Abstract
The clustered prnB, prnC, and prnD genes are repressed by the simultaneous presence of glucose and ammonium. A derepressed mutation inactivating a CreA-binding site acts in cis only on the permease gene (prnB) while derepression of prnD and prnC is largely the result of reversal of inducer exclusion.
All genes involved in the utilization of proline in Aspergillus nidulans are clustered in chromosome VII (Fig. 1). prnB encodes the specific proline permease, prnD encodes proline oxidase, and prnC encodes l-Δ1-pyrroline carboxylate dehydrogenase (11). prnA encodes a Zn-binuclear cluster transcriptional activator mediating proline induction (5, 14). prnX is a gene of unknown function whose inactivation does not affect proline utilization (8).
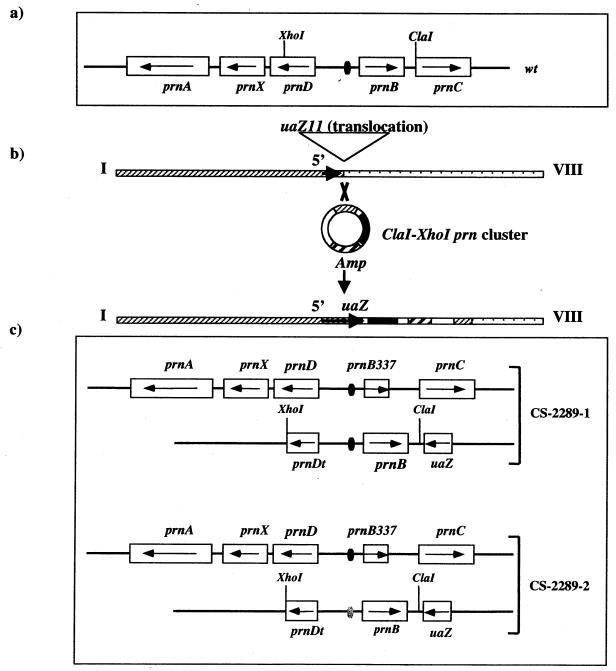
FIG. 1.
(a) Schematic representation of prn cluster. The direction of transcription of the genes (according to references 8, 9, 10, and 16) is indicated by arrows. (b) Strategy used for the construction of strains CS-2289-1 and CS-2289-2. Strain uaZ11 prnB337 pabaA1 riboB2 yA2 was transformed with the plasmids pXCwt and pXCprnd22. Both plasmids contain the 3′ end of the uaZ gene and the XhoI-ClaI region of the prn cluster. pXCwt contains a wild-type region, and pXCprnd22 contains a region carrying a mutation in an essential CreA-binding site (6). (c) Schematic representation of strains CS-2289-1 and CS-2289-2. For each strain, the upper scheme shows the resident prn cluster, and the lower scheme shows the prn sequences integrated at the uaZ locus. The XhoI-ClaI fragment of prn cluster integrated at the uaZ locus contains a truncated copy of prnD gene (prnDt), the prnD-B intergenic region, the prnB gene, and the prnB-C intergenic region. The resident prn locus carries the prnB337 deletion. The CreA-binding sites active in repression are represented as ovals: darks ovals for wild-type sites and a white oval for the prnd-22 mutant site.
The structural genes of the proline utilization cluster are subject to specific induction by proline and to metabolite repression. Carbon repression is mediated by CreA, a negative-acting zinc finger protein (4, 7). Nitrogen derepression is mediated by AreA, a positive-acting transcription factor belonging to the GATA family (1, 12). It was shown many years ago that these genes are repressed significantly only when both repressing carbon and nitrogen sources are present, i.e., in the simultaneous presence of ammonium and glucose (8). We have shown that this process operates at the level of the steady state of the cognate mRNAs (5, 16) (Fig. 2A). Other data indicate that the repression process operates at the level of transcription rather than at the level of mRNA stability (6). We have shown that repression operates directly on the prnB gene and that the expression of the prnA gene is not affected by repression (5, 6, 16). A model accounting for these findings has been presented (9). Mutation in any or both of two specific CreA binding sites (prnd), located between the prnD and prnB intergenic region, results in derepression of prnB, prnC, and prnD expression (5, 6, 16) (Fig. 2B).
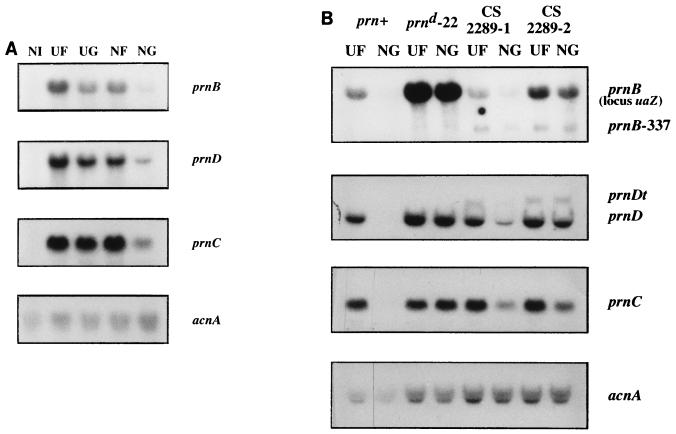
FIG. 2.
Northern blot analysis of prn genes transcription. (A) Expression of the prnB, prnD, and prnC genes in a wild-type strain grown under different conditions. Noninduced levels and all possible combinations of carbon and nitrogen repressing conditions are shown. (B) mRNA levels of the prn genes in a strain carrying prnd22 and in strains CS-2289-1 and CS-2289-2. Only two conditions are shown: induced nonrepression (UF) and induced double repression (NG). mRNAs extracted from a wild-type strain grown under identical conditions are also shown. Less RNA, as shown by the acnA panels, has been loaded for the latter, but the strong repression afforded by the simultaneous presence of glucose and ammonium is clearly visible and can be compared with that of the equivalent mRNAs of panel A. Mycelia were grown for 8 h at 37°C in 0.1% fructose and 5 mM urea and then were either left noninduced (NI) or induced with 20 mM l-proline (UF). Simultaneously, they either were left nonrepressed (UF) or were glucose repressed (1% glucose, 5 mM urea) (UG), nitrogen repressed [20 mM diammonium d-(+)-tartrate, 0.1% fructose] (NF), or carbon and nitrogen repressed [1% glucose, 20 mM diammonium d-(+)-tartrate] (NG) for 2 h at 37°C. The membranes have been probed for prnB, prnD, prnC, and acnA (actin), the latter as a control of RNA loading. The methods used for RNA preparation were those of González et al. (9).
A. nidulans strains lacking areA cannot utilize proline as a nitrogen source in the presence of glucose as the sole carbon source. The prnd mutations suppress this phenotype. Arst and collaborators have taken advantage of this phenotype to show that a mutation in what we now know to be one of the two physiologically essential CreA-binding sites (6) is cis dominant and trans recessive in relation to prnB but is cis and trans dominant in relation to prnC and prnD (3). The cis-trans test was carried out by checking the growth on proline of a homozygous areA diploid mutant carrying a prnd mutation placed either cis or trans to mutations in each of the structural genes. There are two ways to explain the results of the cis-trans test. The first explanation is that there is no direct repression of prnD and/or prnC, but that the apparent repression of these genes is the result of inducer exclusion due to the repression of the prnB-encoded permease by glucose, and their apparent derepression is the result of reversal of inducer exclusion, acting via the derepression of prnB. The second is that the CreA-binding sites in the prnD-prnB intergenic region directly affect the expression of prnD and/or prnC, but that the only limiting step for growth in proline under repressing conditions is the expression of the permease coded by the prnB gene. In this note, we demonstrate that inducer exclusion is largely and perhaps exclusively responsible for prnD and, to a lesser extent, prnC repression.
The strategy we have used to this aim is shown in Fig. 1. In a strain carrying an internal deletion of prnB, prnB337 resulting in a null phenotype for proline uptake (17), we have inserted a fragment of the prn cluster in trans. This fragment includes the whole wild-type prnB gene, the entire prnB-prnD intergenic region, and also a short (902-bp) segment of the prnD gene. Two strains were constructed, one carries the wild-type repressible promoter, the other carries a promoter containing a mutation in one of the essential CreA-binding sites, prnd22 (6, 16). This mutation changes the canonical CreA-binding site 5′CTGGGG into 5′CTGAGG. This base pair change is sufficient to prevent all binding of this site to CreA in vitro (6). uaZ11 is a I/VIII chromosomal translocation, which splits the uaZ gene (13). This null mutation results in the inability to utilize uric acid as the sole nitrogen source. Transformants able to utilize uric acid can be selected by transforming uaZ11 strains with a plasmid containing the 3′ uaZ moiety, including an overlap with the 5′ moiety. Thus, transforming sequences are always targeted to the 5′ moiety of the uaZ gene. The prn transgenes were integrated adjacent to the uaZ locus by transforming a prnB337 uaZ11 strain (see the legend to Fig. 1 for the complete genotype) with plasmids pXCwt and pXCprnd22 (Fig. 1). We can thus directly investigate the levels of expression of the prnD and prnC genes in trans to the promoter region driving the active prnB gene.
The results of this investigation are shown in Fig. 2B. (i) In spite of the presence of the complete prnD-prnB intergenic region, the prnB transgene is expressed less than the corresponding sequences at the prn locus. This remains unexplained. Nevertheless, the ratio between derepressed and repressed levels in the wild-type promoter and the ratios between the levels of the wild type and those of the prnd22 mutant are the same for the gene in the cluster and for the transgene. (ii) The prnd22 mutation in the resident cluster results in derepression of prnB, prnC, and prnD, as previously described by Sophianopoulou et al. (16). The same effect is seen on the prnB transgene. (iii) In the derepressed mutant, the steady-state levels of prnB are higher than in the nonderepressed (wild-type promoter) strain even under conditions of derepression for the latter. It could be said that the prnd22 mutation results also in an “up-promoter” effect. This is true both for the transgene and for the resident locus (16). (iv) We know from classical genetic tests that prnd mutations are cis dominant and trans recessive in relation to prnB (3). However, the level of the short prnB message transcribed from the resident gene carrying a deletion mutation is not repressed when in trans with the prnd22 mutation. This could be expected if overexpression of the prnB transgene resulted in inducer accumulation and if this accumulation could partially bypass carbon and nitrogen metabolite repression. Alternatively or additionally, the small residual message may be more stable than the wild-type prnB message. (v) The crucial observation is that the introduction of the prnd22 mutation results in complete derepression of prnD in trans and a considerable derepression of prnC in trans. The mRNA of the truncated prnD transgene follows the same repression pattern as the wild-type mRNA from the resident cluster. The conclusion is that inducer exclusion accounts for the carbon and nitrogen catabolite repression of prnD and prnC. (vi) For prnC, however, derepression in trans seems less pronounced than derepression in cis. Northern blots provide semiquantitative estimates, and we would not wish to claim that this small difference is significant. However, a clear effect at a distance of sequences in the prnD-prnB intergenic region on the expression of prnC was demonstrated long ago with classical genetics (2). These results have now been confirmed at the level of the expression of the prnC mRNA (15) (D. Gómez and C. Scazzocchio, unpublished results). The region involved has been mapped (D. Gómez and C. Scazzocchio, unpublished results) and coincides with a region involved in the integration of carbon and nitrogen metabolite repression (9). This may suggest that while inducer exclusion is the main parameter involved in the repression of prnD, a moderate cis-acting effect of the prnD-prnB intergenic region might also be exerted on the expression of prnC.
Acknowledgments
This work was supported by European Union grants BIO2-CT93-0147 and BIO-CT96-0535. D.G. received a research studentship from the Ministère de l'Education National, de l'Enseignement Superieur et de la Recherche of the French Government.
REFERENCES
- 1.Arst H N, Jr, Cove D J. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- 2.Arst H N, Jr, MacDonald D W. Reduced expression of a distal gene of the prn cluster in deletion mutants of Aspergillus nidulans: genetics evidence for a dicistronic messenger in an eukaryote. Mol Gen Genet. 1978;163:17–22. doi: 10.1007/BF00268959. [DOI] [PubMed] [Google Scholar]
- 3.Arst H N, Jr, MacDonald D W, Jones S A. Regulation of proline transport in Aspergillus nidulans. J Gen Microbiol. 1980;116:285–294. [Google Scholar]
- 4.Bailey C, Arst H N., Jr Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem. 1975;51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- 5.Cazelle B, Pokorska A, Hull E, Green P M, Stanway G, Scazzocchio C. Sequence, intron-exon organization, transcription and mutational analysis of prnA, the gene encoding the transcriptional activator of the prn gene cluster in Aspergillus nidulans. Mol Microbiol. 1998;28:355–370. doi: 10.1046/j.1365-2958.1998.00801.x. [DOI] [PubMed] [Google Scholar]
- 6.Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CreA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavrias V. Etudes moleculaires sur la régulation et la structure du groupe des gènes prn chez Aspergillus nidulans. Ph.D. thesis. Orsay, France: Université Paris-Sud; 1992. [Google Scholar]
- 9.González R, Gavrias V, Gómez D, Scazzocchio C, Cubero B. The integration of nitrogen and carbon catabolite repression in Aspergillus nidulans requires the GATA factor AreA and an additional positive-acting element, ADA. EMBO J. 1997;16:2937–2944. doi: 10.1093/emboj/16.10.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hull E P, Green P M, Arst H N, Jr, Scazzocchio C. Cloning and physical characterisation of the l-proline catabolism gene cluster of Aspergillus nidulans. Mol Microbiol. 1989;3:553–559. doi: 10.1111/j.1365-2958.1989.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 11.Jones S A, Arst H N, Jr, MacDonald D W. Gene roles in the prn cluster of Aspergillus nidulans. Curr Genet. 1981;3:49–56. doi: 10.1007/BF00419580. [DOI] [PubMed] [Google Scholar]
- 12.Kudla B, Caddick M X, Langdon T, Martínez-Rossi N M, Bennett C F, Sibley S, Davies R W, Arst H N., Jr The regulatory gene areA mediating nitrogen repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oestreicher N, Sealy-Lewis H M, Scazzocchio C. Characterisation, cloning and integrative properties of the gene encoding urate oxidase in Aspergillus nidulans. Gene. 1993;132:185–192. doi: 10.1016/0378-1119(93)90194-8. [DOI] [PubMed] [Google Scholar]
- 14.Sharma K K, Arst H N., Jr The product of the regulatory gene of the proline catabolism gene cluster in Aspergillus nidulans is a positive-acting protein. Curr Genet. 1985;9:299–304. doi: 10.1007/BF00419959. [DOI] [PubMed] [Google Scholar]
- 15.Sophianopoulou V. Le groupe des gènes prn impliqués dans la degradation de la l-proline chez Aspergillus nidulans. Ph.D. thesis. Orsay, France: Université Paris-Sud; 1989. [Google Scholar]
- 16.Sophianopoulou V, Suárez T, Diallinas G, Scazzocchio C. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol Gen Genet. 1993;236:209–213. doi: 10.1007/BF00277114. [DOI] [PubMed] [Google Scholar]
- 17.Tazebay U H, Sophianopoulou V, Cubero B, Scazzocchio C, Diallinas G. Post-transcriptional control and kinetic characterization of proline transport in germinating conidiospores of Aspergillus nidulans. FEMS Microbiol Lett. 1995;132:27–37. doi: 10.1111/j.1574-6968.1995.tb07806.x. [DOI] [PubMed] [Google Scholar]