Abstract
The pR and pRM promoters of bacteriophage lambda direct transcription in divergent directions from start sites separated by 83 phosphodiester bonds. We had previously shown that the presence of an RNA polymerase at pR interfered with open complex formation at pRM and that this effect was alleviated by the deletion of 10 bp between the two promoters. Here we present a detailed characterization of the dependence of the interference on the interpromoter distance. It was found that the reduced interference between the two promoters is unique to the 10-bp deletion. The relief of interference was demonstrated to be due to the facilitation of a step subsequent to RNA polymerase binding to the pRM promoter. A model to explain these observations is proposed. A search of known Escherichia coli promoters identified three pairs of divergent promoters with similar separations to those investigated here.
In the rightward control region of bacteriophage lambda, transcription is initiated in divergent directions from two promoters, pR and pRM, that have start sites separated by 83 phosphodiester bonds (pdb; we are using this designation to avoid ambiguity in the representation of the distance between start sites). These two promoters are among those responsible for implementing the decision as to whether viral development will proceed along the lytic or lysogenic pathways (27). The pR promoter has greater similarity to the promoter consensus sequence than the pRM promoter (27). As a consequence, open complex formation at pR is accomplished in seconds but under the same conditions requires tens of minutes at pRM (15, 27, 34). Therefore, for the wild-type control region, in vitro RNA polymerase (RNAP)-pRM interactions occur almost exclusively in the context of another RNAP already bound to pR. It has been previously shown that this pR-bound RNAP interferes with open complex formation at pRM (16, 17, 21, 34, 37). The effect is not exerted at the initial binding of RNAP to the promoter but rather at a subsequent step (16, 34) that is likely a conformational change in the RNAP (9). Eventually, open complexes do form at pRM and coexist with those at pR (16, 25). The converse of the situation described above has also been shown: when pR has been weakened due to base substitutions, its ability to form open complexes is affected by the presence of pRM on the same DNA fragment (11).
Only 13 pdb separate the start site-distal edges of the −35 regions of the pR and pRM promoters. Given such a short interpromoter distance, it was suggested that the pR-bound RNAP was slowing open complex formation at pRM because of steric hindrance. Consistent with this notion, deletion of 1 bp between the −35 regions was found to further reduce the rate of open complex formation at pRM (40). However, it has also been shown that when the distance between the −35 regions of the promoters is shortened by the deletion of 10 bp (one turn of the DNA helix), unexpectedly the inhibition of open complex formation at pRM is greatly diminished (21). In other phages where the interpromoter distance at pR and pRM is even shorter, such as 434 (66 pdb between start sites) and P22 (52 pdb), concurrent occupancy of the promoters is not observed (8, 41).
To further explore this phenomenon, a series of deletions between the −35 regions of pR and pRM was generated to examine the length dependence of inhibition at the pRM promoter by the presence of RNAP at the pR promoter. DNA constructs lacking 3, 5, 6, 7, 8, 9, 10, 11, and 12 bp between the −35 regions of the two promoters were made (Fig. 1). The distance between the −35 regions of the pRM and pR promoters was deleted, starting from the edge of the −35 region proximal to pRM. The constructs are designated as Dn, where n is the number of base pairs that have been deleted. The promoters were constructed from synthetic oligodeoxyribonucleotides and cloned into the pKK232-8 vector by using BamHI and HindIII restriction sites as described previously (21) and sequenced. The location of the strand-separated region at both promoters was checked by KMnO4 footprinting and found not to be affected by the deletions (data not shown).
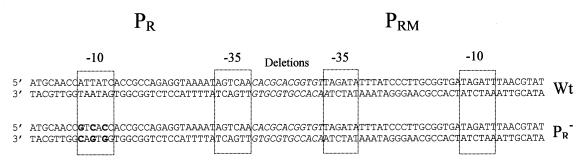
FIG. 1.
Constructs used in this study. The sequences shown were cloned into the pKK232-8 plasmid vector for E. coli at BamHI and HindIII restriction sites. The pR promoter was inactivated by introduction of three base pair substitutions in its −10 region, which are shown in boldface type. The −10 and −35 regions are boxed. The position of the region shortened in the deletion mutants is indicated in italic letters. The deletions start from the upstream edge of the −35 of the pRM promoter and progress towards the pR promoter. Constructs are designated as Dn, with n indicating the number of base pairs deleted. For example, the sequence between the −35 regions is CACGCACGG (top strand in the figure) for the D3 mutant.
Determination of open complex formation by the electrophoretic mobility shift assay.
Open complex formation at the pRM promoter was monitored with an electrophoretic mobility shift assay carried out as described by Mita et al. (21). Approximately 1 to 2 nM 32P-labeled promoter DNA was incubated at 37°C with RNAP (activity, 50% ± 10% [mean ± standard deviation]), at a concentration of active enzyme of 100 nM, in 20 μl of HEPES buffer (30 mM HEPES [pH 7.6], 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol) containing 50 μg of bovine serum albumin per ml. After the addition of 1 μl of a 1-mg/ml solution of heparin to inactivate free RNAP as well as closed complexes and incubation for an additional minute at 37°C, 2 μl of a loading solution (30% glycerol, 0.25% bromophenol blue, 0.25% xylenecyanolphenolfluorine) was added to each reaction mixture prior to loading onto a 4% polyacrylamide gel (29:1 acrylamide-bisacrylamide). The gels were run in TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA) at 6 V/cm for 1.5 h and then exposed to X-ray film to detect the radioactive bands. Two complex bands were observed. On the basis of actual footprinting of the complexes (21), we were able to determine that the faster-moving band represented DNA with an open complex at pR only and the slower one represented a complex of DNA and RNAP bound in open complexes at both pR and pRM (see also Results). Open complexes at both promoters are very stable (reference 28 and our unpublished results); thus, no significant dissociation or redistribution of RNAP is expected to occur during electrophoresis of the complexes.
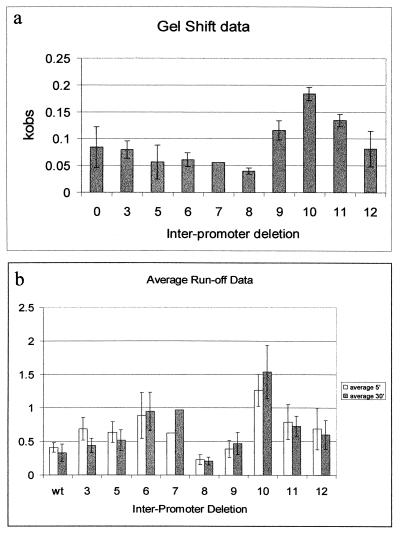
Full saturation of the pR promoter occurs before our first time point (taken at 2 min) and probably within seconds (21, 28). Next, the much slower process of open complex formation at pRM takes place. Our measurements follow the rate of conversion of DNA with one open complex (at pR) to that with two open complexes (at pR and pRM) and thus the rate of open complex formation at pRM. A comparison of the pseudo-first-order rate constants (kobs) for the binding of RNAP to the pRM promoter in the context of the different deletions is graphically shown in Fig. 2a, and the values for kobs for each promoter deletion mutant are given in Table 1. The D10 construct is seen to be unique in the rate with which pRM can form an open complex with RNAP, which was enhanced greater than twofold on this construct. The rate of open complex formation at pRM was slowest for the D8 construct.
FIG. 2.
Activity of the pRM promoter is maximal for a 10-bp deletion in the region separating pRM and pR in both electrophoretic mobility shift and runoff transcription assays. The x-axis represents the number of base pairs deleted between the −35 regions of pR and pRM. (a) Comparison of the average kobs for open complex formation at pRM for each of the promoter variants. The radioactivity in each band, as a percentage of the total in the lane, was plotted against the time of incubation with the RNAP, and the kobs for each DNA was determined by fitting the data to the equation y = Yf · {1 − exp[−(t) · kobs]} + Yo, where y = the percent of open complexes formed, t = time after RNAP mixing, and Yf and Yo are the limiting values for y. (b) Data from runoff transcription assays. The y-axis is the ratio of the band intensity for transcription derived for the pRM promoter compared to the total density of the lane. The empty bars represent the relative amounts of RNA transcribed after incubating the RNAP with promoter for 5 min, while the solid bars represent the relative amounts of RNA synthesized after a 30-min preincubation.
TABLE 1.
Average kobs of the promoter deletion mutants
| Interpromoter deletion no. | Avg kobsa (min−1) |
|---|---|
| 0 | 0.06 ± 0.04 |
| 3 | 0.08 ± 0.02 |
| 5 | 0.06 ± 0.03 |
| 6 | 0.06 ± 0.01 |
| 7 | 0.06b |
| 8 | 0.04 ± 0.01 |
| 9 | 0.12 ± 0.02 |
| 10 | 0.18 ± 0.01 |
| 11 | 0.08 ± 0.01 |
| 12 | 0.09 ± 0.03 |
The kobs for each DNA was determined by fitting the data as described in the legend to Fig. 2. Values are means ± standard deviations based on averaging the results of three independent determinations except where indicated.
Only one determination.
Run-off transcription assays.
The ability of RNAP to form open complexes at pRM for each of the constructs was also determined with a single-round runoff transcription assay. Approximately 5 nM promoter was incubated with 50 nM RNAP in HEPES buffer for either 5 or 30 min, followed by a 1-min incubation with heparin (50 μg/ml). To allow RNA synthesis, ATP, CTP, and GTP were added to 200 μM and UTP (including [32P]UTP) was added to 2 μM. After 10 min, UTP was added to 500 μM and the reaction mixture was incubated for an additional 5 min to ensure complete elongation of all transcripts. Finally, the products were separated on a denaturing gel. Bands apparent after exposure of the gel to Kodak Biomax film were scanned, and the intensities were normalized to the sum of the intensities of the pRM and pR bands.
The amount of runoff product made in this assay is a reflection of the number of open complexes formed during the incubation of RNAP and the promoter, prior to the addition of heparin. The results of these experiments are shown in Fig. 2b. For all constructs, the pRM promoter was found to be competent to initiate RNA synthesis (results not shown). Relative to the other deletion mutants, again a sharp increase is seen in the amount of RNA synthesized from the pRM promoter on the D10 template. In this assay, but not the gel mobility shift experiments, the D6 and D7 constructs also show elevated levels of RNA synthesis, albeit not quite as high as that for D10. We do not understand the underlying cause of this difference between the two assays for these two constructs.
Dependence of kobs on RNAP concentration for the wild type and D10 spacing between pR and pRM.
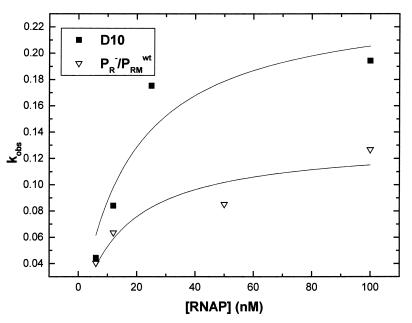
The results described above, as well as those from our previous studies (21, 37), indicate that utilization of the pRM promoter on the construct with the 10-bp deletion was significantly increased in comparison to that on constructs with the wild-type or other spacings between the pRM and pR promoters. To better understand the effect of the 10-bp deletion on open complex formation at pRM, we determined the dependence of kobs on RNAP concentration for two promoter mutants, D10 and pR−/pRM. The rates of open complex formation were determined for each concentration of RNAP as described above. The dependence of kobs on the concentration of RNAP is shown in Fig. 3; the data were fit as described in the figure legend to obtain the values of the association constant for RNAP binding to the promoter in a closed complex (KB) and the first-order rate constant for the conversion of the closed to the open promoter complex (kf). The values of KB (7 × 107 ± 3 × 107 M−1) and kf (0.13 ± 0.02 min−1) for the pR−/pRM construct determined here were similar to those previously reported (16). The fact that mainly kf is increased when pR is inactivated (12, 16) indicates that RNAP binding to pRM is not affected but is rather a subsequent step on the pathway to formation of an open complex. For pRM on D10, similar values for KB (6 × 107 ± 3 × 107 M−1) and kf (0.24 ± 0.04 min−1) are obtained, indicating that on this template the formation of an open complex at pRM takes place as if the pR promoter were not occupied. We routinely observe a slightly greater rate of open complex formation at pRM in the D10 than in the pR− context (reflected here by a twofold-greater kf) (see also references 21 and 37). However, since the effect is quite small, we have not attempted to characterize it further.
FIG. 3.
Dependence of kobs on RNAP concentration for the pR−/pRM and D10 constructs. The kobs were determined as a function of [RNAP], and the data were fit to the equation kobs = (KB · kf) [RNAP]/(KB[RNAP] + 1), where [RNAP] is the concentration of enzyme, KB is the association constant for RNAP binding to the promoter in a closed complex, and kf is the first-order rate constant for the conversion of the closed to the open promoter complex. The curves are the result of the fits. Symbols: ■, promoter with the 10-bp deletion; ▿, wild-type DNA that has had the pR promoter inactivated by the base changes indicated in Fig. 1. The DNA concentration was kept constant (approximately 1 to 2 nM) for all concentrations of RNAP.
Involvement of the α-CTD in the interference of RNAP at pR with open complex formation at pRM.
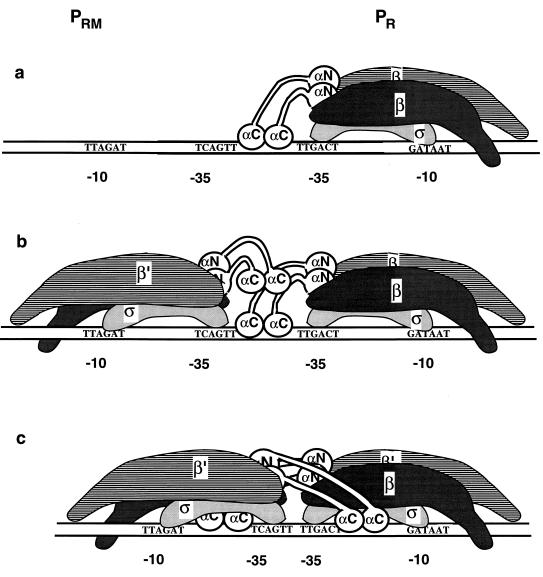
In Fig. 4 we present a model, refined from Tang et al. (37), that takes our results into account and also draws upon recent insights into the interaction of the α subunit of RNAP with upstream DNA sequences. As first shown for the rrnB P1 promoter, the alpha C-terminal domain (α-CTD) binds sequence specifically to an A+T-rich region located between −40 and −60 (the UP element), thereby greatly activating RNA synthesis in vivo and the rate of open complex formation in vitro (30). However, at other promoters, there is also evidence for interactions of the α-CTD with other DNA sequences in upstream regions at similar locations, both in the presence and absence of activator protein (7, 10, 13, 29, 37). The extent of activation that can result from such interactions has not been systematically studied. Based on results with RNAP deleted for the α-CTD, we estimate that at the pR and pRM promoters, the interactions with upstream DNA stimulate open complex formation two- to threefold (37). Without the ability of interaction with upstream sequences, RNAP always exhibited a low level of activity at pRM, even when the pR promoter was inactivated or the template used bore the D10 template (37). These results provide a strong indication that on the D10 template the RNAP at pRM was able to engage in upstream interactions even in the presence of another RNAP at pR.
FIG. 4.
RNAP at pR interferes with open complex formation at pRM for the wild-type (a and b) but not the D10 (c) interpromoter distance. The α subunits of RNAP are shown in white, with the N-terminal domains anchored to the β and β′ subunits of the RNAP (gray and striped regions) and the CTDs and the flexible linkers jutting away from RNAP. The pRM promoter is on the left, and pR is on the right. The sequences of the −10 and −35 regions are indicated for the nontemplate strand of each promoter. The spacer DNAs between the −10 and the −35 regions are shown as devoid of contacts with RNAP. (a) The −35 regions of pR and pRM are separated by 13 pdb. Within seconds of the addition of RNAP, an open complex forms at the pR promoter. Proposed upstream contacts of the α-CTDs of the RNAP are shown. (b) Subsequent interaction of RNAP with pRM in the presence of an RNAP at pR. The RNAP at pR obstructs upstream access by the α-CTDs of the RNAP at pRM. (c) The −35 regions of pR and pRM are separated by 2 bp. This closer-in arrangement allows the spacer DNAs of pR and pRM to be contacted by the α-CTDs of the RNAP at the other promoter, facilitating open complex formation at pRM despite the presence of an RNAP at pR.
In the model presented in Fig. 4, on the template with the wild-type spacing between the two promoters, the interference of pR-bound RNAP with open complex formation at pRM is exerted via obstruction of interactions between the α-CTD and DNA in the −40 to −60 region of pRM. This obstruction would be relieved for the D10 construct. Here the 10-bp deletion between the −35 regions of pRM and pR makes the spacer DNA between the −10 and −35 regions of the pR promoter coincident with bp −44 to −60 with respect to pRM. We envisage that the α-CTD of the RNAP at pRM would be able to reach over the RNAP at pR and contact this region. Few if any contacts have been demonstrated between promoter-bound RNAP and the spacer DNA (1, 35), so that the α-CTD of the RNAP at pRM may well be able to interact with the spacer DNA of pR, even when both promoters are occupied. The steep dependence of promoter activity and the rate of open complex formation on the interpromoter distance may reflect several factors. For shorter deletions (longer interpromoter distances), less of the spacer DNA but more of the −10 region of pR is at −40 to −60 with respect to pRM, leading to obstruction akin to that mentioned above for the wild-type spacer. Conversely, the longer deletions D11 and D12 (with shorter interpromoter distances) would keep the entire spacer DNA within the −40 to −60 region, but steric clashes between the two RNAPs would then become prohibitively severe.
We show that the putative upstream interactions lead to an increase in kf, which is in agreement not only with the mode of pRM activation obtained when the pR promoter is inactivated (12, 16) but also with that observed when pRM is provided with a genuine UP element (36, 37). Thus, the model is consistent with the available experimental evidence indicating that upstream interactions of the α-CTD facilitate a step subsequent to the initial binding of RNAP to the promoter.
Divergent promoters of E. coli.
Divergent promoters are fairly common in E. coli as well. In a 1988 review (3), many instances of divergently transcribed promoters in a back-to-back orientation (i.e., directing the synthesis of nonoverlapping transcripts) similar to that of pR and pRM of phage lambda were recognized. For our current analysis, we focused on promoter pairs that had start sites separated by 120 pdb or fewer. We chose this distance as an upper limit based on the observed 60-bp upstream extension in DNA interactions at promoters containing upstream elements (30). Thus, it is likely that start site separations beyond this distance will allow unimpeded interactions of RNAP at either promoter. Our search of the database RegulonDB (31) for known E. coli promoters satisfying the above criteria identified 13 promoter pairs, five of which were also represented in the earlier compilation (3) (Table 2). Three cases for which the separation between the start sites is in the range of 71 to 83 pdb investigated here were identified. Interestingly, all three have a separation of 78 pdb, similar to that for the D5 deletion (this work and reference 21), where the interference was found to be rather pronounced. The regulatory significance of a separation by this distance has yet to be investigated.
TABLE 2.
Back-to-back divergent promoters in E. coli
| Promoter paira | Transcriptionb | Positionc | Separation (pdb)d | Reference |
|---|---|---|---|---|
| fepA | Reverse | 611892 | 16 | 26 |
| fes | Forward | 611908 | ||
| bioA | Reverse | 808515 | 10 | 24 |
| bioB | Forward | 808525 | ||
| fumA | Reverse | 1686464 | 111 | 20 |
| manA | Forward | 1686575 | ||
| pdx | Reverse | 2435904 | 28 | 33 |
| div | Forward | 2435932 | ||
| udf Px | Reverse | 3208209 | 78 | 6 |
| rpsUp1 | Forward | 3208287 | ||
| dnaAp1 | Reverse | 3881590 | 104 | 23 |
| rpmHp3 | Forward | 3881694 | ||
| asnC | Reverse | 3924656 | 101 | 19 |
| asnA | Forward | 3924757 | ||
| ilvY | Reverse | 3955488 | 45 | 39 |
| ilvC | Forward | 3955533 | ||
| metJp1 | Reverse | 4126138 | 78 | 18 |
| metB | Forward | 4126216 | ||
| trmA | Reverse | 4160874 | 104 | 14 |
| btuB | Forward | 4160978 | 2 | |
| uvrA | Reverse | 4271512 | 78 | 32 |
| ssb | Forward | 4271590 | 5 | |
| IleRp2 | Reverse | 4445871 | 108 | 38 |
| ORF83 | Forward | 4445979 | ||
| smp | Reverse | 4622368 | 64 | 22 |
| serB | Forward | 4622432 |
Only pairs with a distance between start sites of 120 pdb or fewer are shown. All are promoters for RNAP holoenzyme containing ς70.
Forward is in a clockwise direction on the genome (in the direction of increasing numbers on the map of Blattner et al. [4]).
Position of start site.
Separation (in total number of pdb) of the start sites between forward and reverse promoters (map number of forward start site − map number of reverse start site).
ACKNOWLEDGMENT
We thank Alberto Santos for updating the literature references presented in Table 2.
This research was supported by grant GM 31808 from the National Institutes of Health (to P.L.H.). The core facility at Case Western Reserve University (oligonucleotide synthesis) is supported by U.S. Public Health Service grant P30CA43703.
REFERENCES
- 1.Auble D T, Allen T L, deHaseth P L. Promoter recognition by Escherichia coli RNA Polymerase: effects of substitutions in the spacer DNA separating the −10 and −35 regions. J Biol Chem. 1986;261:11202–11206. [PubMed] [Google Scholar]
- 2.Aufrere R, Tempete M, Bohin J P. Regulation of expression of the gene for vitamin B12 receptor cloned on a multicopy plasmid in Escherichia coli. Mol Gen Genet. 1986;205:358–365. doi: 10.1007/BF00430451. [DOI] [PubMed] [Google Scholar]
- 3.Beck C F, Warren R A J. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988;52:318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brandsma J A, Bosch D, de Ruyter M, van de Putte P. Analysis of the regulatory region of the ssb gene of Escherichia coli. Nucleic Acids Res. 1985;13:5095–5109. doi: 10.1093/nar/13.14.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton Z F, Gross C A, Watanabe K K, Burgess R R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- 7.Busby S, Ebright R H. Promoter structure, promoter recognition and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 8.Bushman F D. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J Mol Biol. 1993;230:28–40. doi: 10.1006/jmbi.1993.1123. [DOI] [PubMed] [Google Scholar]
- 9.deHaseth P L, Zupancic M, Record M T., Jr RNA polymerase-promoter interaction: the comings and goings of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove S L, Joung J K, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 11.Fong R S-C, Woody S, Gussin G N. Direct and indirect effects of mutations in lambda PRM on open complex formation at the divergent PR promoter. J Mol Biol. 1994;240:119–126. doi: 10.1006/jmbi.1994.1426. [DOI] [PubMed] [Google Scholar]
- 12.Fong R S-C, Woody S, Gussin G N. Modulation of PRM activity by the lambda PR promoter in both the presence and absence of repressor. J Mol Biol. 1993;232:792–804. doi: 10.1006/jmbi.1993.1432. [DOI] [PubMed] [Google Scholar]
- 13.Giladi H, Murakami K, Ishihama A, Oppenheim A B. Identification of an UP element within the IHF binding site at the PL1-PL2 tandem promoter of bacteriophage λ. J Mol Biol. 1996;260:484–491. doi: 10.1006/jmbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 14.Gustafsson C, Lindström P H R, Hagervall T G, Esberg K B, Bjork G R. The trmA promoter has regulatory features and sequence elements in common with the rRNA P1 promoter family of Escherichia coli. J Bacteriol. 1991;173:1757–1764. doi: 10.1128/jb.173.5.1757-1764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawley D, McClure W R. Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol. 1982;157:493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- 16.Hershberger P A, deHaseth P L. RNA polymerase bound to the PR promoter of bacteriophage lambda inhibits open complex formation at the divergently transcribed PRM promoter: implications for an indirect mechanism of transcriptional activation by lambda repressor. J Mol Biol. 1991;222:479–494. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- 17.Hershberger P A, Mita B C, Tripatara A, deHaseth P L. Interference by PR-bound RNA polymerase with PRM function in vitro. Modulation by the bacteriophage lambda cI protein. J Biol Chem. 1993;268:8943–8948. [PubMed] [Google Scholar]
- 18.Kirby T W, Hindenbach B R, Greene R C. Regulation of in vivo transcription of the Escherichia coli K-12 metJBLF gene cluster. J Bacteriol. 1986;165:671–677. doi: 10.1128/jb.165.3.671-677.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolling R, Lother H. AsnC: an autogenously regulated activator of asparagine synthase A transcription in Escherichia coli. J Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles J S, Guest J R. Nucleotide sequence and transcriptional start point of the phosphomannose isomerase gene (manA) of Escherichia coli. Gene. 1984;32:41–48. doi: 10.1016/0378-1119(84)90030-1. [DOI] [PubMed] [Google Scholar]
- 21.Mita B C, Tang Y, deHaseth P L. Interference of PR-bound RNA polymerase with open complex formation at PRM is relieved by a 10-base pair deletion between the two promoters. J Biol Chem. 1995;270:30428–30433. doi: 10.1074/jbc.270.51.30428. [DOI] [PubMed] [Google Scholar]
- 22.Neuwald A F, Stauffer G V. An Escherichia coli membrane protein with a unique signal sequence. Gene. 1989;82:219–228. doi: 10.1016/0378-1119(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 23.Ohmori H, Kimura M, Nagata T, Sakakibara Y. Structural analysis of the dnaA and dnaN genes of Escherichia coli. Gene. 1984;28:159–170. doi: 10.1016/0378-1119(84)90253-1. [DOI] [PubMed] [Google Scholar]
- 24.Otsuka A, Abelson J. The regulatory region of the biotin operon in Escherichia coli. Nature. 1978;276:689–694. doi: 10.1038/276689a0. [DOI] [PubMed] [Google Scholar]
- 25.Owens E, Gussin G N. Differential binding of RNA polymerase to the PRM and PR promoters of bacteriophage lambda. Gene. 1983;23:157–166. doi: 10.1016/0378-1119(83)90047-1. [DOI] [PubMed] [Google Scholar]
- 26.Pettis G S, Brickman T J, McIntosh M A. Transcriptional mapping and nucleotide sequence of the Escherichia coli fepA-fes enterobactin region. Identification of a unique iron-regulated bidirectional promoter. J Biol Chem. 1988;263:18857–18863. [PubMed] [Google Scholar]
- 27.Ptashne M. A genetic switch. Cambridge, Mass: Cell Press; 1986. [Google Scholar]
- 28.Roe J H, Burgess R R, Record M T., Jr Kinetics and mechanism of the interaction of E. coli RNA polymerase with the λPR promoter. J Mol Biol. 1984;176:495–521. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 29.Ross W, Aiyar S E, Salomon J, Gourse R L. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 31.Salgado H, Santos A, Garza-Ramos U, vanHelden J, Diaz E, Collado-Vides J. RegulonDB (version 2.0): a database on transcriptional regulation in Escherichia coli. Nucleic Acids Res. 1999;27:519–607. doi: 10.1093/nar/27.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancar A, Sancar G B, Rupp W D, Little J W, Mount D W. LexA protein inhibits transcription of the E. coli uvrA gene in vitro. Nature. 1982;298:96–98. doi: 10.1038/298096a0. [DOI] [PubMed] [Google Scholar]
- 33.Schoenlein P V, Roa B B, Winkler M E. Divergent transcription of pdxB and homology between the pdxB and serA gene products in Escherichia coli K-12. J Bacteriol. 1989;171:6084–6092. doi: 10.1128/jb.171.11.6084-6092.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih M-C, Gussin G N. Mutations affecting two different steps in transcription initiation at the phage λ PRM promoter. Proc Natl Acad Sci USA. 1983;80:496–500. doi: 10.1073/pnas.80.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siebenlist U, Simpson R B, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 36.Strainic M G, Jr, Sullivan J J, Velevis A, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase: effects of the UP element on open complex formation and promoter clearance. Biochemistry. 1998;37:18074–18080. doi: 10.1021/bi9813431. [DOI] [PubMed] [Google Scholar]
- 37.Tang Y, Murakami K, Ishihama A, deHaseth P L. Upstream interactions at the lambda PRM promoter are sequence nonspecific and activate the promoter to a lesser extent than an introduced UP element of an rRNA promoter. J Bacteriol. 1996;178:6945–6951. doi: 10.1128/jb.178.23.6945-6951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss D L, Johnson D I, Weith H L, Somerville R L. Structural analysis of the ileR locus of Escherichia coli K12. J Biol Chem. 1986;261:9966–9971. [PubMed] [Google Scholar]
- 39.Wek R C, Hatfield G W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986;261:2441–2450. [PubMed] [Google Scholar]
- 40.Woody S T, Fong R S-C, Gussin G N. Effects of a single base-pair deletion in the bacteriophage lambda PRM promoter. Repression of PRM by repressor bound at OR2 and by RNA polymerase bound at PR. J Mol Biol. 1993;229:37–51. doi: 10.1006/jmbi.1993.1006. [DOI] [PubMed] [Google Scholar]
- 41.Xu J. The role of 434 repressor in regulating transcription initiation at bacteriophage 434 PR and PRM promoters. Ph.D. dissertation. Buffalo: State University of New York at Buffalo; 1999. [Google Scholar]