Africa, from an airplane passenger that arrived in São Paulo State from South Africa, in late November.
Actually, the Omicron (21M) VOC is divided into six subvariants: or BA.1 (21K), BA.2 (21L ), BA.4 (22A), BA.5 (22B), BA.2.12.1 (22C) and BA.2.75 (22D). Since the beginning of the pandemic, the SARS-CoV-2 genome has been rapidly developing, mostly due to the inherent polymerase mistakes and host immune selection factors (Harvey et al., 2021). Different mutations in the spike protein raises concerns and Omicron variant is the most mutated SARS-CoV-2 variant, presenting more than 60 mutations in its genome; of which, 32 were in the receptor binding domain (RBD) of the spike protein (Tsang et al., 2022; Wang and Cheng, 2022).
Compared with previous variants, Omicron is known for decreased hospitalization rates and less severe disease (Maslo et al., 2022). The reduced viral fusogenicity, associated with low pathogenicity in SARS-CoV-2 patients, is probably one of the major reasons but the mechanism is still unclear (Motozono et al., 2021; Rajah et al., 2022). A possible cause of the reduced fusogenicity is related to the S:L452R mutation, present in the Delta variant but absent in Omicron. A recent study reported that a Omicron-L452R mutant generated; displayed increased fusogenicity and infectivity mediated by enhancing the cleavage of the spike protein (Zhang et al., 2022).
About transmissibility, Omicron presents a high rate when compared to Delta, which was linked to its immune evasion properties (Rössler et al., 2021). This resulted in Omicron overtaking Delta in areas where community transmission occurs (Gularte et al., 2022; Torjesen, 2021). Tracking SARS-CoV-2 variants through sequencing becomes a key part of a better understanding of viral evolution, especially after the beginning of the vaccination programs. Therefore, the main goal of this work was to describe the replacement of Delta by Omicron variant in Rio Grande do Sul (RS) state, in southern Brazil. Furthermore, through the genomic viral surveillance, we found, for the first time to our knowledge, a new Omicron-related lineage that is circulating in south Brazil, the Omicron-L452R variant.
2. Material and methods
A total of 152 SARS-CoV-2 complete genomes were sequenced through Illumina MiSeq platform at Laboratório de Microbiologia Molecular - Feevale University, one of the institutions linked to the Corona-ômica.BR-MCTI Network, a relevant genomic surveillance initiative in Brazil. Naso-oropharyngeal swab samples were received from suspected patients of COVID-19 at Laboratório de Microbiologia Molecular of Universidade Feevale. After the diagnosis confirmation, a pre-selection was carried out, choosing samples that presented cycle threshold value (Ct) below 25. The commercial MagMAX™ CORE Nucleic Acid Purification Kit (Applied biosystems™, Thermo Fisher Scientific, Waltham, MA, USA) kit was used to perform viral RNA extraction using the automated equipment KingFisher™ Duo Prime (Thermo Fisher Scientific™). As previously described (da Silva et al., 2021), viral genome sequencing and phylogenetic analysis were carried out. Briefly, reverse transcription reaction was carried out in RNA extracted by SuperScript IV reverse transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA). Following the manufacturer instructions (QIAGEN, Hilden, Germany), viral genome library preparation was carried out using the QIAseq SARS-CoV-2 Primer Panel paired for library enrichment and QIAseq FX DNA Library UDI Kit. Also, Illumina MiSeq platform (Foster City, CA, USA), using MiSeq Reagent Kit v3 (600-cycle) was used.
FASTQ files were input into the Illumina BaseSpace and consensus sequences were assembled using DRAGEN software with alignment to the reference SARS-CoV-2 sequence (NC_045512.2), minimum depth 10, minimum allele frequency 0.5, and genome at 5X coverage. The Geneious PrimeTM suite was used for genome annotation, editing, and mapping the sequences against the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124) available in the EpiCoV database from Global Initiative on Sharing Avian Influenza Data (GISAID) (Shu and Mccauley, 2017). PANGO and Nextrain lineage assignments were applied to characterize the consensus sequences. Phylogenetic tree was constructed including all SARS-CoV-2 complete genomes available from GISAID through the Nextclade tool on Nextstrain server (Hadfield et al., 2018). Mutational sequence profiles and signatures were also analyzed in order to better understand the lineage sequences. RDP v.4.101 software was used to evaluate the presence of recombinants among the sequences obtained (Martin et al., 2015).
3. Results
3.1. Epidemiological data
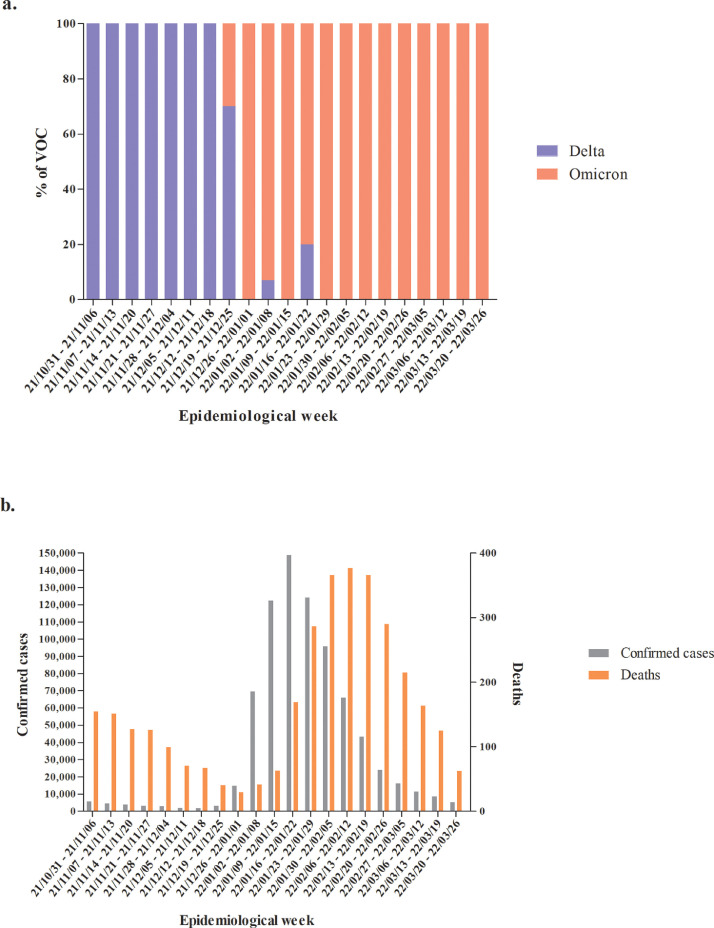
Performing an epidemiological analysis of the sequences generated by our laboratory Delta variant represented 100% of frequency (between October and November 2021). In December of the same year, the first Omicron variant detections were performed and less than a month later (January 2022), the scenario reversed. Delta variant detection considerably decreased (to 6% of all samples analyzed) while Omicron represented 94% of the samples. In February 2022, Delta was not detected anymore, and Omicron represented 100% of the sequences until now (March, 2022) (Fig. 1 a).
Fig. 1.
Graphical display of (a) the proportion of SARS-CoV-2 lineages identified in sequencing data and (b) the epidemiological data on COVID-19 cases and deaths in RS state by epidemiological week for the period October 2021 to March 2022, RS state, Brazil.
Considering these variant fluctuations, we compared the number of cases and deaths according to the same epidemiological weeks. Between October and December 2021, when the Delta variant was dominant, the number of cases was low and stable with decreasing the number of deaths. From the introduction of the Omicron variant (in the end of December), the number of cases and deaths started to exponentially increase, and a major COVID-19 wave was observed in RS state. After the peak, the number of the cases and deaths started to decline in January and February 2022, respectively (Fig. 1b).
3.2. Genetic analysis
A total of 152 SARS-CoV-2 complete genomes were retrieved through Illumina MiSeq. The samples were collected between late November, 2021 and mid-March, 2022 from patients in RS state, Brazil. Regarding genetic characterization, the sequences were aligned with complete SARS-CoV-2 genomes of different lineages through the NextClade online tool (https://clades.nextstrain.org/) and the phylogenetic tree was inferred (Fig. 1a). Of the total samples, 17% (26/152) were classified as Delta variant (21J) and 83% (126/152) as Omicron (21K and 21L). All sequences were uploaded at GISAID, and the accession numbers could be observed at Supplementary Tables 1 and 2.
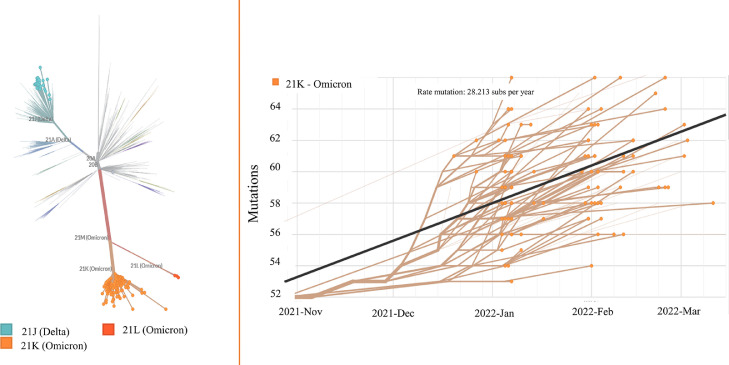
Subsequently, the sequences were analyzed through the Pangolin online tool (https://github.com/hCoV-2019/pangolin) and characterized as belonging to the following: i) Delta (AY.99.2, AY.101, AY.122, AY.43, AY.112, AY.25, AY.43.2, AY.46) and ii) Omicron (BA.1, BA.1.1, BA.1.14, BA.1.15, BA.1.17, BA.1.9 and BA.2) sublineages. The most prevalent were from the major Omicron clade (21K); BA.1.1 (41%) and B.A.1 (23%). The other 21K frequencies were: BA.1.14 (0.6%), BA.1.15 (14%), BA.1.17 (0.7%), BA.1.9 (0.4%). The newest Omicron B.2 (21L) sublineage was detected in only 3 sequences from February, 2022. The Nexstrain unrooted phylogenetic tree corroborated previous analysis and showed that the sequences clustered in two major clusters, belonging to Delta and Omicron (Fig. 2 a).
Fig. 2.
Phylogenetic and phylodynamic of SARS-CoV-2 variant analysis in Rio Grande do Sul State. Phylogenetic tree of all circulating lineages, developed through the Nextstrain server (Nextclade tool), using GISAID data. The sequences generated herein are clustered into a Delta (21J) and Omicron lineage (21K and 21L), marked in green and orange, respectively (left). Phylogenetic tree of 126 SARS-CoV-2 Omicron variants. The phylogeny is embedded as a root-to-tip plot, in which the x axis represents the date of sample collection, and the y axis represents the number of genome wide mutations that have occurred since the phylogeny root (right). Gisaid accession numbers available at Supplementary Material 1 (Omicron) and 2 (Delta).
Omicron sequences, through root-to-tip regression, showed an estimated high rate of 28,123 substitutions per year, suggested through molecular clock analysis (Fig. 2b). A wide range in quantification of genome nucleotide mutations compared to the reference strain is observed among the Omicron clade sequences, and a sub cluster above the regression line suggests a mutation rate above the average.
3.3. Mutational profile
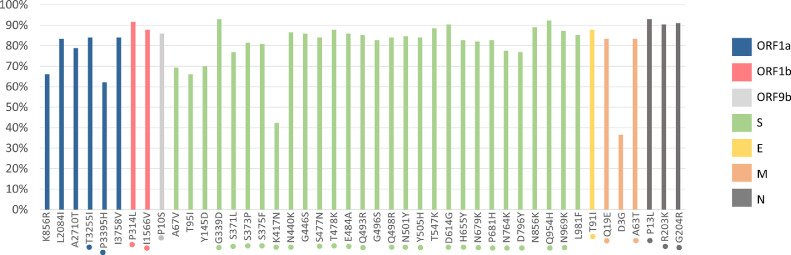
A complete mutational profile was performed, describing the amino acid mutations found in complete SARS-CoV-2 coding regions and comparing the Omicron sublineages. Mutation analysis at the Spike (S) protein revealed the presence of the Omicron signatures. The most frequent S protein mutations shared between the two Omicron major clades 21K (BA.1, BA.1.1, BA.1.14, BA.1.15, BA.1.17, BA.1.9) and 21L (BA.2) includes: G339D, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H and N969K. Analyzing only 21K cluster, this group shared additional mutations not present in 21L clade: A67V, T95I, Y145D, L212I, S371L, S373P, S375F, G446S, G496S, T547K, N856K and L981F (Fig. 3 ). Mutations in other viral proteins and their prevalence are also shown in Fig. 3. The major difference between BA.1 and BA.1.1, is that the BA.1.1 carried an additional S:R346K mutation, also described in our sequences. Additional acquired mutations among the sequences were restricted to few sequences and complete analysis could be observed in Table 1 .
Fig. 3.
Frequency of non-synonymous mutations across the genomes of Omicron sublineages sequenced during October 2021 to March 2022, RS state, Brazil. The x axis represents the mutations along the genome and y axis represents each mutation's frequencies. The analysis was performed in all coding regions, as observed. The mutations marked with dot were also present in BA.2. Only the mutations with more than 40% at the same sub lineage were shown.
Table 1.
Mutational profile of 126 Omicron whole-genome sequences. The first column presents the common mutations between Omicron 21K sub lineages (BA.1, BA.1.1, BA.1.1.14, BA.1.15, BA.1.17 and BA.1.9) and in bold the mutations also find in 21L cluster (BA.2). The unique mutations are present in at least one sequence, and the mutations marked with * represent more than 50% at the same sub lineage.
| Region | Common Omicron mutations | Unique mutations on BA.1 | Unique mutations on BA.1.1 | Unique mutations on BA.1.1.14 | Unique mutations on BA.1.1.15 | Unique mutations on BA.1.1.17 | Unique mutations on BA.1.9 | Unique mutations on BA.2 |
|---|---|---|---|---|---|---|---|---|
| S | A67V, T95I, Y145D, L212I, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F | |||||||
| ORF1a | K856R, L2084I, A2710T, T3255I, P3395H, I3758V | R24C, T333M, V675A, G1100S, H1113Y, G1307S, L1334V, F1501S, V1535A, T1682I, T1854I, E2904D, L3027F, T3090I, L3116F, L3201F, L3919I | G379E, Y621C, E743K, T1605S, T1788M, S1857L, P2079S, R3066K, L3606F, L3612S, A3615V, F3624L, M3626T, F3628P | T4174I*, E622D, R175C, A3456V | L3116F*, R1628S*, T2906I, T2153I, E1245K, E932G, I3758V, E311G* | N1324S, P1803S, P3395H*, S2114F, A3235V | L3606F | S135R*, T842I*, G1307S*, L3027F*, T3090I*, L3201F* |
| ORF1b | P314L,I1566V | P105S*, A1302Y, K2421N, P1570S, F2283L | T11A | C130S, D1735Y | G1014S, R1315C* | |||
| ORF7a | S83Y | |||||||
| ORF3a | L106F*, S165F*, T32I | Y154N* | ||||||
| ORF9b | P10S | |||||||
| E | ET9I | |||||||
| M | MQ19E, MD3G, MA63T | A85S* | I82T | |||||
| N | NP13L, NR203K, NG204R | P67S, E378D* | R68P, T135A, D402Y | Q163K | D343G | S413R |
3.4. Omicron-L452R
Two sequences, classified as BA.1 Omicron sub lineage and named as hCoV-19/Brazil/LMM69880 (Gisaid accession number EPI_ISL_11514450; collection date: 2022/01/18) and hCoV-19/Brazil/LMM71052 (Gisaid accession number EPI_ISL_11514436; collection date 2022/02/07), draw attention to the presence of a Delta signature S:L452R mutation. Both sequences presented Omicron signatures mutations sharing ORF1a:L2084I, ORF1a:I3758V, ORF1b:P314L, ORF1b:I1566V, ORF9b:P10S, N:P13L, N:R203K, N:G204R, S:G339D, S:L452R, S:T478K, S:D614G, S:H655Y, S:N856K, S:Q954H and S:N764K. Additional uncommon mutations were also found in hCoV-19/Brazil/LMM69880/2022 (ORF1b:P1570S and ORF7a:S83Y) and hCoV-19/Brazil/LMM71052/2022 (ORF1a:F1501S, ORF1a:V1535A, ORF1a:F3624, ORF1a:M3626T, ORF1a:F3628P and ORF1b:F2283L). No evidence of recombination was found for the analyzed sequences.
These patients presented different clinical presentations. The patient LMM69880, 60 years old, body mass index (BMI) 34 with incomplete vaccine protocol (single Oxford/AstraZeneca Covid-19 vaccine in 2021/06/18) dose presented mild symptoms with no complications. The patient LMM71052, 43 years old, BMI 37 and two Pfizer vaccine doses (last dose: 2021/10/18) had severe symptoms and was hospitalized requiring mechanical ventilation. These patients were respectively infected approximately six and four months after vaccination. Our virus genome surveillance effort found a new variant of concern (VOC), the Omicron-L452R, which should be closely monitored.
4. Discussion and conclusion
Brazil faced the fourth wave of COVID-19 cases, with the highest peak since the beginning of the pandemic. Analyzing data from the RS health authorities, comparing December 2021 and January 2022, the confirmed case numbers increased 2.572,4% (SES-RS, 2022). It's possible to observe that the increase COVID-19 cases and deaths from the beginning of December 2021 is synchronous with the entry and predominance of the Omicron variant. Omicron has triggered a massive wave of new infections and re-emergent outbreaks worldwide. This variant spreads more easily than the original SARS-CoV-2 virus and other VOCs, with an expressively higher transmissibility; and also can evade natural and vaccine-induced immunity better than its predecessor variants (Mohapatra et al., 2022; Rössler et al., 2021; Torjesen, 2021). The variant established in a short period of time, which was also observed in other countries, contaminating an expressive number of individuals in a short period of time. Even the apparently milder severity, especially in fully vaccinated individuals, the public health impact should be strongly considerable since the cases increased exponentially.
The SARS-CoV-2 variant Omicron sublineages has spread rapidly in RS state, and a diversification was reported herein, with BA.1 and BA.1.1 (21K) being the most frequent ones. As observed in our study, according to covariants.org, Omicron has overtaken the Delta and has spread rapidly to in Europe, Asia, North and South America. The ability of 21K Omicron sublineages to replace the previously predominant Delta variant has been credited to immune escape rather than a higher intrinsic transmissibility (Eggink et al., 2022; Lyngse et al., 2021), but BA.2 has been shown to be even more transmissible than BA.1 (Lyngse et al., 2022). Despite this, the presence of BA.2 was very low in the samples analyzed in this study, maybe due to the still recent entrance into the RS state.
Mutational SARS-CoV-2 analysis have been important since the beginning of the pandemic, especially for the S glycoprotein, which mediates virus attachment to ACE2 receptor, membrane fusion, and entry into the host cell, and also acts as a primary target for neutralizing antibodies elicited by the host immune response (Walls et al., 2020). Omicron sublineages have mutations in common and also specific signatures, as shown in the results section. SARS-CoV-2 is constantly evolving, new mutations are reported daily, and some overlap with other variants. The following mutations S:K417N, S:T478K, S:N501Y, S:D614G, S:H655Y and S:P681H, detected in all Omicron sublineages analyzed herein, were also described in other VOCs (Alpha, Beta, Gamma and Delta) and have been previously related to increase viral binding, immune evasion and high transmissibility (Arora et al., 2021; Greaney et al., 2021; Harvey et al., 2021). L452R is related to increased SARS-CoV-2 fusogenicity and infectivity (Motozono et al., 2021). In our study, we report a natural Omicron-L452R in two patients. Through genetic engineering, a recent reported that an Omicron-L452R mutant showed enhanced fusion activity and increased ability to infect lung tissues of humanized mice (Zhang et al., 2022) and SARS-CoV-2 pathogenicity in patients is strongly related to fusogenic capability (Motozono et al., 2021). This could be the reason that the big COVID-19 wave associated with Omicron infections, less hospitalization rates and clinical severity were observed (Maslo et al., 2022). The clinical signs of the infected Omicron-L52R reported in this study were divergent, one of them presented serious COVID-19 complications, and the other, only mild clinical signs. Despite this, it is important to note that the severity of the disease is multifactorial, and, to link L452R with specific clinical signs it's not possible without further studies as only two Omicron-L453R variants were identified in this study.
The BA.4 and BA.5 Omicron sublineages, that emerged after the sequences reported herein, also presented the S:L452R mutation (Tegally et al., 2022). Despite this, their classification also depends on the presence of other synonymous and non-synonymous mutations, including the S:F486V mutation (reported as BA.4 and BA.5 signature), which was not observed in our sequences. Thus, our sequences probably represented a transition point between lineages, with a mutation that later stabilized in new Omicron BA.4 and BA.5.
SARS-CoV-2 overall rate of evolution, estimated by preliminary molecular clock in 2020 was 8×10-4 substitutions/site/year, which equates to 24 substitutions per year. The current global estimate including multiple variants of concern/interest suggests a similar rate of approximately 25.941 substitutions per year (derived from a global Nextstrain build, https://nextstrain.org/ncov/gisaid/global, accessed April 1st, 2022). Our Omicron analysis presented a higher clock rate of 28.125 substitutions per year, showing a higher substitution rate than the majority of other sequences.
In summary, we described the replacement of the Delta lineage by Omicron in a short period of time and the presence of seven sublineages (BA.1, BA.1.1, BA.1.14, BA.1.15, BA.1.17, BA.1.9 and BA.2), mapping their mutational profile. Furthermore, we report the first natural infections with Omicron-L452R variant, an important S protein mutation previously characteristic only of the Delta VOC. Our study has provided a comprehensive investigation concerning the epidemiological and genetic characteristics of the major wave caused by Omicron in RS state, Southern Brazil, and contributed to important scientific findings enriching the knowledge on SARS-CoV-2.
Ethical aspects
Project approved by the Research Ethics Committee (CEP) at Feevale University. Process number: CAAE: 33202820.7.1001.5348.
CRediT authorship contribution statement
Mariana Soares da Silva: Conceptualization, Formal analysis, Investigation, Writing – original draft. Juliana Schons Gularte: Conceptualization, Formal analysis, Investigation. Micheli Filippi: Conceptualization, Formal analysis, Investigation. Meriane Demoliner: Conceptualization, Formal analysis, Investigation. Viviane Girardi: . Ana Cristina Sbaraini Mosena: Formal analysis, Investigation. Vyctoria Malayhka de Abreu Góes Pereira: Formal analysis, Investigation. Alana Witt Hansen: Formal analysis, Investigation. Matheus Nunes Weber: Formal analysis, Investigation, Resources. Paula Rodrigues de Almeida: Formal analysis, Investigation. Juliane Deise Fleck: Funding acquisition, Resources, Supervision. Andrea Gurgel Batista Leite Dal Bó: Writing – review & editing. Marcus Herbert Jones: Writing – review & editing. Frederico Friedrich: Writing – review & editing. Luiz Amorim Filho: Writing – review & editing. Fábio Klamt: Writing – review & editing. Fernando Rosado Spilki: Conceptualization, Writing – review & editing, Funding acquisition, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Brazilian Coordination for the Improvement of Higher-Level Personnel (CAPES), Foundation for Research Support of the State of Rio Grande do Sul (FAPERGS), and Brazilian National Council for Scientific Development (CNPq) for scholarships. This work is an initiative of Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP = 01.20.0029.000462/20, CNPq = 404096/2020-4), and FAPERGS (grant 21/2551-0000081-3). IMMUNESHARE – MCTI Trial (SEFEP/MCTI/CNPq #401558/2021-5). We thank the researchers Mariene Amorim and José Luiz Proença-Módena from Laboratório de Estudos de Vírus Emergentes – Unicamp for their collaboration.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198907.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Arora P., Pöhlmann S., Hoffmann M. Mutation D614G increases SARS-CoV-2 transmission. Signal Transduct. Target. Ther. 2021;6:101. doi: 10.1038/s41392-021-00502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva M.S., Demoliner M., Hansen A.W., Gularte J.S., Silveira F., Heldt F.H., Filippi M., Pereira V.M., de A.G., da Silva F.P., Mallmann L., Fink P., da Silva L.L., Weber M.N., de Almeida P.R., Fleck J.D., Spilki F.R. Early detection of SARS-CoV-2 P.1 variant in southern brazil and reinfection of the same patient by p.2. Rev. Inst. Med. Trop. Sao Paulo. 2021;63:1–8. doi: 10.1590/S1678-9946202163058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D., Andeweg S.P., Vennema H., van Maarseveen N., Vermaas K., Vlaemynck B., Schepers R., van Gageldonk-Lafeber A.B., van den Hof S., Reusken C.B.E.M., Knol M.J. Increased risk of infection with SARS-CoV-2 Omicron BA.1 compared with Delta in vaccinated and previously infected individuals, the Netherlands, 22 November 2021 to 19 January 2022. Eurosurveillance. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.4.2101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E.J., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gularte J.S., da Silva M.S., Mosena A.C.S., Demoliner M., Hansen A.W., Filippi M., Pereira V.M., de A.G., Heldt F.H., Weber M.N., de Almeida P.R., Hoffmann A.T., Valim A.R., de M., Possuelo L.G., Fleck J.D., Spilki F.R. Early introduction, dispersal and evolution of Delta SARS-CoV-2 in Southern Brazil, late predominance of AY.99.2 and AY.101 related lineages. Virus Res. 2022;311 doi: 10.1016/j.virusres.2022.198702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngse, F.P., Kirkeby, C.T., Denwood, M., Christiansen, L.E., Mølbak, K., Møller, C.H., Skov, R.L., Krause, T.G., Rasmussen, M., Sieber, R.N., Johannesen, T.B., Lillebaek, T., Fonager, J., Fomsgaard, A., Møller, F.T., Stegger, M., Overvad, M., Spiess, K., Mortensen, L.H., 2022. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish Households. medRxiv 2022.01.28.22270044. doi: 10.1101/2022.01.28.22270044. [DOI]
- Lyngse, F.P., Mortensen, L.H., Denwood, M.J., Christiansen, L.E., Møller, C.H., Skov, R.L., Spiess, K., Fomsgaard, A., Lassaunière, M.M., Rasmussen, M., Stegger, M., Nielsen, C., Sieber, R.N., Cohen, A.S., Møller, F.T., Overvad, M., Mølbak, K., Krause, T.G., Kirkeby, C.T., 2021. SARS-CoV-2 Omicron VOC Transmission in Danish Households. medRxiv 2021.12.27.21268278. doi: 10.1101/2021.12.27.21268278. [DOI]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4 : detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:1–5. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 omicron wave compared with previous waves. JAMA. 2022;327:583–584. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra R.K., Sarangi A.K., Kandi V., Azam M., Tiwari R., Dhama K. Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: current global scenario. J. Med. Virol. 2022;94:1780–1783. doi: 10.1002/jmv.27561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., Yue Y., Shimizu R., Ito J., Torii S., Yonekawa A., Shimono N., Nagasaki Y., Minami R., Toya T., Sekiya N., Fukuhara T., Matsuura Y., Schreiber G., Ikeda T., Nakagawa S., Ueno T., Sato K. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136. doi: 10.1016/j.chom.2021.06.006. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajah M.M., Bernier A., Buchrieser J., Schwartz O. The mechanism and consequences of SARS-CoV-2 spike-mediated fusion and syncytia formation. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler, A., Riepler, L., Bante, D., Laer, D. von, Kimpel, J., 2021. SARS-CoV-2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv 2021.12.08.21267491. doi: 10.1101/2021.12.08.21267491. [DOI]
- Secretaria da Saúde do Rio Grande do Sul (SES-RS), 2022. Boletim Epidemiológico. Disponível em: https://coronavirus.rs.gov.br/boletins-epidemiologicos-2022. Acesso realizado em: 03/08/2022.
- Shu Y, McCauley J.GISAID. Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. pii=30494. DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., Baxter C., Althaus C.L., Anyaneji U.J., Kekana D., Viana R., Giandhari J., Lessells R.J., Maponga T., Maruapula D., Choga W., Matshaba M., Mbulawa M.B., Msomi N., Naidoo Y., Pillay S., Sanko T.J., San J.E., Scott L., Singh L., Magini N.A., Smith-Lawrence P., Stevens W., Dor G., Tshiabuila D., Wolter N., Preiser W., Treurnicht F.K., Venter M., Chiloane G., McIntyre C., O'Toole A., Ruis C., Peacock T.P., Roemer C., Pond S.L.K., Williamson C., Pybus O.G., Bhiman J.N., Glass A., Martin D.P., Jackson B., Rambaut A., Laguda-Akingba O., Gaseitsiwe S., von Gottberg A., de Oliveira T. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;2021 doi: 10.1038/s41591-022-01911-2. NGS-SA consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- Tsang A.K.L., Cheng P.K.C., Mak G.C.K., Leung P.K.L., Yip P.C.W., Lam E.T.K., Ng K.H.L., Chan R.C.W. Unusual high number of spike protein mutations for the SARS-CoV-2 strains detected in Hong Kong. J. Clin. Virol. 2022;148 doi: 10.1016/j.jcv.2022.105081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cheng G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2022;94:1728–1733. doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- WHO, 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. [WWW Document]. World Heal. Organ. 2021. URL https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed 3.31.22).
- Zhang Y., Zhang T., Fang Y., Liu J., Ye Q., Ding L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct. Target. Ther. 2022;7:76. doi: 10.1038/s41392-022-00941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.