Abstract
Work presented here establishes a connection between cellular coenzyme A (CoA) levels and thiamine biosynthesis in Salmonella enterica serovar Typhimurium. Prior work showed that panE mutants (panE encodes ketopantoate reductase) had a conditional requirement for thiamine or pantothenate. Data presented herein show that the nutritional requirement of panE mutants for either thiamine or pantothenate is manifest only when flux through the purine biosynthetic pathway is reduced. Further, the data show that under the above conditions it is the lack of thiamine pyrophosphate, and not decreased CoA levels, that directly prevents growth.
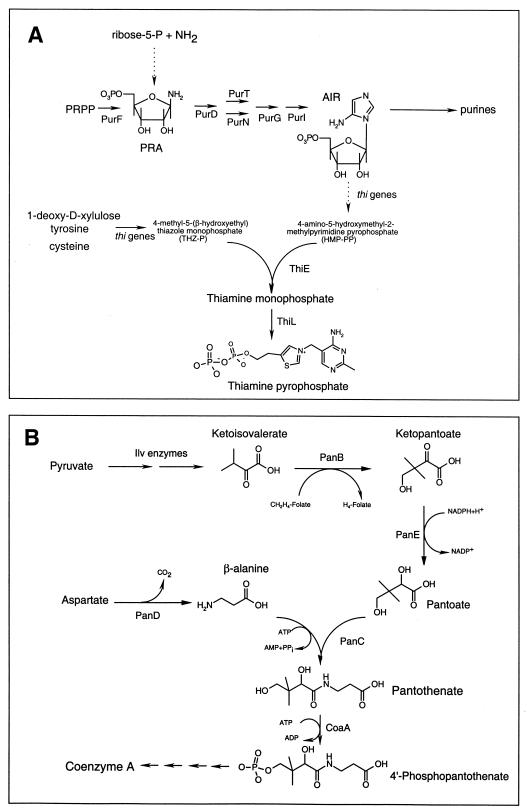
Thiamine pyrophosphate (TPP) is a required cofactor in many reactions, including those involving the removal or transfer of C2 units. TPP is synthesized de novo through the condensation of two independently synthesized compounds, 4-methyl-5-(β-hydroxyethyl)thiazole phosphate and 4-amino-5-hydroxymethyl-2-methyl-pyrimidine pyrophosphate (HMP-PP) (4). Our current understanding of thiamine biosynthesis is outlined in Fig. 1. Relevant to this work is that HMP-PP is synthesized from aminoimidazole ribotide (AIR), an intermediate in de novo purine biosynthesis. AIR is synthesized by using the first five enzymes in purine biosynthesis (20–22) or, in the absence of the first enzyme (PurF), by an uncharacterized phosphoribosylamine (PRA)-forming activity (10, 12, 14). In the latter case, sufficient PRA is generated to allow thiamine but not purine synthesis. The mechanism of PRA formation in a purF mutant has been refractile to genetic and biochemical analyses and has been proposed to involve contributions by several enzymes with primary roles elsewhere in metabolism.
FIG. 1.
Relevant metabolic pathways. (A) Our present understanding of thiamine biosynthesis is illustrated. Where known, the enzyme responsible for each step is indicated. Dotted arrows represent biochemical reactions that have not been characterized. (B) The biosynthetic pathways contributing to coenzyme A synthesis are illustrated. Where relevant and known, the enzyme catalyzing each reaction is indicated.
Lesions in several loci result in inability, or reduced ability, of the cell to synthesize thiamine in the absence of PurF (2, 3, 6, 11, 23, 24). Characterization of several of these loci has resulted in the conclusion that with the exception of PRA generation, thiamine synthesis is mechanistically similar with and without PurF (2, 6, 14, 16, 23). Therefore, to explain the finding that several mutant loci prevent thiamine synthesis in a purF strain but not in wild-type strains, we suggest that the metabolic status of a purF mutant (i.e., low flux through purine biosynthesis) renders thiamine synthesis more sensitive to disruption.
Mutations in the pantothenate biosynthetic gene panE (which encodes ketopantoate reductase) were recently shown to cause a requirement for either thiamine or pantothenate under some growth conditions (11, 17; J. Zilles, J. Kappock, J. Stubbe, and D. M. Downs, unpublished data). It is important to point out that panE mutants have a wild-type growth rate on minimal medium. This growth is possible because an enzyme in isoleucine biosynthesis, acetohydroxy acid isomeroreductase (IlvC), can reduce sufficient ketopantoate for pantothenate biosynthesis (Fig. 1B) (25).
The work presented here was pursued in order to understand the connection between pantothenate, coenzyme A (CoA), and thiamine synthesis. In the literature, several references have been made to a phenotypic connection between pantothenate and thiamine (9, 11, 19). Results presented herein demonstrate that the nutritional requirement of panE mutants results from reduced flux through the purine biosynthetic pathway. Further, the data show that under conditions of low purine flux, there is an increased requirement for CoA (or an intermediate between pantothenate and CoA) in thiamine synthesis.
Levels of CoA are reduced in panE mutants.
Table 1 contains data showing growth rates on different media, and the resulting CoA and TPP levels, in a panE mutant and a wild-type strain. Significant aspects of these data are discussed below. The TPP concentrations of cells grown in exogenous thiamine are not reported but were routinely >100-fold higher than those measured in cells grown in medium lacking exogenous thiamine. As expected, exogenous pantothenate resulted in up to a twofold increase of CoA levels even in the wild-type strain (7).
TABLE 1.
TPP and CoA pools in wild-type and panE mutant strains
| Strain | Addition(s)a | Level (avg ± SD) ofb:
|
Growth rate (μ)c | |
|---|---|---|---|---|
| TPP | CoA | |||
| Wild type (LT2) | ||||
| 1 | None | 62.4 ± 22.4 | 0.189 ± .003 | 0.48 ± 0.0 |
| 2 | Thi | NA | 0.186 ± .005 | 0.48 ± 0.01 |
| 3 | Pan | 53.8 ± 1.0 | 0.211 ± .023 | 0.46 ± 0.01 |
| 4 | Ade | 38.7 ± 13.0 | 0.149 ± .017 | 0.49 ± 0.01 |
| 5 | Ade-Thi | NA | 0.148 ± .002 | 0.46 ± 0.02 |
| 6 | Ade-Pan | 43.2 ± 15.1 | 0.240 ± .024 | 0.47 ± 0.01 |
| panE1::MudJ (DM63) | ||||
| 7 | None | 58.0 ± 13.6 | 0.023 ± .004 | 0.45 ± 0.04 |
| 8 | Thi | NA | 0.044 ± .017 | 0.48 ± 0.01 |
| 9 | Pan | 44.9 ± 9.8 | 0.164 ± .014 | 0.44 ± 0.02 |
| 10 | Ade | 14.5 ± 3.6 | 0.032 ± .011 | 0.25 ± 0.01 |
| 11 | Ade-Thi | NA | 0.023 ± .005 | 0.43 ± 0.05 |
| 12 | Ade-Pan | 40.1 ± 10.0 | 0.166 ± .016 | 0.49 ± 0.05 |
Abbreviations: Ade, adenine; Thi, thiamine; Pan, pantothenate.
TPP levels are expressed as picomoles per milligram (dry weight) of cells (29), and CoA levels are expressed as nanomoles per milligram (dry weight) of cells (1, 15). Data are averages of three independent experiments. NA, not applicable.
Specific growth rates were determined as μ = ln(X/Xo)/T, as previously described (24). Growth curves were performed with three independent innocula, and the rates were averaged.
(i) Minimal medium.
When grown on minimal medium, panE mutants have no detectable growth defect, since the IlvC enzyme is able to reduce ketopantoate for pantothenate biosynthesis (25). However, the contribution of PanE to pantothenate and/or CoA biosynthesis is reflected by the eightfold reduction in total CoA levels in panE mutants compared to the isogenic wild-type strain (Table 1, lines 1 and 7). This result indicates that the cell maintains a CoA level in excess of that required for full growth in unsupplemented medium and represents the first report of an upper limit of the CoA level required for efficient prototrophic growth in Salmonella enterica serovar Typhimurium.
(ii) Minimal adenine medium.
Recent work has shown that the presence of adenine in the medium reduces the cellular pool of TPP (14). Phenotypic analyses and labeling studies suggest that this reduction is due primarily to a decrease in flux through the purine biosynthetic pathway (26; Zilles et al., unpublished data). A purF panE double mutant was unable to grow in the absence of adenine and either pantothenate or thiamine (11, 17). Consistent with previous work, we found that mutants defective in panE had a significantly reduced growth rate in medium supplemented with adenine containing gluconate as the carbon and energy source (Table 1, line 10). Taken together, these results suggest that reduced flux through the purine biosynthetic pathway is responsible for generating the nutritional requirement in a panE mutant.
Growth of a panE strain depends on adequate TPP levels.
On medium containing adenine, panE mutants had lowered levels of both CoA and TPP compared to a wild-type strain (Table 1, lines 4 and 10). Since the nutritional requirement of a panE mutant could be satisfied by either pantothenate or thiamine, we considered two possibilities. Either one, but not both, of the respective metabolite pools could be reduced without affecting growth, or the reduction in one pool could lead to a reduction in the other. If the former possibility was correct, addition of either supplement would restore its respective metabolite pool as well as the growth rate. If the latter possibility was true, we expected that addition of the primary metabolite would restore both the growth rate and the levels of both metabolites.
The data shown in Table 1 are consistent with the second possibility above. Although addition of thiamine to the growth medium of a panE mutant restored a wild-type growth rate and elevated the TPP levels, it failed to result in increased CoA levels. In contrast, the addition of pantothenate restored the wild-type growth rate and increased the levels of both relevant metabolites. From this result, we conclude that the decreased CoA levels caused by a panE mutation prevented thiamine synthesis when flux through the purine pathway was reduced. This conclusion is consistent with the ability of a panE mutant to grow prototrophically when flux through the purine pathway is elevated and CoA levels remain reduced.
Conditions that decrease the rate of pantothenate biosynthesis cause a conditional requirement for TPP.
If the conclusion in the previous section is correct, any strain with lowered pantothenate biosynthesis should display a thiamine requirement when there is low flux through the purine pathway. Since a panE mutant is only partially blocked in pantothenate biosynthesis, testing of this hypothesis required conditions that would also partially block pantoate biosynthesis. A temperature-sensitive mutation in panB (which encodes ketopantoate hydroxymethyltransferase [Fig. 1B]) was utilized to generate such a block. Since the CoA level in strain DM374 [panB628(Ts)] was inversely proportional to growth temperature (Table 2) while the isogenic wild-type strain had constant CoA levels, we concluded that the panB628(Ts) allele resulted in a partial block of pantothenate biosynthesis at intermediate temperatures.
TABLE 2.
CoA levels change as a function of growth temperature in a mutant carrying allele panB628(Ts)
| Temp (°C) | Strain | panB allele | CoA levela | Ratio of CoA levels [wild type/ panB628(Ts)] |
|---|---|---|---|---|
| 30 | LT2 | Wild type | 0.227 ± 0.062 | |
| DM374 | panB628(Ts) | 0.068 ± 0.002 | 3.33 | |
| 32 | LT2 | Wild type | 0.231 ± 0.027 | |
| DM374 | panB628(Ts) | 0.053 ± 0.005 | 4.35 | |
| 35 | LT2 | Wild type | 0.258 ± 0.084 | |
| DM374 | panB628(Ts) | 0.029 ± 0.016 | 8.90 |
Strain DM4961 [purF2085 panB628(Ts)] was constructed to test if thiamine synthesis was prevented when flux through the purine pathway was reduced. Growth of this strain was monitored over a temperature range from 30 to 37°C in gluconate medium containing adenine, adenine and pantothenate, or adenine and thiamine. Representative growth rates from these experiments are shown in Table 3. These data demonstrate that an intermediate temperature (32°C) could generate the anticipated phenotype in the panB(Ts) purF mutant. At 30°C (Table 3, line 1) this strain showed wild-type growth behavior, while at 32°C (line 2) a clear requirement for thiamine or pantothenate was detected, similar to that described for the purF panE mutant (11). As expected, at higher temperatures only pantothenate restored the full growth of strain DM4961, consistent with other metabolic requirements becoming unmasked by the progressive reduction of CoA levels. These results confirm that a partial block in pantothenate biosynthesis generates a thiamine requirement when flux through the purine biosynthetic pathway is reduced.
TABLE 3.
panB628(Ts) generates a temperature-conditional thiamine auxotrophy
| Temp (°C) | Specific growth rate (μ) on minimal medium witha:
|
||
|---|---|---|---|
| Ade | Ade-Thi | Ade-Pan | |
| 30 | 0.34 | 0.45 | 0.46 |
| 32 | 0.12 | 0.32 | 0.50 |
| 35 | 0.07 | 0.12 | 0.42 |
| 37 | 0.03 | 0.13 | 0.50 |
Medium contained gluconate (22 mM) as the sole carbon and energy source plus the indicated additions. Data are for growth of strain DM4961 [purF2085 panE628(Ts)] at the indicated temperatures and are from a representative experiment. Growth curves were performed as described previously (24) with the specific growth rate determined as μ = ln (X/Xo)/T. Abbreviations: Ade, adenine; Thi, thiamine; Pan, pantothenate.
Low CoA, not pantothenate, levels affect thiamine synthesis.
Results in the previous sections were consistent with the hypothesis that low CoA levels prevent thiamine synthesis when flux through the purine pathway is significantly reduced. However, it remained formally possible that pantothenate played a role in thiamine synthesis in addition to elevating CoA levels. This was an important distinction, since the above experiments utilized exogenous pantothenate to alter CoA levels.
To investigate the effect of decreased CoA levels on thiamine biosynthesis, we constructed a strain that had reduced CoA levels independently of pantothenate biosynthesis. Pantothenate kinase, the product of the coaA gene, is an essential enzyme and is the first dedicated step in CoA biosynthesis (Fig. 1B). Past work with Escherichia coli characterized conditional coaA mutants and showed that these mutations decrease the levels of CoA and simultaneously elevate the levels of endogenous pantothenate (28). We isolated a conditional (temperature-sensitive) coaA mutant in serovar Typhimurium. This mutant was determined to be defective in the coaA gene based on the following criteria: (i) the mutant displayed wild-type growth on minimal or rich medium at 30°C and lack of growth on either medium at 42°C, (ii) the mutant had fourfold-reduced pantothenate kinase activity in cell extracts at 40°C (33 versus 145 pmol/min/mg of protein) (13), and (iii) the lesions mapped to the expected position in the chromosome and were complemented by a plasmid that carried only the wild-type allele of coaA from E. coli (27) (data not shown).
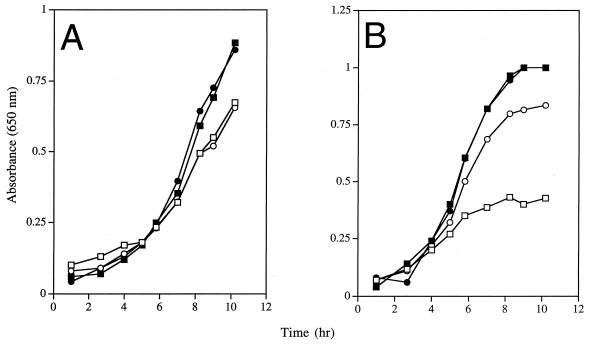
The coaA(Ts) mutation was transduced into DM1936 (purF2085), generating the isogenic strains DM5242 [purF2085 zii-8038::Tn10d(Tc) coaA1(Ts)] and DM5243 [purF2085 zii-8038::Tn10d(Tc)]. The thiamine requirements of these two strains were determined at 30 and 34°C, and representative results are shown in Fig. 2. Two points could be made from these data. First, the growth rates of the two strains were indistinguishable (μ ≅ 0.29) at either temperature when thiamine was present in the medium, demonstrating that the coaA mutation did not adversely affect normal metabolism at these temperatures. Second, the purF coaA+ mutant was able to grow in the absence of thiamine at both temperatures (μ = 0.28) due to the PurF-independent thiamine synthesis known to occur (24). Significantly, the purF coaA(Ts) mutant became a thiamine auxotroph at 34°C (μ = 0.14), exhibiting a requirement for thiamine that could not be satisfied by pantothenate. This experiment effectively uncoupled exogenous pantothenate from endogenous CoA levels. Thus, this result allows us to conclude that a reduced level of CoA, or a metabolite between pantothenate and CoA, is responsible for the thiamine requirement of panE mutants when flux through the purine pathway is reduced.
FIG. 2.
Low CoA levels result in thiamine auxotrophy in a purF background. Growth curves were performed as described previously (24). Cultures depicted in panel A were grown at 30°C; those in panel B were grown at 34°C. Squares represent strain DM5342 [purF2085 zii-8038::Tn10d(Tc) coaA1(Ts)]; circles represent strain DM5343 [purF2085 zii-8038::Tn10d(Tc)]. Strains were grown in medium with gluconate as the sole carbon and energy source plus adenine in the absence (open symbols) or presence (solid symbols) of exogenous thiamine.
Summary.
The work presented here has made three contributions to our understanding of metabolism in Salmonella serovar Typhimurium. First, experiments herein have defined an upper limit for the CoA levels required for prototrophic growth, since panE mutants with an eightfold reduction in CoA levels had no apparent growth defect on minimal medium. Second, data from this work have validated the hypothesis that the metabolic requirements for thiamine synthesis differ depending on the level of flux through the purine biosynthetic pathway. We have demonstrated that while a reduction in cellular CoA levels does not normally interfere with thiamine synthesis, the combination of low CoA levels plus reduced flux through the purine pathway generates a defect in thiamine synthesis. We suggest that this result implicates CoA (or an intermediate between pantothenate and CoA) in thiamine synthesis, at least under conditions of low purine flux. There are various ways to incorporate this finding into a hypothesis. The model we favor is that a CoA thioester is involved in modifying (e.g., succinylating or acetylating) an intermediate in thiamine synthesis and that this modification increases the efficiency of the respective enzymatic reaction. There is precedent for an enzyme favoring a modified substrate while being able to function poorly with an unmodified substrate (e.g., ornithine transaminase [5, 8]). Such a model would predict that modification is not critical when substrate (AIR) levels are high but becomes critical for efficient conversion of AIR to HMP when substrate levels are limiting due to reduced flux through the purine biosynthetic pathway. Work is under way to characterize ThiC, the protein predicted to catalyze the conversion of AIR to HMP, and to begin addressing the hypothesis presented by this work.
Finally, we have found that the first metabolic defect resulting from a reduction in CoA levels is the conditional thiamine requirement. This result differs from a previous report for E. coli, which indicated that growth stasis in CoA-depleted strains was due to the inability to synthesize amino acids and proteins (18). Two points should be made about the difference in experimental conditions: in the earlier experiments, thiamine was routinely added to the medium and, since flux through the purine pathway was at wild-type levels, no thiamine requirement would be expected.
Acknowledgments
This work was supported by competitive grant GM47296 from the NIH. A.R. is supported by NIH Predoctoral Individual National Research Service Award F31 AI09638.
We acknowledge Chris Herring, who first characterized the coaA(Ts) mutant, and thank J. C. Escalante for helpful discussions.
REFERENCES
- 1.Allred J B, Guy D G. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal Biochem. 1969;29:293–299. doi: 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- 2.Beck B J, Connolly L E, de las Penas A, Downs D M. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1997;179:6504–6508. doi: 10.1128/jb.179.20.6504-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck B J, Downs D M. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1998;180:885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley T P, Downs D M, Ealick S, McLafferty F, van Loon D, Taylor S, Chiu H, Kinsland C, Reddick J, Xi J, Campobasso N. Thiamin synthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 5.Billheimer J T, Carnevale H N, Leisinger T, Eckhardt T, Jones E E. Ornithine δ-transaminase activity in Escherichia coli: its identity with acetylornithine δ-transaminase. J Bacteriol. 1976;127:1315–1323. doi: 10.1128/jb.127.3.1315-1323.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claas K, Weber S, Downs D M. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:228–232. doi: 10.1128/jb.182.1.228-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan J E, Jr, Littel K J, Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1982;149:916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunn R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalal F R, Gots R E, Gots J E. Mechanism of adenine inhibition in adenine-sensitive mutants of Salmonella typhimurium. J Bacteriol. 1966;91:507–513. doi: 10.1128/jb.91.2.507-513.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downs D M. Evidence for a new, oxygen-regulated biosynthetic pathway for the pyrimidine moiety of thiamine in Salmonella typhimurium. J Bacteriol. 1992;174:1515–1521. doi: 10.1128/jb.174.5.1515-1521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downs D M, Petersen L. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1994;176:4858–4864. doi: 10.1128/jb.176.16.4858-4864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downs D M, Roth J R. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J Bacteriol. 1991;173:6597–6604. doi: 10.1128/jb.173.20.6597-6604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn S D, Snell E E. Isolation of temperature-sensitive pantothenate kinase mutants of Salmonella typhimurium and mapping of the coaA gene. J Bacteriol. 1979;140:805–808. doi: 10.1128/jb.140.3.805-808.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enos-Berlage J, Downs D M. Biosynthesis of the pyrimidine moiety of thiamine independent of the PurF enzyme (phosphoribosylpyrophosphate amidotransferase) in Salmonella typhimurium: incorporation of stable isotope-labeled glycine and formate. J Bacteriol. 1999;181:841–848. doi: 10.1128/jb.181.3.841-848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enos-Berlage J E. Ph.D. thesis. University of Wisconsin, Madison; 1998. [Google Scholar]
- 16.Enos-Berlage J L, Downs D M. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J Bacteriol. 1997;179:3989–3996. doi: 10.1128/jb.179.12.3989-3996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frodyma M, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackowski S, Rock C O. Consequences of reduced intracellular coenzyme A content in Escherichia coli. J Bacteriol. 1986;166:866–871. doi: 10.1128/jb.166.3.866-871.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaRossa R A, Van Dyk T K. Leaky pantothenate and thiamine mutations of Salmonella typhimurium conferring sulphometuron methyl sensitivity. J Gen Microbiol. 1989;135:2209–2222. doi: 10.1099/00221287-135-8-2209. [DOI] [PubMed] [Google Scholar]
- 20.Newell P C, Tucker R G. Biosynthesis of the pyrimidine moiety of thiamine. Biochem J. 1968;106:279–287. doi: 10.1042/bj1060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newell P C, Tucker R G. New pyrimidine pathway involved in the biosynthesis of the pyrimidine of thiamine. Nature (London) 1967;215:1384–1385. doi: 10.1038/2151384a0. [DOI] [PubMed] [Google Scholar]
- 22.Newell P C, Tucker R G. Precursors of the pyrimidine moiety of thiamine. Biochem J. 1968;106:271–277. doi: 10.1042/bj1060271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen L, Downs D M. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol. 1996;178:5676–5682. doi: 10.1128/jb.178.19.5676-5682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen L A, Enos-Berlage J E, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primerano D A, Burns R O. Role of acetohydroxyacid isomeroreductase in biosynthesis of pantothenic acid in Salmonella typhimurium. J Bacteriol. 1983;153:259–269. doi: 10.1128/jb.153.1.259-269.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts R B, Dowie D B, Abelson P H, Bolton E T, Britten R J. Studies of biosynthesis in Escherichia coli. Washington, D.C.: Carnegie Institution of Washington; 1955. [Google Scholar]
- 27.Song W-J, Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J Bacteriol. 1992;174:6411–6417. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallari D S, Rock C O. Isolation and characterization of temperature-sensitive pantothenate kinase (coaA) mutants of Escherichia coli. J Bacteriol. 1987;169:5795–5800. doi: 10.1128/jb.169.12.5795-5800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb E, Downs D M. Characterization of thiL, encoding thiamine monophosphate kinase, in Salmonella typhimurium. J Biol Chem. 1997;272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]