Abstract
Dendritic cells (DCs) bridge the connection between innate and adaptive immunity. DCs present antigens to T cells and stimulate potent cytotoxic T‐cell responses. Metabolic reprogramming is critical for DC development and activation; however, metabolic adaptations and regulation in DC subsets remains largely uncharacterized. Here, we mapped metabolomic and lipidomic signatures associated with the activation phenotype of human conventional DC type 1, a DC subset specialized in cross‐presentation and therefore of major importance for the stimulation of CD8+ T cells. Our metabolomics and lipidomic analyses showed that Toll‐like receptor (TLR) stimulation altered glycerolipids and amino acids in cDC1. Poly I:C or pRNA stimulation reduced triglycerides and cholesterol esters, as well as various amino acids. Moreover, TLR stimulation reduced expression of glycolysis‐regulating genes and did not induce glycolysis. Conversely, cDC1 exhibited increased mitochondrial content and oxidative phosphorylation (OXPHOS) upon TLR3 or TLR7/8 stimulation. Our findings highlight the metabolic adaptations required for cDC1 maturation.
Keywords: BDCA3, dendritic cell activation, lipid mediators, metabolomics
Our findings highlight the metabolic adaptations required for maturation of cDC1 dendritic cells (DC). Toll like receptor stimulation alters glycerolipids and amino acids and increases mitochondrial content and oxidative phosphorylation in cDC1 DC.

Abbreviations
- ECAR
extracellular acidification rate
- LC‐MS
liquid chromatography coupled to high‐resolution mass spectrometry
- LDs
lipid droplets
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- pRNA
protamine RNA
- TLR
Toll‐like receptor
INTRODUCTION
Dendritic cells (DCs) are professional antigen‐presenting cells, which serve as a key connection between innate and adaptive immunity [1, 2]. DCs comprise a family of different subsets that vary in ontogeny, localization, phenotype and specialized immune functions. All DC subsets share the capacity to activate naïve T cells [3, 4], which can be harnessed to generate tumour‐specific immune response. DCs are divided into three major subsets in humans, that is cDC1 (BDCA3+), cDC2 (BDCA1+) and plasmacytoid DC (pDC). These subsets originate from distinct DC precursors, exist in the steady state and have specialized functions [5]. Analysis of gene expression profiling identified cDC1 as a distinct subset among human myeloid DCs, equivalent to mouse CD8α+ and CD103+ DC [6, 7] and with a monocytic morphology [8]. Blood cDC1s are a very rare subset expressing myeloid lineage markers (e.g. CD13 and CD33), CLEC9A, XCR1, Necl2 and TLR3 [9, 10]. cDC1s are found in blood, lymph nodes, tonsil, bone marrow and spleen [10, 11] as well as in peripheral tissues, for example skin, liver and lung [7, 12, 13]. cDC1s have a more activated phenotype in peripheral and lymphoid tissues compared with blood [13] and have an innate resistance to broad range of viral infections [14].
To detect antigen and subsequently activate adaptive immune responses, DCs contain pattern recognition receptors to recognize danger‐associated molecular patterns which upon binding induce DC maturation. There are several groups of pattern recognition receptors, the first and most studied are the TLRs. These are transmembrane molecules expressed in hetero‐ or homodimers, either on the cell surface or in the endosomal compartment [15]. cDC1s express TLR3 and TLR8 but do not express TLR4, 7 or 9 [16]. Upon stimulation with TLR3 ligand poly I:C and its analogs, cDC1s upregulate co‐stimulatory molecules, produce IFN‐α, IFN‐β and CXCL10 and increase their capacity to cross‐present [10, 17]. In contrast to blood cDC1s which require Toll‐like receptor (TLR) stimulation for Ag cross‐presentation [7, 18], peripheral cDC1s can cross‐present antigens (Ag) even in the absence of a stimulus [19]. Blood cDC1s express higher levels of TLR3 and lower levels of CCR7 expression compared with peripheral tissues [7]. TLR stimulation of cDC1s results in production of high IFN‐λ [20], IFN‐α, IFN‐β, IL‐6, IL‐8 and TNF‐α [21]. Evidence shows that blood cDC1s are potent inducers of Th1 response, while lymph node‐derived cDC1s are potent inducers of T cells producing both Th1 [10] and Th2 cytokines [18].
Recent data indicate that upon TLR stimulation, changes in DC metabolism are indispensable for the maturation process [22, 23]. However, at present, there is no knowledge about the metabolic phenotype of human cDC1 which dictates their activation and immunogenicity. Targeting suitable DC subset(s) in combination with optimal adjuvant is crucial for expression of appropriate maturation markers and secretion of the beneficial cytokines is central to the efficacy of DC‐based vaccines. Previously, we have performed immunotherapies with naturally occurring CD1c+ DC and pDC [24, 25]. Furthermore, we identified metabolic adaptations associated with activation of CD1c+ DC and pDC [23]. Considering the intricate interplay between biochemical events that govern DC activation and the distinct ontogeny of cDC1s, we here identified biochemical and bioenergetic changes in cDC1s following stimulation with poly I:C and pRNA, the latter acting via TLR7/8.
MATERIALS AND METHODS
Chemicals
Antimycin A (#A8674), FCCP (#C2920), oligomycin A (#O4876), rotenone (#R8875), A 922500 and poly I:C were obtained from Sigma‐Aldrich. MitoTracker™ Green FM (#M7514) was obtained from Thermo Fisher Scientific.
Cell isolation and stimulation
For lipidomic and metabolomic analysis, cDC1s were isolated from aphaeresis product using a fully automated CliniMACS prodigy isolation and culture system (Miltenyi Biotec, Bergisch‐Gladbach, Germany). Vials with antibodies or beads were connected to the CliniMACS prodigy tubing set with a vial adaptor. For CliniMACS isolation, the aphaeresis product was connected to a CliniMACS tubing set (Miltenyi Biotec, Bergisch‐Gladbach, Germany) using a sterile tubing welder. B cells, T cells and monocytes were depleted using GMP‐grade magnetic bead‐coupled CD19 antibodies, CD3 antibodies and CD14 antibodies, respectively, using program CD141 PreDepletion of the CliniMACS prodigy isolation system. In the next step, cDC1s were positively selected with GMP‐grade magnetic bead‐coupled anti‐CD141 antibodies. Magnetically labelled cDC1s were positively selected using program CD141 enrichment of the CliniMACS prodigy isolation system. This procedure results in purified cDC1s. One third of the cDC1 were spun down, snap‐frozen and stored at −80℃ in RNA‐free tubes. The other cDC1s were cultured for 12 h at a concentration of 1–2 × 106 cells/ml in X‐VIVO15 medium supplemented with 2% human serum and, respectively, stimulated with protamine RNA (15µg/ml) or poly‐IC (10µg/ml). The next day the cells were harvested, spun down, snap‐frozen and stored at −80℃ in RNA‐free tubes.
For RNA‐seq, cDC1s were isolated from buffy coats of healthy volunteers (Sanquin, Nijmegen, the Netherlands) after obtaining written informed consent as per the Declaration of Helsinki and according to institutional guidelines. Peripheral blood mononuclear cells (PBMCs) were isolated by using Ficoll density centrifugation (Lymphoprep; Axis‐Shield PoC AS, Oslo, Norway). Anti‐human lineage cocktail 1 in FITC (LIN1) (BD Bioscience Pharmingen, San Jose, CA) containing antibodies for CD3, CD14, CD16, CD19, CD20, CD56 receptors, together with the anti‐FITC‐conjugated magnetic microbeads of Miltenyi Biotec (Bergisch‐Gladbach, Germany), was used to deplete the LIN1+ cell fraction, by following the manufacturer's instructions. Next, cDC1s were further purified by sorting (flow cytometry) using anti‐BDCA3‐APC combined with anti‐HLA‐DR‐PE‐Vio770 (Miltenyi Biotec) to a purity of 99·9%. cDC1s were cultured in X‐VIVO‐15 medium (Lonza, Basel, Switzerland) supplemented with 2% human serum (Sanquin).
Cytokine detection
Supernatant was taken from each sample after incubation, and TNF‐α, IFN‐α and IL‐6 were measured with the MACSPlex cytokine 12 Kit (Miltenyi Biotech), according to the manufacturer's instructions.
Metabolism assays
An XF‐96 Extracellular Flux Analyzer (Seahorse Bioscience) was used for extracellular flux analyses of cDC1 (30 000 cells/well), as described previously [26]. For mitochondrial fitness tests, OCR was measured sequentially at basal and following the addition of 1 µM oligomycin, 3 µM FCCP (fluorocarbonyl cyanide phenylhydrazone) and 1 µM ROT+1 µM AA.
Metabolomics
Extraction of metabolites from cells was performed by addition of 0·5 ml of a pre‐chilled at −80℃ methanol/chloroform/water (8·1:0·9:1 v/v/v) mixture to each cell sample, vortex for 10 s, incubate on dry ice for 15 min and sonicate for 1 min. The supernatant containing the polar metabolites was collected after centrifugation at 18 000 g for 20 min at −4℃. Extracts were subsequently dried and reconstituted in 0·15 M phosphate buffer in 2H2O for NMR analysis. The latter was carried on a 14·1 T NMR (600·13 MHz for 1H) Bruker Avance II, under standardized conditions as described [27]. Chenomx NMR suite 8·2 (Chenomx Inc) was used for quantification of metabolites from NMR spectra.
Lipidomics
Extracts used for the NMR metabolomics analysis were dried and reconstituted in 50:50 isopropanol:water (v/v). A Shimadzu Nexera LC‐30 (Shimadzu, ‘s‐Hertogenbosch, the Netherlands) was used to deliver a gradient of water: acetonitrile 80:20 (v/v) (eluent A) and water: acetonitrile: 2‐propanol 1:9:90 (v/v/v) (eluent B). Both eluents also contained 5 mM ammonium formate and 0·05% formic acid. The applied gradient, with a column flow of 300 µl/min, was as follows: 0 min 30% B, 1 min 30% B, 10 min 50% B, 10·5 min 70% B, 12 min 70% B, 12·5 min 30% B, 16 min 30% B. A Phenomenex Kinetex C18, 2·7 µm particles, 50 × 2·1 mm (Phenomenex, Utrecht, the Netherlands) was used as the column. The injection volume was 5 µl. The MS was a Sciex TripleTOF 6600 (AB Sciex Netherlands B.V., Nieuwerkerk aan den IJssel, the Netherlands) operated in positive (ESI+) and negative (ESI‐) ESI mode, with the following conditions: ion source gas 1, 2 and curtain gas 30 psi, temperature 350°C, acquisition range m/z 100–1800, IonSpray voltage 5500 V (ESI+) and −4500 V (ESI−), declustering potential 80 V (ESI+) and −80 V (ESI−). An information‐dependent acquisition (IDA) method was used to identify lipids, with the following conditions for MS analysis: collision energy ±10, acquisition time 250 ms and for MS/MS analysis: collision energy ±45, collision energy spread 25, ion release delay 30, ion release width 14, acquisition time 40 ms. The IDA switching criteria where set as: for ions greater than m/z 300, which exceeds 200 cps, exclude former target for 2 s, exclude isotopes within 1·5 Da, max. candidate ions 20. Before data analysis, raw MS data files were converted with the Reifycs Abf Converter (v1·1) to the Abf file. MS‐DIAL (v3·30), with the FiehnO (VS39) database, was used to align the data and identify the different lipids [28].
Protamine RNA (pRNA) complexes
pRNA complexes were made fresh before adding to the cells. Protamine (protaminehydrochloride MPH 5000 IE/ml; Meda Pharma BV Amstelveen, the Netherlands) was diluted to 0·5 mg/ml in RNase‐free water and mixed with 2‐kbp‐long single‐stranded mRNA (coding for human gp100 protein). It was extensively mixed and incubated for 5–10 min at room temperature, before being added to the cells.
RNA sequencing
cDC1s were isolated as described above, and total RNA was extracted using Trizol (Invitrogen, MA, USA), following the standard protocol. The quality control of the isolated RNA (concentration, RIN, 28S/18S and size) was performed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). RNA sequencing and read alignment were performed by BGI TECH SOLUTIONS (Hong Kong). Reads were aligned to human genome version 19.
RESULTS
cDC1 activation is accompanied by upregulation in oxidative phosphorylation (OXPHOS)
TLR stimulation induces the release of proinflammatory cytokines, which are necessary to facilitate the induction of potent immune responses by DCs [29]. The release of cytokines is, besides the upregulation of antigen‐presenting molecules and co‐stimulatory molecules, a major factor of DC maturation [30]. To determine whether the TLR3 agonist poly I:C or the TLR7/8 agonist pRNA was able to activate cDC1, expression of co‐stimulatory molecule CD86 and changes in secreted cytokines were evaluated. TLR stimulation of blood cDC1 with poly I:C or pRNA, for 12 h, increased CD86 surface expression (Figure 1a) and release of TNF‐α, IL‐6 and IFN‐α (Figure 1b‐d). TLR stimulation of blood cDC1 with poly I:C or pRNA, for 12 h did not affect cell viability (Figure S1A). The culture of cDC1 did not increase CD86 surface expression (Figure S1B). We questioned whether activation of cDC1 in response to poly I:C and pRNA was accompanied by metabolic changes. A hallmark of DC activation is induction of glycolysis [31, 32]. To investigate whether TLR stimulation induces glycolysis in cDC1, we analysed the expression of glycolysis related genes. Among the glycolysis‐regulating genes, pRNA stimulation significantly reduced expression of G6PD, PGAM2, PDPR, PCK2, ENO3 and SLC2A5 while PDK3, SLC2A6 and SLC2A13 were increased (Figure 2a). Mitochondrial mass and mitochondrial activity are intricately linked [33]. To explore the effect of TLR stimulation on cDC1s, cells were stained with MitoTracker™ Green FM, a fluorescent dye that localizes to mitochondria, in a mitochondrial membrane potential independent manner, where it covalently binds to mitochondrial proteins by reacting with free thiol groups of cysteine residues [34]. TLR stimulation with poly I:C and pRNA increased mitochondrial content of cDC1 (Figure 2b). Investigation of extracellular flux analysis (EFA) showed that poly I:C and pRNA increased basal oxygen consumption rate (OCR), ATP‐linked OCR but reduced spare respiratory capacity (SRC) in cDC1 (Figure 2c‐d). Poly I:C or pRNA did not increase extracellular acidification rate (ECAR) (Figure 2e). Collectively, these data suggest that TLR stimulation increases OXPHOS but not glycolysis in cDC1s.
FIGURE 1.
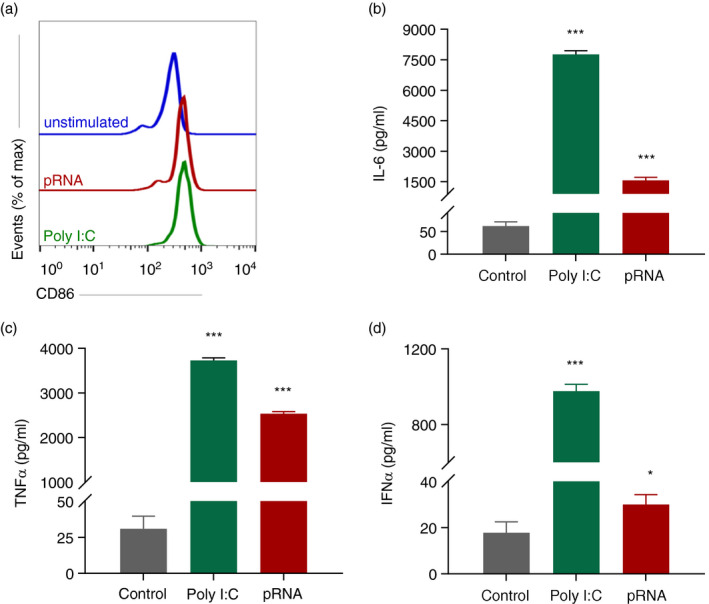
Poly I:C and pRNA activate cDC1. (a) Flow cytometry histograms of CD86 cDC1 stimulated cells for 12 h. (b–d) Cytokines levels on protein level were measured in the supernatant of the cDC1 stimulated for 12 h. Data represent mean ± SEM of three independent experiments. *p < 0·05; ***p < 0·001 (Student's t‐test)
FIGURE 2.
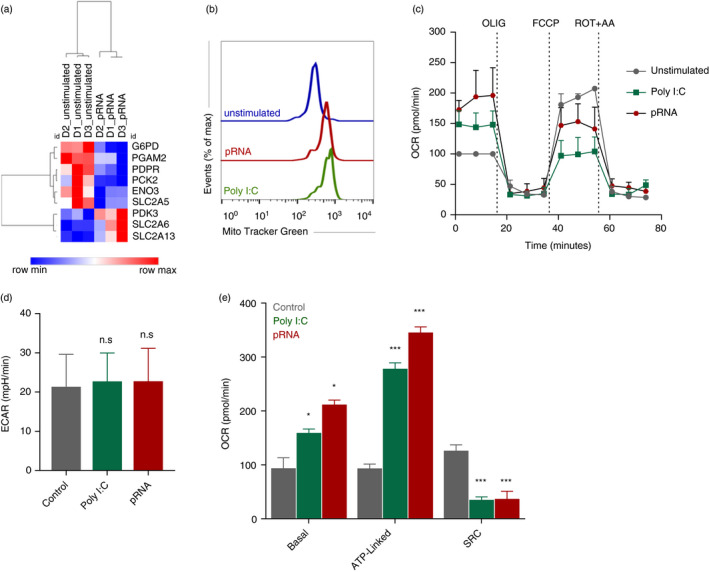
TLR stimulation induces OXPHOS in cDC1. (a) Heatmap showing expression of significantly changed genes which regulate glycolysis in cDC1 upon pRNA‐stimulation for 12 h. Red colour indicates increased expression while blue colour shows decreased expression. (b) Flow cytometry histograms of cDC1 cells stained with MitoTracker Green FM and stimulated with poly I:C or pRNA for 12 h. (c) Mitochondrial fitness test of cDC1 stimulated with poly I:C or pRNA for 12 h. Data represent mean ± SEM of three independent experiments. (d–e) Data were collected within same experiments as c, but are shown separately for better understanding. Data represent mean ± SEM of three independent experiments. *p < 0·05; ***p < 0·001 (Student's t‐test) [Colour figure can be viewed at wileyonlinelibrary.com]
Lipidome signature associated with cDC1 activation
Lipid metabolism is crucial for innate immune functions of DC [35]. To identify how TLR ligands modify lipid metabolism, we analysed the effect of poly I:C and pRNA on the cDC1 lipidome. Untargeted lipidomic analysis by liquid chromatography coupled to high‐resolution mass spectrometry (LC‐MS) was performed in both negative and positive electrospray ionization modes. Lipids were annotated using the MS‐DIAL based on their MS/MS spectra, which allowed us to identify 8 distinct lipid classes, including phospholipids (phosphatidylcholine, phosphatidylethanolamine), etherphospholipids, lyso‐phospholipids (lyso‐phosphatidylcholine, lyso‐phosphatidylethanolamine), sphingomyelins, ceramides, fatty acids, glycerolipids and cholesterol esters. Multidimensional scaling (MDS1 and MDS2) revealed a clear separation between poly I:C or pRNA‐stimulated and unstimulated cDC1 (Figure 3a). To identify lipid species, which changed upon poly I:C or pRNA stimulation we analysed all identified lipids by computing pairwise correlation coefficients and organizing the resulting matrix by hierarchical clustering (Figure 3b). Unsupervised clustering of lipid species, presenting a more than twofold change, discriminated between unstimulated cDC1 and poly I:C or pRNA‐stimulated cDC1s are shown (Figure 3b). Univariate analysis of detected lipids with a p‐value <0·05 based on ANOVA revealed that 25 distinct lipid species were significantly decreased in poly I:C‐ and pRNA‐matured cDC1s (Figure 3c), whereas 12 and 13 lipid species were significantly increased in poly I:C and pRNA‐stimulated cDC1, respectively (Figure 3c). The majority of these altered lipids belong to the subclass of glycerolipids, especially triacylglycerides (TAG), while few belong to the cholesterol esters (CE) (Figure 3c). To determine the functional significance of TAG in immune response, we used A 922500, a selective DGAT‐1 inhibitor (DGAT1i) to suppress TG accumulation. A 922500 significantly reduced pRNA‐stimulated release of TNF‐α and IL‐6 (Figure 3d–e). These data suggest the involvement of triglycerides and cholesterol esters in regulating cDC1 innate immune function.
FIGURE 3.
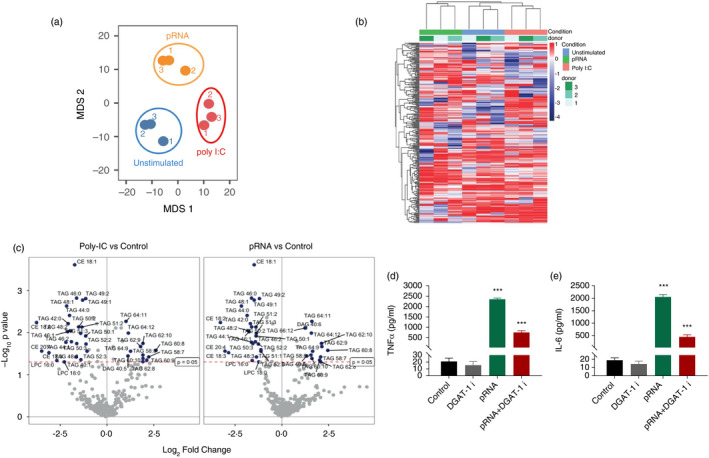
TLR stimulation affects glycerolipids in cDC1. (a) Non‐metric multidimensional scaling (MDS1 and MDS2) ordination plotting to test the differences between unstimulated cDC1 and/or poly I:C or pRNA stimulated. (b) Heatmap showing expression of log2 fold change lipids in cDC1 upon poly I:C or pRNA‐stimulation after 12 h. (c) Volcano plots for the differential lipid expression following poly I:C or pRNA stimulation of cDC1. The x‐axis describes the log2 fold change in expression levels between poly I:C or pRNA‐stimulated cDC1 relative to unstimulated cells. The y‐axis shows the statistical significance expressed as −log10(p‐value) with p ≤ 0·05. (d–e) Cytokine levels on protein level were measured in the supernatant of the cDC1 stimulated for 12 h. Data represent mean ± SEM of three independent experiments. ***p < 0·001 (Student's t‐test) [Colour figure can be viewed at wileyonlinelibrary.com]
TLR stimulation alters amino acid metabolism in cDC1
To identify the pathways, which are altered upon poly I:C or pRNA stimulation in cDC1s, the metabolite profile was analysed using MetaboAnalyst 2.0 software. MetaboAnalyst 2.0 is a web‐based software, which utilizes the KEGG metabolic pathway database to derive its predictive ability. The software employs pathway enrichment and topology analysis to identify significantly altered pathways under the given experimental setting [36]. In cDC1s, stimulation with poly I:C or pRNA altered aminoacyl‐tRNA biosynthesis, alanine, aspartate and glutamate metabolism, glycine, serine and threonine metabolism, pyruvate metabolism and pantothenate and CoA biosynthesis (Figure 4a,b). The results of the pathway analysis are shown in Tables 1 and 2. Differential expression analysis of stimulated versus unstimulated (p‐value <0·05) showed that both poly I:C and pRNA result in reduced amino acids and its derivatives (Figure 4c,d).
FIGURE 4.
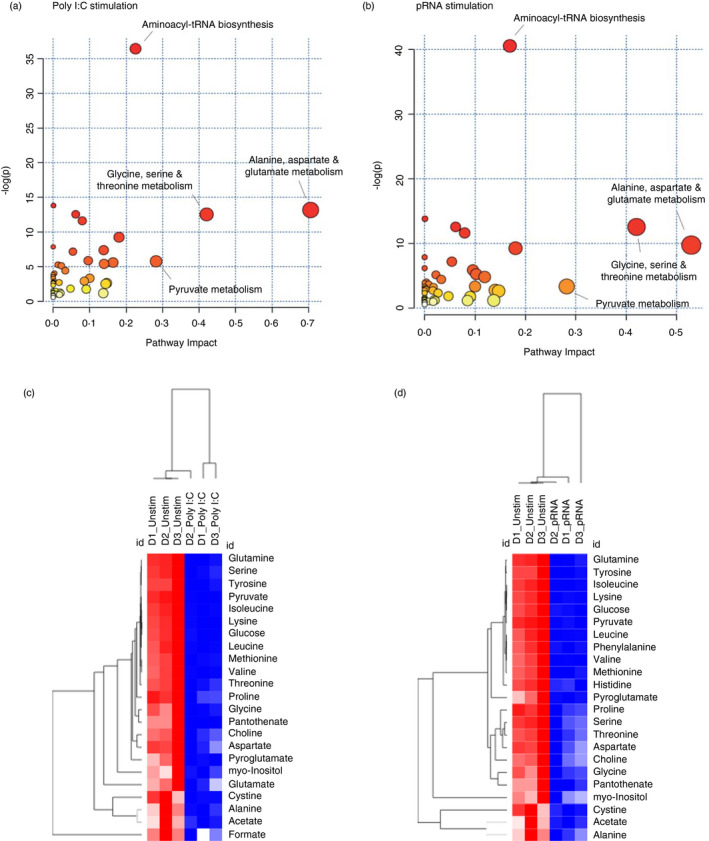
TLR stimulation alters amino acid metabolism in cDC1. (a) Metabolome view from pathway analysis performed using MetaboAnalyst in cDC1 upon poly I:C stimulation. Select pathways with high pathway impact and/or high p‐value are labelled. (b) Metabolome view from pathway analysis performed using MetaboAnalyst in cDC1 upon pRNA stimulation. Select pathways with high pathway impact and/or high p‐value are labelled. (c) Heatmap showing levels of significantly changed metabolites which regulate amino acid metabolism in cDC1 upon poly I:C stimulation for 12 h. Red colour indicates higher levels while blue colour shows lower levels. (d) Heatmap showing levels of significantly changed metabolites which regulate amino acid metabolism in cDC1 upon pRNA‐stimulation for 12 h. Red colour indicates increased levels while blue colour shows decreased levels [Colour figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
List of selected pathways identified by pathway analysis in poly I:C stimulated cDC1 using MetaboAnalyst
| No | Pathway name | Total cmpda | Hitsb | Expected | Raw p c | Holm adjust | Impactd |
|---|---|---|---|---|---|---|---|
| 1 | Aminoacyl‐tRNA biosynthesis | 75 | 14 | 0·71666 | 1·52E−16 | 1·22E−14 | 0·22536 |
| 2 | Nitrogen metabolism | 39 | 6 | 0·37266 | 1·00E−06 | 7·92E−05 | 0·00067 |
| 3 | Alanine, aspartate and glutamate metabolism | 24 | 5 | 0·22933 | 1·89E−06 | 0·00014768 | 0·70561 |
| 4 | Valine, leucine and isoleucine biosynthesis | 27 | 5 | 0·258 | 3·53E−06 | 0·00027173 | 0·06148 |
| 5 | Glycine, serine and threonine metabolism | 48 | 6 | 0·45866 | 3·57E−06 | 0·00027173 | 0·42039 |
| 6 | Cysteine and methionine metabolism | 56 | 6 | 0·53511 | 8·99E−06 | 0·0006743 | 0·07941 |
| 7 | Pantothenate and CoA biosynthesis | 27 | 6 | 0·258 | 9·63E−05 | 0·0071228 | 0·18014 |
| 8 | Cyanoamino acid metabolism | 16 | 3 | 0·15289 | 0·000394 | 0·028752 | 0 |
| 9 | Arginine and proline metabolism | 77 | 5 | 0·73577 | 0·00063 | 0·045351 | 0·13813 |
| 10 | Taurine and hypotaurine metabolism | 20 | 3 | 0·19111 | 0·000782 | 0·055522 | 0·05395 |
Total cmpd is the total number of compounds in the pathway.
Hits is the actual matched number from the uploaded data.
Raw p is the p‐value calculated from the pathway analysis.
Impact is the pathway impact value calculated from pathway topology analysis.
TABLE 2.
List of selected pathways identified by pathway analysis in pRNA‐stimulated cDC1 using MetaboAnalyst
| No | Pathway name | Total cmpda | Hitsb | Expected | Raw p c | Holm adjust | Impactd |
|---|---|---|---|---|---|---|---|
| 1 | Aminoacyl‐tRNA biosynthesis | 75 | 15 | 0·71666 | 2·39E−18 | 1·91E−16 | 0·16902 |
| 2 | Nitrogen metabolism | 39 | 6 | 0·37266 | 1·00E−06 | 7·92E−05 | 0·00067 |
| 3 | Valine, leucine and isoleucine biosynthesis | 27 | 5 | 0·258 | 3·53E−06 | 0·00027526 | 0·06148 |
| 4 | Glycine, serine and threonine metabolism | 48 | 6 | 0·45866 | 3·57E−06 | 0·00027526 | 0·42039 |
| 5 | Cysteine and methionine metabolism | 56 | 6 | 0·53511 | 8·99E−06 | 0·00068329 | 0·07941 |
| 6 | Alanine, aspartate and glutamate metabolism | 24 | 4 | 0·22933 | 5·94E−05 | 0·004455 | 0·52897 |
| 7 | Pantothenate and CoA biosynthesis | 27 | 4 | 0·258 | 9·63E−05 | 0·0071228 | 0·18014 |
| 8 | Cyanoamino acid metabolism | 16 | 3 | 0·15289 | 0·00039387 | 0·028752 | 0 |
| 9 | Taurine and hypotaurine metabolism | 20 | 3 | 0·19111 | 0·00078201 | 0·056304 | 0·05395 |
| 10 | beta‐Alanine metabolism | 28 | 3 | 0·26755 | 0·0021377 | 0·15178 | 0 |
Total cmpd is the total number of compounds in the pathway.
Hits is the actual matched number from the uploaded data.
Raw p is the p‐value calculated from the pathway analysis.
Impact is the pathway impact value calculated from pathway topology analysis.
DISCUSSION
In this study, a combined use of mass spectrometry and nuclear magnetic resonance spectroscopy was used to comparatively map metabolomic and lipidomic molecular species associated with the activation of cDC1s. Our untargeted approach yielded a distinct pattern of lipidomic and metabolomic alterations upon TLR3 and TLR7/8 stimulation in cDC1s.
Lipidomic data show that TLR stimulation of cDC1 significantly modulated TAG and CEs. Both, TAG and CE, are stored in lipid droplets (LD) [37] and their accumulation is indicator of inflammatory activation [38]. Interestingly, bone marrow‐derived DCs, stimulated with LPS or IL‐4, exhibit accumulation of lipid droplets (LDs) [39]. This suggests that TLR stimulation of cDC1 increases TAG and CE synthesis, which are sequestered in LDs. de novo TAG synthesis is catalysed by DGAT1 and DGAT2, via esterification of a fatty acyl moiety to a DAG [40] and DGAT1 inhibition reduces TAG levels [41]. Our data showing that DGAT1 inhibition reduces TNF‐α and IL‐6 levels in stimulated cDC1 suggest that production of TAG is reduced, which in turn reduces cDC1 activation. We did not test effect of DGAT2 inhibition on cDC1 activation; however, marked reduction in TNF‐α and IL‐6 suggests that DGAT1 plays a major role in TAG synthesis in cDC1 and DGAT2 might play a minor role. These data are in line with role of DGAT1 in the production of TAGs and LDs in macrophages [42]. Together, these data pose a scenario in which TLR stimulation increases DGAT1‐mediated TAG and CE production, which are stored in LDs and are used in activation process.
Metabolomic analysis showed that aminoacyl‐tRNA biosynthesis is one of the pathways affected upon TLR stimulation of cDC1s. Upon pathogen infection, aminoacyl‐tRNA synthase (ARS) is secreted from monocytes to activate macrophages to induce innate immune responses [19]. Reduction of metabolites in cDC1 upon poly I:C and pRNA suggests that intracellular ARS release is reduced, which in turn induces Th responses. Furthermore, metabolomics analysis shows the alanine, aspartate and glutamate metabolism pathway is involved in cDC1 activation upon poly I:C and pRNA stimulation. Interestingly, excessive glutamate in DCs favours the development of Treg cells, whereas glutamate‐deficient DCs upregulate both Th1‐specific (IL‐12 and IL‐27) and Th17‐specific (IL‐6 and IL‐23) cytokines [43]. Moreover, glutamate released by DCs enhances T‐cell proliferation and secretion of Th1 and proinflammatory cytokines [44]. Our data show that poly I:C and pRNA‐stimulated cDC1 have lower levels of glutamate. Together these data suggest that TLR stimulation results in release of glutamate from cDC1, which results in potent Th responses. We also observed reduction in amino acids (AA) in cDC1s after stimulation suggest. Amino acids fuel the TCA cycle after conversion into to either acetyl‐CoA or α‐keto acid intermediates, that is pyruvate, oxaloacetate and succinyl‐CoA. Therefore, decreases in amino acids suggest that these are consumed to support OXPHOS as a source of ATP in cDC1.
Our data show that poly I:C or pRNA stimulation increased basal oxygen consumption rate and ATP‐linked respiration but decreased the spare respiratory capacity in cDC1s. Interestingly, Du et al performed ex vivo analysis of cDC1s and identified that cDC1 retains higher levels of both glycolysis and mitochondrial metabolism, which are crucial for priming of CD8+ T cells [45]. In contrast, we did not observe glycolysis induction upon TLR stimulation in cDC1s. Gene expression analysis showed that 12h TLR stimulation reduced the majority of glycolysis‐regulating genes, for example G6PD, PGAM2, PDPR, PCK2, ENO3 and SLC2A5. No functional data on relevance of SLC2A13 for OXPHOS or glycolysis have been found. SLC2A6 and PDK3 have been shown to be required for glycolytic changes in inflammatory macrophages [46, 47]. However, SLC2A6‐mediated glycolysis is dispensable for inflammatory responses in macrophages, while PDK activity is required only for glycolytic changes. The increased OXPHOS in cDC1 upon TLR stimulation indicates that increased SLC2A6 and PDK3 do not play role to induce glycolysis in cDC1. Furthermore, metabolomics analysis showed that 12h poly I:C or pRNA stimulation reduced pyruvate and acetate levels in cDC1. During glycolysis, glucose is converted to pyruvate which then enters TCA cycle [48]. Reduction in ENO3, which regulates the penultimate step of glycolysis [49], together with reduced lactate, pyruvate and acetate levels suggests the observed disabled glycolysis after 12h.
Understanding the metabolic regulation of DC subsets and functions can significantly impact our understanding of DC biology and immune regulation. Our data provide new insight into the metabolic regulation in cDC1s by identifying different metabolites and metabolic adaptation, which orchestrate the described functions. These findings may help improve DC‐based immunotherapies.
CONFLICT OF INTEREST
The authors have no competing interests to declare.
AUTHOR CONTRIBUTIONS
FB and IJMdV conceived the research. FB, SK and RJED performed the experiments. FB, JvB and SK, RJED, MG analysed the data. FB and IJMdV wrote the manuscript. IJMdV supervised the research.
Supporting information
Fig S1
ACKNOWLEDGEMENT
Authors would like to thank Jasper J P van Beek and Georgina Flórez‐Grau for critical discussions.
Basit F, van Oorschot T, van Buggenum J, Derks RJE, Kostidis S, Giera M, et al. Metabolomic and lipidomic signatures associated with activation of human cDC1 (BDCA3+/CD141+) dendritic cells. Immunology.2022;165:99–109. 10.1111/imm.13409
Funding information
This work was supported by Vici grant 918.146.55 from the Dutch research council (NOW) and the Oncode Institute.
REFERENCES
- 1. Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y‐J, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. [DOI] [PubMed] [Google Scholar]
- 2. Sittig SP, de Vries IJM, Schreibelt G. Primary human blood dendritic cells for cancer immunotherapy‐tailoring the immune response by dendritic cell maturation. Biomedicines. 2015;3:282–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allan RS, Waithman J, Bedoui S, Jones CM, Villadangos JA, Zhan Y, et al. Migratory dendritic cells transfer antigen to a lymph node‐resident dendritic cell population for efficient CTL priming. Immunity. 2006;25:153–62. [DOI] [PubMed] [Google Scholar]
- 4. Flinsenberg TW, Compeer EB, Koning D, Klein M, Amelung FJ, van Baarle D, et al. Fcgamma receptor antigen targeting potentiates cross‐presentation by human blood and lymphoid tissue BDCA‐3+ dendritic cells. Blood. 2012;120:5163–72. [DOI] [PubMed] [Google Scholar]
- 5. Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154:3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robbins SH, Walzer T, Dembélé D, Thibault C, Defays A, Bessou G, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome‐wide expression profiling. Genome Biol. 2008;9:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, et al. Human tissues contain CD141hi cross‐presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, et al. BDCA‐2, BDCA‐3, and BDCA‐4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–46. [DOI] [PubMed] [Google Scholar]
- 9. Contreras V, Urien C, Guiton R, Alexandre Y, Vu Manh TP, Andrieu T, et al. Existence of CD8alpha‐like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J Immunol. 2010;185:3313–25. [DOI] [PubMed] [Google Scholar]
- 10. Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, et al. Human CD141+ (BDCA‐3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross‐presents necrotic cell antigens. J Exp Med. 2010;207:1247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poulin LF, Salio M, Griessinger E, Anjos‐Afonso F, Craciun L, Chen JL, et al. Characterization of human DNGR‐1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207:1261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu C‐C, Ali N, Karagiannis P, Di Meglio P, Skowera A, Napolitano L, et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL‐10 and induce regulatory T cells that suppress skin inflammation. J Exp Med. 2012;209:935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, et al. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon‐lambda in response to hepatitis C virus. Hepatology. 2013;57:1705–15. [DOI] [PubMed] [Google Scholar]
- 14. Silvin A, Yu CI, Lahaye X, Imperatore F, Brault JB, Cardinaud S, et al. Constitutive resistance to viral infection in human CD141(+) dendritic cells. Sci Immunol. 2017;2(13):eaai8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Botos I, Segal DM, Davies DR. The structural biology of Toll‐like receptors. Structure. 2011;19:447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pearson FE, Chang K, Minoda Y, Rojas IML, Haigh OL, Daraj G, et al. Activation of human CD141(+) and CD1c(+) dendritic cells in vivo with combined TLR3 and TLR7/8 ligation. Immunol Cell Biol. 2018;96:390–400. [DOI] [PubMed] [Google Scholar]
- 17. Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, et al. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–81. [DOI] [PubMed] [Google Scholar]
- 18. Segura E, Valladeau‐Guilemond J, Donnadieu MH, Sastre‐Garau X, Soumelis V, Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med. 2012;209:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahn YH, Park S, Choi JJ, Park B‐K, Rhee KH, Kang E, et al. Secreted tryptophanyl‐tRNA synthetase as a primary defence system against infection. Nat Microbiol. 2016;2:16191. [DOI] [PubMed] [Google Scholar]
- 20. Zhang S, Kodys K, Li K, Szabo G. Human type 2 myeloid dendritic cells produce interferon‐lambda and amplify interferon‐alpha in response to hepatitis C virus infection. Gastroenterology. 2013;144:414–25 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meixlsperger S, Leung CS, Ramer PC, Pack M, Vanoaica LD, Breton G, et al. CD141+ dendritic cells produce prominent amounts of IFN‐alpha after dsRNA recognition and can be targeted via DEC‐205 in humanized mice. Blood. 2013;121:5034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wculek SK, Khouili SC, Priego E, Heras‐Murillo I, Sancho D. Metabolic control of dendritic cell functions: digesting information. Front Immunol. 2019;10:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basit F, Mathan T, Sancho D, de Vries IJM. Human dendritic cell subsets undergo distinct metabolic reprogramming for immune response. Front Immunol. 2018;9:2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schreibelt G, Bol KF, Westdorp H, Wimmers F, Aarntzen EHJG, Duiveman‐de Boer T, et al. Effective clinical responses in metastatic melanoma patients after vaccination with primary myeloid dendritic cells. Clin Cancer Res. 2016;22:2155–66. [DOI] [PubMed] [Google Scholar]
- 25. Tel J, Aarntzen EHJG, Baba T, Schreibelt G, Schulte BM, Benitez‐Ribas D, et al. Natural human plasmacytoid dendritic cells induce antigen‐specific T‐cell responses in melanoma patients. Cancer Res. 2013;73:1063–75. [DOI] [PubMed] [Google Scholar]
- 26. Pelgrom LR, van der Ham AJ, Everts B. Analysis of TLR‐induced metabolic changes in dendritic cells using the seahorse XF(e)96 extracellular flux analyzer. Methods Mol Biol. 2016;1390:273–85. [DOI] [PubMed] [Google Scholar]
- 27. Kostidis S, Addie RD, Morreau H, Mayboroda OA, Giera M. Quantitative NMR analysis of intra‐ and extracellular metabolism of mammalian cells: a tutorial. Anal Chim Acta. 2017;980:1–24. [DOI] [PubMed] [Google Scholar]
- 28. Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, et al. MS‐DIAL: data‐independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12:523–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fumagalli S, Torri A, Papagna A, Citterio S, Mainoldi F, Foti M. IL‐22 is rapidly induced by pathogen recognition receptors stimulation in bone‐marrow‐derived dendritic cells in the absence of IL‐23. Sci Rep. 2016;6:33900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Said A, Weindl G. Regulation of dendritic cell function in inflammation. J Immunol Res. 2015;2015:743169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, et al. TLR‐driven early glycolytic reprogramming via the kinases TBK1‐IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Everts B, Amiel E, van der Windt GJW, Freitas TC, Chott R, Yarasheski KE, et al. Commitment to glycolysis sustains survival of NO‐producing inflammatory dendritic cells. Blood. 2012;120:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bratic I, Trifunovic A. Mitochondrial energy metabolism and ageing. Biochim Biophys Acta. 2010;1797(6–7):961–7. [DOI] [PubMed] [Google Scholar]
- 34. Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser‐induced fluorescence detection. J Chromatogr B. 2003;793:141–50. [DOI] [PubMed] [Google Scholar]
- 35. Saas P, Varin A, Perruche S, Ceroi A. Recent insights into the implications of metabolism in plasmacytoid dendritic cell innate functions: potential ways to control these functions. F1000Res. 2017;6:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chong J, Yamamoto M, Xia J. MetaboAnalystR 2.0: from raw spectra to biological insights. Metabolites. 2019;9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerrini V, Gennaro ML. Foam cells: one size doesn't fit all. Trends Immunol. 2019;40:1163–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quiroga IY, Pellon‐Maison M, Gonzalez MC, Coleman RA, Gonzalez‐Baro MR. Triacylglycerol synthesis directed by glycerol‐3‐phosphate acyltransferases ‐3 and ‐4 is required for lipid droplet formation and the modulation of the inflammatory response during macrophage to foam cell transition. Atherosclerosis. 2021;316:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maroof A, English NR, Bedford PA, Gabrilovich DI, Knight SC. Developing dendritic cells become ‘lacy’ cells packed with fat and glycogen. Immunology. 2005;115:473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhatt‐Wessel B, Jordan TW, Miller JH, Peng L. Role of DGAT enzymes in triacylglycerol metabolism. Arch Biochem Biophys. 2018;655:1–11. [DOI] [PubMed] [Google Scholar]
- 41. Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, et al. DGAT1‐dependent lipid droplet biogenesis protects mitochondrial function during starvation‐induced autophagy. Dev Cell. 2017;42:9–21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castoldi A, Monteiro LB, van Teijlingen Bakker N, Sanin DE, Rana N, Corrado M, et al. Triacylglycerol synthesis enhances macrophage inflammatory function. Nat Commun. 2020;11:4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen AM, Caspi RR. Glutamate joins the ranks of immunomodulators. Nat Med. 2010;16:856–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pacheco R, Oliva H, Martinez‐Navío JM, Climent N, Ciruela F, Gatell JM, et al. Glutamate released by dendritic cells as a novel modulator of T cell activation. J Immunol. 2006;177:6695–704. [DOI] [PubMed] [Google Scholar]
- 45. Du X, Wen J, Wang Y, Karmaus PWF, Khatamian A, Tan H, et al. Hippo/Mst signalling couples metabolic state and immune function of CD8alpha(+) dendritic cells. Nature. 2018;558:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maedera S, Mizuno T, Ishiguro H, Ito T, Soga T, Kusuhara H. GLUT6 is a lysosomal transporter that is regulated by inflammatory stimuli and modulates glycolysis in macrophages. FEBS Lett. 2019;593:195–208. [DOI] [PubMed] [Google Scholar]
- 47. Na YR, Jung D, Song J, Park JW, Hong JJ, Seok SH. Pyruvate dehydrogenase kinase is a negative regulator of interleukin‐10 production in macrophages. J Mol Cell Biol. 2020;12:543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams NC, O'Neill LAJ. A Role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol. 2018;9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim AY, Lim B, Choi J, Kim J. The TFG‐TEC oncoprotein induces transcriptional activation of the human beta‐enolase gene via chromatin modification of the promoter region. Mol Carcinog. 2016;55:1411–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1


