Abstract
Aim
To assess the pathobiological and translational importance of whole-blood transcriptomic analysis in inflammatory bowel disease [IBD].
Methods
We analysed whole-blood expression profiles from paired-end sequencing in a discovery cohort of 590 Europeans recruited across six countries in the IBD Character initiative (newly diagnosed patients with Crohn’s disease [CD; n = 156], ulcerative colitis [UC; n = 167], and controls [n = 267]), exploring differential expression [DESeq2], co-expression networks [WGCNA], and transcription factor involvement [EPEE, ChEA, DoRothEA]. Findings were validated by analysis of an independent replication cohort [99 CD, 100 UC, 95 controls]. In the discovery cohort, we also defined baseline expression correlates of future treatment escalation using cross-validated elastic-net and random forest modelling, along with a pragmatic ratio detection procedure.
Results
Disease-specific transcriptomes were defined in IBD [8697 transcripts], CD [7152], and UC [8521], with the most highly significant changes in single genes, including CD177 (log2-fold change [LFC] = 4.63, p = 4.05 × 10-118), MCEMP1 [LFC = 2.45, p = 7.37 × 10-109], and S100A12 [LFC = 2.31, p = 2.15 × 10-93]. Significantly over-represented pathways included IL-1 [p = 1.58 × 10-11], IL-4, and IL-13 [p = 8.96 × 10-9]. Highly concordant results were obtained using multiple regulatory activity inference tools applied to the discovery and replication cohorts. These analyses demonstrated central roles in IBD for the transcription factors NFE2, SPI1 [PU.1], CEBPB, and IRF2, all regulators of cytokine signalling, based on a consistent signal across cohorts and transcription factor ranking methods. A number of simple transcriptome-based models were associated with the need for treatment escalation, including the binary CLEC5A/CDH2 expression ratio in UC (hazard ratio = 23.4, 95% confidence interval [CI] 5.3–102.0).
Conclusions
Transcriptomic analysis has allowed for a detailed characterisation of IBD pathobiology, with important potential translational implications.
Keywords: Inflammatory bowel disease, transcriptome, Crohn’s disease, ulcerative colitis, transcription factor
Graphical Abstract
Graphical Abstract.
SIGNIFICANCE OF THIS STUDY.
What is already known about this subject?
Transcriptomic studies in IBD have identified susceptibility genes that represent new biological pathways and potential treatment targets, including PAI1.
Blood expression profiles associate with the need for treatment escalation in IBD. Mucosal transcriptomic signatures of IBD may predict response to biologic agents.
Dynamic growth of knowledge about regulatory networks has enabled inference of transcription factor activity from transcriptomic data.
Despite a decade of expression profiling research, whole-blood transcriptome in IBD remains underexplored.
What are the new findings?
We demonstrate that an IBD-specific whole-blood transcriptomic profile can be defined and replicated with similarities in expression profiles between Crohn’s disease and ulcerative colitis.
New-onset IBD is characterised by overexpression of bactericidal neutrophil glycoprotein CD177 and an intestinal stem cell marker OLFM4.
Smoking in IBD is associated with greater expression of GPR15, which encodes a T cell colon-homing receptor.
Dysregulation of IL-1, IL-4, and IL-13 pathways are present at diagnosis.
Comparative transcription factor activity in the IBD Character and independent replication cohorts revealed consistently increased expression footprints of NFE2, SPI1 [PU.1], CEBPB, and IRF2 in IBD compared with controls.
A number of relatively simple oligo-marker expression models are associated with treatment escalation in UC and include two-parameter models—notably the proportion of expression of CLEC5A to CDH2 [C-type lectin-like protein to cadherin 2].
How might it impact on clinical practice in the foreseeable future?
The identified transcription factors may reveal targets for novel therapeutic interventions.
Whole-blood transcription analysis may allow for the assessment of disease course and outcome: a critical step towards precision medicine in IBD.
1. Introduction
The inflammatory bowel diseases [IBD], Crohn’s disease [CD] and ulcerative colitis [UC], are common causes of chronic illness worldwide, affecting 0.5–1.0% of the population in Europe and North America.1 IBD is now a global health burden with rising incidence and prevalence, particularly among newly industrialised nations.1 Although mortality is low, morbidity associated with IBD remains high and health care costs continue to increase.
IBD is characterised by intestinal and circulatory inflammatory changes, and much of the progress in drug development has been fuelled by our improved understanding of the molecular processes involved in disease pathogenesis. Although this has transformed the medical management of IBD,2,3 the pathogenesis of this disease is still not completely understood. Furthermore, given the heterogeneity within IBD, treatment response differs widely between patients. With the advancements in ‘-omic’ technologies, there has been progress in our understanding of the molecular profiles of disease onset, drug resistance, and disease course.4
Among these, transcriptomic analyses have contributed to addressing these challenges by identifying signatures correlated with response to therapy5,6 and a T cell-derived signature that associates with the need for treatment escalation.7 The reproducibility of these signals has not been established and the field has been confounded by concerns regarding inconsistencies in analytical techniques and in the criteria for treatment escalation.8 Most recently, analysis of the whole blood-derived transcriptome has shown promise in a study by Biasci et al., and has led to a multicentre clinical trial in the UK to validate this signature as a predictor of treatment escalation.9
The applications of ‘-omic’ technologies to define disease pathogenesis and to identify new targets for therapeutic interventions are high on the research agenda for IBD.10 Here, we provide the first report of the circulating transcriptome in the extended IBD Character inception cohort, presenting with suspected IBD across six European centres. We describe the gene expression profiles and, using a series of recently developed analytical tools, define expression modules and novel transcription factor [TF] involvement. We confirm these key findings in an independent, publicly available dataset, setting the scene for subsequent mechanistic and interventional studies.
2. Materials and Methods
2.1. Discovery cohort
Patients with suspected or established IBD and healthy controls were recruited across six European centres over 2013–2016 [see Supplementary Methods, including a schematic overview of the study]. Whole blood was collected in Paxgene tubes. Ion AmpliSeq Human Gene Expression Core Panels [20 802 amplicons] were run with the Ion AmpliSeq Library Kit Plus in the Wellcome Trust Clinical Research Facility in Edinburgh.
2.2. Replication cohort
The replication cohort11 from Poland included 96 adult and 103 paediatric patients with IBD [CD, n = 99, and UC, n = 100] and 95 controls of whom 52 were children. Moderate-to-severe disease was present in 36 participants with CD [29 children] and 38 with UC [31 children]. The controls included adults undergoing cancer screening and children with surgical, orthopaedic, or ophthalmological conditions without inflammation. Details of paired-end sequencing of whole-blood RNA in the replication cohort and clinical characteristics are given by Ostrowski et al.11
2.3. Transcriptomic analysis
After alignment, filtering, and quality check, differential expression was assessed with DESeq2. Genes expressed differentially with log-fold change [LFC] >1, which also retained significance after Bonferroni correction, were subject to ontology investigations. DESeq2-normalised expression data were used in downstream applications. A weighted gene co-expression network analysis was done with the R package WGCNA. Traits correlated with modules included: diagnosis, gender, age, C-reactive protein [CRP], smoking, Montreal classification, and the need for treatment escalation. Three methods were used for the inference of TF activity from the expression data: Effector and Perturbation Estimation Engine [EPEE],12 ChIP-X Enrichment Analysis 3 [ChEA3],13 and Discriminant Regulon Expression Analysis [DoRothEA2] v2.14 In order to maximise the true-positive rate, the EPEE and ChEA3 results were intersected [Supplementary Methods]. Immunity-focused differential ratio analysis with intermediary inference [DRAIMI] was conducted. Similarities of individual transcription profiles to LM22 cell type signatures were assessed using CIBERSORT. To determine how these results compare with the mucosal expression profiles, we used a meta-analysis of intestinal biopsy microarray studies performed by Granlund et al.15
2.4. Treatment escalation
This sub-study aimed to differentiate between patients who required an escalation of therapy within the first year following diagnosis and those who remained escalation-free throughout and at the end of the follow-up period lasting not less than 1 year [patients with escalation within the first week following diagnosis were excluded]. Treatment escalation was defined as the need for a biologic, ciclosporin, and/or surgery, instituted for a disease flare after initial remission. In UC, the definition of treatment escalation also included any patient requiring colectomy during the index admission. We built models predicting the need for escalation using analyses of whole-blood transcriptomes and models based on: [a] the elastic-net regression, [b] the random forest, and [c] a custom procedure exploiting differences of unrelated transcript ratios. Additionally, we applied elastic-net and random forest classifications to a set of transcripts matching the ones used by Biasci et al.16 in order to confirm their predictive potential. Details, including escalation criteria from Biasci et al., are given in the Supplementary Methods.
3. Results
3.1. Overview of results section
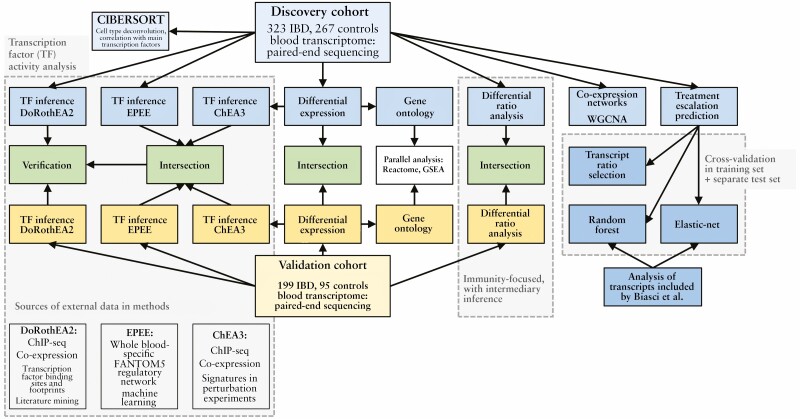
This section starts with the presentation of the differential expression analysis in the discovery and replication cohorts, and the related gene ontology investigations. These data are followed by a description of modules and hub genes demonstrated by weighted gene correlation network analysis [WGCNA]. Next, outcomes from a new type of gene expression ratio-based analysis are presented. Subsequently,TF inference using the three methods across the discovery and replication cohorts is described, and the results are compared and analysed in the contexts of gene expression and deconvoluted cell type abundance. Finally, the transcriptomic data are used to build models predicting treatment escalation. We demonstrate that the best-performing predictive model in UC was related to the key identified TFs. A graphical overview of the study is also presented in Figure 1.
Figure 1.
Overview of the study.
3.2. Clinical characteristics of the discovery cohort [IBD Character]
In total, 590 participants were recruited, including 156 patients with CD, 167 diagnosed with UC, and 267 controls, which included 54 healthy controls and 213 symptomatic controls with no evidence of IBD at follow-up. The baseline clinical characteristics are summarised in Table 1 in Supplementary Methods by centre. Of note, UC patients were older [p = 2.4 × 10-5] and less likely to smoke [p = 1.0 × 10-8] compared with controls, but no differences in smoking patterns or demographics were observed between CD patients and controls.
Table 1.
Basic characteristics of the discovery [IBD Character] transcriptomics cohort. Values are expressed as a median [1st–3rd quartile] or a percentage, and also as mean ± standard deviation for endoscopic scores.
| CD n = 156 [27%] |
UC n = 167 [28%] |
Non-IBD n = 267 [45%] |
|
|---|---|---|---|
| Age, years | 26 [21–37] | 35 [27–46] | 30 [23–40] |
| Sex, % female | 77 [49%] | 67 [40%] | 145 [54%] |
| CRP, mg/L | 7 [2–28] | 3 [1–14] | 2 [1–5] |
| Current smoker | 43/132 [33%] | 10/163 [6%] | 47/245 [19%] |
| CD location | |||
| L1/ L1+L4 | 49 [31%]/ 1 [1%] | ||
| L2/ L2+L4 | 41 [26%]/ 5 [3%] | ||
| L3/ L3+L4 | 47 [30%]/ 10 [6%] | ||
| L4 | 3 [2%] | ||
| CD behaviour | |||
| B1 | 127 [84%] | ||
| B2 | 13 [8%] | ||
| B3 | 12 [8%] | ||
| Froslie score [n = 121] | 4 [2–7] mean 5.3 ± 4.6 |
||
| Harvey–Bradshaw Index [n = 99] | 6 [2.5–7.75] | ||
| UC extent | |||
| E1 | 42 [25%] | ||
| E2 | 54 [32%] | ||
| E3 | 72 [43%] | ||
| Mayo endoscopic subscore [n = 122] | 4 [2-7] mean 4.6 ± 3.0 |
||
| Treatment-naive | 38 [75%] | 104 [63%] | |
| Most common medications at recruitment | Oral prednisone 16 [10%] Intravenous steroids 11 [7%] Oral budesonide 8 [5%] Oral 5-ASA 6 [4%] |
Rectal 5-ASA 26 [16%] Oral 5-ASA 20 [12%] Oral prednisone 15 [9%] Intravenous steroids 13 [8%] |
|
| Froslie score in patients on treatment | 3 [2.75–7.0] mean 4.5 ± 3.4 |
||
| Mayo endoscopic subscore in patients on treatment | 4.0 [2.0–5.0] mean 4.0 ± 2.9 |
CD, Crohn’s disease; hsCRP. high-sensitivity C-reactive protein; IBD, inflammatory bowel disease; UC, ulcerative colitis; 5-ASA, 5-aminosalicylate.
3.3. Discovery cohort: differential expression analysis highlights inflammation, neutrophils, and antimicrobial peptides
After filtering, 14 182 whole-blood transcripts were included in the differential expression analysis in the discovery cohort. The number of differentially expressed transcripts was 8697 in IBD vs. controls, 7152 in CD vs. controls, 8521 in UC vs. controls, and 1664 in CD vs. UC [Supplementary Table 1]. The split between the over- and under-expressed genes was almost equal. However, the absolute value of the log2-fold change [LFC] in comparisons against controls exceeded unity only for over-expression [1.4% for IBD vs. controls]. Colonic CD and UC appeared transcriptomically similar. An increased expression of alpha defensins was found in colonic vs. ileal CD. The genes with the most significant expression differences are presented in volcano plots [Figure 2; Supplementary Table 1]. Results of a subanalysis focusing on smoking, which implicate GPR15 and LRRN3, are presented in Supplementary Table 2.
Figure 2.
Volcano plots showing differential expression of whole-blood transcripts between inflammatory bowel disease [IBD] and subtypes of IBD vs. controls [A–C] and CD vs. UC [D] in the discovery cohort. Only transcripts with a false-discovery rate <0.05 are shown. Genes with log2-fold change [LFC] >2 are indicated in red. Genes with the top 10 most significant p-values and top 10 LFC are labelled [the two lists may overlap]. The genes labelled in panel D may be considered overexpressed in CD relative to UC. CD, Crohn’s disease; UC, ulcerative colitis.
3.4. Replication cohort: differential expression analysis and integration confirm involvement of neutrophil marker CD177, alpha-defensins, and OLFM4
The results of an analogous DESeq2 analysis of pooled IBD and control data from the replication cohort are presented in Supplementary Table 1 and Supplementary Figure 1. Briefly, 13 264 genes were included and the number of entities with a false-discovery ratio [FDR] <0.05 was 5402 in IBD, 4200 in CD, 5491 in UC, and 385 in an additional CD vs. UC comparison. From the highest results for both datasets [absolute LFC >1], genes with significant Bonferroni-corrected p-values were extracted to provide a consensus shortlist [Figure 3].
Figure 3.
The inflammatory bowel disease [IBD] differential expression consensus shortlist obtained by intersecting data from the discovery [IBD Character] and replication cohorts [Ostrowski et al.]. Flowchart illustrating the intersection of differential expressed transcripts in the discovery and the replication data. Mean log2-fold changes [LFC] and Bonferroni-corrected p-values [pBonf.] for genes overexpressed in the discovery and replication cohorts. Only genes with absolute LFC >1 and pBonf. <0.05 in both datasets were selected. This selection was performed separately in the IBD, Crohn’s disease [CD], and ulcerative colitis [UC] data.
3.5. Gene ontology brings focus to neutrophils, antimicrobial peptides, and metalloproteinases
To identify common ontology terms between the discovery and replication cohort, gene ontology analyses were performed. Primary gene ontology results were concordant in CD and UC, providing a high level of significance for IBD. The main ontology terms in the discovery and the replication cohort overlapped [Figure 4]. Myeloid leukocyte function and immune responses best described the differences between IBD and controls. Neutrophil degranulation [p = 1.1 × 10-16], antimicrobial peptides [p = 9.8 × 10-8], and metalloproteinase activity [p = 5.5 × 10-4Figure 4] were implicated. The complete results of the gene ontology and Reactome analyses can be found in Supplementary Tables 3 and 4.
Figure 4.
Most significant gene ontology sets [A] and reactome pathways [B] enriched by genes overexpressed in transcriptomes from the discovery [IBD Character] and replication [Ostrowski et al.] cohorts. Ratio represents the fraction of genes from the target dataset which were identified among the overexpressed genes. FDR, false-discovery ratio; IBD, inflammatory bowel disease.
3.6. Weighted correlation network analysis modules link inflammation and clinical traits, and feature toll-like receptor and neutrophil cytosolic factor genes
WGCNA of the discovery dataset identified 20 modules in IBD, 19 in CD, and 16 in UC. From these, we selected modules that were at least moderately associated with any of the traits [r >0.4, p <0.05]: three in IBD, two in CD, and five in UC. The pro-inflammatory modules were similar in all analyses and comprised genes associated with leukocyte activation and immunity. Conversely, the identified anti-inflammatory modules related to T cell differentiation and activation. The only module to associate with the need for treatment escalation [r = -0.50, p <0.0001] was found in UC and included genes with low module membership scores [hub gene AMPD2]. The analysis of modules related to clinical traits of IBD using protein-protein interaction networks revealed the involvement of TLR4, NCF2, and SPI1 across IBD, and integrins in UC [ITGA2B, ITGB3]. A more detailed description of WGCNA results along with a complete list of hub genes are presented in Supplementary File 1. A description of the modules and a full list of genes with module membership scores and tables of module-trait associations are presented in Supplementary File 1 and Supplementary Table 5.
3.7. Immunity-focused differential ratio analysis with intermediary inference implicates regulation in pattern recognition receptors and NfkB pathways
In the discovery dataset, DRAIMI unequivocally implicated pathways linked to pattern recognition receptors. The top result was RELA [also known as p65; 67.2% of all protein-protein interactions], followed by TLR4 [55.3%], and MYD88 [54.2%]. High involvement was noted also for three dual-specificity phosphatases [DUSP7, DUSP4, DUSP6] and natural cytotoxicity receptors 1 and 3 [NCR1, NCR3]. In terms of the number of interactions, these were supplemented by NFKB1, TRIM21, and MAPK3. The top results also included SYK. Almost all the listed genes [RELA, TLR4, MYD88, DUSP4, DUSP6, DUSP7, NFKB1, TRIM21, MAPK3] belong to pattern recognition pathways and specifically highlight the TLR4-MYD88-RELA[p65]-NfkB axis. Within the 304 implicated genes [Supplementary Table 6], notable results also include HLA and IRF, NFATC2, IL4 and its receptor, IL13, STAT3, ITGAV1, ITGAM, JAK1, IL2 and its receptors, FYN, TYK2, TNF, VCAM1, and other genes that fit well with prior knowledge. The list also includes CEBPB, which is one of the key genes identified by the TF activity inference analysis.
In the validation cohort, SYK, MAPK1, and MAPK9 were among the top results, emphasising the role of pattern-recognition receptor pathways, and RELA was replicated, albeit with less significance [Supplementary Table 6]. The two most important results in the validation analysis were STAT4 and PTPN6, and the results’ gene ontology was strongly linked to cell activation in the immune context and also included protein degradation. Overall, DRAIMI underscored the importance of regulation in pattern recognition receptors and NfkB pathways in the whole blood of patients with IBD.
3.8. Transcription factor activity inference across cohorts and methods exposes the relevance of NFE2, SPI1, CEBPB, and IRF2 to IBD
TF activity inference was then conducted, and the obtained results were intersected across methods and cohorts. Both EPEE and ChEA3 identified TFs whose activity differed between patients from the IBD groups and the controls. The number of TFs ordered according to EPEE was 302, and in the case of ChEA3 it equalled 1632. Here, we initially focused on TFs most consistently appearing in the most important EPEE and ChEA3 results from the discovery [IBD Character] and replication [Ostrowski et al.] datasets [Table 2]. All the EPEE regulator scores and ChEA3 and DoRothEA2 results are presented in the supplementary data [Supplementary Tables 7–9].
Table 2.
Key transcription factor [TF] activity differences in inflammatory bowel disease [IBD], Crohn’s disease [CD], and ulcerative colitis [UC] as measured vs. controls in two datasets using two different approaches [EPEE and ChEA3]. TFs inferred from both datasets [discovery: IBD Character, replication: Ostrowski et al.], using both methods, and confirmed by DoRothEA2, are printed in bold and highlighted. Each set of results lists TFs in decreasing order of importance. SPI1 encodes PU.1
| Discovery cohort | Replication cohort | |||||
|---|---|---|---|---|---|---|
| IBD | CD | UC | IBD | CD | UC | |
| EPEE – overactive | SPI1, NFE2, FOXO3, ETS2, ITGB2, RXRA, IRF1, ETV6, IRF2, RARA | ARID3A, CEBPB, PKNOX1, PLAGL1, MAX, CEBPA, MAFB, CUX1, E2F3, LMO2 | ARID3A, NFE2, CEBPB, CEBPA, ETS2, PLAGL1, RFX2, MAX, E2F3, TAL1 | SPI1, FOXO3, NFE2, ITGB2, STAT6, IRF1, KLF13, EWSR1, IRF2, ETV6 | SPI1, IRF1, NFE2, ETV6, FOXO3, FOXO4, STAT6, FLI1, CEBPD, KLF13 | SPI1, FOXO3, NFE2, KLF13, ITGB2, STAT6, ETS2, FOXO4, XBP1, ELF4 |
| EPEE – deficient | LEF1, RUNX3, KLF12, SPIB, IRF8, SP4, FOXP1, TP53, SMAD3, FOXJ3 | EWSR1, LEF1, ELK1, STAT6, RUNX3, SP1, IRF1, ELF1, ZFX, YY1 | LEF1, IRF1, EWSR1, STAT6, RUNX3, ZFX, SP1, ELK4, ELK1, SP4 | SREBF1, POU2F2, GATA3, SPIB, TFCP2, ZNF740, TBP, IRF8, TBX21, ZNF350 | TBX21, POU2F1, NFKB2, EWSR1, SPIB, SP1, SMAD3, REST, FOXO1, BHLHE40 | ELF1, SP1, CREB1, TBX21, POU2F1, GABPA, NFKB2, REST, BHLHE40, FOXO1 |
| ChEA3 – altered activity | SPI1, NFE2, NFE4, ZNF746, CEBPB, ZNF438, MTF1, BATF2, HLX, GATA1, CEBPE, ELF4, IRF2, MXD1, LYL1, ZNF467, SP110, IRF1, BORCS8-MEF2B, TRAFD1 | SPI1, NFE4, BATF2, CEBPB, NFE2, HLX, IRF1, MTF1, ZNF438, ZNF746, ZNF467, IRF2, PLSCR1, SP110, MXD1, ELF4, CEBPE, TRAFD1, IRF9, TFEC | SPI1, NFE2, NFE4, ZNF746, ZNF438, CEBPB, MTF1, GATA1, HLX, CEBPE, ELF4, MXD1, BORCS8-MEF2B, LYL1, ZNF467, TAL1, SP110, TET2, STAT5B, TFE3 | DNTTIP1, NFE2, CEBPE, ZNF524, ZNF787, SPI1, NFE4, GLMP, TFE3, LTF, BORCS8-MEF2B, ELF4, ZNF581, CEBPB, KLF1, RARA, TFEB, ZNF746, ZNF672, LYL1 | ZNF524, NFE2, CEBPE, HLX, ZNF787, SPI1, DNTTIP1, LTF, GLMP, BORCS8-MEF2B, TFE3, ZNF467, FLI1, ZNF581, RARA, CEBPA, KLF1, CEBPB, NR1H2, LYL1 | ZNF524, ZNF787, DNTTIP1, LTF, GLMP, TFE3, ZNF581, ZNF408, BORCS8-MEF2B, CEBPE, KLF1, NFE2, THAP7, SPI1, GMEB2, ZBTB45, NFE4, ZNF672, ZBTB17, CEBPB |
3.9. Intersection of results from TF inference uncovers patterns constant across diverse methods and two IBD cohorts
Four TFs were identified using the intersection procedure: NFE2, SPI1 [PU.1], CEBPB, and IRF2 [Figure 5]. DoRothEA2 predicted the activity of all four TFs to be increased in IBD. Given the transcriptome similarity between CD and UC—and insignificant differences in the Montreal subgroup analyses—attempts at identifying CD- or UC-specific TF activity yielded inconsistent results. EPEE suggested that IRF9, STAT1, and ELF1 activity in CD was lower than in UC. On the other hand, the transcriptional footprints of SPI1 [discovery] and IRF4 [replication] were clearer in CD than in UC, where C-reactive protein [CRP] was lower. In a supplementary analysis, ChEA3 highly ranked NFE2, SPI1, CEBPB, and IRF2 in IBD mucosal transcriptomes.
Figure 5.
The intersection of EPEE and ChEA3 results in the discovery and replication data. Rectangles filled in blue indicate the presence of a transcription factor among the most significant results in the given analysis. Contrasts between inflammatory bowel disease [IBD], Crohn’s disease [CD], ulcerative colitis [UC], and controls were explored. DoRothEA2 was used to confirm the findings [thus filtering out IRF1, which is not shown]. SPI1 encodes PU.1.
3.10. Correlation between key TFs and other transcripts unveils inflammatory context
Correlations of NFE2, SPI1 [PU.1], CEBPB, and IRF2 with other transcripts are presented in Supplementary Table 10. NFE2 correlated strongly with OSM [rho = 0.79, p = 4.75 × 10-69], TLR4, NCF4, and ITGAM. SPI1 [PU.1] correlated with the kinase RPS6KA1 [rho = 0.86, p = 3.75 × 10-97] and matrix metalloproteinase inhibitor TIMP2, as well as with NCF2 and NCF4. CEBPB expression associated with STX3 [rho = 0.74, p = 1.73 × 10-56], whose loss causes microvillus inclusion disease,17 INFGR2, inflammasome-triggering NLRC4, and TLR5. Last, IRF2 correlated with mitochondrial superoxide dismutase SOD2 [rho = 0.75, p = 2.1 × 10-60], the IL-1 antagonist ILR1N, the immune signal transducer MYD88, and IRF9.
3.11. Analysis of TFs and cellular composition hint at a possible role of NFE2 and SPI1 in macrophage polarisation in IBD
The expression of the four selected TFs in individual subjects was correlated with the predictions of cell type abundance obtained using LM22 signatures in the discovery cohort. Correlations in IBD and controls appeared different with regard to B cells [in IBD, stronger correlation with TFs in naïve and weaker in memory B cells], γδ T cells [stronger correlation with CEBPB in IBD], and M0 macrophages [stronger correlation with NFE2 and SPI1/PU.1 in IBD, Figure 6]. Of note, stronger correlations imply a more systematic association between cell type abundance and gene expression, not necessarily higher expression. Similarities to LM22 signatures by group can be accessed in Supplementary Table 11.
Figure 6.
Heatmap illustrating Pearson’s correlations between transcription factor expression and LM22-predicted cell type abundance in the whole blood of patients with inflammatory bowel disease [IBD] and in controls, in the discovery cohort. A stronger absolute correlation suggests more consistent expression in the cell type. Apart from B cells, γδ T cells, and M0 macrophages, the absolute differences between the two groups were small [<0.3].
3.12. Prediction of treatment escalation using gene expression data
Data on escalation of treatment were available for 204 IBD patients from the discovery cohort [mean follow-up 580 days, 24.0% required escalation], including 89 with CD [mean follow-up length 502 days, escalation in 34.8%] and 115 with UC [mean follow-up length 641 days, escalation in 15.6%]; 151 patients have not been exposed to anti-inflammatory or immunosuppressive medication at baseline [74%; Table 3]. Before escalation in the training cohort, patients with CD most commonly received the following treatments: azathioprine [n = 9], 6-mercaptopurine [n = 4], azathioprine and prednisolone [n = 2], azathioprine and budesonide [n = 2], prednisolone [n = 2], budesonide [n = 2], intravenous methylprednisolone [n = 2]. In UC, the following treatments were most frequently used before escalation: prednisolone and 5-aminosalicylates [n = 6], azathioprine [n = 4], 5-aminosalicylates alone [n = 2].
Table 3.
Basic characteristics of randomly split subgroups of the discovery cohort [IBD-Character] used for building predictive models and assessing their performance. Values are expressed as a median [1st-3rd quartile] or a percentage.
| CD | UC | |||
|---|---|---|---|---|
| Training | Testing | Training | Testing | |
| n | 71 | 18 | 92 | 23 |
| Sex, % female | 38 [53%] | 10 [56%] | 39 [42%] | 9 [39%] |
| Age, years | 27.0 [24.0-34.0] | 32.0 [24.0-52.0] | 36.5 [27.0-44.0] | 32.0 [24.0-46.0] |
| CRP, mg/L | 8.75 [2.2-43.0] | 9.4 [2.2-73.0] | 3.7 [1.2-14.0] | 2.2 [1.5-9.1] |
| Treatment-naive | 56 [79%] | 14 [78%] | 64 [70%] | 17 [74%] |
CRP, C-reactive protein.
3.13. Machine learning methods achieve transcriptome-only prognostication
We applied machine learning to build prognostic models differentiating between patients who would require treatment escalation within the first year or remain escalation-free for at least 1 year. Elastic-net models had moderate performance in cross-validation (area under the curve [AUC] 0.70–0.79), which was confirmed in test samples of IBD and UC but not in CD cases [AUC 0.80–0.83 and in CD 0.41; Supplementary File 2]. Random forest classifiers had cross-validation AUCs ranging from 0.68 in IBD to 0.82 in UC [in test sets 0.74–0.76], indicating that the transcriptomic data could serve to predict escalation, albeit not precisely. Analysis of variable importance in random forest models hints at the potential role of C14orf28 in IBD overall, and of FZD6 and ARRDC2 in CD, as well as of MPZL2 and CSNK2A1 in UC [Supplementary File 2]. Gene ontology did not reveal specific pathway enrichment.
3.14. CLEC5A-to-CDH2 gene expression ratio enables prognostication in UC and appears related to macrophage polarisation
The custom search for predictive transcript ratios yielded some of the highest AUC values in cross-validation [IBD 0.76, CD 0.87, UC 0.90]. The top-performing ratios discovered by the custom procedure in the training sets were CLEC5A/CDH2 in UC [AUC in test set 0.85], HIST1H3H/GPR162 in CD [AUC 0.74], and STAB1/GPR162 in IBD [AUC 0.69]. The CLEC5A/CDH2 ratio achieved performance superior to complex models, with a hazard ratio for treatment escalation of 23.4 [95% CI 5.3–102.0; Figure 7]. In UC patients with available high-sensitivity [hs] CRP, AUC for the CLEC5A/CDH2 ratio was higher compared with hsCRP: 0.90 [0.84–0.96] vs. 0.69 [0.53–0.85; p = 0.016] and not affected by prior exposure to IBD medications. In UC, the ratio with the second-best discriminatory power was ANPEP/SEC61A2. The top quotients are listed in Supplementary Table 12.
Figure 7.
Ulcerative colitis treatment escalation depending on the CLEC5A/CDH2 ratio [low-risk <3]. The log-rank test p-value is shown. Hazard ratio: 23.4 (95% confidence interval [CI] 5.3–102.0). Patients with escalation after 1 year and censored within the first year [excluded from modelling] are also included for illustration.
3.15. Genes belonging to a model by Biasci et al. are validated to have predictive potential
Of the 15 informative [non-housekeeping] transcripts used in the prediction score proposed by Biasci et al., 12 could be extracted from the IBD Character dataset [Supplementary Methods]. The optimal elastic-net and random forest models performed well despite the limited number of predictors. A cross-validation AUC of 0.65–0.82 was achieved for IBD Character escalation criteria and 0.60–0.79 using the original escalation criteria from Biasci et al. The random forest model for UC reached AUC 0.87 in the test set using the discovery cohort [IBD Character] escalation criteria. Variable importance analysis of IBD Character random forest models revealed that the following transcripts from Biasci et al. were the most informative: NUDT7, GZMH, and IL18RAP in IBD overall, GZMK and LY96 in CD [dominating the remaining genes], and IL18RAP in UC [exceeding other transcripts in UC].
4. Discussion
This combined dataset represents a comprehensive study of the whole-blood transcriptome in IBD, and successfully identifies and replicates genes of interest in differential expression profiles. We also are able to describe co-expression networks and, in a novel detailed analysis, we identify and replicate TFs that may be driving the disease. To strengthen the TF analyses, we employed two complementary methods and examined two independent datasets of IBD cases and controls. New correlates of treatment escalation in UC are proposed. The results highlight underexplored areas of IBD biology, which provide new insights into pathogenesis and possible therapeutic interventions.
4.1. Patients and controls
The IBD Character cohort comprised comparable numbers of patients with CD, UC, and controls across Europe. This was an inception cohort at presentation, which is reflected by a slightly higher upper quartile of CRP in CD. The location of CD and the extent of UC mirror the general structure in the population.18 The controls included both symptomatic controls undergoing investigations for IBD, with no evidence of IBD at follow-up, and young, healthy Swedish volunteers. The access to and analysis of the dataset described by Ostrowski et al. provided replication of the findings from the discovery cohort.
4.2. Differential expression
The overriding finding in this context was that CD and UC expression profiles were remarkably similar. CD177, OLFM4 [olfactomedin 4], DEFA4 [defensin A4], GALNT14, and ELANE exhibited the greatest differences in the joint analysis. CD177 encodes human neutrophil alloantigen 2a, which is present on cells exhibiting bactericidal activity in IBD,19 and impairs neutrophil migration.20 Interestingly, CD177+ neutrophils producing IL-17 are increased in allergic asthma.21 Olfactomedin 4 is a marker of intestinal stem cells and is known to be increased in the IBD mucosal proteome.22,23 The copy number of defensins A1–3 correlates with the diagnosis of CD and its colonic location.24 The product of GALNT14—a glycosylase—is enriched in neutrophils, and ELANE is a neutrophil elastase. Increased expression of alpha defensins in colonic vs. ileal CD is in line with a current hypothesis on defensin deficiency in ileal CD.25
Smoking is considered the most significant modifiable risk factor for CD and its differential effects in CD and UC are of particular interest. The only transcript that clearly distinguished itself in the comparisons of current vs. never smokers was the G-protein coupled receptor GPR15, which encodes a colon-homing molecule for T cells.26 GPR15+ T cells secrete less IFN-γ, but more IL-17 after stimulation, suggesting a Th17-like phenotype.27 It has been shown that regulatory T cell GPR15 expression in UC patients is increased28 and, most recently, the dependence of GPR15 transcription on aryl hydrocarbon receptor signalling has been demonstrated.29
4.3. Weighted gene co-expression network analysis
WGCNA yielded modules correlated with IBD and CRP. Yet, none of the modules associating with the diagnosis were independent of inflammation. Although the resulting gene lists were too extensive to clearly illustrate specific functions, they seem informative with regard to the actors of the anti-inflammatory response: CD3E, which is a subunit of the CD3 T cell co-receptor [otelixizumab target], CARD11, which induces apoptosis and NF-κB, and SMAD3, which is crucial for TGFβ signalling.30 Some of the genes with high anti-inflammatory module membership cause very early-onset IBD when deficient [ZAP70, LRBA].31
4.4. Transcription factors implicated in analysis
A key novel discovery in our study lies in the replication of multiple TFs that are dysregulated in IBD [Table 4].
Table 4.
Transcription factors implied in inflammatory bowel disease by this study point towards involvement of the Th9 immune response, monocytes/macrophages, and IFN-ϒ.
| Biological significance | |
|---|---|
| NFE2 | Implied in megakaryocyte maturation and development of erythroid colonies.32,33 May bind IL-8 promoter.34 In late-stage megakaryocytes, NFE2 is upregulated by IRF2 [see below], but suppressed by IFNα 35,36 |
| SPI1 [PU.1] | Key regulator of Th9 immunity, marker of Th9 cells. Inhibits uncontrolled neutrophil activation.37 Together with CEBPB [see below] enables IL1B transcription38 |
| CEBPB | Involved in monocyte survival,39 expression of IL-17-regulated genes,40 and development of Th2 cells. May form homo- or heterodimers with other CEBP proteins |
| IRF2 | Induced by IFNϒ, competes with IRF1 to deactivate the expression of IFNα and IFNβ. Activates IL-7 and belongs to non-canonical inflammasome detecting cytosolic lipopolysaccharide41,42 |
NFE2, nuclear factor erythroid 2; SPI1, Spi-1 proto-oncogene; CEBPB. CCAAT enhancer binding protein beta; IRF2, interferon regulatory factor 2.
NFE2 is underexplored in the context of mucosal inflammation and immunity in general. Both NFE2 and SPI1 [PU.1] are overexpressed and undermethylated in systemic sclerosis in comparison with controls.43
SPI1 [PU.1] governs Th9 immunity, which in IBD is still insufficiently understood, but appears predominantly pro-inflammatory.44 In animal models, interleukin-9 deficiency protected mice against trinitrobenzene sulphonic acid [TNBS]- and oxazolone-induced colitis.45 The IL-9 receptor is also overexpressed in the intestinal epithelium of patients with IBD. Consequently, IL-9 appears as a therapeutic target in UC. Enokizumab [MEDI-528], a humanised anti-IL-9 monoclonal antibody, was well tolerated in asthmatic patients.46 To the best of our knowledge, there are no current trials examining anti-IL-9 for the treatment of IBD. Moreover, strategies against PU.1 [SPI1 product] are currently being developed in oncology.47
CEBPB was found to lie 150 kb downstream from an IBD risk locus.48 Interestingly, profiling of transcription start sites in IBD biopsies also highlighted CEBPB as potentially involved in the disease.49 It is overexpressed in the mucosa of patients with UC50 and strategically positioned at the interface of myeloid and epithelial inflammation clusters in a network analysis of mucosal IBD transcriptomes.51 A recent study by Sudhakar et al., employing ChEA3 in transcriptomic modules from CD4+ and CD14+ cells in 33 patients with CD, hinted at the involvement of both SPI1 and CEBPB.52 Some research has indicated a role for fibrates in reducing CRP [IL-6] in response to IL-1 via CEBPB.53
Finally, IRF2 mediates the functions of IFNγ. Fontolizumab, a monoclonal antibody against IFNγ, was not efficacious in CD trials, a result that might have been influenced by a large placebo effect as well as the anti-inflammatory effects of IFNγ through downregulation of IL-23.54
4.5. Transcription factor activity inference
In brief, the two main TF inference methods employed in this study—EPEE and ChEA3—are complementary in terms of accepted transcriptomics data, computational approaches, regulon tissue-specificity, and regulatory network sources. Other notable TF inference tools include BART, DoRothEA v2, iRegulon [a Cytoscape application], TFEA.ChIP, and oPOSSUM. We chose DoRothEA v2 for reasons that included an up-to-date regulon library [Supplementary Methods]. Our concordance-based approach to filtering is arbitrary, but motivated by the way TF inference tools are developed and aimed at indicating only TFs unequivocally supported by diverse methods in independent data sources.
4.6. Predicting treatment escalation
The translational application of transcriptional profiling of IBD in practice is of immediate clinical interest. A whole blood-derived 17-gene classifier proposed by Biasci et al. has been shown to be highly predictive of disease course. This forms the basis of the current biomarker-stratified PROFILE trial in CD.16 By applying machine learning to our dataset using 12 out of 15 informative genes, we provide confirmatory evidence of such a signature predicting treatment escalation with an AUC of up to 0.87. This is important as the first independent confirmation of this signature. Moreover, we report that simpler models, involving two transcripts only, may have comparable utility. In the strongest binary model we derive, the CLEC5A/CDH2 signature [Box 1] strongly associates with the need for treatment escalation in UC and makes a promising candidate for further external validation [Box 1].
Box 1. Functions of genes in the ulcerative colitis [UC] treatment escalation-associated CLEC5A/CDH2 ratio.
CLEC5A [C-type lectin-like protein] is expressed in leukocytes and remains under the control of SPI1 [PU.1] and CEBPB,55,56 which we show herein to be critical IBD transcription factors. Although CLEC5A is chiefly known for being a dengue virus receptor, it was recently implicated in IBD as a marker for M1 macrophages, which express TNF.57,58 The CLEC5A signalling cascade includes IBD-related proteins such as Zap-70, TREM-2, and SYK. CDH2 [cadherin 2, N-cadherin] is a calcium-dependent cell adhesion glycoprotein that enables dendritic cell migration59 and was proposed as a marker for the epithelial-to-mesenchymal transition in circulating tumour cells.60 CDH2 may associate with immune tolerance as it predicts breast cancer recurrence, shorter survival in multiple myeloma,61 and Ewing’s sarcoma.62 Increased CDH2 levels are also found in CD stenotic tissue.63 In mice, CDH1 [but not CDH2] is a marker of M2 macrophages; establishing a similar role for CDH2 would suggest that the CLEC5A/CDH2 ratio is chiefly related to polarisation of these cells.
The predictive accuracy of the signature proposed by Biasci and colleagues is worthy of discussion. This was assessed using two sets of criteria: our more stringent criteria, predefined for the discovery cohort, and the broader criteria, originally applied by Biasci et al., which included azathioprine. Both demonstrate that the signature may predict the need for an escalation of therapy. However, there are some caveats. First, we note that our analysis is approximated, as the original equation is not available and could not be assessed. We did not use separate criteria for CD and UC and no data on the number of escalation events were available. Furthermore, not all transcripts used by Biasci et al. were expressed in the discovery cohort at sufficient levels. Nevertheless, these favourable results highlight the potential for generalisation of this specific gene signature, and the value of prognostic transcriptomic markers in IBD in general.
4.7. Limitations and future directions
The prognostication part of the study, albeit based on a large, international cohort, would require independent validation, and a quantitative polymerase chain reaction [qPCR] assay would need to be developed. Alternatively, flow cytometry or immunohistochemistry could also be helpful in further investigation of the role of CLEC5A in IBD, with the latter potentially bringing the method close to the bedside by prospectively studying blood smears or retrospectively analysing archived UC intestinal biopsy material. A larger sample size of any future investigations in this domain would allow for the study of the relationship between disease activity on treatment and the accuracy of prediction of the disease course.
The presented analyses implicating NFE2, SPI1, CEBPB, IRF2, and CLEC5A, CDH2 would all benefit from functional validation. Further study of the transcription factors’ importance could involve chromatin immunoprecipitation-sequencing. With regard to differentially expressed genes, discriminating between IBD-related transcriptomic changes that are protective and those that are pathogenic would also require further mechanistic study. These limitations are balanced by the large sample size, as well as by a significant overlap of the main results between different methods and in an independent replication cohort.
4.8. Conclusion
Detailed whole-blood transcriptomic analyses in two large European cohorts has progressed our understanding of several aspects of IBD. The most overexpressed genes may represent important molecules in the pathogenesis of this disease and are potentially druggable targets—in this context we highlight CD177, OLFM4, and GPR15. Moreover, a number of TFs are implicated in our discovery and validation datasets as having an important role in inflammation. In particular, NFE2, SPI1 [PU.1], CEBPB, and IRF2 are the most involved and warrant further study. Importantly, we also provide the first validation of the predictive signature proposed by Biasci et al. to be of translational use, and provide evidence that simpler models, such as the CLEC5A/CDH2 expression ratio, constitute promising candidates for validation in the prediction of treatment escalation in IBD.
Overall, our study provides the foundation for future work focusing on the mechanistic and clinical roles of the TFs implicated in IBD, including single-cell sequencing studies of blood leukocytes in IBD, with the aim of pinpointing targetable pathways and establishing the role of genes overexpressed in IBD. Further scaling up of transcriptomic research in terms of cohort sizes will be necessary to reveal the disease biology in finer detail and to establish practical classifiers with a range of diagnostic, subtyping, and prognostic applications.
Supplementary Material
Acknowledgements
The authors would like to thank the creators of EPEE, Murat Çobanoğlu and Viren Amin, for their assistance in the study, as well as all the patients and volunteers for their participation.
The data underlying this article are available in ArrayExpress (accession E-MTAB-11349).
Contributor Information
Jan K Nowak, Translational Gastroenterology Unit, Nuffield Department of Medicine, Experimental Medicine Division, University of Oxford, John Radcliffe Hospital, Oxford, UK; Department of Pediatric Gastroenterology and Metabolic Diseases, Poznan University of Medical Sciences, Poznan, Poland.
Alex T Adams, Translational Gastroenterology Unit, Nuffield Department of Medicine, Experimental Medicine Division, University of Oxford, John Radcliffe Hospital, Oxford, UK.
Rahul Kalla, MRC Centre for Inflammation Research, Queens Medical Research Institute, University of Edinburgh, Edinburgh, UK.
Jonas C Lindstrøm, Health Services Research Unit, Akershus University Hospital, Lørenskog, Norway; Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Simen Vatn, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Gastroenterology, Akershus University Hospital, Lørenskog, Norway.
Daniel Bergemalm, Department of Gastroenterology, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Åsa V Keita, Department of Biomedical and Clinical Sciences, Linköping University, Linköping, Sweden.
Fernando Gomollón, HCU ‘Lozano Blesa’, IIS Aragón, Zaragoza, Spain.
Jørgen Jahnsen, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Gastroenterology, Akershus University Hospital, Lørenskog, Norway.
Morten H Vatn, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; EpiGen Institute, Akershus University Hospital, University of Oslo, Oslo, Norway.
Petr Ricanek, Department of Gastroenterology, Akershus University Hospital, Lørenskog, Norway.
Jerzy Ostrowski, Department of Genetics, Maria Skłodowska-Curie National Research Institute of Oncology, Warsaw, Poland; Department of Gastroenterology, Hepatology and Clinical Oncology, Centre for Postgraduate Medical Education, Warsaw, Poland.
Jaroslaw Walkowiak, Department of Pediatric Gastroenterology and Metabolic Diseases, Poznan University of Medical Sciences, Poznan, Poland.
Jonas Halfvarson, Department of Gastroenterology, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Jack Satsangi, Translational Gastroenterology Unit, Nuffield Department of Medicine, Experimental Medicine Division, University of Oxford, John Radcliffe Hospital, Oxford, UK; Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK.
IBD Character Consortium:
Erik Andersson, Ian D Arnott, Monica Bayes, Ferdinando Bonfiglio, Ray K Boyapati, Adam Carstens, Christina Casén, Ewa Ciemniejewska, Mauro D’Amato, Fredrik A. Dahl, Trond Espen Detlie, Hazel E Drummond, Gunn S Ekeland, Daniel Ekman, Anna B Frengen, Mats Gullberg, Ivo G Gut, Marta Gut, Simon C Heath, Fredrik Hjelm, Henrik Hjortswang, Gwo-Tzer Ho, Daisy Jonkers, Nicholas A Kennedy, Charles W Lees, Torbjørn Lindahl, Mårten Lindqvist, Angelika Merkel, Eddie Modig, Aina E F Moen, Hilde Nilsen, Elaine R Nimmo, Colin L Noble, Niklas Nordberg, Kate R O’Leary, Anette Ocklind, Christine Olbjørn, Erik Pettersson, Marieke Pierik, Dominique Poncelet, Dirk Repsilber, Céline Sabatel, Renaud Schoemans, Alan G Shand, Johan D Söderholm, Janne Sølvernes, Mikael Sundell, Tone M Tannæs, Leif Törkvist, Anne-Clémence Veillard, Nicholas T Ventham, David C Wilson, and Panpan You
Funding
This work was supported by the European Union’s Seventh Framework Programme [FP7] grant IBD Character [2858546] and the Polish National Science Centre [2017/25/B/NZ5/02783 to JW].
Conflict of Interest
JKN reports personal fees from Norsa Pharma, a grant from Biocodex Microbiota Foundation, and non-financial support from Nutricia outside of the submitted work. RK has served as a speaker for Ferring and has received support for research from IBD-Character [EU FP7 2858546]. DB has received personal fees as speaker and/or advisory board member for Ferring, Janssen, Pfizer, and Takeda. JW reports personal fees and non-financial support from Biocodex, BGP Products, Chiesi, Hipp, Humana, Mead Johnson Nutrition, Merck Sharp & Dohme, Nestle, Norsa Pharma, Nutricia, Roche, Sequoia Pharmaceuticals, and Vitis Pharma, outside of the submitted work, and also grants, personal fees, and non-financial support from Nutricia Research Foundation Poland, also outside of the submitted work. JJH has received personal fees as speaker, consultant, and/or advisory board member for AbbVie, Aqilion AB, Celgene, Celltrion, Dr Falk Pharma and the Falk Foundation, Ferring, Hospira, Janssen, MEDA, Medivir, MSD, Olink Proteomics, Pfizer, Prometheus Laboratories, Sandoz/Novartis, Shire, Takeda, Thermo Fisher Scientific, Tillotts Pharma, Vifor Pharma, UCB, and has received grant support from Janssen, MSD, and Takeda, outside of the submitted work. JS has served as a speaker, a consultant, and an advisory board member for MSD, Ferring, Abbvie, and Shire, a consultant with Takeda, has received speaking fees from MSD, travel support from Shire, and has received research funding from Abbvie, Wellcome, CSO, MRC, and the EC grant IBDBIOM.
Author Contributions
Study design: JKN, ATA, RK, MHV, PR, JH, JS. Patient recruitment and sample processing: ATA, RK, JCL, SV, DB, AVK, FG, MHV, PR, JO, JW, JH, JS. Data analysis: JKN, ATA, RK, JS. Drafting the manuscript: JKN, ATA, RK. Study coordination: JH, JS. All authors were involved in critical review, editing, revision and approval of the final manuscript.
Conference presentation: Digestive Disease Week 2020.
IBD Character Consortium: Erik Andersson, Ian D. Arnott, MD, Monica Bayes, PhD, Ferdinando Bonfiglio, PhD, Ray K. Boyapati, MD, Adam Carstens, MD, Christina Casén, MSc, Ewa Ciemniejewska, MSc, Mauro D’Amato, PhD, Fredrik A. Dahl, PhD, Trond Espen Detlie, MD, Hazel E. Drummond, BSc, Gunn S. Ekeland, MSc, Daniel Ekman, MSc, Anna B. Frengen, PhD, Mats Gullberg, PhD, Ivo G. Gut, PhD, Marta Gut, PhD, Simon C. Heath, PhD, Fredrik Hjelm, PhD, Henrik Hjortswang, MD, PhD, Gwo-Tzer Ho, PhD, Daisy Jonkers, PhD, Nicholas A. Kennedy, MBBS, PhD, FRACP, Charles W. Lees, PhD, Torbjørn Lindahl, MSc, Mårten Lindqvist, PhD, Angelika Merkel, PhD, Eddie Modig, BSc, Aina E. F. Moen, PhD, Hilde Nilsen, PhD, Elaine R. Nimmo, PhD, Colin L. Noble, MD, Niklas Nordberg, PhD, Kate R. O’Leary, MSc, Anette Ocklind, PhD, Christine Olbjørn, MD, Erik Pettersson, PhD, Marieke Pierik, MD, PhD, Dominique Poncelet, PhD, Dirk Repsilber, PhD, Céline Sabatel, PhD, Renaud Schoemans, PhD, Alan G. Shand, MD, Johan D. Söderholm, MD, PhD, Janne Sølvernes, MS, Mikael Sundell, BSc, Tone M. Tannæs, PhD, Leif Törkvist, MD, PhD, Anne-Clémence Veillard, PhD, Nicholas T. Ventham, MRCS[Eng], PhD, MBBS, David C. Wilson, MD, MRCPCH, Panpan You, MS.
References
- 1. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018;390:2769–78. [DOI] [PubMed] [Google Scholar]
- 2. Sandborn WJ, Su C, Sands BE, et al. ; OCTAVE Induction 1, OCTAVE Induction 2, and OCTAVE Sustain Investigators. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N Engl J Med 2017;376:1723–36. [DOI] [PubMed] [Google Scholar]
- 3. Sands BE, Sandborn WJ, Panaccione R, et al. ; UNIFI Study Group. Ustekinumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med 2019;381:1201–14. [DOI] [PubMed] [Google Scholar]
- 4. Borg-Bartolo SP, Boyapati RK, Satsangi J, Kalla R. Precision medicine in inflammatory bowel disease: concept, progress and challenges. F1000Res 2020;9. Doi: 10.12688/f1000research.20928.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haberman Y, Karns R, Dexheimer PJ, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun 2019;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Czarnewski P, Parigi SM, Sorini C, et al. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun 2019;10:2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gasparetto M, Payne F, Nayak K, et al. Transcription and DNA methylation patterns of blood-derived CD8+ T cells are associated with age and inflammatory bowel disease but do not predict prognosis. Gastroenterology 2021;160:232–44.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parkes M, Noor NM, Dowling F, et al. PRedicting Outcomes For Crohn’s dIsease using a moLecular biomarkEr [PROFILE]: protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open 2018;8:e026767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weersma RK, Xavier RJ, Vermeire S, Barrett JC; IBD Multi Omics Consortium. Multiomics analyses to deliver the most effective treatment to every patient with inflammatory bowel disease. Gastroenterology 2018;155:e1–4. [DOI] [PubMed] [Google Scholar]
- 11. Ostrowski J, Dabrowska M, Lazowska I, et al. Redefining the practical utility of blood transcriptome biomarkers in inflammatory bowel diseases. J Crohns Colitis 2019;13:626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amin V, Ağaç D, Barnes SD, Çobanoğlu MC. Accurate differential analysis of transcription factor activity from gene expression. Bioinformatics 2019;35:5018–29. [DOI] [PubMed] [Google Scholar]
- 13. Keenan AB, Torre D, Lachmann A, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res 2019;47:W212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 2019;29:1363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Granlund Av, Flatberg A, Østvik AE, et al. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One 2013;8:e56818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut 2019;68:1386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wiegerinck CL, Janecke AR, Schneeberger K, et al. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology 2014;147:65–8.e10. [DOI] [PubMed] [Google Scholar]
- 18. Ng SC, Tang W, Ching JY, et al. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-Pacific Crohn’s and colitis epidemiology study. Gastroenterology 2013;145:158–65.e2. [DOI] [PubMed] [Google Scholar]
- 19. Zhou G, Yu L, Fang L, et al. CD177+ neutrophils as functionally activated neutrophils negatively regulate IBD. Gut 2018;67:1052–63. [DOI] [PubMed] [Google Scholar]
- 20. Bai M, Grieshaber-Bouyer R, Wang J, et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood 2017;130:2092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramirez-Velazquez C, Castillo EC, Guido-Bayardo L, Ortiz-Navarrete V. IL-17-producing peripheral blood CD177+ neutrophils increase in allergic asthmatic subjects. Allergy Asthma Clin Immunol 2013;9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ning L, Shan G, Sun Z, et al. Quantitative proteomic analysis reveals the deregulation of nicotinamide adenine dinucleotide metabolism and CD38 in inflammatory bowel disease. Biomed Res Int 2019;2019:3950628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neyazi M, Bharadwaj SS, Bullers S, et al. ; Oxford IBD Cohort Study Investigators. Overexpression of cancer-associated stem cell gene OLFM4 in the colonic epithelium of patients with primary sclerosing cholangitis. Inflamm Bowel Dis 2021;27:1316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jespersgaard C, Fode P, Dybdahl M, et al. Alpha-defensin DEFA1A3 gene copy number elevation in Danish Crohn’s disease patients. Dig Dis Sci 2011;56:3517–24. [DOI] [PubMed] [Google Scholar]
- 25. Wehkamp J, Stange EF. An update review on the paneth cell as key to ileal Crohn’s disease. Front Immunol 2020;11:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim SV, Xiang WV, Kwak C, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 2013;340:1456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen LP, Pan J, Dinh TT, et al. Role and species-specific expression of colon T cell homing receptor GPR15 in colitis. Nat Immunol 2015;16:207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adamczyk A, Gageik D, Frede A, et al. Differential expression of GPR15 on T cells during ulcerative colitis. JCI Insight 2017; 2:e90585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swaminathan G, Nguyen LP, Namkoong H, et al. The aryl hydrocarbon receptor regulates expression of mucosal trafficking receptor GPR15. Mucosal Immunol 2021;14:852–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benahmed M, Meresse B, Arnulf B, et al. Inhibition of TGF-beta signaling by IL-15: a new role for IL-15 in the loss of immune homeostasis in celiac disease. Gastroenterology 2007;132:994–1008. [DOI] [PubMed] [Google Scholar]
- 31. Charbit-Henrion F, Parlato M, Hanein S, et al. Diagnostic yield of next-generation sequencing in very early-onset inflammatory bowel diseases: a multicentre study. J Crohns Colitis 2018;12:1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zang C, Luyten A, Chen J, Liu XS, Shivdasani RA. NF-E2, FLI1 and RUNX1 collaborate at areas of dynamic chromatin to activate transcription in mature mouse megakaryocytes. Sci Rep 2016;6:30255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rheinemann L, Seeger TS, Wehrle J, Pahl HL. NFE2 regulates transcription of multiple enzymes in the heme biosynthesis pathway. Haematologica 2014;99:e208–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wehrle J, Seeger TS, Schwemmers S, Pfeifer D, Bulashevska A, Pahl HL. Transcription factor nuclear factor erythroid-2 mediates expression of the cytokine interleukin 8, a known predictor of inferior outcome in patients with myeloproliferative neoplasms. Haematologica 2013;98:1073–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamane A, Nakamura T, Suzuki H, et al. Interferon-alpha 2b-induced thrombocytopenia is caused by inhibition of platelet production but not proliferation and endomitosis in human megakaryocytes. Blood 2008;112:542–50. [DOI] [PubMed] [Google Scholar]
- 36. Stellacci E, Testa U, Petrucci E, et al. Interferon regulatory factor-2 drives megakaryocytic differentiation. Biochem J 2004;377:367–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fischer J, Walter C, Tönges A, et al. Safeguard function of PU.1 shapes the inflammatory epigenome of neutrophils. Nat Immunol 2019;20:546–58. [DOI] [PubMed] [Google Scholar]
- 38. Pulugulla SH, Workman R, Rutter NW, et al. A combined computational and experimental approach reveals the structure of a C/EBPβ-Spi1 interaction required for IL1B gene transcription. J Biol Chem 2018;293:19942–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura A, Hirai H, Yokota A, et al. Accelerated apoptosis of peripheral blood monocytes in Cebpb-deficient mice. Biochem Biophys Res Commun 2015;464:654–8. [DOI] [PubMed] [Google Scholar]
- 40. Chiricozzi A, Nograles KE, Johnson-Huang LM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PLoS One 2014;9:e90284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benaoudia S, Martin A, Puig Gamez M, et al. A genome-wide screen identifies IRF2 as a key regulator of caspase-4 in human cells. EMBO Rep 2019;20:e48235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kayagaki N, Lee BL, Stowe IB, et al. IRF2 transcriptionally induces GSDMD expression for pyroptosis. Sci Signal 2019;12. Doi: 10.1126/scisignal.aax4917. [DOI] [PubMed] [Google Scholar]
- 43. Zhu H, Zhu C, Mi W, et al. Integration of genome-wide DNA methylation and transcription uncovered aberrant methylation-regulated genes and pathways in the peripheral blood mononuclear cells of systemic sclerosis. Int J Rheumatol 2018;2018:7342472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gerlach K, McKenzie AN, Neurath MF, Weigmann B. IL-9 regulates intestinal barrier function in experimental T cell-mediated colitis. Tissue Barriers 2015;3:e983777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerlach K, Hwang Y, Nikolaev A, et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol 2014;15:676–86. [DOI] [PubMed] [Google Scholar]
- 46. Oh CK, Leigh R, McLaurin KK, Kim K, Hultquist M, Molfino NA. A randomized, controlled trial to evaluate the effect of an anti-interleukin-9 monoclonal antibody in adults with uncontrolled asthma. Respir Res 2013;14:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antony-Debré I, Paul A, Leite J, et al. Pharmacological inhibition of the transcription factor PU.1 in leukemia. J Clin Invest 2017;127:4297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC]. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boyd M, Thodberg M, Vitezic M, et al. Characterisation of the enhancer and promoter landscape of inflammatory bowel disease from human colon biopsies. Nat Commun 2018;9:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ufer M, Häsler R, Jacobs G, et al. Decreased sigmoidal ABCB1 [P-glycoprotein] expression in ulcerative colitis is associated with disease activity. Pharmacogenomics 2009;10:1941–53. [DOI] [PubMed] [Google Scholar]
- 51. Kaiko GE, Chen F, Lai CW, et al. PAI-1 augments mucosal damage in colitis. Sci Transl Med 2019;11. Doi: 10.1126/scitranslmed.aat0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sudhakar P, Verstockt B, Cremer J, et al. Understanding the molecular drivers of disease heterogeneity in Crohn’s disease using multi-omic data integration and network analysis. Inflamm Bowel Dis 2020. Doi: 10.1093/ibd/izaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kleemann R, Gervois PP, Verschuren L, Staels B, Princen HM, Kooistra T. Fibrates down-regulate IL-1-stimulated C-reactive protein gene expression in hepatocytes by reducing nuclear p50-NFkappa B-C/EBP-beta complex formation. Blood 2003;101:545–51. [DOI] [PubMed] [Google Scholar]
- 54. Abraham C, Dulai PS, Vermeire S, Sandborn WJ. Lessons learned from trials targeting cytokine pathways in patients with inflammatory bowel diseases. Gastroenterology 2017;152:374–88.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Batliner J, Mancarelli MM, Jenal M, et al. CLEC5A [MDL-1] is a novel PU.1 transcriptional target during myeloid differentiation. Mol Immunol 2011;48:714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu J, Chen W, Liu H, Yang H, Liu T. Transcription factor CEBPB inhibits the proliferation of osteosarcoma by regulating downstream target gene CLEC5A. J Clin Lab Anal 2019;33:e22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. González-Domínguez É, Samaniego R, Flores-Sevilla JL, et al. CD163L1 and CLEC5A discriminate subsets of human resident and inflammatory macrophages in vivo. J Leukoc Biol 2015;98:453–66. [DOI] [PubMed] [Google Scholar]
- 58. Elleisy N, Rohde S, Huth A, et al. Genetic association analysis of CLEC5A and CLEC7A gene single-nucleotide polymorphisms and Crohn’s disease. World J Gastroenterol 2020;26:2194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konradi S, Yasmin N, Haslwanter D, et al. Langerhans cell maturation is accompanied by induction of N-cadherin and the transcriptional regulators of epithelial-mesenchymal transition ZEB1/2. Eur J Immunol 2014;44:553–60. [DOI] [PubMed] [Google Scholar]
- 60. Blassl C, Kuhlmann JD, Webers A, Wimberger P, Fehm T, Neubauer H. Gene expression profiling of single circulating tumor cells in ovarian cancer: establishment of a multi-marker gene panel. Mol Oncol 2016;10:1030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vandyke K, Chow AW, Williams SA, To LB, Zannettino AC. Circulating N-cadherin levels are a negative prognostic indicator in patients with multiple myeloma. Br J Haematol 2013;161:499–507. [DOI] [PubMed] [Google Scholar]
- 62. Przybyl J, Kozak K, Kosela H, et al. Gene expression profiling of peripheral blood cells: new insights into Ewing sarcoma biology and clinical applications. Med Oncol 2014;31:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kurahara LH, Sumiyoshi M, Aoyagi K, et al. Intestinal myofibroblast TRPC6 channel may contribute to stenotic fibrosis in Crohn’s disease. Inflamm Bowel Dis 2015;21:496–506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.