Abstract
While antibodies garner the lion’s share of attention in SARS-CoV-2 immunity, cellular immunity (T cells) may be equally, if not more important, in controlling infection. Both CD8+ and CD4+ T cells are elicited earlier and are associated with milder disease, than antibodies, and T-cell activation appears to be necessary for control of infection. Variants of concern (VOCs) such as Omicron have escaped the neutralizing antibody responses after two mRNA vaccine doses, but T-cell immunity is largely intact. The breadth and patient-specific nature of the latter offers a formidable line of defense that can limit the severity of illness, and are likely to be responsible for most of the protection from natural infection or vaccination against VOCs which have evaded the antibody response. Comprehensive searches for T-cell epitopes, T-cell activation from infection and vaccination of specific patient groups, and elicitation of cellular immunity by various alternative vaccine modalities are here reviewed. Development of vaccines that specifically target T cells is called for, to meet the needs of patient groups for whom cellular immunity is weaker, such as the elderly and the immunosuppressed. While VOCs have not yet fully escaped T-cell immunity elicited by natural infection and vaccines, some early reports of partial escape suggest that future VOCs may achieve the dreaded result, dislodging a substantial proportion of cellular immunity, enough to cause a grave public health burden. A proactive, rather than reactive, solution which identifies and targets immutable sequences in SARS-CoV-2, not just those which are conserved, may be the only recourse humankind has to disarm these future VOCs before they disarm us.
Keywords: alternative vaccine modalities, cellular immunity, COVID, SARS-CoV-2, special patient groups, T-cell epitopes
Introduction
In late 2019, a novel coronavirus infected humans, and rapidly spread, resulting in a global pandemic. Subsequently called SARS-CoV-2 (severe acute respiratory syndrome coronavirus type 2), it is the causative agent of Coronavirus disease of 2019 (COVID-19). As of 8 June 2022, there have been 87.0 million cases in the United States and 1.04 million deaths, and 538 million cases worldwide and 6.33 million deaths. 1 COVID-19 symptoms include fever, chills, cough, difficulty breathing, fatigue, body aches, headache, loss of taste or smell, sore throat, runny nose, nausea or vomiting, and diarrhea. 2 A substantial number of those infected with SARS-CoV-2 go on to develop severe disease, which can result in hospitalization or death. The elderly and those with other preexisting conditions are particularly susceptible. The majority of young people who are infected are asymptomatic or experience mild disease. However, 10–30% of COVID-19 patients, including even those who had mild disease, go on to develop ‘long-COVID-19’, and are called ‘long-haulers’, 3 which can manifest as, besides the symptoms of acute COVID-19, joint pain, chest pain, memory loss, concentration or sleep problems, rapid heartbeat, depression or anxiety, and dizziness. The virus can damage organs other than the respiratory tract and lungs, such as heart, kidney, and brain. 4
The drive to develop a vaccine for COVID-19 resulted in a wholly unprecedented speed from concept to Food and Drug Administration (FDA) authorization, with the whole process taking a little over a year. There are currently three vaccines approved for use in the United States: BNT162b2 from Pfizer/BioNTech (mRNA); mRNA-1273 from Moderna; and Ad26.COV2.S (adenovirus) from J&J, all three consisting entirely of SARS-CoV-2 spike protein. Outside the United States, the most widely used vaccines include ChAdOx1-S (adenovirus) from AstraZeneca; GAM-COVID-Vac (adenovirus), or Sputnik, from the Gamaleya Institute in Russia; and CoronaVac (inactivated virus) from Sinovac in China.
SARS-CoV-2 is an RNA virus which infects cells of the upper and lower respiratory tract via the receptor human angiotensin converting enzyme-2 (hACE2). 5 Its RNA polymerase has proofreading capability, 6 and thus the mutation rate is lower than that of, for example, influenza virus and human immunodeficiency virus-1 (HIV);7,8 however, the sheer volume of virus in an infected individual, and the extent of infection in the global population, results in the continual emergence of new variants. When a variant rises to the level of demanding special public health action, it is classified as a Variant of Concern (VOC) by the U.S. government SARS-CoV-2 Interagency Group. Currently, Delta and Omicron are VOCs, and past VOCs include Alpha, Beta, and Gamma. 9 At the cellular level, some VOCs can infect their target cells more efficiently, resulting in increased contagiousness (although some are less transmissible, such as the Beta VOC). In some cases, they can cause more severe disease and, perhaps most significantly, can evade natural or vaccine-mediated immunity to some extent. 10 Delta outpaced the existing strains so efficiently that in a matter of months it constituted virtually 100% of new infections worldwide. Since then, Omicron has superseded even Delta, such that now it makes up 100% of new infections in the United States, 11 since it emerged in November 2021.
Antiviral immunity
When the body detects an invading virus, the first line of immunological defense is the innate immune system. This arm is initiated immediately and includes neutrophils, macrophages, dendritic cells, and natural killer cells. Innate immunity activates inflammation, recruits immune cells to the infection site, activates complement, phagocytoses foreign bodies, and activates the adaptive immune response. 12 The adaptive immune response is antigen specific and takes longer to develop. It consists of humoral immunity (antibodies) and cell-mediated immunity (T cells). In the humoral response, B cells are activated to secrete antibodies, which can bind to extracellular virus and neutralize it, that is, prevent it from infecting target cells. There is also a non-neutralizing antibody response, where binding of antibody to antigen can result in enhanced phagocytosis or activation of the complement cascade. T cells respond to intracellular virus and largely consist of CD4+ T cells and CD8+ T cells. Viral protein that is produced in the cytosol, or that is phagocytosed into lysosomes, is digested by the proteasome, and short peptides are translocated to the endoplasmic reticulum where they bind to major histocompatibility complex (MHC) molecules. Cytosolic-derived peptides bind to MHC Class I and are presented on the cell surface to CD8+ T cells. Virus phagocytosed by professional antigen-presenting cells (APCs) produces peptides that bind to MHC Class II, which are presented on the cell surface to CD4+ T cells. 13
CD8+ T cells, also known as cytotoxic T lymphocytes (CTLs), recognize virus-infected cells presenting the same peptide-MHCs that activated them in the first place and destroy the cell via granzyme B or perforin, limiting viral spread. 14 CD4+ T cells, which are activated by professional APCs, include several specific groups with different functions. Broadly speaking, T helper 1 (Th1) cells enhance the CTL response; Th2 cells enhance the antibody response and anti-parasitic immunity; T follicular cells (Tfh) enhance the antibody response; Th17 cells promote inflammation but can also cause autoimmunity; and regulatory T cells (Tregs) act to maintain homeostasis (a balance between immunogenicity and immune tolerance) and inhibit autoimmunity (Figures 1 and 2). 15
Figure 1.
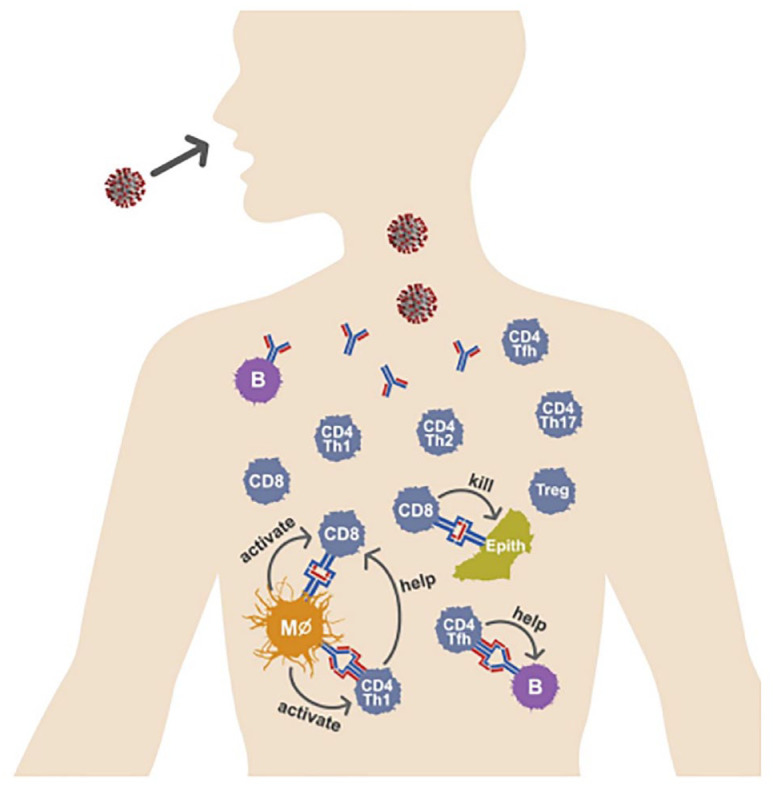
The adaptive immune response to viral infection. Circulating antibody; B cells; CD8+ T cells; and CD4+ T follicular helper (Tfh), Th1, Th2, Th17, and regulatory T (Treg) cells. Shown are Th1 cells being activated by antigen-presenting cells (APC; here a macrophage), which in turn ‘help’ a CD8+ T cell also being activated by the same APC; a Tfh cell helping an antibody-producing B cell; and a CD8+ cytotoxic T cell killing a virally infected epithelial cell.
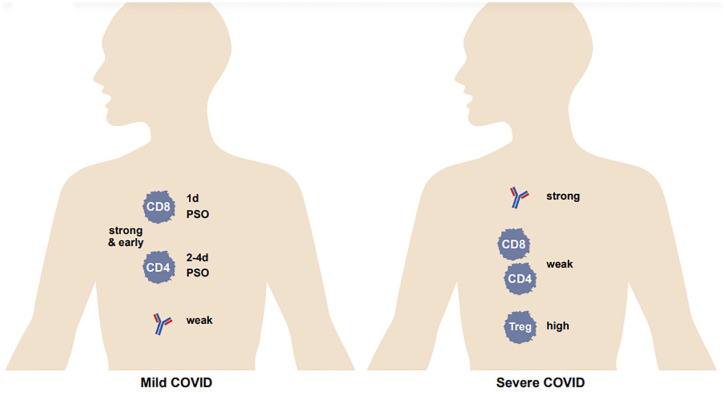
Figure 2.
T-cell and antibody activity in mild versus severe COVID-19. T-cell activity is high and B-cell activity is low in mild COVID-19, whereas T-cell activity is low and B-cell activity is high in severe COVID-19. Treg activity is high in severe COVID-19. PSO, post-symptom onset.
T-cell control of SARS-CoV-2 infection
Antibodies and T cells in mild versus severe disease
It has been widely recognized that a strong antibody response is associated with severe COVID-19, 16 whereas for T cells it is the opposite – a strong and early response is associated with mild disease.17,18 On the other hand, there is minimal induction of T cells in moderate to severe disease, 19 and an impairment in the T-cell response in death. 20 (However, some reports have observed opposite results: for example, higher activation of CD4+ and CD8+ T cells in severe disease; 21 also see below on conflicting evidence from studies on T-cell exhaustion.) A high level of Tregs (suppressive T-cell response) was associated with severe COVID. 22 One cannot immediately conclude from this, however, that antibodies are failing in their control of infection, or that they actually enhance infection, and that T cells are able to control infection on their own. To our knowledge, which is the cause and which is the effect has not been unraveled (nor would this be easy to do): is a robust antibody response generated proportionate to a high viral load, or is a strong antibody response generated in a subset of individuals and yet cannot control infection? However, in contrast, high T-cell activity is associated with milder disease, and a poor T-cell response with severe disease. Because it is difficult to imagine that high viral loads induce less T cells than low viral loads, one might hypothesize that individuals who are able to generate a potent T-cell response to SARS-CoV-2 infection are able to dampen viral replication by this means. Still, antibodies appear to be of substantial importance for controlling infection, as evidenced by the high (at least initial) effectiveness of the current COVID-19 vaccines, which elicit both antibody and T-cell responses. This is further underscored by the effectiveness of monoclonal antibody therapy for treatment of COVID-19. 23
Kinetics of antibodies versus T cells
While one might think that antibodies are critical for an immediate response to SARS-CoV-2 infection, and T cells control the progression of infection as a slower response (given that they only react to infected cells), the evidence may suggest otherwise. T cells are induced in natural SARS-CoV-2 infection extremely rapidly: CD4+ T cells can be detected at 2–4 days post-symptom onset (PSO), 18 and CD8+ T cells as early as 1 day PSO, 24 whereas seroconversion (generation of antibodies) occurs between 5 and 15 days PSO. 25 Many studies have examined the duration of T-cell immunity after natural infection, as well as that of antibodies and memory B cells. CD4+ and CD8+ T cells have a half-life of 200 days. 26 Another study observed a decline in CD4+ and CD8+ T cells after natural infection, with a half-life of 3–5 months. 27 Chen et al. 28 report that the level of spike-specific CD4+ T cells does not change by 7 months after infection. A detectable T-cell response to SARS-CoV-2 antigens was present in 93% and 92% of patients at 6 months and 12 months after disease onset, respectively. 29 And Jung et al. 30 find that stem cell-like memory T cells actually increase over the course of convalescence, with a peak at 120 days PSO. The neutralizing antibody (NAb) response is also of similar duration: NAbs have a half-life of >200 days; however, this is likely attributable to long-lived plasma (antibody-producing) cells rather than the persistence of an antibody in circulation. 26 About 95% of COVID-19 convalescents had detectable NAbs from 6 months to 12 months after the onset of disease. 29 Changes in the status of long-lived CD8+ T cells develop over time: a study on CD8+ T cells specific for nucleocapsid 322-331 found that there was a decrease in the activation state and polyfunctionality of these T cells, but an increase in the homeostatic proliferation potential and in their lymph node-homing. 31
T-cell subsets in SARS-CoV-2 infection
Virus-specific CD4+ T cells dominate the T-cell response, with a smaller contribution from virus-specific CD8+ T cells. In 100% and 70% of convalescent patients, SARS-CoV-2-specific CD4+ T cells and CD8+ T cells could be detected, respectively. 32 Within the CD4+ T-cell population, Th1 and Tfh subpopulations predominate.33,34 Th1 cells produce interferon-γ (IFN-γ) and other cytokines which help control viral infection, as well as help CD8+ T cells. Tfh cells help elicit virus-specific NAbs. 35 There is evidence that skewing toward a Th2-dominant response is associated with poor viral control. 36 In an experiment where Tfh cells were perturbed after spike protein vaccination in mice, it was shown that these cells promote the frequency and somatic hypermutation (random mutation of the B-cell receptor/antibody which improves affinity) of germinal center B cells. 37 A study of patients with severe COVID-19 found Tfh-independent antibodies induced by vaccination or infection; while somatic hypermutation was reduced, the antibodies were still of high affinity and durable, and the clonal diversity was actually higher (Table 1). 38
Table 1.
Alternative vaccine platforms against SARS-CoV-2 in development.
| Format | Antigen(s) | Species tested | T-cell immunity? | Special considerations | Reference |
|---|---|---|---|---|---|
| Mesoporous silica rods | S and N | Mouse | Yes | Can be lyophilized | 131 |
| Protein fused to alpaca-derived nanobody | Spike RBD | Mouse | Yes | Binds to MHC Class II | 132 |
| Peptide with XS15 and Montanide ISA51VG adjuv | T-cell epitopes from various prots | Human | Yes | Multifunctional CD4+ and CD8+ T cells exceeding natural infection and vaccination | 133 |
| Codon-optimized DNA | Spike | Mouse | Yes | Lasted at least 6 months | 134 |
| DNA + protein | Spike | Macaques | Yes | Delivered by electroporation | 135 |
| Ferritin nanoparticle with Alhydrogel and Army Liposome adjuv | Spike | Mouse | Yes | Polyfunctional memory CD4+ and long-lived memory CD8+ | 136 |
| Intradermal skin patch | Nucleoprot. | Mouse | Yes | Dissolvable microneedles | 137 |
| N,N,N-trimethyl chitosan particles | Spike | Mouse | Yes | Peritoneal delivery | 138 |
| Peptide w/Freund’s adjuv | Spike 446-480 | HLA-transgenic mouse | Yes | Detected multiple human T-cell epitopes | 139 |
| Protein encapsulated in polymersome | Spike | Mouse | Yes | Self-assembling nanoscale vesicle | 140 |
| Polymeric glyco-adjuvant | Spike | Mouse | Yes | 141 | |
| mRNA fused to nanoparticles | Mouse | Yes | Th1-biased | 142 | |
| 20 peptides | S and N | Mouse & hamster | Yes | RNA adjuvant | 143 |
| Modified vaccinia virus Ankara | Spike | Hamster | Yes | Prime-boost (i.m.-i.n.) | 144 |
| MVA | Spike | Mouse | Yes | Prime-boost | 145 |
| Adenovirus serotype 5 | Spike RBD | Mouse | Yes | Lasted over 6 months with 1 dose | 146 |
| Ad5 | S and N | Macaques | Yes | Enhanced T-cell Stimulation Domain to target MHC Class II to elicit Th1-biased response | 147 |
| Ad5 | N | Mouse & hamster | Yes | Rapid T-cell recall response in respiratory mucosa | 148 |
| Inactivated vaccine | Whole virus | Human | Yes | Equivalent T-cell response to approved vaccines | 149 |
| Inactivated vaccine | Whole virus | Human | Yes | Immunogenicity in PBMC ELISPOT against S, N, and E | 150 |
| Fusion protein Spike-RBD-Rotavirus VP6 | Spike RBD | Mouse | Yes | High quantity T cells and no Abs | 151 |
Functional experiments: T cells necessary and/or sufficient in SARS-CoV-2 infection?
In animal models, adoptive transfer of viral-specific T cells was able to control SARS-CoV-1 viral infection in mice. 39 In rhesus macaques, prior depletion of CD8+ T cells abrogated control of SARS-CoV-2 viral infection. 40 Intriguing data were reported on human patients lacking antibodies, due to a genetic deficiency such as agammaglobulinemia, with these patients being able to fully control infection. 41 These cumulative observations provocatively suggest that in natural SARS-CoV-2 infection, T cells shoulder the bulk of the antiviral adaptive immune response, while antibodies play a lesser role. However, patients receiving B-cell–depleting anti-CD20 therapy for B-cell lymphoma had a more severe and protracted clinical course of COVID-19. 42 Also, antibodies may be critical for the prevention of reinfection. 43
T-cell exhaustion
In persistent viral infection, T cells can undergo a state of exhaustion. CD8+ T-cell exhaustion involves the loss of interleukin-2 (IL-2), tumor necrosis factor-α (TNF-α), and IFN-γ production and a gradual loss of proliferative capacity. CD4+ T cells can also undergo exhaustion, exemplified by a loss in effector function. A high level of inhibitory receptors are expressed, including lymphocyte-activation protein 3 (LAG3), cytotoxic T lymphocyte-activation antigen 4 (CTLA-4), and T-cell immunoglobulin and mucin-domain containing protein 3 (TIM-3). 44 Interestingly, the frequency of exhausted SARS-CoV-2-reactive CD8+ T cells was increased in COVID-19 patients with mild disease as compared with those with severe disease, and this subset displayed less cytotoxicity and inflammatory features. This study also demonstrated an impaired IFN response, and the generation of a robust CD8+ T-cell memory response with pro-survival transcriptomic markers, in those with severe disease. It is unclear how these associations can be reconciled with earlier observations on the association of a strong T-cell response with mild COVID-19 and a weak one with severe disease; Kusnadi et al., 45 at least, propose that a robust memory CD8+ T-cell response in severe COVID-19 may be contributory toward prevention against re-exposure. It may also be that in certain patients T cells may themselves promote disease pathogenesis, 46 which might convolute association data. In contrast, Zheng et al. 47 found the frequency of the non-exhausted CD8+ T-cell subset to be reduced in those with severe infection. Lymphopenia is a common feature of severe COVID-19 and is associated with poor outcomes for these patients. 48 Nonetheless, it should be pointed out that most studies on T-cell exhaustion have been with chronic viruses like HIV; 49 the vast majority of COVID-19 patients clear their infection, so T-cell exhaustion must not usually be due to persistent virus, except for those who are immunocompromised in whom virus can exist for a long duration.
Preexisting immunity to SARS-CoV-2 from immunity to other coronaviruses
SARS-CoV-2 belongs to the beta group of coronaviruses, which also includes SARS-CoV, MERS-CoV, and four endemic coronaviruses causing colds in humans (some alpha coronaviruses also cause colds in humans). Surprisingly, although SARS-CoV-2 is a relatively new human coronavirus, many studies have found preexisting T cells to the endemic cold coronaviruses which also cross-reacted to SARS-CoV-2 in a subset of individuals. Fifty-two COVID-19 household contacts were sampled at the earliest timepoint after exposure, and those who remained polymerase chain reaction (PCR)-negative for virus had statistically significantly higher T cells cross-reactive to both human endemic coronavirus and SARS-CoV-2 nucleocapsid (but not to spike) than those who became PCR-positive. 50 Sagar et al. 51 found that individuals who had a recent history of human coronavirus infection had lower rates of intensive care unit admissions and better survival after acquiring SARS-CoV-2 infection. In fact, SARS-CoV-2 cross-reactive CD4+ T cells were found in as many as 66% of healthy, unexposed individuals. 52 Richards et al. 53 took circulating CD4+ T cells from prior to 2020 and tested them for human coronavirus (other than SARS-CoV-2) spike, and then examined positive cells for reactivity to SARS-CoV-2 spike, nucleoprotein, and membrane/envelope, and found a highly variable functional potential, with nucleocapsid-specific CD4+ T cells producing the highest levels of granzyme. Preexisting polymerase-specific T cells were detected in health-care workers who were able to abort infection and stay seronegative, and those individuals with the highest early transcribed replication-transcription complex-reactive T cells had an increase in IFI27, a robust early innate signature of SARS-CoV-2. 54
T cells after SARS-CoV-2 vaccination
Comparison between the three U.S. vaccines
A comparison of the three vaccines approved in the United States found that mRNA-1273 (Moderna) and BNT162b2 (Pfizer) yielded similar antibody and neutralization titers after one dose, which were higher than Ad26.COV2.S (J&J). The two mRNA vaccines also induced higher bulk and cytotoxic T-cell responses than Ad26.COV2.S. All three elicited CD8+ T-cell responses in less than 50% of vaccinees after one dose. 55 Collier et al. 56 found that all three vaccines elicited similar kinetics of humoral and cellular immune responses and that at 8 months post-vaccination, the two mRNA vaccines (two doses) had slightly higher CD8+ T-cell responses than Ad26.COV2.S (0.016%, 0.017%, and 0.012% for BNT162b2, mRNA-1273, and Ad26COV2.S, respectively).
A study by Ivanova et al. found that while COVID-19 patients and vaccinees both induced robust innate and adaptive immune responses, there were qualitative differences in the character of these responses. Patients had a much stronger interferon response, which likely led to more dramatic upregulation of cytotoxic genes in T cells. The majority of B and T cells in patients were effector cells, whereas in vaccinees most were memory cells. 57 Sahin et al. reported that most recipients of BNT162b2 had a strong IFN-γ+ or IL-2+ CD8+ and CD4 + Th1 response. An examination of several epitopes presented by frequent MHC alleles revealed that epitope-specific CD8+ T cells of an early-differentiated effector memory phenotype were 0.02–2.92% of CD8+ T cells 1 week after boost. 58 A study on T-cell immunity after AZD1222 (Astra Zeneca’s ChAdOx1 nCoV-19) showed that the Th1 and CD8+ T cells that were induced showed a high degree of polyfunctionality, with extensive breadth and depth across spike protein. 59
Kinetics and characteristics of T-cell induction by vaccines
Using the activation-induced marker (AIM) assay on peripheral blood, with spike peptide megapools, Painter et al. found that for mRNA vaccinees, CD4+ T cells were induced in all individuals surveyed. CD8+ T cells, however, were more gradual in their response, with lower numbers of individuals having a detectable response (71% after prime, rising to 88% after boost). 60 In contrast, Oberhardt et al., testing for three spike epitopes [restricted to three different human leukocyte antigen (HLA) alleles], observed a rapid and substantial induction of CD8+ T cells in 9 of 13 individuals at 6–8 days post-prime, and peaking for most individuals as early as 9–12 days post-prime. Measurement of a spike-specific CD4+ T-cell epitope revealed only a weakly detectable CD4+ T-cell level, as was the case also for neutralizing antibody levels, at these times. 61 The disparity in these two publications may owe to the different assays utilized, and/or the epitope restrictions tested. Gil-Manso et al. 62 found that the maximum levels of both the humoral response (as measured by circulating spike-specific IgG) and the cellular response (as measured by SARS-CoV-2-reactive, cytokine-releasing T cells) were not reached until 14 days after the second vaccine dose.
Guerrera et al. 63 reported that anti-RBD (receptor-binding domain of spike) antibodies did not reach significant levels until after the BNT162b2 boost, and by 6 months post-boost, were down overall about 3.5-fold. AIM+ CD4+ T cells, on the other hand, were hardly reduced at 6 months post-boost; and AIM + CD8 + T cells, which were present in 87% of vaccinees after the first dose, and which underwent further expansion after the second dose, were still present in 88% of vaccinees 6 months post-boost. In another study, low-dose mRNA-1273 elicited spike-specific CD8+ T cells in 88% of vaccinees and were maintained in 67% of vaccinees at 6 months post-boost; notably, both spike-specific CD4+ and CD8+ T cells were induced to just as high levels in older individuals as in the 18- to 55-year-old age group. 64 Barouch et al., 65 examining Ad26.COV2.S, found that spike-specific IFN-γ+ CD4 + and CD8 + T cells, as measured by intracellular cytokine staining (ICS), were not diminished out to 8 months post-vaccination. Fine-needle aspiration of draining lymph nodes in patients who had received the BNT162b2 vaccine detected spike-specific T cells being maintained at constant frequencies out to 6 months post-vaccination. 66 Interestingly, in neuro-COVID long-haulers, that is, patients with post-acute sequelae of SARS-CoV-2 infection (PASC), there were decreased spike-specific but increased nucleocapsid- and membrane-specific T cells compared with healthy convalescents. 67
Numerous studies reported that convalescents who received their first mRNA vaccine dose had similar levels of T cells induced as naïve patients who received their second dose, and that the second dose does not further induce T cells in convalescent patients.68–71 Heterologous prime-boost with the UK ChAdOx1 (a chimpanzee adenoviral-vectored spike vaccine) and mRNA vaccine induced higher frequencies of spike-specific CD4+ and CD8+ T cells than homologous prime-boost (with either ChAdOx1, BNT162b2, or mRNA-1273).72,73
T-cell immunity against variants of concern induced by the vaccines
Much work has been done analyzing T-cell immunity against VOCs elicited by the vaccines. While the VOCs have significant reductions in susceptibility to the vaccine-induced humoral response, it appears that the cellular response is less affected. A study on B.1.1.7 (Alpha) and B.1.351 (Beta) variants found no observable differences in CD4+ T-cell activation in comparison with the vaccine strain. 74 Another study observed that the neutralizing antibody response against B.1.351 was more reduced than that against B.1.1.7, but that the T-cell response was directed against epitopes that were conserved between the vaccine strain and these two VOCs. 75 Neidleman et al. 76 reported that the overall T-cell response to the ancestral (vaccine) strain, B.1.1.7, and B.1.351 were similar; but spike-specific T cells from convalescent vaccinees had more long-term persistence and better homing to the respiratory tract, including the nasopharynx, than naïve vaccinees. Later studies incorporated the Delta (B.1.617.2) variant. Both Woldemeskal et al. 77 and Jordan et al. 78 found that the overall T-cell response and the CD4+ and CD8+ T-cell response to BNT162b2, respectively, were virtually the same against Delta as the ancestral strain. In contrast, Keeton et al. 79 looked at Ad26.COV2.S and found some reduction in the CD8+ T-cell response to Delta, with 8/15 vaccinees having a 2-fold or greater reduction, and five in the group having complete loss of a T-cell response to Delta. Data on Omicron are more sparse, since this VOC is newer, but preliminary observations are that most T-cell epitopes against the ancestral strain are conserved in Omicron, suggesting that even this variant might not have escaped the T-cell arm of the immune response to the vaccine and to prior infection. 80 Still, a recent report found that 20% of individuals examined had a T-cell activity against Omicron that was reduced by more than 50%. 81
Why would convalescent patients and vaccinees have a less affected cell-mediated response than humoral response to variants? It has been argued that T-cell epitopes tend to be more conserved than antibody epitopes, 82 but it would be helpful to inquire into this hypothesis more deeply. There is no biochemical reason why a given individual T-cell epitope would be more conserved than a given antibody epitope. We have noted, in our studies on influenza virus, that there is a qualitative and substantial difference between sequence conservation and functional invariance.83,84 The former indicates the degree to which a residue, sequence stretch, or whole protein is the same among most or all circulating strains, while the latter is a measure of the degree of attenuation or lethality of a given mutation on viral replication, in quantifiable terms. The two are neither theoretically nor experimentally equivalent, as our genomic data on influenza virus has demonstrated (unpublished observations). It has been offered that T-cell epitopes are, in general, more conserved than antibody epitopes, and that they are more functionally invariant (from a mutational standpoint), in the same breath. Mutations arise from errors in replication, and on the nucleotide level, there should be no bias from one protein to the next; even though some nucleotide conversions are more common than others, this will average out over the whole protein or a subdomain of it. It is true, indeed, that T-cell epitopes can sample among all viral proteins, even those that are internal in the viral particle, whereas antibodies can only react to external proteins, namely those that are structural and exposed. And, it is true that for influenza virus, internal proteins are more conserved than the two surface proteins, hemagglutinin and neuraminidase; 85 similarly, SARS-CoV-2 nucleocapsid is well-conserved among the human coronaviruses. 86 One might in fact think that a surface protein that mediates attachment and internalization should be more conserved than internal proteins, which need not necessarily interact with cellular proteins.
Alternative explanations for the more intact T-cell response
Preferable hypotheses have been offered by other virologists as to why the overall T-cell response to SARS-CoV-2 seems to be less affected by variants than the antibody response. First, there are more potential T-cell epitopes than antibody epitopes, because the former can sample across all viral proteins, not just external ones. Greater breadth means less chance for a variant to escape all of them – and even any of them, because mutation away from one cannot be selected for unless it can mutate away from, not necessarily all, but enough comprising the overall T-cell response. 87 Second, T-cell epitopes are highly diverse from individual-to-individual, because they are restricted by the MHC (HLA) haplotype. Even if, in a hypothetical scenario, a variant arises within the quasispecies (the diverse population of minor or unique variants within a single individual) to escape that patient’s T-cell immunity, it is likely to be fully sensitive to the T-cell response of the next individual, and thus would be stopped dead in its tracks. We offer another, not mutually exclusive, hypothesis: viruses may have evolved a ‘flexibility’ in amino acid sequence in external proteins, so that they can better evade antibody immunity. This is a rather complicated notion, as it implies that viruses have evolved evolvability; and how exactly they do that on a molecular level is difficult to envision. One prominent example, however, is influenza virus: the bulk of the antibody response to the virus, either from natural infection or vaccination, is directed against the head domain of hemagglutinin, which is the most mutable portion of the most mutable protein in the viral proteome. 81 Somehow, the virus has managed to devise a structure that both binds the cellular receptor and can do so in more ways than one, so that if it ever has to mutate to avoid antibody binding, it can still maintain its ability to bind the receptor.
All of these observations underscore a valuable lesson: it perhaps will prove to be highly important, especially in the long run, to design vaccines that specifically and potently target T cells. We do not suggest that antibody vaccines or antibody components to vaccines should be abandoned, but rather supplemented. There are ways to directly target T cells, such as epitope vaccines, vaccines that incorporate internal proteins, or the use of T cell-activating adjuvants. T cells often get short shrift in the vaccine world, and antibodies often not only take center stage but can even lead people to lose sight of T cells; this is likely to be a grievous mistake, but one that is certainly correctable.
Anti-SARS-CoV-2 T-cell epitope analyses
All that being said, there is evidence of relative T-cell escape among SARS-CoV-2 variants, both on an epitope level and in overall reactivity. Tarke et al. found that the total reactivity of CD4+ and CD8+ T cells in convalescents and vaccinees against B.1.351 (Beta) was reduced by 14% and 22%, respectively, and the reactivity of CD8+ T cells against CAL.20 C (a Southern California variant) dropped by 10%. It must be acknowledged that a drop in overall T-cell reactivity could partly owe to epitope-independent aspects, such as T-cell exhaustion, as mentioned above. Only 7% and 3% of previously identified CD4+ and CD8+ T-cell epitopes, respectively, were mutated in various VOCs in this study. 88 de Silva et al. surveyed 360 experimentally verified T-cell epitopes and identified seven immunodominant epitopes that had variants in global SARS-CoV-2 sequence data. They made polyclonal CD4+ and CD8+ T-cell lines against each of these wild-type sequence epitopes and found that for six of the variants (five in nucleocapsid and one in ORF3a), there was complete loss of responsiveness by the T-cell lines. 89 Agerer et al. 90 deep-sequenced 747 virus isolates and identified mutations in CD8+ T-cell epitopes that they then tested against CD8+ T cells isolated from HLA-matched COVID-19 patients and observed reduced IFN-γ production and cytotoxic activity. Zhang et al. 91 analyzed 13,432 SARS-CoV-2 strains harboring 4420 mutations and found mutations in cross-reactive T-cell epitopes between SARS-CoV-2 and seasonal human coronavirus. A method called T-Scan was used to identify CD8+ T-cell epitopes from convalescent patients with four major HLA alleles (HLA-A*02:01; 02:07; 11:01; and 24:02). Four epitopes that were mutated in VOCs, including two from Delta, elicited a decreased activation of T cells from at least two patients (3–6 patients per HLA allele were tested); and T cells from nearly all vaccinees tested responded with a decreased activation to the mutated forms of these four epitopes. 92 A more recent study reported that the overall CD4+ T-cell response in peripheral blood mononuclear cells (PBMCs) to B.1.1.529 (Omicron) in convalescent and vaccinated individuals, dropped by 16% and 9%, relative to the response to wild-type virus, respectively; and that of CD8+ T cells dropped by 30% and 8%, respectively. 93 We also reiterate the provocative recent study in which 20% of individuals studied (previously infected and/or vaccinated) had a >50% reduction in T-cell reactivity to Omicron. 79 In light of the T-cell reactivity to VOCs, a review by Ferrari et al., while more confident that overall T-cell protection should be relatively impervious to overall mutational escape, nonetheless counseled that, to maximize the effectiveness conveyed by the T-cell arm, one should target conserved and immunodominant epitopes that cannot be mutated due to functional and/or structural constraints. However, assessment of this so-called ‘invariance’ was theoretical, based on structural considerations of spike, as there was no discussion of mutations made or tested. 94
It is also our contention that more T-cell escape mutations will likely arise in nature as the pandemic continues on. The antibody response that vaccinees mount against natural infection effectively ‘masks’ the T-cell response, because it can neutralize virus before T cells see it; take away that early neutralization, as many variants seem to be doing, and more and more T-cell escape mutants should arise over time. Also, it may take longer for T-cell escape mutants to arise because of the diversity of HLA haplotypes; but taking longer does not mean never happening. Despite the seeming individuality of peptide-MHC restrictions, many peptides can bind multiple HLA supertypes. In addition, certain HLAs are widely prevalent among certain populations, such as HLA-A2 among Caucasians, 95 and HLA-A26 among Japanese. 96 SARS-CoV-2 also mutates less rapidly, on a per nucleotide per replication cycle level, than influenza virus and HIV, 8 because unlike the latter two viruses, its RNA polymerase has proofreading capability. 6 It may require a long incubation time, for example, in an immunocompromised person, for T-cell escape mutants to arise. Nonetheless, given the sheer bulk of both the viral load in individuals and the global disease burden, these mutants are likely to eventually emerge, just as a variant like Omicron, with its 30 mutations in spike and 20 mutations elsewhere, 97 demonstrates. It would be surprising to us if eventually variants with a similar number of new mutations do not arise that escape much of the overall T-cell response – even if not fully, enough to cause a serious public health concern. Underscoring these predictions, it is well known that influenza virus, which has obviously been around for much longer than SARS-CoV-2, has strongly selected for numerous T-cell epitope escape mutants.98,99 Even if the diversity of the T-cell response allows the host to still target other epitopes than the one(s) the virus mutates, a partial escape could potentially ‘tip the balance’, to the point where the virus can then transmit to another host, which ultimately is the selectable trait of relevance. In the context of immunodominance, this becomes even more plausible: for example, for influenza virus, the M1 58-66 HLA-A2-restricted epitope can make up >75% of the T-cell response in these individuals; 100 and the NP366 epitope in C57Bl6 mice is so dominant that the Cal09 strain, which has a single mutation in this epitope, entirely evades control by an NP vaccine. 101 All this being said, if indeed this hypothesis bears out, one might expect to see more evidence of T-cell escape by SARS-CoV-2 prior to the issuance of the vaccines, when the T-cell response was engaged without a robust masking from a vaccine-induced antibody response (see above on the kinetics of the antibody response versus that of T cells in natural SARS-CoV-2 infection); however, to the author’s knowledge evidence on this topic is sparse. Also, vaccination does not always prevent transmission, which necessitates infected cells and a (putative) engaged T-cell response, so that pressure for T-cell escape could have been substantially present before ample escape from the antibody response induced by the vaccine. Thus, it must be acknowledged that theoretical considerations for T-cell and antibody escape from infection and vaccination remain a complicated issue.
The most recent meta-analysis of papers reporting SARS-CoV-2 epitopes was published in July 2021, which collated 382 distinct CD4+ and 1052 CD8+ T-cell epitopes from 25 studies. 102 The majority of these were from a comprehensive analysis with the sensitive AIM assay by Tarke et al. 103 Some more recent studies used less conventional approaches. A novel reverse epitope discovery platform started with large public single-cell gene expression and T-cell receptor (TCR) data sets and identified >1200 highly public anti-SARS-CoV-2 CD4+ T-cell clonotypes. 104 Another study utilized mass spectrometry of two SARS-CoV-2-infected cell lines to derive an HLA-I immunopeptidome; interestingly, several peptides from out-of-frame open reading frames (ORFs) were recovered and were shown to elicit a T-cell response in COVID-19 patients which exceeded those of in-frame ORFs. 105
One study compared the TCR repertoire of infected versus vaccinated individuals and found a largely identical repertoire, with similar magnitudes of response, memory phenotypes, and TCR motifs, providing mechanistic insight on the ability of mRNA vaccines to boost the recall T-cell response in those who had previously been infected with SARS-CoV-2. 106 More evidence of preexisting T cells cross-reactive to common cold coronaviruses in SARS-CoV-2-unexposed individuals has been uncovered by specific epitope analysis: 10 SARS-CoV-2 HLA-A*02:01 epitopes were able to induce activation of CD8+ T cells from unexposed donors; 107 and eight peptides from spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins were able to activate naïve T cells from within PBMCs isolated from unexposed donors. 108
Most vaccines in wide use, and all three approved for use in the United States, consist exclusively of spike. However, infection presents all viral antigens; and many epitopes from non-spike proteins have been identified in various studies. A survey of 34 acute and recovered COVID-19 patients against peptides spanning the full-length of E, M, and N found 10 peptides containing CD4+ T-cell epitopes that were positive in more than 36% of patients, and three in more than 55% of patients. 109 The distribution of epitope-reactivity may be important as well. Fazolo et al. 110 reported that, in a set of South Brazil pediatric COVID-19 patients, CD8+ T-cell responses to S were weaker, while those to M and N were stronger, than those in COVID-19 adults.
T-cell immunity of special patient groups
Since the vast majority of COVID-19 deaths are among the elderly, it is of substantial interest to understand how the T cells of this age group respond to the vaccine. After vaccination with BNT162b2, individuals over 80 years of age had lower IFN-γ and IL-2 production by spike-specific CD4+ T cells than younger people. 111 Those in long-term care facilities also responded with a lower frequency of viral-specific CD8+ T cells. 112 Underlying these phenotypes was a more restricted TCR repertoire diversity, increased self-reactive T cells, and more active Treg cells. 113
About 2.7% of Americans are immunosuppressed for various reasons, 114 so it is an important question how the vaccine elicits immunity in this patient group. Five of 21 patients with inborn errors of immunity, following vaccination with BNT162b2, did not develop any specific CD4+ CD40 L+ T cells 115 (CD40 L is primarily expressed on activated T cells). Collier et al. 116 found that CD4+ and CD8+ T-cell IFN-γ responses, following mRNA vaccination, were reduced in patients receiving immunosuppressive therapy. In contrast, a study with a small (n = 5) cohort of autoimmune patients receiving immunosuppressive therapy observed high IFN-γ levels in all five patients following BNT162b2 vaccination. 117 For those people living with HIV, inactivated vaccines induced lower levels of neutralizing antibodies, possibly due to lower CD4 + T-cell counts; 118 however, another study found that HIV patients infected with SARS-CoV-2 mounted a T-cell response comparable to that of HIV-negative patients which persisted for 5–7 months, with CD4+ T cells greater in number than CD8+ T cells, and the response largely against S, M, and N. 119
People in whom B cells are depleted can still activate the cellular immune response. All multiple sclerosis patients receiving anti-CD20 therapy tested mounted strong, antigen-specific CD4+ and CD8+ T-cell responses to mRNA vaccination; however, there was a relative deficit in Tfh cells, while CD8+ T cells were stronger. 120 The induction of higher spike-specific CD8+ T cells in anti-CD20-treated patients after mRNA vaccination was corroborated by Madelon et al., 121 who also found an equivalent percentage of these patients with detectable spike-specific CD4+ T cells as compared with non-anti-CD20 patients.
Immunosuppressive therapy is also administered to transplant patients, and many studies have been conducted analyzing the response to vaccination or infection in various types of transplantation. The percentage of viral-specific CD4+ and CD8+ T cells in solid organ transplant recipients (SOTR) following BNT162b2 vaccination was lower than in healthy controls. 122 Infection with SARS-CoV-2 in SOTRs induced a greater T-cell response than those who were vaccinated. 123 Soberingly, 90% of cardiothoracic transplant recipients vaccinated with two doses of BNT162b2 in another study did not mount a detectable T-cell response. 124 In patients receiving allogeneic stem cell transplants, 82% of patients who received two doses of BNT162b2 mounted a T-cell response, 71% a CD4+ T-cell response, and 52% a CD8+ T-cell response. 125
Cancer patients unexposed to SARS-CoV-2 had lower prevalences of preexisting SARS-CoV-2 cross-reactive CD4+ T cells and those with COVID-19 mounted a profoundly reduced intensity, expandability, and diversity of SARS-CoV-2 T-cell responses. 126 Scurr et al. 127 found that while IFN-γ and/or IL-2 virus-specific T-cell responses were induced in 91% of healthy donors after one dose of vaccine, they were only induced in 48% of solid organ cancer patients. As for hematologic cancers, while 77% of these patients who were hospitalized for COVID-19 had detectable SARS-CoV-2-specific T-cell responses, this group had higher mortality than patients with solid cancer; those in whom a greater number of CD8+ T cells were induced had improved survival. 128 T-cell responses to SARS-CoV-2 were detected in 41–88% of convalescents who did not have detectable anti-S antibodies, but this number dropped to 35% for CD4+ T cells in multiple myeloma convalescents without anti-S antibodies and just 28% for CD8+ T cells. 129
Alternative vaccine approaches
Besides the mRNA and Ad26 or ChAdOx1 vaccines, numerous other vaccine modalities have been or are being attempted either preclinically or in clinical trials. A biomaterial vaccine based on a mesoporous silica rods platform was able to induce robust and durable cellular immunity in mice and could be lyophilized. 130 A protein vaccine comprised by the spike RBD fused to an alpaca-derived nanobody that binds to MHC Class II elicited robust cellular immunity in mice. 131 A peptide vaccine consisting of T-cell epitopes from various viral proteins, given with the Toll-like receptor 1/2 agonist XS15 adjuvant emulsified in Montanide ISA51VG, induced multifuctional Th1 CD4+ and CD8+ T cells in a Phase 1 trial, which exceeded those from natural infection and from the approved vaccines. 132 Administration of a codon-optimized DNA vaccine with S produced high levels of a Th1 response as well as memory CD8+ and CD4+ T cells in mice that lasted for at least 6 months. 133 A spike DNA plus protein vaccine was electroporated into rhesus macaques and induced a robust T-cell response. 134 A spike-ferritin nanoparticle vaccine with Alhydrogel and Army Liposome Formulation adjuvants given to mice produced a polyfunctional memory CD4+ T cell as well as a long-lived memory CD8+ T-cell response. 135 Intradermal delivery of N with a dissolvable microneedle skin patch induced a significant T-cell response in mice. 136 Peritoneal delivery of spike encapsulated in N,N,N-trimethyl chitosan particles to mice induced strong T-cell responses. 137 A synthetic peptide vaccine with Freund’s adjuvant consisting of spike 446-480 protected mice from a lethal dose of SARS-CoV-2. 138 Spike protein given along with CpG adjuvant, together encapsulated in an artificial cell membrane polymersome, which produces a self-assembling nanoscale vesicle, induced functional memory CD4+ and CD8+ T cells in mice. 139 A polymeric glyco-adjuvant with spike activated Tfh cells and polyfunctional CD4+ and CD8+ T cells in mice. 140 An mRNA vaccine fused to ferritin-formed nanoparticles elicited a Th1-biased cellular response that protected mice challenged with SARS-CoV-2. 141 A 20-peptide vaccine from S and N with an RNA adjuvant induced effector memory T cells in hamsters and mice. 142
For viral vectors, delivery of a Modified vaccinia virus Ankara (MVA) with spike in a prime-boost (intramuscular-intranasal) regimen induced a massive expansion of tissue-resident CD8+ T cells that protected hamsters from SARS-CoV-2 lung infection and pathology. 143 Another MVA vaccine with S given in a prime-boost regimen elicited CD8+ T cells and protected mice from SARS-CoV-2 challenge. 144 An Ad5 vaccine with spike RBD induced strong CD4+ and CD8+ T-cell responses in mice that lasted for over 6 months, with just a single dose. 145 Another Ad5-vectored vaccine expressing both S and N with an Enhanced T-cell Stimulation Domain to target MHC Class II elicited a Th1-biased response to S and N and was protective in rhesus macaques. 146 Still another Ad5 vaccine, expressing N, protected hamsters and mice from weight loss and reduced viral load in hamsters and mice and induced a rapid T-cell recall response in the respiratory mucosa. 147
An inactivated SARS-CoV-2 vaccine elicited IFN-γ-secreting T cells that were equivalent to those in approved vaccine recipients. 148 Another inactivated vaccine in a small cohort of individuals produced IFN-γ-secreting T cells against S, N, and E in a PBMC enzyme-linked immune absorbent spot (ELISpot) assay. 149 A fusion protein between spike RBD and rotavirus VP6 induced a high quantity of T cells in mice, with no antibodies detected. 150
Conclusion
Antibodies against SARS-CoV-2, as with many other respiratory viruses, are afforded an outsized emphasis, both by the lay public and by virologists, as exemplified by, for example, the near-immediate analysis of the humoral response when a new variant arises. This is not just due to the relative ease with which neutralization assays can be implemented, but also to the dogma that antibodies stand first in line to prevent infection altogether. However, as mentioned above, T cells arise much earlier after natural SARS-CoV-2 infection than do antibodies; and high T-cell activity is associated with controlled viral infection, whereas the opposite has proven to be the case for antibodies. Indeed, that the current vaccines are seemingly unable to neutralize Omicron (the neutralizing antibody titer is down 22-fold after two doses), 151 but can still greatly reduce severe disease, and often even symptomatic infection, is likely attributable in large part to the cellular immune response. Also of note is the few patients with agammaglobulinemia who have been studied who fully control SARS-CoV-2 infection. Given these observations, it will be prudent to research vaccine modalities that can specifically elicit a T-cell response, which will be especially important for those groups who tend to mount weaker T-cell responses, like the elderly and otherwise immunocompromised. In particular, vaccines that present epitopes that are functionally invariant (as determined by mutational analysis, not by sequence analysis or structural inferences) to prevent mutational escape could figure as important players in our ongoing battle with this amazingly devious pathogen.
Acknowledgments
None
Footnotes
ORCID iD: Arthur Young  https://orcid.org/0000-0001-8690-6361
https://orcid.org/0000-0001-8690-6361
Declarations
Ethics approval and consent to participate: Not applicable
Consent for publication: Not applicable
Author contributions: Arthur Young: Conceptualization; Writing – original draft; Writing – review & editing.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Competing interests: The author declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: AY is President of InvVax.
Availability of data and materials: Not applicable
References
- 1. COVID worldometer, https://www.worldometers.info/coronavirus/ (accessed 8 June 2022).
- 2. Centers for Disease Control and Prevention. Symptoms of COVID-19, https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed 28 January 2022).
- 3. Allen Media Broadcasting. Mayo Clinic, https://www.wxow.com/coronavirus/mayo-says-10-30-of-their-patients-deal-with-long-haul-covid-19-symptoms/article_f0f19a62-27a6-11ec-a811-6326ec151e09.html (2022, accessed 28 January 2022).
- 4. Monroe I, Dale M, Schwabe M, et al. The COVID-19 patient in the surgical intensive care unit. Surg Clin North Am 2022; 102: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta D, Sharma P, Singh M, et al. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell Mol Life Sci 2021; 78: 7967–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romano M, Ruggeiro A, Squeglia F, et al. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020; 9: 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung PP, Rogozin IB, Choy KT, et al. Comparative mutational analyses of influenza A viruses. RNA 2015; 21: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fischer W, Giogi EE, Chakraborty S, et al. HIV-1 and SARS-CoV-2: patterns in the evolution of two pandemic pathogens. Cell Host Microbe 2021; 29: 1093–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aleem A, Samad ABA, Slenker AK. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). 1st ed. Treasure Island, FL: StatPearls Publishing, 2022. [PubMed] [Google Scholar]
- 10. Thye AYK, Law JWF, Pusparajah P, et al. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines 2021; 9: 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. COVID data tracker weekly review, https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html (accessed 8 June 2022).
- 12. O’Neill LAJ, Bowie AG. Sensing and signaling in antiviral innate immunity. Curr Biol 2010; 20: R328–R333. [DOI] [PubMed] [Google Scholar]
- 13. Pishesha N, Harmand TJ, Ploegh HL. A guide to antigen processing and presentation. Nat Rev Immunol. 13 April 2022. DOI: 10.1038/s41577-022-00707-2. [DOI] [PubMed] [Google Scholar]
- 14. Hay ZLZ, Slansky JE. Granzymes: the molecular executors of immune-mediated cytotoxicity. Int J Mol Sci 2022; 23: 1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy KM, Weaver C. Janeway’s immunobiology. 9th ed. New York: Norton & Company, 2016. [Google Scholar]
- 16. Imai K, Kitagawa Y, Tabata S, et al. Antibody response patterns in COVID-19 patients with different levels of disease severity in Japan. J Med Virol 2021; 93: 3211–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lafon E, Diem G, Witting C, et al. Potent SARS-CoV-2-specific T cell immunity and low anaphylatoxin levels correlate with mild disease progression in COVID-19 patients. Front Immunol 2021; 12: 684014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan AT, Linster M, Tan CW, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 2021; 34: 108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao L, Zhou J, Yang S, et al. The dichotomous and incomplete adaptive immunity in COVID-19 patients with different disease severity. Signal Transduct Target Ther 2021; 6: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Li R, Song G, et al. Impairment of T cells’ antiviral and anti-inflammation immunities may be critical to death from COVID-19. R Soc Open Sci 2021; 8: 211606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zenarruzabeitia O, Astarloa-Pando G, Terrén I, et al. T cell activation, highly armed cytotoxic cells and a shift in monocytes CD300 receptors expression is characteristic of patients with severe COVID-19. Front Immunol 2021; 12: 655934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neidleman J, Luo X, George AF, et al. Distinctive features of SARS-CoV-2-specific T cells predict recovery from severe COVID-19. Cell Rep 2021; 36: 109414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miguez-Rey E, Choi D, Kim S, et al. Monoclonal antibody therapies in the management of SARS-CoV-2 infection. Expert Opin Investig Drugs 2022; 31: 41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schulien I, Kemming J, Oberhardt V, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med 2021; 27: 78–85. [DOI] [PubMed] [Google Scholar]
- 25. Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26: 845–848. [DOI] [PubMed] [Google Scholar]
- 26. Cohen KW, Linderman SL, Moodie Z, et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med 2021; 2: 100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection. Science 2021; 371: eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Liu X, Zhang X, et al. Decline in neutralising antibody responses, but sustained T-cell immunity, in COVID-19 patients at 7 months post-infection. Clin Transl Immunology 2021; 10: e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J, Lin H, Ye B, et al. One-year sustained cellular and humoral immunities of COVID-19 convalescents. Clin Infect Dis 2021; 1: ciab884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jung JH, Rha M-S, Sa M, et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat Commun 2021; 12: 4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma T, Ryu H, McGregor M, et al. Protracted yet coordinated differentiation of long-lived SARS-CoV-2- specific CD8+ T cells during convalescence. J Immunol 2021; 207: 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181: 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tincati C, Cannizzo ES, Giacomelli M, et al. Heightened circulating interferon-inducible chemokines, and activated pro-cytolytic Th1-cell phenotype features Covid-19 aggravation in the second week of illness. Front Immunol 2020; 11: 580987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui D, Tang Y, Jiang Q, et al. Follicular helper T cells in the immunopathogenesis of SARS-CoV-2 infection. Front Immunol 2021; 12: 731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baumjohann D, Fazilleau N. Antigen-dependent multistep differentiation of T follicular helper cells and its role in SARS-CoV-2 infection and vaccination. Eur J Immunol 2021; 51: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Golovkin A, Kalinina O, Bezrukikh V, et al. Imbalanced immune response of T-cell and B-cell subsets in patients with moderate and severe COVID-19. Viruses 2021; 13: 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cavazzoni CB, Hanson BL, Podestà MA, et al. Follicular T cells optimize the germinal center response to SARS-CoV-2 protein vaccination in mice. Cell Rep 2022; 38: 110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen JS, Chow RD, Song E, et al. High-affinity, neutralizing antibodies to SARS-CoV-2 can be made without T follicular helper cells. Sci Immunol 2022; 7: eabl5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol 2010; 84: 9318–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021; 590: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soresina A, Moratto D, Chiarini M, et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol 2020; 31: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gaitzsch E, Passerini V, Khatamzas E, et al. COVID-19 in patients receiving CD20-depleting immunochemotherapy for B-cell lymphoma. Hemasphere 2021; 5: e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Townsend JP, Hassler HB, Wang Z, et al. The durability of immunity against reinfection by SARS-CoV-2: a comparative evolutionary study. Lancet Microbe 2021; 2: e666–e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mishra KP, Singh M, Saraswat D, et al. Dysfunctional state of T cells or exhaustion during chronic viral infections and COVID-19: a review. Viral Immunol 2022; 35: 284–290. [DOI] [PubMed] [Google Scholar]
- 45. Kusnadi A, Ramírez-Suástegui C, Fajardo V, et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8+ T cells. Sci Immunol 2021; 6: eabe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khanolkar A. Elucidating T cell and B cell responses to SARS-CoV-2 in humans: gaining insights into protective immunity and immunopathology. Cells 2021; 11: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020; 17: 541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang I, Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J Intensive Care 2020; 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng K, Zheng X, Yang W. The role of metabolic dysfunction in T-cell exhaustion during chronic viral infection. Front Immunol 2022; 13: 843242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kundu R, Narean JS, Wang L, et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun 2022; 13: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sagar M, Reifler K, Rossi M, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest 2021; 131: e143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ansari A, Arya R, Sachan S, et al. Immune memory in mild COVID-19 patients and unexposed donors from India reveals persistent T cell responses after SARS-CoV-2 infection. Front Immunol 2021; 12: 636768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Richards KA, Glover M, Crawford JC, et al. Circulating CD4 T cells elicited by endemic coronaviruses display vast disparities in abundance and functional potential linked to antigen specificity and age. J Infect Dis 2021; 223: 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Swadling L, Diniz MO, Schmidt NM, et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022; 601: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Naranbhai V, Garcia-Beltran WF, Chang CC, et al. Comparative immunogenicity and effectiveness of mRNA-1273, BNT162b2 and Ad26.COV2.S COVID-19 vaccines. J Infect Dis 2021; 1: jiab593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Collier AY, Yu J, McMachan K, et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N Engl J Med 2021; 385: 2010–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ivanova EN, Devlin JC, Buus TB, et al. Discrete immune response signature to SARS-CoV-2 mRNA vaccination versus infection. medRxiv 2021, https://www.medrxiv.org/content/10.1101/2021.04.20.21255677v1
- 58. Sahin U, Muik A, Vogler I, et al. BNT162b2 vaccine induces neutralising antibodies and poly-specific T cells in humans. Nature 2021; 595: 572–577. [DOI] [PubMed] [Google Scholar]
- 59. Swanson PAII, Padilla M, Hoyland W, et al. T-cell mediated immunity after AZD1222 vaccination: a polyfunctional spike-specific Th1 response with a diverse TCR repertoire. Sci Transl Med 2021; 13: eabj7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Painter MM, Mathew D, Goel RR, et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 2021; 54: 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oberhardt V, Luxenburger J, Kemming J, et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature 2021; 597: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gil-Manso S, Carbonell D, López-Fernández L, et al. Induction of high levels of specific humoral and cellular responses to SARS-CoV-2 after the administration of Covid-19 mRNA vaccines requires several days. Front Immunol 2021; 12: 726960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guerrera G, Picozza MD’, Orso S, et al. BNT162b2 vaccination induces durable SARS-CoV-2 specific T cells with a stem cell memory phenotype. Sci Immunol 2021; 6: eabl5344. [DOI] [PubMed] [Google Scholar]
- 64. Mateus J, Dan JM, Zhang Z, et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 2021; 374: eabj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barouch DH, Stephenson KE, Sadoff J, et al. Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N Engl J Med 2021; 385: 951–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mudd PA, Minervina AA, Pogorelyy MV, et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T cell follicular helper cell response in humans. Cell 2022; 185: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Visvabharathy L, Hanson B, Orban Z, et al. Neuro-COVID long-haulers exhibit broad dysfunction in T cell memory generation and responses to vaccination. medRxiv 2021, https://www.medrxiv.org/content/10.1101/2021.08.08.21261763v3
- 68. Vizcarra P, Haemmerle J, Velasco H, et al. BNT 162b2 mRNA COVID-19 vaccine reactogenicity: the key role of immunity. Vaccine 2021; 39: 7367–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Payne RP, Longet S, Austin JA, et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell 2021; 184: 5699–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zollner A, Watschinger C, Rssler A, et al. B and T cell response to SARS-CoV-2 vaccination in health care professional with and without previous COVID-19. eBioMedicine 2021; 70: 103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Casado JI, Haemmerle J, Vizcarra P, et al. T-cell response after first dose of BNT162b2 SARS-CoV-2 vaccine among healthcare workers with previous infection or cross-reactive immunity. Clin Transl Immunology 2021; 10: e1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Barros-Martins J, Hammerschmidt SI, Cossmann A, et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat Med 2021; 27: 1525–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schmidt T, Klemis V, Schub D, et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat Med 2021; 27: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Geers D, Shamier MC, Bogers S, et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol 2021; 6: eabj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Skelly DT, Harding AC, Gilbert-Jaramillo J, et al. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat Commun 2021; 12: 5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Neidleman J, Luo X, McGregor M, et al. mRNA vaccine-induced T cells respond identically to SARS-CoV-2 variants of concern but differ in longevity and homing properties depending on prior infection status. eLife 2021; 10: e72619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Woldemeskel BA, Garliss CC, Blankson JN. mRNA vaccine-elicited severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific T cells persist at 6 months and recognize the Delta variant. Clin Infect Dis 2021; 1: ciab915. [DOI] [PubMed] [Google Scholar]
- 78. Jordan SC, Shin B-H, Gadsden T-AM, et al. T cell immune responses to SAS-CoV-2 and variants of concern (Alpha and Delta) in infected and vaccinated individuals. Cell Mol Immunol 2021; 18: 2554–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Keeton R, Richardson SI, Moyo-Gwete T, et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe 2021; 29: 1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tarke A, Coelho CH, Zhang Z, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022; 185: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Naranbhai V, Nathan N, Kaseke C, et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell 2022; 185: 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Choi SJ, Kim DU, Noh JY, et al. T cell epitopes in SARS-CoV-2 proteins are substantially conserved in the Omicron variant. Cell Mol Immunol 2022; 19: 447–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu NC, Young AP, Al-Mawsawi LQ, et al. High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci Rep 2014; 4: 4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang M, Wang C, Otto TD, et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 2018; 360: eaap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tchilian E, Holzer B. Harnessing local immunity for an effective universal swine influenza vaccine. Viruses 2017; 9: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yewdell J, Lopez-Munoz A, Kosik I, et al. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Res Sq. Epub ahead of print 13 December 2021. DOI: 10.21203/rs.3.rs-1162804/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021; 184: 861–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tarke A, Sidney J, Methot N, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021; 2: 100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. de Silva TI, Liu G, Lindsey BB, et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. Iscience 2021; 24: 103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Agerer B, Koblischke M, Gudipati V, et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci Immunol 2021; 6: eabg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang C, Jin X, Chen X, et al. Antigenic evolution on a global scale reveals the potential natural selection of severe acute respiratory syndrome-coronavirus 2 by pre-existing cross-reactive T-cell immunity. Front Microbiol 2021; 12: 599562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang H, Deng S, Ren L, et al. Profiling CD8+ T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants. Cell Rep 2021; 36: 109708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gao Y, Cai C, Grifoni A, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med 2022; 28: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Boni C, Cavazzini D, Bolchi A, et al. Degenerate CD8 epitopes mapping to structurally constrained regions of the spike protein: a T cell-based way-out from the SARS-CoV-2 variants storm. Front Immunol 2021; 12: 730051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marescotti D, Destro F, Baldisserotto A, et al. Characterization of a human leucocyte antigen A2-restricted Epstein-Barr virus nuclear antigen-1-derived cytotoxic T-lymphocyte epitope. Immunology 2010; 129: 386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Niu Y, Terasaki Y, Komatsu N, et al. Identification of peptides applicable as vaccines for HLA-A26-positive cancer patients. Cancer Sci 2009; 100: 2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. American Society for Microbiology. How ominous is the Omicron variant? https://asm.org/Articles/2021/December/How-Ominous-is-the-Omicron-Variant-B-1-1-529 (accessed 8 June 2022).
- 98. Valkenburg SA, Quiñones-Parra S, Gras S, et al. Acute emergence and reversion of influenza A virus quasispecies within CD8+ T cell antigenic peptides. Nat Commun 2013; 4: 2663. [DOI] [PubMed] [Google Scholar]
- 99. Machkovech HM, Bedford T, Suchard MA, et al. Positive selection in CD8+ T cell epitopes of influenza virus nucleoprotein revealed by a comparative analysis of human and swine viral lineages. J Virol 2015; 89: 11275–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tan ACL, La Gruta NL, Zeng W, et al. Precursor frequency and competition dictate the HLA-A2-restricted CD8+ T cell responses to influenza A infection and vaccination in HLA-A2.1 transgenic mice. J Immunol 2011; 187: 1895–1902. [DOI] [PubMed] [Google Scholar]
- 101. Uddback IEM, Pedersen LMI, Pedersen SR, et al. Combined local and systemic immunization is essential for durable T-cell mediated heterosubtypic immunity against influenza A virus. Sci Rep 2016; 6: 20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Grifoni A, Sidney J, Vita R, et al. SARS-COV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 2021; 29: 1076–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tarke A, Sidney J, Kidd CK, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med 2021; 2: 100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rosati E, Pogorelyy MV, Minervina AA, et al. Characterization of SARS-CoV-2 public CD4+ ab T cell clonotypes through reverse epitope discovery. bioRxiv 2021, https://www.biorxiv.org/content/10.1101/2021.11.19.469229v1.full
- 105. Weingarten-Gabbay S, Klaeger S, Sarzikova S, et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell 2021; 184: 3962–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Minervina AA, Pogorelyy MV, Kirk AM. Convergent epitope-specific T cell responses after SARS-CoV-2 infection and vaccination. medRxiv 2021, https://www.medrxiv.org/content/10.1101/2021.07.12.21260227v3
- 107. Quiros-Fernandez I, Poorebrahim M, Fakhr E, et al. Immunogenic T cell epitopes of SARS-CoV-2 are recognized by circulating memory and naïve CD8 T cells of unexposed individuals. eBioMedicine 2021; 72: 103610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hu W, He M, Wang X, et al. Specific CD8+ T cell repertoire recognizing conserved antigens of SARS-CoV-2 in unexposed population: a prerequisite for broad-spectrum CD8+ T cell immunity. Vaccines 2021; 9: 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Heide J, Schulte S, Kohsar M, et al. Broadly directed SARS-CoV-2-specific CD4+ T cell response includes frequently detected peptide specificities within the membrane and nucleoprotein in patients with acute and resolved COVID-19. PLoS Pathog 2021; 17: e1009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Fazolo T, Lima K, Fontoura JC, et al. Pediatric COVID-19 patients in South Brazil show abundant viral mRNA and strong specific anti-viral responses. Nat Commun 2021; 12: 6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Collier DA, Ferreira IATM, Kotagiri P, et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature 2021; 596: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Demaret J, Corroyer-Simovic B, Alidjinou EK, et al. Impaired functional T-cell response to SARS-CoV-2 after two doses of BNT 162b2 mRNA vaccine in older people. Front Immunol 2021; 12: 778679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang W, Thomas R, Oh J, et al. Thymic aging may be associated with COVID-19 pathophysiology in the elderly. Cells 2021; 10: 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Harpaz R, Dahl RM, Dooling KL. Prevalence of immunosuppression among US adults, 2013. JAMA 2016; 316: 2547–2548. [DOI] [PubMed] [Google Scholar]
- 115. Amodio D, Ruggiero A, Sgrulletti M, et al. Humoral and cellular response following vaccination with the BNT162b2 mRNA COVID-19 vaccine in patients affected by primary immunodeficiencies. Front Immunol 2021; 12: 727850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Collier AY, Yu J, McMahan K, et al. Coronavirus disease 2019 messenger RNA vaccine immunogenicity in immunosuppressed individuals. J Infect Dis 2021; 1: jiab569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Malipiero G, Moratto A, Infantino M, et al. Assessment of humoral and cellular immunity induced by the BNT162b2 SARS-CoV-2 vaccine in healthcare workers, elderly people, and immunosuppressed patients with autoimmune disease. Immunol Res 2021; 69: 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lv Z, Li Q, Feng Z, et al. Inactivated SARS-CoV-2 vaccines elicit immunogenicity and T-cell responses in people living with HIV. Int Immunopharmacol 2022; 102: 108383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Alrubayyi A, Gea-Mallorquí E, Touizer E, et al. Characterization of humoral and SARS-CoV-2 specific T cell responses in people living with HIV. Nat Commun 2021; 12: 5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Apostolidis SA, Kakara M, Painter MM, et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat Med 2021; 27: 1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Madelon N, Lauper K, Breville G, et al. Robust T cell responses in anti-CD20 treated patients following COVID-19 vaccination: a prospective cohort study. Clin Infect Dis 2021; 1: ciab954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yanis A, Haddadin Z, Spieker AJ, et al. Humoral and cellular immune responses to the SARS-CoV-2 BNT162be vaccine among a cohort of solid organ transplant recipients and healthy controls. Transpl Infect Dis 2022; 24: e13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ferreira VH, Marinelli T, Ierullo M, et al. Severe acute respiratory syndrome coronavirus 2 infection induces greater T-cell responses compared to vaccination in solid organ transplant recipients. J Infect Dis 2021; 224: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schramm R, Costard-Jäckle A, Rivinius R, et al. Poor humoral and T-cell response to two-dose SARS-CoV-2 messenger RNA vaccine BNT162b2 in cardiothoracic transplant recipients. Clin Res Cardiol 2021; 110: 1142–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Harrington P, Doores KJ, Saha C, et al. Repeated vaccination against SARS-CoV-2 elicits robust polyfunctional T cell response in allogeneic stem cell transplantation recipients. Cancer Cell 2021; 39: 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bilich T, Roerden M, Maringer Y, et al. Preexisting and post-COVID-19 immune responses to SARS-CoV-2 in patients with cancer. Cancer Discov 2021; 11: 1982–1995. [DOI] [PubMed] [Google Scholar]
- 127. Scurr MJ, Zelek WM, Lippiatt G, et al. Whole blood-based measurement of SARS-CoV-2-specific T cells reveals asymptomatic infection and vaccine immunogenicity in healthy subjects and patients with solid-organ cancers. Immunology 2022; 165: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bange EM, Han NA, Wileyto P, et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med 2021; 27: 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Aleman A, Upadhyaya B, Tuballes K, et al. Variable cellular responses to SARS-CoV-2 in fully vaccinated patients with multiple myeloma. Cancer Cell 2021; 39: 1442–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Langellotto F, Dellacherie MO, Yeager C, et al. A modular biomaterial scaffold-based vaccine elicits durable adaptive immunity to subunit SARS-CoV-2 antigens. Adv Healthc Mater 2021; 10: e2101370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pishesha N, Harmand TJ, Rothlauf PW, et al. A class II MHC-targeted vaccine elicits immunity against SARS-CoV-2 and its variants. Proc Natl Acad Sci USA 2021; 118: e2116147118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Heitmann JS, Bilich T, Tandler C, et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022; 601: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Alamri SS, Alluhaybi KA, Alhabbab RY, et al. Synthetic SARS-CoV-2 spike-based DNA vaccine elicits robust and long-lasting Th1 humoral and cellular immunity in mice. Front Microbiol 2021; 12: 727455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rosati M, Agarwal M, Hu X, et al. Control of SARS-CoV-2 infection after Spike DNA or Spike DNA+Protein co-immunization in rhesus macaques. PLoS Pathog 2021; 17: e1009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Carmen JM, Shrivastava S, Lu Z, et al. SARS-CoV-2 ferritin nanoparticle vaccine induces robust innate immune activity driving polyfunctional spike-specific T cell responses. NPJ Vaccines 2021; 6: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kuwentrai C, Yu J, Zhang BZ, et al. Induction of humoral and cellular immunity by intradermal delivery of SARS-CoV-2 nucleocapsid protein using dissolvable microneedles. J Immunol Res 2021; 2021: 5531220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jearanaiwitayakul T, Apichirapokey S, Chawengkirttikul R, et al. Peritoneal administration of a subunit vaccine encapsulated in a nanodelivery system not only augments systemic responses against SARS-CoV-2 but also stimulates responses in the respiratory tract. Viruses 2021; 13: 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Aparicio B, Casares N, Egea J, et al. Preclinical evaluation of a synthetic peptide vaccine against SARS-CoV-2 inducing multiepitopic and cross-reactive humoral neutralizing and cellular CD4 and CD8 responses. Emerg Microbes Infect 2021; 10: 1931–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lam JH, Khan AK, Cornell TA, et al. Polymersomes as stable nanocarriers for a highly immunogenic and durable SARS-CoV-2 spike protein subunit vaccine. ACS Nano 2021; 15: 15754–15770. [DOI] [PubMed] [Google Scholar]
- 140. Gray LT, Raczy MM, Briquez PS, et al. Generation of potent cellular and humoral immunity against SARS-CoV-2 antigens via conjugation to a polymeric glyco-adjuvant. Biomaterials 2021; 278: 121159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sun W, He L, Zhang H, et al. The self-assembled nanoparticle-based trimeric RBD mRNA vaccine elicits robust and durable protective immunity against SARS-CoV-2 in mice. Signal Transduct Target Ther 2021; 6: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lee YS, Hong SH, Park HJ, et al. Peptides derived from S and N proteins of severe acute respiratory syndrome coronavirus 2 induce T cell responses: a proof of concept for T cell vaccines. Front Microbiol 2021; 12: 732450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bošnjak B, Odak I, Barros-Martins J, et al. Intranasal delivery of MVA vector vaccine induces effective pulmonary immunity against SARS-CoV-2 in rodents. Front Immunol 2021; 12: 772240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tscherne A, Schwarz JH, Rohde C, et al. Immunogenicity and efficacy of the COVID-19 candidate vector vaccine MVA-SARS-2-S in preclinical vaccination. Proc Natl Acad Sci USA 2021; 118: e2026207118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. King RG, Silva-Sanchez A, Peel JN, et al. Single-dose intranasal administration of AdCOVID elicits systemic and mucosal immunity against SARS-CoV-2 and fully protects mice from lethal challenge. Vaccines 2021; 9: 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Gabitzsch E, Safrit JT, Verma M, et al. Dual-antigen COVID-19 vaccine subcutaneous prime delivery with oral boosts protects NHP against SARS-CoV-2 challenge. Front Immunol 2021; 12: 729837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Matchett WE, Joag V, Stolley JM, et al. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J Immunol 2021; 207: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Melo-González F, Soto JA, González LA, et al. Recognition of variants of concern by antibodies and T cells induced by a SARS-CoV-2 inactivated vaccine. Front Immunol 2021; 12: 747830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Deng Y, Li Y, Yang R, et al. SARS-CoV-2- specific T cell immunity to structural proteins in inactivated COVID-19 vaccine recipients. Cell Mol Immunol 2021; 18: 2040–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Tamminen K, Heinimäki S, Gröhn S, et al. Fusion protein of rotavirus VP6 and SARS-CoV-2 receptor binding domain induces T cell responses. Vaccines 2021; 9: 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Muik A, Lui BG, Wallisch A-K, et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 2022; 375: eabn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]