Abstract
It is difficult to identify people with nonalcoholic fatty liver disease (NAFLD) who are at high risk for developing hepatocellular carcinoma (HCC). A polygenic risk score (PRS) for hepatic fat (HFC‐PRS) derived from non‐Asians has been reported to be associated with HCC risk in European populations. However, population‐level data of this risk in Asian populations are lacking. Utilizing resources from 24,333 participants of the Singapore Chinese Health Study (SCHS), we examined the relationship between the HFC‐PRS and HCC risk. In addition, we constructed and evaluated a NAFLD‐related PRS (NAFLD‐PRS) with HCC risk in the SCHS. Cox proportional hazards models were used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) of HCC incidence with both HFC‐PRS and NAFLD‐PRS. The HFC‐PRS and NAFLD‐PRS were highly correlated (Spearman r = 0.79, p < 0.001). The highest quartiles of both the HFC‐PRS and the NAFLD‐PRS were associated with significantly increased risk of HCC with HR of 2.39 (95% CI 1.51, 3.78) and 1.77 (95% CI 1.15, 2.73), respectively, compared with their respective lowest quartile. Conclusion: The PRS for hepatic fat content or NAFLD may be useful for assessing HCC risk in both Asian and European populations. The findings of this and prior studies support a potential causal role of genetically determined NAFLD in HCC development.
It is difficult to identify people with non‐alcoholic fatty liver disease (NAFLD) who are at high risk for developing hepatocellular carcinoma (HCC). We examined if two polygenic risk scores: HFC‐PRS (hepatic fat), derived in a mixed ancestry American population and previously validated in Europeans; and NAFLD‐PRS, which we calculated utilizing East Asian populations, were associated with HCC risk in our East Asian population in the Singapore Chinese Health Study. Higher levels of both HFC‐PRS and NAFLD‐PRS were associated with higher risk of HCC, and both may be useful assessing HCC risk in Asian and European populations.
![]()
INTRODUCTION
Liver cancer, most of which is hepatocellular carcinoma (HCC), is now the second‐leading cause of cancer death globally behind lung cancer.[ 1 ] The increasing prevalence of nonalcoholic fatty liver disease (NAFLD) along with obesity and type 2 diabetes has elevated NAFLD as a prominent risk factor for HCC.[ 2 ] NAFLD has become the most common cause of chronic liver disease, affecting about 25% of the global population.[ 2 ] NAFLD is estimated to contribute to 10%–12% of HCC burden in Europe and North America and 1%–6% of HCC burden in Asian countries.[ 2 ] In Asian countries, NAFLD‐related HCC is expected to increase due to increasing NAFLD and obesity prevalence on account of globalization and changing dietary and lifestyle habits.[ 2 ]
While NAFLD in Europe and North America is often linked to type 2 diabetes and obesity, approximately 8%–19% of people in Asian countries with body mass index (BMI) less than 25 kg/m2 have NAFLD, which is referred to as lean NAFLD.[ 3 ] Genetic predisposition may be a particularly important factor for NAFLD development in this nonobese population that may not otherwise be screened for HCC. For example, patatin‐like phospholipase domain containing 3 (PNPLA3), a gene strongly associated with NAFLD, has previously been shown to have a stronger impact on hepatic steatosis, fatty liver disease, in people of Chinese ancestry without metabolic syndrome.[ 3 , 4 ] Genetic risk profiling for other diseases such as coronary artery disease and breast cancer have resulted in targeted lifestyle interventions, better prediction of age of disease onset, and more streamlined screening.[ 5 ] Genetic predisposition, calculated as a polygenic risk score (PRS), may be an effective noninvasive biomarker to improve identification of individuals at high risk for NAFLD‐related HCC.
Recently, Bianco et al. used a previously developed PRS that sums the number of steatosis‐predisposing alleles in four genes, PNPLA3, transmembrane 6 superfamily member 2 (TM6SF2), membrane Bound O‐Acyltransferase Domain Containing 7 (MBOAT7) and glucokinase regulatory protein (GCKR), weighed by their effect size on hepatic fat content in Europeans, African Americans, Hispanics, and non‐Hispanic Whites in the United States[ 6 ] (referred to in this paper as the hepatic fat content– related PRS [HFC‐PRS]). Bianco et al. examined genetically determined hepatic fat content in relation to risk of HCC in patients with NAFLD and normal controls in Italy and the United Kingdom as well as in the general population in the United Kingdom.[ 7 ] They reported that the HFC‐PRS was significantly associated with increased risk of HCC in the NAFLD cohort and in the UK general population. However, their study is limited by the cross‐sectional design of the NAFLD cohort and generalizability to European‐only populations.[ 7 ]
To extend these findings to other populations, we investigated whether the established HFC‐PRS based on non‐Asian populations was associated with HCC in an Asian population. In addition, we constructed a PRS for NAFLD (NAFLD‐PRS) using publicly available genome‐wide association study (GWAS) data generated in East Asian samples and examined the association for this NAFLD‐PRS with HFC‐PRS and with the risk of developing HCC in East Asians in Singapore. Finally, we applied Mendelian randomization (MR) methods to assess the causality of the relationship between NAFLD and risk of HCC in East Asians. The overall goal of this work was to investigate a causal link between NAFLD and HCC in Asian populations, and to broaden applicability of PRS beyond typically studied European populations.
METHODS
Study population
The current study was conducted within the Singapore Chinese Health Study (SCHS), a population‐based prospective cohort study for which participants were recruited in Singapore between April 1993 and December 1998.[ 8 ] In total, 63,257 Han Chinese men and women aged 45–74 at baseline were enrolled into the study. Eligible subjects were required to be permanent residents of Singapore residing in government‐built housing and belong to one of two major Chinese dialect groups, either the Hokkien or Cantonese. At enrollment, trained interviewers administered an in‐person interview using a structured questionnaire for information on participants’ demographic and lifestyle characteristics. Interviewers also administered a validated semi‐quantitative food frequency questionnaire to collect information on habitual dietary intake, including alcohol intake.[ 9 ] The SCHS is approved by the institutional review board at the National University of Singapore and the University of Pittsburgh, and all subjects provided written informed consent.
Sample collection and storage for the SCHS have been described previously.[ 10 ] Briefly, 32,535 participants donated blood, buccal, and/or urine samples for research. A subset of HCC cases and their matched controls were tested previously for serologic status of hepatitis B surface antigen (HBsAg) and antibodies for hepatitis C virus (anti‐HCV), which are established risk factors for HCC.[ 11 , 12 ] Among the 208 HCC cases with PRS data, 16 did not donate serum or plasma samples. The remaining 192 cases were tested for HBsAg, in which four cases failed testing. Therefore, 188 cases had available HBsAg‐positivity status. The presence of HBsAg was determined using a standard radioimmunoassay (AUSRIA; Abbott Laboratories, North Chicago, IL, USA), and anti‐HCV using the ELISA version 2.0 kit (Ortho Diagnostic Systems, Raritan, NJ, USA), with confirmation of positive samples using the RIBA version 2.0 (Chiron, Emeryville, CA, USA). Given the low prevalence of anti‐HCV positivity in the 366 SCHS participants evaluated (2%),[ 11 ] anti‐HCV testing was stopped to preserve samples for future studies.
All study participants were followed annually for incidence of cancer and death. Incident cancer cases were identified through linkage analysis with the nationwide Singapore Cancer Registry, and deaths were ascertained via the Singapore Birth and Death Registry. The Singapore Cancer Registry has collected comprehensive information on cancer diagnoses since 1968.[ 13 ] To date, 56 participants (<0.1%) have been cumulatively lost to follow‐up. HCC cases were defined by the International Classification of Diseases–Oncology, 2nd Edition code C22.0.[ 13 ] The present analysis includes 208 incident cases of HCC in the genotyped subcohort.
Genotyping and PRS
Methods for genotyping SCHS samples have been published previously.[ 14 , 15 , 16 ] Briefly, a total of 23,600 SCHS samples, genotyped on the Illumina Global Screening Array, and 2003 SCHS samples, genotyped on the Illumina HumanOmni ZhongHua‐8 Bead Chip, passed standard GWAS quality control procedures.[ 14 , 15 , 16 ] An additional 156 samples identified to be duplicate or likely first degree–related samples between data sets were excluded from the study. After excluding individuals with prevalent cancer at baseline, related individuals, and those with poor genotyping quality, all participants with available genotyping were included in the present analysis.
Single‐nucleotide polymorphisms (SNPs) included in the HFC‐PRS were developed for predicting hepatic fat content in an American mixed‐ancestry cohort[ 6 , 7 ]; descriptive information about variants and weights are provided in Table S1. The HFC‐PRS included four SNPs: rs1260326 (GCKR), rs58542926 (TM6SF2), rs641738 (MBOAT7), and rs738409 (PNPLA3). The SNPs rs58542926 (TM6SF2), rs641738 (MBOAT7), and rs738409 (PNPLA3) were genotyped, and rs1260326 (GCKR) was imputed with an imputation INFO score of 0.9882. The East Asian NAFLD‐PRS was calculated by extracting summary statistics information on SNP‐NAFLD associations from the Phenoscanner[ 17 , 18 ] database at study start on July 13, 2021. All SNPs associated with the phenotype “non‐alcoholic fatty liver disease” in East Asian ancestry were included in the search. After the initial search, 13 records for a total of 12 SNPs had beta (β) and p values available in East Asians as defined by Phenoscanner for the association with NAFLD (Table S2). SNPs were pruned based on p value with NAFLD and linkage disequilibrium (LD) with each other, in which SNPs were included with p values of genome‐wide significance (<5 × 10−8) and independent SNPs with r 2 ≤ 0.2. LD was determined through correlations in our data as well as a comparison using the LDMatrix tool provided by the National Cancer Institute based on the population of Han Chinese in Beijing, China (https://ldlink.nci.nih.gov). If two SNPs were in LD, the SNP with a larger effect size and smaller p value for association with NAFLD was chosen for inclusion in the PRS. The SNPs selected for the East Asian NAFLD‐PRS and additional information provided by Phenoscanner are provided in Table S1. After pruning, three SNPs remained that were associated with NAFLD among those with East Asian ancestry: rs1260326 (GCKR), rs4808199 (GATA zinc finger domain containing 2A [GATAD2A]), and rs2896019 (PNPLA3). The rs2896019 (PNPLA3) SNP was genotyped, and rs4808199 (GATAD2A) was imputed with an imputation INFO score of 0.9998. A weighted score was calculated by summing the three SNP effect alleles associated with higher log odds of NAFLD weighted by their respective beta (β) values.[ 19 ]
We first examined the relationship between HFC‐PRS and risk of HCC in our study population. We then calculated the East Asian–based NAFLD‐PRS to determine whether the association was consistent when the same ancestral population was used for the exposure and outcome associations. The SNPs used in the HFC‐PRS and included in our East Asian NAFLD‐PRS were extracted from the SCHS for analysis. A total of 24,333 individuals (including 208 HCC cases) had complete data for the HFC‐PRS. Among those individuals, 24,294 (including 206 HCC cases) had complete data for the three SNPs included in our East Asian NAFLD PRS.
Statistical analysis
Person‐years of follow‐up for each subject were calculated from the date of enrollment into the study to the date of HCC diagnosis, death, migration out of Singapore, or December 31, 2015, whichever occurred first. Follow‐up was calculated from the date of enrollment as opposed to date of blood draw, because alleles are static regardless of measurement date. Allele frequency differences between HCC cases and non‐cases were determined by chi‐square tests. Quartiles of NAFLD PRSs were determined among all subjects who had complete data for each genetic score. Participants were compared by PRS quartile or HCC status for covariate differences by chi‐square tests for categorical variables and either Wilcoxon two‐sample test (for comparison by HCC status) or Kruskal‐Wallis test (for comparison by PRS quartile) for continuous variables.
Cox proportional hazards regression models were used to estimate hazard ratio (HR) and 95% confidence interval (CI) for the association between NAFLD PRSs and HCC risk with adjustment for potential confounding variables. Initial models were adjusted for only age (years) and sex (male, female). Additional covariates measured at baseline included in the second model were dialect group (e.g., Cantonese, Hokkien), BMI (kg/m2; continuous), education (i.e., no formal education/primary, secondary or higher), smoking status (i.e., never, light, heavy), alcohol intake (i.e., one or more alcoholic drinks per day, less than one alcoholic drink per day), year of enrollment (i.e., 1993–1995, 1996–1998), and diabetes status (yes, no). Heavy smokers were defined as those who began smoking before 15 years of age and smoked 13 or more cigarettes per day, whereas all other smokers were considered as light smokers.[ 20 ] To account for potential population stratification, a third model adjusted for all previous covariates as well as the top three principal components (PCs) of population stratification (PCs 1–3). Quartiles of NAFLD PRSs were used to evaluate a linear relationship between NAFLD risk and HCC. Continuous NAFLD PRSs were scaled to mean = zero and SD = 1 to compare the two different scores. Heterogeneity in the NAFLD polygenic scores–HCC risk association by traditional NAFLD risk factors (BMI, diabetes, dietary fat intake, total energy) was tested by including a product term of the scaled continuous NAFLD genetic score and a binary indicator of the covariate in the model. To examine the association between the two PRSs and HCC risk without the influence of hepatitis B or C chronic infection nor heavy alcohol consumption, we conducted a sensitivity analysis excluding patients with HCC who were HBsAg or anti‐HCV positive or heavy drinkers. Heavy drinking status was defined as those who consumed ≥15 drinks/week for men and ≥8 drinks/week for women, following definitions from the US Center for Disease Control and Prevention (https://www.cdc.gov/alcohol/pdfs/excessive_alcohol_use.pdf).
To estimate the causal association between NAFLD and risk of HCC in those of East Asian ancestry, we used SNPs included in the East Asian NAFLD‐PRS as an instrumental variable: rs1260326 (GCKR), rs4808199 (GATAD2A), and rs2896019 (PNPLA3). The causal effect of NAFLD in East Asians on HCC risk was estimated using the MR package in R[ 21 ]. NAFLD was our explanatory variable and HCC the outcome, in which the inverse‐variance weighted (IVW) method—an estimator that combines ratio estimates for each variant[ 22 ]—was our primary result. To account for possible pleiotropy or invalid instruments, we also conducted sensitivity analyses including the weighted median,[ 23 ] MR Egger,[ 24 ] contamination mixture,[ 25 ] and maximum likelihood estimation[ 22 ] methods. To verify that a potentially pleiotropic SNP (rs1260326) did not affect our overall association, we used the IVW method, dropping rs1260326 (GCKR) as an instrumental variable.
To determine the PRS thresholds able to identify individuals at higher risk of HCC and assess whether adding the PRS to a clinical model improved its performance in identifying high‐risk individuals, we used area under the curve (AUC) statistics including cases diagnosed within 10 years and all non‐cases, and identified the optimal cutoff point for the HFC‐PRS maximizing both sensitivity and specificity. The corresponding cutoff point for NAFLD‐PRS was determined based on sensitivity similar to the HFC‐PRS cutoff point. Positive test (greater than or equal to the PRS cutoff point; yes/no) was added as a predictor to clinical models among all participants to determine whether adding PRS significantly improved HCC prediction compared with clinical variables alone (age, sex, BMI).
All statistical analyses were performed using R version 4.04 (https://www.r‐project.org/). All p values presented are two‐sided, and p values < 0.05 were considered statistically significant.
RESULTS
The present analysis included 24,333 SCHS participants with valid data on the HFC‐PRS and 24,294 participants with valid data on the East Asian NAFLD‐PRS. The HFC‐PRS and East Asian NAFLD‐PRS overlapped on three chromosomes (chromosomes 2, 19, and 22) and two genes: GCKR and PNPLA3. The GCKR SNP is the same for both PRSs (rs1260326). The GATAD2A SNP of the East Asian NAFLD‐PRS on chromosome 19 was not in LD with the TM6SF2 SNP (r 2 = 0.13) or the MBOAT7 SNP (r 2 < 0.001), both from the HFC‐PRS and located on chromosome 19. Rs738409 (HFC‐PRS) and rs2896019 (East Asian NAFLD‐PRS), both in the PNPLA3 gene, were in high LD (r 2 = 0.87). Overall, the HFC‐PRS and East Asian NAFLD‐PRS had a Spearman correlation of 0.79 (p < 0.001).
HFC‐PRS association with risk of HCC
After extracting SNP data from the SCHS, 24,333 individuals including 208 HCC cases had valid data for the four SNPs included in the HFC‐PRS. Among those with valid HFC‐PRS data, the average (SD) follow‐up time was 18.9 (3.6) years. Participants who developed HCC were older at baseline and had slightly higher BMI (Table 1). HCC cases were less likely to be female or speak Cantonese dialect and were more likely to be heavy smokers and diabetic at baseline (Table 1). There were no significant differences between cases and non‐cases for education or alcoholic drink intake. Two of the four SNPs included in the HFC‐PRS had significantly different minor allele frequencies (MAFs) between HCC cases and non‐cases: rs58542926 (TM6SF2, p < 0.001) and rs738409 (PNPLA3, p = 0.002) (Table 2). Participants in higher quartiles of HFC‐PRS were more likely to speak Hokkien dialect compared with Cantonese and slightly more likely to be diabetic or a current smoker (Table S3).
TABLE 1.
Baseline characteristics by HCC status in the SCHS HFC‐PRS population (n = 24,333)
| Characteristics | HCC cases | Non‐cases | p value |
|---|---|---|---|
| n | 208 | 24,125 | |
| Age, years, median (25th, 75th percentiles) | 59 (54, 64) | 54 (49, 61) | <0.001 |
| Female sex, n (%) | 62 (30%) | 13,225 (55%) | <0.001 |
| Cantonese dialect, n (%) | 81 (39%) | 11,890 (49%) | 0.004 |
| Education, secondary school or higher, n (%) | 62 (30%) | 8069 (33%) | 0.299 |
| BMI, kg/m2, median (25th, 75th percentiles) | 23.5 (22.5, 26.3) | 23.1 (21.1, 24.8) | <0.001 |
| Smoking status a (%) | |||
| Never smoker | 120 (58%) | 17,152 (71%) | <0.001 |
| Light smoker | 71 (34%) | 6102 (25%) | |
| Heavy smoker | 17 (8%) | 871 (4%) | |
| One or more alcoholic drinks per day, n (%) | 11 (5.2%) | 1114 (4.6%) | 0.770 |
| Heavy drinkers b , n (%) | 5 (2.4%) | 367 (1.5%) | 0.454 |
| Diabetes, n (%) | 36 (17%) | 1848 (8%) | <0.001 |
Note: Chi‐square test was used for categorical variables; Wilcoxon two‐sample test was used for continuous variables.
Cigarette smoking: The “heavy” smokers were those who began smoking before 15 years of age and smoked 13 or more cigarettes; all remaining ever smokers were defined as light smokers.
Heavy drinkers were defined as those who consumed ≥15 drinks/week for men and ≥8 drinks/week for women, following definitions from the US Center for Disease Control and Prevention (https://www.cdc.gov/alcohol/pdfs/excessive_alcohol_use.pdf).
TABLE 2.
Characteristics of SNPs in HFC‐PRS and East Asian NAFLD‐PRS in the SCHS
| CHR | SNP | Position (hg19) | Gene | Minor allele | Major allele | MAF Overall | MAF among cases | MAF among non‐cases | CHISQ | p |
|---|---|---|---|---|---|---|---|---|---|---|
| HFC‐PRS (n = 24,333, 208 HCC cases) | ||||||||||
| 2 | rs1260326 | chr2:27730940 | GCKR | T | C | 0.47 | 0.51 | 0.47 | 3.86 | 0.145 |
| 19 | rs58542926 | chr19:19379549 | TM6SF2 | T | C | 0.07 | 0.12 | 0.07 | 63.65 | <0.001 |
| 19 | rs641738 | chr19:54676763 | MBOAT7 | T | C | 0.25 | 0.27 | 0.25 | 1.46 | 0.483 |
| 22 | rs738409 | chr22:44324727 | PNPLA3 | G | C | 0.37 | 0.45 | 0.37 | 12.41 | 0.002 |
| East Asian NAFLD‐PRS (n = 24,294, 206 HCC cases) | ||||||||||
| 2 | rs1260326 | chr2:27730940 | GCKR | T | C | 0.47 | 0.51 | 0.47 | 3.57 | 0.167 |
| 19 | rs4808199 | chr19:19545099 | GATAD2A | A | G | 0.31 | 0.34 | 0.31 | 2.07 | 0.355 |
| 22 | rs2896019 | chr22:44333694 | PNPLA3 | G | T | 0.38 | 0.45 | 0.38 | 9.35 | 0.009 |
Note: p value significance is for the chi‐square statistical test.
Abbreviations: CHR, chromosome; CHISQ, chi‐square statistic; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Higher quartile and levels of the HFC‐PRS were associated with a statistically significant higher risk of HCC (p trend < 0.001). Compared with the lowest quartile, HR (95% CI) of HCC for the fourth quartile of HFC‐PRS was 2.39 (1.51, 3.78) after adjustment for age and sex. Estimates did not materially change when adjusted for further covariates, including dialect, BMI, education, smoking status, alcohol intake, year of enrollment, diabetes status, and PCs 1–3 (Table 3). Each SD increment in HFC‐PRS was associated with a statistically significant 38% increase in HCC risk (Table 3). When stratified by NAFLD risk factors including BMI, history of diabetes, daily intake of fat or total energy, the HFC‐PRS–HCC risk associations were robust across all subgroups (Figure 1). After excluding 60 HBsAg‐positive, 4 anti‐HCV‐positive, and 3 heavy‐drinker cases, the strength of the association between HFC‐PRS and risk of HCC increased (fourth quartile compared with first quartile HR = 2.78, 95% CI 1.57, 4.93) (Table S4).
TABLE 3.
Quartiles and continuous hazard ratios for risk of HCC by HFC‐PRS and East Asian NAFLD‐PRS in the SCHS
| Weighted score quartile | Persons | Person‐years | Cases | HR a (95% CI) | p a | HR b (95% CI) | p b | HR c (95% CI) | p c |
|---|---|---|---|---|---|---|---|---|---|
| HFC‐PRS (n = 24,333, 208 HCC Cases) | |||||||||
| First quartile (<0.128) | 4730 | 88,859 | 24 | 1.00 | 1.00 | 1.00 | |||
| Second quartile (0.128 to <0.331) | 6337 | 119,466 | 50 | 1.59 (0.98, 2.58) | 0.063 | 1.61 (0.99, 2.61) | 0.056 | 1.62 (1.00, 2.64) | 0.051 |
| Third quartile (0.331 to <0.459) | 7157 | 135,516 | 60 | 1.64 (1.02, 2.63) | 0.041 | 1.60 (1.00, 2.58) | 0.050 | 1.62 (1.01, 2.60) | 0.046 |
| Fourth quartile (≥0.459) | 6109 | 114,865 | 74 | 2.39 (1.51, 3.78) | 0.0002 | 2.31 (1.46, 3.66) | 0.0004 | 2.35 (1.48, 3.73) | 0.0003 |
| p trend | 0.0002 | 0.0004 | 0.0003 | ||||||
| Continuous, HR for 1‐SD increase (SD = 0.22) | 1.38 (1.21, 1.57) | <0.001 | 1.37 (1.20, 1.55) | <0.001 | 1.37 (1.20, 1.56) | <0.001 | |||
| East Asian NAFLD‐PRS (n = 24,294, 206 HCC cases) | |||||||||
| First quartile (<0.615) | 4594 | 85,933 | 29 | 1.00 | 1.00 | 1.00 | |||
| Second quartile (0.615 to <0.937) | 6121 | 115,712 | 43 | 1.13 (0.71, 1.82) | 0.601 | 1.14 (0.71, 1.82) | 0.598 | 1.15 (0.72, 1.84) | 0.569 |
| Third quartile (0.937 to <1.259) | 7310 | 137,843 | 63 | 1.37 (0.88, 2.13) | 0.161 | 1.34 (0.86, 2.09) | 0.189 | 1.35 (0.87, 2.10) | 0.178 |
| Fourth quartile (≥1.259) | 6269 | 118,489 | 71 | 1.77 (1.15, 2.73) | 0.009 | 1.68 (1.09, 2.60) | 0.018 | 1.70 (1.10, 2.62) | 0.017 |
| p trend | 0.003 | 0.008 | 0.007 | ||||||
| Continuous, HR for 1‐SD increase (SD = 0.52) | 1.26 (1.10, 1.44) | 0.0006 | 1.24 (1.09, 1.42) | 0.002 | 1.24 (1.09, 1.42) | 0.001 | |||
Abbreviation: HR, hazard ratio.
Adjusted for age and sex only.
Adjusted for age, sex, dialect, BMI, education, smoking status, alcohol intake, year of enrollment, and diabetes status.
Adjusted for all covariates in model b with additional adjustment for principal components 1–3.
FIGURE 1.
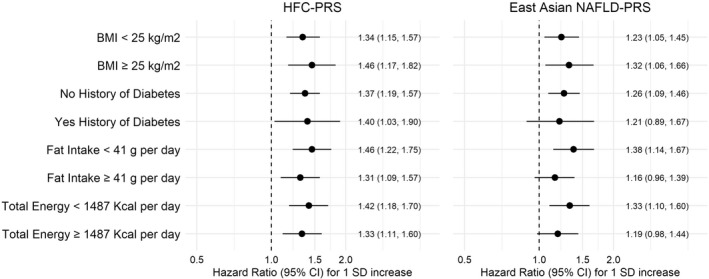
Association of hepatic fat content polygenic risk score (HFC‐PRS) and East Asian nonalcoholic fatty liver disease PRS (NAFLD‐PRS) with risk of hepatocellular carcinoma (HCC) stratified by NAFLD risk factors in the Singapore Chinese Health Study (SCHS). Fat intake and total energy cutoffs were determined by the median of the distribution. All models were adjusted for age and sex. BMI, body mass index; CI, confidence interval
East Asian NAFLD‐PRS association with risk of HCC
Among those with valid data for the HFC‐PRS, 24,294 individuals including 206 HCC cases in the SCHS also had valid data for the three SNPs included in the East Asian NAFLD‐PRS. The average and SD follow‐up time was the same for those with East Asian NAFLD‐PRS data as compared with the HFC‐PRS. Baseline characteristics between cases and non‐cases were nearly identical to the PRS‐HRC data given in Table 1 (Table S5). One SNP included in the East Asian NAFLD‐PRS, rs2896019 (PNPLA3), had a significantly different MAF between HCC cases and non‐cases (p = 0.009), in which the G allele frequency was 0.45 among cases and 0.38 among non‐cases (Table 2). Participants in higher quartiles of East Asian NAFLD‐PRS were more likely to speak Hokkien dialect compared with Cantonese and slightly more likely to be diabetic (Table S3).
Higher quartile and levels of the East Asian NAFLD‐PRS were associated with a statistically significant higher risk of HCC, showing similar results as compared with the HFC‐PRS (p trend = 0.003). Compared with the lowest quartile, HR (95% CI) of HCC for the fourth quartile of East Asian NAFLD‐PRS was 1.77 (1.15, 2.73) after adjustment for age and sex. Estimates were not materially changed when further adjusted for additional covariates including dialect, BMI, education, smoking status, alcohol intake, year of enrollment, diabetes status, and PCs 1–3 (Table 3). A 1‐SD increase in East Asian NAFLD‐PRS was associated with a 26% increase in HCC risk (Table 3). When stratified by NAFLD risk factors, the East Asian NAFLD‐PRS was consistently associated with higher risk of HCC among all subgroups; however, some stratified subgroups did not remain statistically significant (Figure 1). After excluding 60 HBsAg‐positive, 4 anti‐HCV‐positive, and 3 heavy‐drinker cases, the strength of the association between East Asian NAFLD‐PRS and risk of HCC increased (fourth quartile compared with first quartile HR = 1.98, 95% CI 1.14, 3.44) (Table S4).
Causal relationship between NAFLD and HCC among East Asians
MR analysis showed that NAFLD was causally associated with development of HCC, in which those with NAFLD have 1.58‐fold higher risk of HCC compared to those without NAFLD (IVW estimate; p < 0.001) (Table 4). All sensitivity analyses conducted to robustly assess MR results were consistent with the IVW estimate (Figure 2), in which the causal HR estimate ranged from 1.53 to 1.58. All sensitivity analyses in which methods allow for some invalid instruments or account for possible pleiotropy, and one gene affects multiple traits, showed a significant causal effect except for one. The p value for MR Egger was well above the threshold for significance (p = 0.281). However, the robust MR Egger estimate, which may be less sensitive to outliers or influential points, was highly significant (p < 0.001). The intercept terms for the MR Egger and robust MR Egger were both not significantly different from zero, indicating minimal pleiotropic effects (Table 4). To further verify that our results were not affected by pleiotropic effects, we calculated the IVW estimate dropping the potentially pleiotropic GCKR SNP, rs1260326. After dropping rs1260326, the IVW estimate remained consistent for a causal effect of NAFLD on HCC risk (IVW estimated beta = 0.456; 95% CI 0.171, 0.741; p = 0.002).
TABLE 4.
Causal estimates of NAFLD on HCC by multiple MR approaches using gene variants included in the East Asian NAFLD‐PRS as instruments
| Method | Intercept a | Beta | HR (95% CI) | p |
|---|---|---|---|---|
| IVW | — | 0.457 | 1.58 (1.22, 2.04) | <0.001 |
| Simple median | — | 0.462 | 1.59 (1.12, 2.25) | 0.01 |
| Weighted median | — | 0.457 | 1.58 (1.19, 2.1) | 0.002 |
| Contamination mixture method | — | 0.46 | 1.58 (1.22, 2.03) | <0.001 |
| Maximum‐likelihood method | — | 0.457 | 1.58 (1.21, 2.06) | 0.001 |
| MR‐Egger | 0.006 | 0.445 | 1.56 (0.69, 3.51) | 0.281 |
| Robust MR‐Egger | 0.006 | 0.445 | 1.56 (1.24, 1.97) | <0.001 |
p value for intercept terms: MR‐Egger = 0.976; robust MR‐Egger = 0.938.
FIGURE 2.
Visual comparison of causality estimates of NAFLD on HCC by multiple Mendelian randomization (MR) approaches using variants included in the East Asian NAFLD‐PRS as instruments. IVW, inverse‐variance weighted
Optimal PRS thresholds and clinical utility
The AUC for each PRS was calculated using cases diagnosed within 10 years and all non‐cases. The AUC for HFC‐PRS was 0.59 (49 cases among 24,174 participants) and only 0.57 for NAFLD‐PRS (49 cases among 24,137 participants). The optimal cutoff for HFC‐PRS was 0.6 (greater than highest quartile cutoff point), with a sensitivity of 33% and specificity of 87%. Using this cutoff, the HFC‐PRS was strongly associated with HCC risk among all participants after adjustment for covariates (HR = 2.18, 95% CI 1.59, 2.99, p < 0.001) (Table S6). The corresponding cutoff for NAFLD‐PRS, similar sensitivity (33%), was 1.26 (greater than highest quartile cutoff point) with a specificity of 74%. Using this cutoff, the NAFLD‐PRS was associated with HCC risk among all participants after adjustment for covariates (HR = 1.51; 95% CI 1.12, 2.04; p = 0.007) (Table S6). Adding HFC‐PRS or NAFLD‐PRS (above or equal to the cutoff) significantly improved the performance; the AUC increased from 0.717 to 0.734 (p = 0.021) and from 0.716 to 0.727 (p = 0.040), respectively, compared to a model with clinical variables including age, sex, and BMI only (data not shown). Both PRS were more strongly associated with risk of HCC compared to single variants alone (Table S6).
DISCUSSION
In this study, the HFC‐PRS and the East Asian NAFLD‐PRS were both associated with HCC risk before and after adjustment for nongenetic factors and were highly correlated with each other in a Singaporean sample. Risk scores for NAFLD/hepatic fat derived in specific ancestral populations may yield consistent results when applied in different populations. PRS may be a cost‐effective way to identify individuals at high risk for HCC that goes beyond lifestyle factors to incorporate genetic predisposition. In addition, our instrumental variable analysis suggests that beyond association to classify individuals into different risk strata, NAFLD may be a causal mechanism for HCC risk in East Asian populations. In summary, the two risk scores evaluated here may prove useful (1) in establishing causes of HCC in East Asian populations, and (2) in the identification/stratification of people with NAFLD for whom preventive and surveillance programs may be administered to reduce the risk and/or improve early detection of HCC.
Bianco et al. determined that a PRS for hepatic fat was associated with increased risk of HCC in Europeans. We extended their findings by applying their PRS and an NAFLD‐PRS in an East Asian population using weights derived from East Asian ancestry. We used a prospective study design, allowing us to include incident HCC cases and time‐to‐event analyses. The point estimates for the East Asian NAFLD‐PRS were lower in magnitude than the HFC‐PRS. The NAFLD‐PRS may more accurately reflect unbiased risk of HCC from NAFLD in East Asians compared with the HFC‐PRS, which was derived using data from a mixed‐ancestry study population. Our point estimates may also differ due to differently defined exposures: continuous hepatic fat content compared with binary NAFLD diagnosis. The PRS for hepatic fat content and the PRS for NAFLD contained the same gene regions except one additional SNP in the HFC‐PRS, indicating that each exposure is associated with very similar genetic predisposition. This study increases the generalizability of both PRS for genetically predicted hepatic fat/NAFLD in association with HCC.
Currently, the prevalence of NAFLD in Asian countries is about 25%, similar to countries in Europe and North America, and has been increasing in the past two decades.[ 3 ] Although obesity, metabolic disease, and other lifestyle factors may not be as prevalent in Asia as in Europe and North America, the similar prevalence of NAFLD in Asia as Europe and North America is partially due to lean NAFLD.[ 3 ] Another reason for similar NAFLD prevalence despite differing risk factors may be genetic predisposition. One of the SNPs in the HFC‐PRS, rs738409, is highly correlated with a SNP in the East Asian NAFLD‐PRS and is a missense mutation in the PNPLA3 gene that has been well established to increase risk of NAFLD, deposition of liver lipids, and NAFLD progression.[ 26 ] The risk (GG) genotype is found in 13%–19% of the general population in Asian studies compared with 4% in Europeans. This difference in genetic susceptibility may be responsible for similar prevalence despite different risk factors. Prevalence of lean NAFLD may also be related to different obesity cutoffs. Many studies recommend lower cutoff points for defining overweight and obese in Asian populations.[ 27 ] Given that lean NAFLD is prevalent in Asia, identifying those with genetically predicted higher risk of HCC may help identify those who could benefit from early detection in this subgroup to reduce incidence and/or mortality of HCC.
Our East Asian NAFLD‐PRS was developed using all available GWAS results in East Asians with appropriate significance and LD cutoffs in the Phenoscanner database at the time of study start. The three SNPs that were included in the score were from the same study published by Kawaguchi et al. that examined these SNPs in association with NAFLD risk, in which those in the highest quintile of the PRS had 5 times the risk of NAFLD compared with the lowest quintile.[ 28 ] The investigators used a stepwise model to determine which SNPs to include in their model with initially four SNPs, then further refining by switching SNPs for those that had been studied previously (including PNPLA3 rs738409 and TM6SF2 rs58542926) and adding 14 additional SNPs. When included in the model, the additional SNPs did not increase the AUC for classifying NAFLD cases from controls.[ 28 ] These results increase our confidence that the East Asian NAFLD‐PRS, while consisting of only three SNPs, broadly covers genetic susceptibility to NAFLD in this population.
Results from our sensitivity analyses suggest that these PRS may be associated specifically with HCC caused by NAFLD. The magnitude for both PRS‐HCC risk associations was strengthened after excluding HCC cases infected with hepatitis B and/or C virus or heavy consumption of alcohol, suggesting that NAFLD (as determined by PRS) had a greater impact on HCC risk. We measured HBsAg positivity for all cases with available blood except 4 cases, and while we only have anti‐HCV measurements for 366 participants, anti‐HCV positivity in HCC cases in our previous study was only 5%,[ 11 ] suggesting that HCV infection has limited impact on HCC risk in our population. We also excluded HCC cases who were defined as heavy drinkers according to the US Center for Disease Control and Prevention guidelines. After exclusion of HCC cases associated with these established risk factors, the only remaining strong risk factor for HCC was NAFLD. Thus, we conclude to the best of our ability that most of the remaining HCC cases were NAFLD‐related. Both PRS were more strongly associated with HCC after exclusion of cases for exposure to other risk factors, suggesting that genetically determined NAFLD plays a more significant role in NAFLD‐related HCC.
MR is an instrumental variable analysis method in which genetic variants are used in observational analyses to make causal inferences about the effect of an exposure on an outcome.[ 29 ] Accordingly, the basic assumptions for MR are that (1) the variant is associated with the exposure (in our case, NAFLD); (2) the variant is not associated with the outcome (i.e., HCC) via a confounding pathway; and (3) the variant does not affect the outcome directly, but purely through the exposure. One potential violation of MR results is pleiotropy, in which genetic variants are associated with multiple risk factors on different causal pathways. A sensitivity analysis designed to test this is the MR Egger method, where if the intercept term is significantly different from zero, there is evidence of either a pleiotropic effect or the InSIDE (INstrument Strength Independent of Direct Effect) assumption is violated, or both.[ 24 ] In our study, there appears to be a slight association between the different PRS and age, dialect, smoking status, and diabetes status. However, dialect may be reflective of underlying genetic or lifestyle differences, and the differences between PRS quartiles by age, smoking, and diabetes were materially very small. Finally, both the MR Egger and robust MR Egger intercept terms were not statistically significant, indicating that pleiotropy may not be a violating factor. To further verify that pleiotropic effects were not impacting our estimates, we conducted a sensitivity analysis in which we calculated the IVW estimate by dropping the GCKR SNP, rs1260326. The minor T‐allele of rs1260326 has previously been associated with lower fasting glucose and insulin levels and a protective effect against type 2 diabetes,[ 30 , 31 , 32 ] and the C‐allele has previously been associated with number of alcoholic drinks consumed per week.[ 33 ] After removing this SNP from our analysis, the IVW estimate was consistent with a causal effect of NAFLD on HCC risk.
Although the AUC for both PRSs alone were poor, using the optimal cutoff, both scores significantly improved the diagnostic accuracy of a clinical model for the entire cohort. These PRSs may be useful in future studies when combined with other genetic variants to produce a score with stronger clinical relevance. Additionally, our analysis expands on the work of Bianco et al.[ 7 ] by showing that HFC‐PRS improved diagnostic accuracy among non‐European populations, expanding its utility.
Our study has multiple strengths. As previously mentioned, we were able to use a prospective study design, collect incident HCC outcomes, and conduct time‐to‐event analyses. Our study is a population‐based cohort representative of a general population; therefore, our findings may be more generalizable to other East Asian populations. We estimated associations for both HFC‐PRS and East Asian NAFLD‐PRS with HCC risk, allowing us to compare PRSs developed in different ancestral populations. Our study also has several limitations. While we overcame the limitation of different ancestral populations by calculating the East Asian NAFLD‐PRS, the published genetic associations we used to calculate the PRS came from a Japanese population.[ 28 ] While we do not expect significant differences in these weights between Han Chinese and Japanese, some differences may exist. Because our study design uses NAFLD exposures from other cohorts, this limits our ability to account for liver‐disease risk factors such as fibrosis. The standard errors of the SNPs included in the HFC‐PRS were not publicly available; therefore, the MR analysis was performed only for the NAFLD‐PRS. We did not have reliable data available on the HSD17B13 SNP, rs72613567, which was studied as part of the PRS‐5 score in Bianco et al. Therefore, we were unable to examine the association between PRS‐5 and HCC risk in the present study. Additionally, while we have shown that different PRSs are associated with HCC in East Asians, more research is needed to determine whether this finding is applicable to other ancestral groups.
In conclusion, we found that PRSs for either hepatic fat or NAFLD based on either American or East Asian populations were associated with HCC incidence. An MR analysis, in which key assumptions were met, allows for the causal inference of a relationship between NAFLD and HCC in East Asians. NAFLD or hepatic fat PRSs may improve HCC risk stratification and be applicable across different populations.
CONFLICT OF INTEREST
Nothing to report.
AUTHOR CONTRIBUTIONS
Study concept: Claire E. Thomas, Brenda Diergaarde, Allison L. Kuipers, Jennifer J. Adibi, Hung N. Luu, and Jian‐Min Yuan. Formal analysis: Claire E. Thomas, Renwei Wang, Aizhen Jin, and Jian‐Min Yuan. Visualization: Claire E. Thomas. Manuscript draft, reviewing, and editing: Claire E. Thomas, Brenda Diergaarde, Allison L. Kuipers, Hung N. Luu, Xuling Chang, Rajkumar Dorajoo, Chew‐Kiat Heng, Chiea‐Chuen Khor, Woon‐Puay Koh, and Jian‐Min Yuan. Methodology: Brenda Diergaarde, Allison L. Kuipers, and Jennifer J. Adibi. Data curation: Xuling Chang, Rajkumar Dorajoo, Chew‐Kiat Heng, Chiea‐Chuen Khor, Woon‐Puay Koh, and Jian‐Min Yuan. Resources: Woon‐Puay Koh and Jian‐Min Yuan. Funding acquisition and study supervision: Jian‐Min Yuan.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The Singapore Chinese Health Study has been approved by the institutional review boards of the National University of Singapore and the University of Pittsburgh. The present study was approved by the institutional review board of the University of Pittsburgh. Informed consent was obtained from all participants, and this study was performed in accordance with the Declaration of Helsinki.
Supporting information
Appendix S1 Supporting Information
ACKNOWLEDGMENT
We thank the Singapore Cancer Registry for the identification of incident cancer cases among participants of the Singapore Chinese Health Study, and Siew‐Hong Low of the National University of Singapore for supervising the fieldwork of the Singapore Chinese Health Study. This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided.
Thomas CE, Diergaarde B, Kuipers AL, Adibi JJ, Luu HN, Chang X, NAFLD polygenic risk score and risk of hepatocellular carcinoma in an East Asian population. Hepatol Commun. 2022;6:2310–2321. 10.1002/hep4.1976
Funding informationThe Singapore Chinese Health Study was supported by grants from the National Medical Research Council, Singapore (NMRC/CIRG/1456/2016); the National Research Foundation, Singapore (Project Number 370062002); and the U.S. National Institutes of Health (NIH) (R01 CA144034 and UM1 CA182876). C.E.T. is supported through an NIH training grant (T32 CA186873). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institutes or the National Institutes of Health. This research was supported in part by Cancer Center Support Grant from the National Cancer Institute (P30 CA047904; Robert L. Ferris, principal investigator).
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Pineros M, Anaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149(4):778–89. 10.1002/ijc.33588. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2. Wong S, Ting Y, Chan W. Epidemiology of non‐alcoholic fatty liver disease‐related hepatocellular carcinoma and its implications. JGH Open. 2018;2(5):235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fan JG, Kim SU, Wong VWS. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67(4):862–73. [DOI] [PubMed] [Google Scholar]
- 4. Shen J, Wong GLH, Chan HLY, Chan HY, Yeung DKW, Chan RSM, et al. PNPLA3 gene polymorphism accounts for fatty liver in community subjects without metabolic syndrome. Aliment Pharmacol Ther. 2014;39(5):532–9. [DOI] [PubMed] [Google Scholar]
- 5. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–90. [DOI] [PubMed] [Google Scholar]
- 6. Dongiovanni P, Stender S, Pietrelli A, Mancina RM, Cespiati A, Petta S, et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018;283(4):356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bianco C, Jamialahmadi O, Pelusi S, Baselli G, Dongiovanni P, Zanoni I, et al. Non‐invasive stratification of hepatocellular carcinoma risk in non‐alcoholic fatty liver using polygenic risk scores. J Hepatol. 2021;74(4):775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan JM, Stram DO, Arakawa K, Lee HP, Yu MC. Dietary cryptoxanthin and reduced risk of lung cancer: the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2003;12(9):890–8. [PubMed] [Google Scholar]
- 9. Hankin JH, Stram DO, Arakawa K, Park S, Low SH, Lee HP, et al. Singapore Chinese Health Study: development, validation, and calibration of the quantitative food frequency questionnaire. Nutr Cancer. 2001;39(2):187–95. [DOI] [PubMed] [Google Scholar]
- 10. Thomas CE, Luu HN, Wang R, Xie G, Adams‐Haduch J, Jin A, et al. Association between pre‐diagnostic serum bile acids and hepatocellular carcinoma: the Singapore Chinese Health Study. Cancers (Basel). 2021;13(11):2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koh WP, Robien K, Wang R, Govindarajan S, Yuan JM, Yu MC. Smoking as an independent risk factor for hepatocellular carcinoma: the Singapore Chinese Health Study. Br J Cancer. 2011;105(9):1430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan JM, Ross RK, Stanczyk FZ, Govindarajan S, Gao YT, Henderson BE, et al. A cohort study of serum testosterone and hepatocellular carcinoma in Shanghai, China. Int J Cancer. 1995;63(4):491–3. [DOI] [PubMed] [Google Scholar]
- 13. Parkin D, Whelan S, Ferlay J, Teppo L, Thomas D. Cancer Incidence in Five continents, Volume 8. Lyon: IARC Press; 2002. [Google Scholar]
- 14. Chang X, Chua KY, Wang L, Liu J, Yuan JM, Khor CC, et al. Midlife leukocyte telomere length as an indicator for handgrip strength in late life. J Gerontol A Biol Sci Med Sci. 2021;76(1):172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang X, Gurung RL, Wang L, Jin A, Li Z, Wang R, et al. Low frequency variants associated with leukocyte telomere length in the Singapore Chinese population. Commun Biol. 2021;4(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dorajoo R, Sun Y, Han Y, Ke T, Burger A, Chang X, et al. A genome‐wide association study of n‐3 and n‐6 plasma fatty acids in a Singaporean Chinese population. Genes Nutr. 2015;10(6):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics. 2016;32(20):3207–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype‐phenotype associations. Bioinformatics. 2019;35(22):4851–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42(4):1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsong WH, Koh WP, Yuan JM, Wang R, Sun CL, Yu MC. Cigarettes and alcohol in relation to colorectal cancer: the Singapore Chinese Health Study. Br J Cancer. 2007;96(5):821–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR‐Egger method. Eur J Epidemiol. 2017;32(5):377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11(1):376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai G, Liu P, Li X, Zhou X, He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: a meta‐analysis. Medicine (Baltimore). 2019;98(7):e14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Low S, Chin MC, Ma S, Heng D, Deurenberg‐Yap M. Rationale for redefining obesity in Asians. Ann Acad Med Singapore. 2009;38(1):66–9. [PubMed] [Google Scholar]
- 28. Kawaguchi T, Shima T, Mizuno M, Mitsumoto Y, Umemura A, Kanbara Y, et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS One. 2018;13(1):e0185490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgess S, Thompson SG. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. Boca Raton, Florida: CRC Press; 2015. [Google Scholar]
- 30. Orho‐Melander M, Melander O, Guiducci C, Perez‐Martinez P, Corella D, Roos C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C‐reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57(11):3112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaxillaire M, Cavalcanti‐Proença C, Dechaume A, Tichet J, Marre M, Balkau B, et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57(8):2253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beer NL, Tribble ND, McCulloch LJ, Roos C, PRV J, Orho‐Melander M, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18(21):4081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


