Abstract
Plants use specialized root outgrowths, termed root hairs, to enhance acquisition of nutrients and water, help secure anchorage, and facilitate interactions with soil microbiome. One of the major regulators of this process is GLABRA2 (GL2), a transcriptional repressor of root hair differentiation. However, regulation of the GL2-function is relatively well characterized, it remains completely unknown whether GL2 itself functions in complex with other transcriptional regulators. We identified GIR1 and GIR2, a plant-specific two-member family of closely related proteins that interact with GL2. Loss-of-function mutants of GIR1 and GIR2 enhanced development of root hair whereas gain-of-function mutants repressed it. Thus, GIR1 and GIR2 might function as adaptor proteins that associate with GL2 and participate in control of root hair formation.
Keywords: Root hair development, GL2-Interacting proteins, Arabidopsis
1. Introduction
Root hairs enhance acquisition of nutrients and water, help secure anchorage, and facilitate interactions with soil microbiome. In Arabidopsis, single-layered root epidermal cells differentiate into hair and non-hair cells based on the position-dependent cues: epidermal cells, termed H cells, located between underlying cortical cells differentiate into root hair cells whereas epidermal cells, termed N cells, positioned directly over single cortical cells differentiate into non-hair cells [1]. One of the major regulators of this process is GLABRA2 (GL2), a transcriptional repressor that functions to suppress root hair differentiation as well as to initiate leaf trichome development [2–4]. To date, most of our knowledge about the GL2-mediated pathway for regulation of root hair development is derived mainly from the events that precede the actual involvement of GL2 and regulate its activity [4]. Thus, whereas theGL2-regulating multiprotein complexes are relatively well characterized, it remains completely unknown whether GL2 itself functions in complex with other transcriptional regulators. Here, we report identification of GIR1 and GIR2, two closely-related proteins that are specific for plants, interact with the GL2 repressor, and act as negative regulators of root hair development.
2. Materials and methods
2.1. Plant material, growth conditions, and root hair density measurements
Seeds of the wild-type Arabidopsis thaliana (ecotype Col-0) plants, the gl2-1 mutant, and the SALK_089888 and SALK_048394 lines, corresponding to the gir1-1 and gir2-1 T-DNA insertion mutants of GIR1 and GIR2, respectively, were obtained from the Arabidopsis Biological Resource Center (abrc.osu.edu). The insertion position of the left T-DNA border in the gir1-1 and gir2-1 mutants was confirmed by DNA sequencing [5]. Arabidopsis plants were grown on half-strength Murashige and Skoog (MS) medium [6], pH 5.7 with 0.6% (w/v) phytoagar, containing 1% sucrose, or on soil in the greenhouse under long-day conditions (16-h light/8-h dark cycle) at 22–23 °C. For production of the gir1-1 gir2-1 line, the pollen of the gir1-1 plants was cross-pollinated onto the gir2-1 plants, and the homozygous double mutant progeny plants were identified by PCR.
For BiFC and GIR1 transcriptional activity assays, Nicotiana benthamiana plants were grown in soil at 22–23 °C under long day conditions. Root hair linear density per millimeter root length was determined by counting the number of hairs from one side of a 2-mm segment from the mature zone of at least 10 roots under a Leica MZ FLIII stereoscope.
2.2. RT-qPCR and gene expression assays
Total RNA was extracted from frozen material using TRIzol reagent and then treated with RNase-free DNase to eliminate genomic DNA (Thermo Fisher Scientific). Total RNA was reverse-transcribed, using the Superscript II RT kit (Invitrogen), and the cDNA was then used as template for real-time PCR. The reactions were performed as described [7] using a MiniOpticon Real-Time PCR System with SYBR Green Realtime PCR Master Mix (BioRad) and primer pairs specific for ACT8 (5′TCCAGGCATTGTCCACAGAA3’/5′ACCTGCTCCTCCTTAGACAT3′), GIR1 (5′GACGAGCCATCTGTGAGATA3’/5′TTTGTGGCGTTTTCATGGAG3′), or GIR2 (5′TTCTCAAGCCAGCCAGATGA 3’/5′GTCTGGTATTGGCAGCGGTA 3′). Tested gene expression values were standardized to the expression levels of the constitutive housekeeping ACT8 gene. For each sample, at least three replications were performed in one experiment. The relative expression level of each gene was calculated using the cycle threshold (CT) 2−ΔΔCT method [8].
2.3. Plasmid construction and production of transgenic plants
The coding sequence of GIR1 was amplified from Arabidopsis cDNA using the primer pair (5′AAGATGAGTCGAAGAAGTCC3’/5′AGTTCCTTCGAGTCTTGCGA 3′), and the GIR1 promoter was amplified from Arabidopsis genomic DNA with the primer pair 5′AGACACCTATACGCTATGCA3′/5′CTTTGGACTTCTTCGACTCA3′. The amplified products were cloned into the pGEM®-T Easy Vector (Promega), producing pGEM-AtGIR1 and pGEM-ProAtGIR1, respectively, and confirmed by DNA sequencing. pGEM-GIR1-mutEAR and pGEM-GIR1-delEAR were produced using primer pairs 5′GAAGTCGAATCTCTCGCCGCCTA3′/5′GATTCCGACTTTGGACTTCTTCGACT3′ and 5′TCGCCGCCTACGTCAAGC3′/5′TTCCAGCTTTGGACTTCTTCGAC3′, respectively, and the New England BioLabs site-directed mutagenesis protocol (https://www.neb.com/applications/cloning-and-synthetic-biology/site-directed-mutagenesis). The GIR1 over-expression construct was generated by ligation of the XhoI-BamHI fragment from pGEM-GIR1 into the same sites of pSAT5A [9]. To generate construct that expresses GUS from the GIR1 promoter, we first amplified the promoter fragment from pGEM-ProAtGIR1 with primers 5′TCCACCGGTAGACACCTATACGCTATGCA3’/5′CAGGTCGACCTTTGGACTTCTTCGACTCA3′ and inserted it into the AgeI-SalI sites of pSAT5A, replacing the 35S promoter and producing pSAT5A-GIR1pro. The GUS reporter gene was then cloned into the KpnI-BamHI sites of pSAT5A-GIR1pro downstream of the GIR1 promoter. To produce construct that expresses the wild-type GIR1 from the GIR1 promoter, the coding sequence of GIR1 was inserted into the SalI-BamHI site of pSAT5A-GIR1pro. The resulting expression cassettes were excised with I-CeuI and inserted into the pPZP-RCS2 binary vector [9]. Transgenic Arabidopsis plants were generated using these binary vectors and the Agrobacterium tumefaciens strain EHA105 as described previously [10].
2.4. Yeast two-hybrid assay
The sequence encoding N-terminal 140 amino acid residues of GL2 was amplified using the primer pair 5′CTGAGAATTCATGAAGTCGATCGATGGCTG3’/5′TGTCGGATCCGATCGGTGGTGTGACGATGA3′, cloned into the EcoRI-BamHI sites of a LexA plasmid pSTT91 (TRP1+) [11], and introduced into the Saccharomyces cerevisiae strain TAT7 (L40-ura3) [12]. The resulting yeast cells were transformed with our normalized cDNA library prepared from Arabidopsis seedlings in pGAD424 (LEU2+) (Clontech) [13]. Transformants were selected on a triple dropout medium that lacked tryptophan, leucine, and histidine and contained 2 mM of 3-amino-1,2,4-triazole. cDNA clones encoding putative GL2 interactors were isolated from these HIS3+ LacZ + yeast clones and analyzed by DNA sequencing. For interaction in the yeast two-hybrid system, the GIR1 and GIR2 cDNAs amplified with the primer pairs 5′CGTAGAATTCAAGATGAGTCGAAGAAGTCC3’/5′GTCCGGATCCAGTTCCTTCGAGTCTTGCGA3′ and 5′GAGGAATTCATGAGTCGGAGGAACAAGAAC3’/5′TAATGGATCCAGTTCCACCAGGTCTTCTT3′, respectively, were cloned into the EcoRI-BamHI sites of pSTT91. The GL2 cDNA was amplified with the primer pair 5′CAGGGAATTCTGTATGTCAATGGCCGTCGAC3’/5′ACTTGGATCCAGCAATCTTCGATTTGTAGACT3′ and cloned into the EcoRI-BamHI site of pGAD424 (Clontech). Protein interactions were determined by the histidine prototrophy as well as by β-galactosidase assay on a nitrocellulose filter as described [13].
2.5. BiFC
The GIR1 and GIR2 coding sequences amplified with the primer pairs 5′CGTAGAATTCAAGATGAGTCGAAGAAGTCC3’/5′GTCCGGATCCAGTTCCTTCGAGTCTTGCGA3′ and 5′GAGGAATTCATGAGTCGGAGGAACAAGAAC3’/5′TAATGGATCCAGTTCCACCAGGTCTTCTT3′, respectively, were cloned into the EcoRI-BamHI sites of pSAT1-nEYFP-C1 [14], and the GL2 cDNA was cloned into the EcoRI-BamHI sites of pSAT6-cEYFP-C1 [14], respectively. The resulting expression cassettes were excised with AscI or PI-PspI from pSAT1-or pSAT6-based vectors, respectively, and inserted into pPZP-RCS2. pRCS2-nEYFP and pRCS2-cEYFP plasmids, produced by inserting expression cassettes from pSAT1-nEYFP-C1 and pSAT6-cEYFP-C1 into pPZP-RCS2, respectively, were used as negative controls. The tested combinations of these constructs were transiently co-expressed in N. benthamiana leaves by agroinfiltration, and the expressing tissues were analyzed using a Zeiss LSM 5 Pascal confocal microscope. All experiments were repeated at least three times.
2.6. Agroinfiltration
Agrobacterium EHA105 strain carrying the tested expression constructs was grown in LB medium supplemented with spectinomycin (100 μg/ml) overnight at 28 °C. Cells were harvested by centrifugation, resuspended to optical density of A600 = 0.1 in infiltration buffer [10 mM MgCl2, 10 mM MES (pH 5.5), 100 μM acetosyringone], incubated for 2 h at room temperature, and infiltrated into the abaxial sides of 3-4-week-old intact N. benthamiana leaves with a 1-ml needleless syringe. Plants were grown for 24 h in the dark and 72 h in the light before being harvested for analyses.
3. Results
3.1. GIR1 and GIR2 interact with GL2
To identify potential factors that function in complex with the GL2 repressor, we employed a yeast two-hybrid screen [12] of our normalized cDNA library prepared from Arabidopsis seedlings [13]. Because the full-length GL2 exhibited strong self-activation when fused to the binding domain (DBD) in the two-hybrid assay, we identified one of its largest fragments, i.e., the N-terminal 140 amino acid residues, GL2-140 (Fig. 1A), that does not self-activate; furthermore, GL2-140 lacks the conserved homeodomain and START domain of GL2, which is expected to minimize interactions with proteins that recognize these conserved domains in a fashion not specific for GL2. Using GL2-140 as bait, we screened 2.77 × 106 transformants and isolated seven different cDNA clones, e.g., At2g27200, At5g06270, etc., encoding potential GL2-interacting repressors (GIRs). Here, we focus on one such clones designated GIR1, which was isolated in two independent screening experiments and corresponded to the Arabidopsis locus At5g06270. The GIR1 gene encodes a protein of 123 amino acid residues with an unknown function based on the annotation of the TAIR database.
Fig. 1.
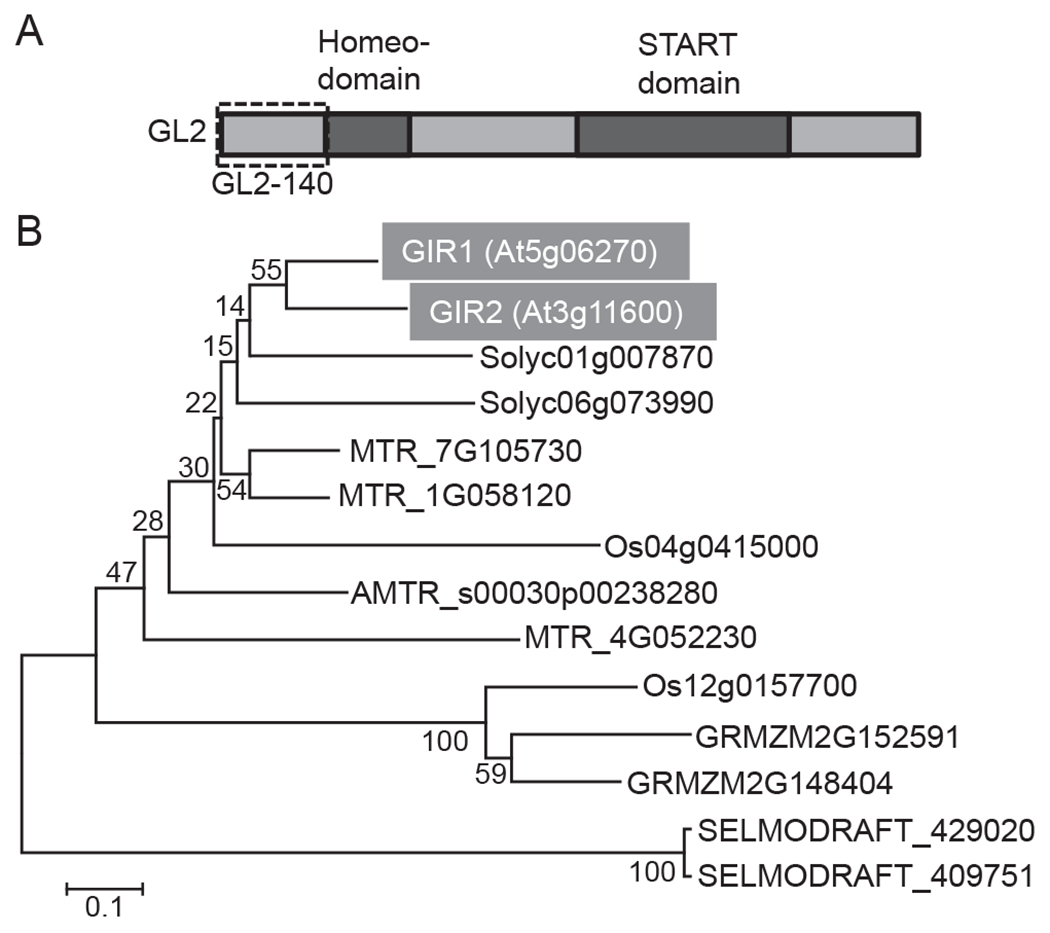
GL2 structure and phylogenetic analysis of GIR1 and GIR2. (A) Schematic representation of the GL2 domain structure. The GL2 N-terminal region, GL2-140, used for identification of GIR1 is delineated with a dashed box. (B) Phylogenetic tree of GIR1 and GIR2 homologs in different vascular plant species: Arabidopsis thaliana (GIR1, At), Zea mays (GRMZM), Oryza sativa (Os), Solanum lycopersicum (Solyc), Medicago truncatula (MTR), Amborella trichopoda (AMTR), and Selaginella moellendorffii (SELMODRAFT1). GIR1 and GIR2 are highlighted by shaded boxes and white letters. The evolutionary history was inferred using the Neighbor-Joining method [27]. The optimal tree with the sum of branch length of 3.86591361 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [28]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [29] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 79 positions in the final dataset. Evolutionary analyses were conducted using the Molecular Evolutionary Genetics Analysis tool (MEGA, version 6.0.5 for Mac OS) (http://www.megasoftware.net), which also generated this description of the analysis. Bar, 0.1 amino acid substitutions per site.
GIR1 is plant-specific, and, whereas it has no close homologs in non-plant genomes, GIR-like proteins are found in many diverse vascular plant species, from dicotyledonous to monocotyledonous plants (Fig. 1B). In Arabidopsis, GIR1 belongs to a small, two-member family (Fig. 1B) with the second member, designated GIR2 (At3g11600), that exhibits 65% identity and 71% similarity to GIR1. Whereas the biological function of GIR1/2 homologs in other plant species remains unknown, one of them, SGN-U226950/SlCycB2 of tomato, has been reported to interact with Wo, a tomato homolog of GL2, and regulate trichome development [15].
Next, we tested both GIR1 and GIR2 for interaction with the full-length GL2 in yeast and in planta. Fig. 2A shows that GIR1 fused to DBD interacted with GL2 fused to the activation domain (AD) in the yeast two-hybrid assay, in which protein interaction is indicated by histidine prototrophy and confirmed by a β-galactosidase assay independent on prototrophy. This interaction of GIR1 was specific because it was not observed with an unrelated control protein, the large T-antigen of SV40. Under the non-selective conditions, all combinations of the tested proteins resulted in the efficient cell growth, indicating that none of the constructs interfered with the cell viability (Fig. 2A). Essentially similar results were observed with GIR2 binding to GL2 in the yeast two-hybrid system (Fig. 2A).
Fig. 2.
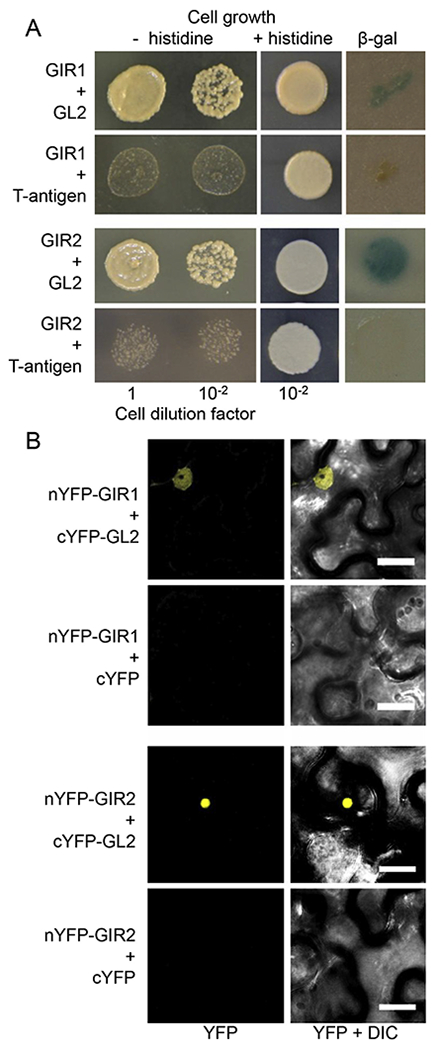
Interaction between GIR1 or GIR2 and GL2. (A) GIR1 and GIR2 interact with GL2 in the yeast two-hybrid system. The indicated dilutions of yeast cells were grown in the absence or presence of histidine as shown. Protein interaction was detected as cell growth on histidine-deficient medium and as β-galactosidase (β-gal) activity on a nitrocellulose filter. (B) GIR1 and GIR2 interact with GL2 in planta, and the interacting proteins accumulate in the cell nucleus. Protein interaction was detected by BiFC. YFP signal is in yellow. Fluorescence images are single confocal sections. Scale bars = 20 μm.
Finally, the GIR1-GL2 and GIR2-GL2 interactions were detected and subcellular localization of the GIR1-GL2 and GIR2-GL2 complexes was determined using bimolecular fluorescence complementation (BiFC) within living plant cells [14,16]. GIR1 and GIR2 were tagged with the N-terminal fragment of YFP (nYFP), and GL2 was tagged with the C-terminal fragment of YFP (cYFP). Then, nYFP-GIR1 and cYFP-GL2 or nYFP-GIR2 and cYFP-GL2 were transiently co-expressed in Nicotiana benthamiana leaves. Fig. 2B shows that nYFP-GIR1 and cYFP-GL2 as well as nYFP-GIR2 and cYFP-GL2 interacted in plant cells, producing the BiFC signal, and that the interacting proteins accumulated predominantly in the cell nucleus. As expected, co-expression of nYFP-GIR1 or nYFP-GIR2 with free cYFP failed to reconstitute the BiFC fluorescence (Fig. 2B).
3.2. GIR1 and GIR2 are functionally redundant and repress development of root hair
To understand the function of GIR1 and GIR2, we employed reverse genetics, using both loss-of-function and gain-of-function lines. For loss-of-function, we utilized two homozygous T-DNA insertional mutant lines gir1-1 (SALK_089888) and gir2-1 (SALK_048394) with mutagenic T-DNA insertions within the single exon of GIR1 and GIR2 (Fig. 3A). We used quantitative RT-PCR (RT-qPCR) analysis to examine expression levels of both genes in the homozygous mutant lines in comparison to the wild-type plants. Fig. 3B shows that, in the gir1-1 line, the GIR1 expression was suppressed by 10 fold whereas the expression level of GIR2 was not affected, and that the gir2-1 line showed a 10-fold reduction in the GIR2 expression, with the expression of GIR1 remained virtually unchanged. Because GIR1 and GIR2 are closely related, and thus may be functionally redundant, we also produced a homozygous double gir1-1 gir2-1 mutant and confirmed that expression of both GIR1 and GIR2 genes was suppressed in this mutant also by 10 fold relative to the wild-type plants (Fig. 3B). For gain-of-function, we focused on GIR1 and generated 10 transgenic lines that constitutively expressed the GIR1 cDNA from a CaMV 35S promoter and analyzed them by RT-qPCR; based on this analysis, we selected two of the gain-of-function alleles, a high expresser and a moderate expresser, designated GIR1-OE1 and GIR1-OE3, that accumulated 70- and 20-fold higher levels of GIR1 transcripts, respectively, than the wild-type plants (Fig. 3C).
Fig. 3.
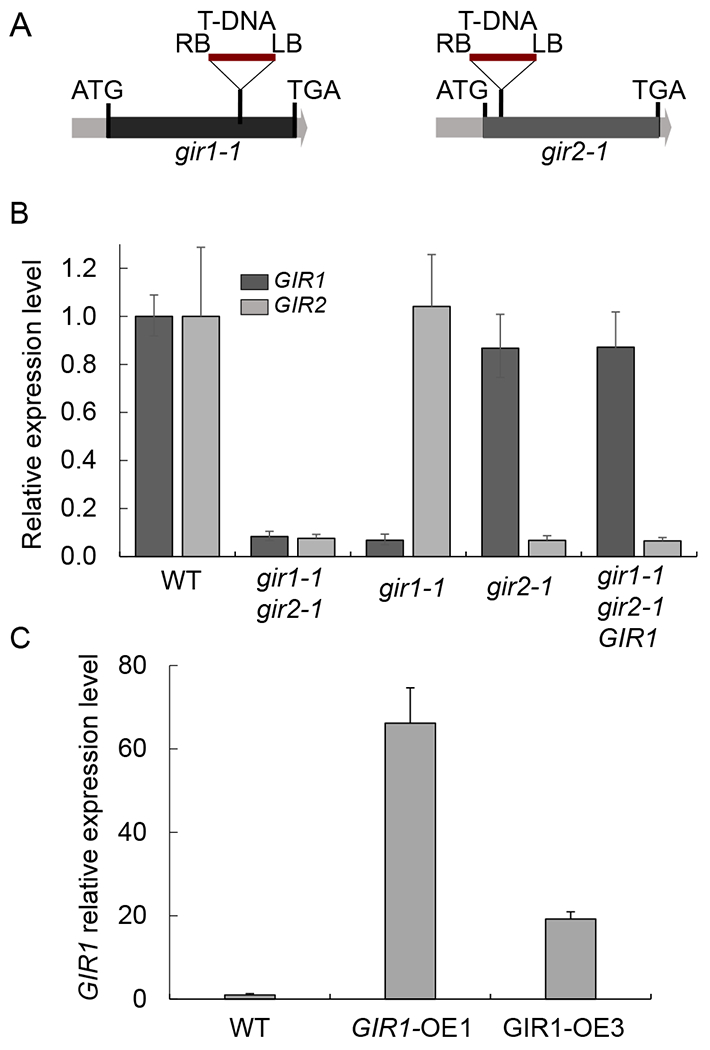
Quantitative RT-PCR analysis of GIR1 gene expression in mutant gir1-1 and over-expressing GIR1-OE plants. (A) Schematic representation of the locations of the mutagenic T-DNA integration sites in the gir1-1 and gir2-1 mutants. LB and RB, T-DNA left and right borders. Black boxes delineate the single exons of GIR1 and GIR2 between their translation initiation (ATG) and termination codons (TGA). (B) Relative expression levels of the GIR1 (dark gray boxes) and GIR2 genes (light gray boxes) in 6-week-old wild-type (WT) plants and the indicated loss-of-function and genetically complemented mutant lines. The expression level of each of the genes in the WT plants is set to 1.0, and error bars represent SEM of N = 3 independent biological replicates. (C) Relative expression levels of GIR1 in WT plants and the indicated gain-of-function lines. The expression level in the WT plants is set to 1.0, and error bars represent SEM of N = 3 independent biological replicates.
Next, we examined gir1-1, gir2-1, gir1-1 gir2-1, GIR1-OE1, and GIR1-OE3 plants for development of their root hair and trichomes, the known hallmarks of the GL2 function [2–4]. Fig. S1 shows that gir1-1 gir2-1 plants exhibited no discernable alterations in the size, shape, or surface density of their trichomes as compared to the wild-type plants while, as expected, the gl2-1 mutant produced virtually no trichomes. Furthermore, gir1-1 and gir2-1 also did not show changes in their root hair formation in comparison to the wild-type plants (Fig. 4A). However, the double mutant gir1-1 gir2-1 plants exhibited excessive root hair formation, comparable to that observed in the gl2-1 mutant, whereas, in the GIR1-OE1, and GIR1-OE3 plants, root hair was sparse (Fig. 4A). These phenotypes were then quantified by measuring linear density of root hairs along the entire seedling root. Fig. 4B shows that, the wild-type plants and the gir1-1 and gir2-1 single mutants developed ca. 50–60 root hairs/mm, but the gir1-1 gir2-1 double mutant plants developed more than twice as many root hairs (130 root hairs/mm), and this elevated root hair frequency phenotype was comparable to that exhibited by the gl2-1 mutant (120 root hairs/mm). These data indicate that GIR1 and GIR2 act as functionally redundant negative regulators of root hair development. Thus, we focused our subsequent studies of the GIR1/2 function on GIR1.
Fig. 4.
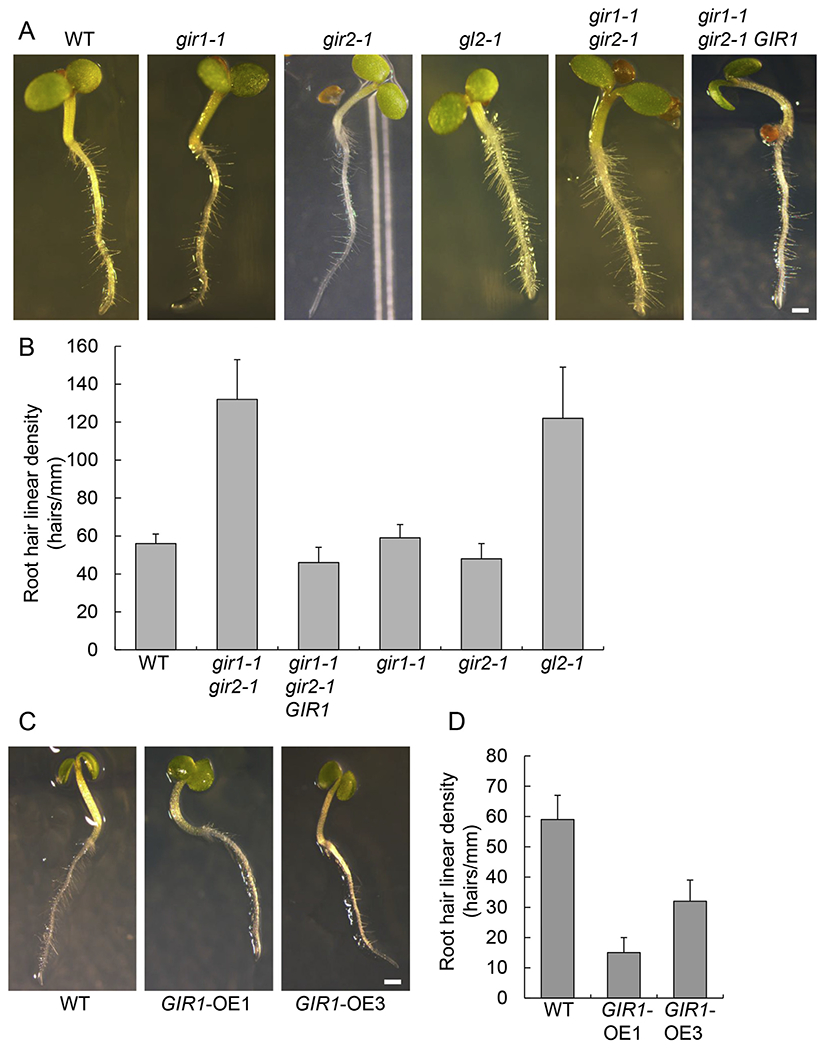
Effects of GIR1 and GIR2 on root hair development. (A) Root hair phenotype in 6-day-old seedlings of the wild-type (WT) plants and the indicated loss-of-function and genetically complemented mutant lines. Scale bar = 1 mm. (B) Root hair linear density in plants shown in panel A. Error bars represent SEM of N = 3 independent biological replicates. (C) Root hair phenotype in 6-day-old seedlings of the WT plants and the indicated gain-of-function lines. Scale bar = 1 mm. (D) Root hair linear density in plants shown in panel C. Error bars represent SEM of N = 3 independent biological replicates.
Gain-of-function of GIR1 in the GIR1-OE1 high expresser plants confirmed the negative regulatory effect of GIR1 on root hair development (Fig. 4C), reducing the average linear density of root hair to approximately one quarter of that observed in the wild-type plants, i.e., 15 root hairs/mm vs. 60 root hairs/mm (Fig. 4D). The moderate expresser GIR1-OE3 plants produced 35 root hairs/mm (Fig. 4D), suggesting correlation of the GIR1 effect with the extent of its over-expression.
Based on the functional redundancy of GIR1 and GIR2, transgenic expression of GIR1 should genetically complement the gir1-1 gir2-1 mutant to a near wild-type phenotype. We expressed the GIR1 genomic sequence from its putative native promoter in the gir1-1 gir2-1 plants; specifically, we utilized 2.1 kb upstream of the GIR1 translation initiation codon, an intergenic region which, in Arabidopsis, often encompasses promoter sequences [17]. The resulting gir1-1 gir2-1/GIR1 plants developed the wild-type root hair phenotype (Fig. 4A), with linear density of ca. 55 root hairs/mm (Fig. 4B).
To gain additional insights into the function of GIR1, we examined its pattern of expression in the plant. First, we used RT-qPCR to analyze GIR1 expression in the whole wild-type seedlings and in different organs of the wild-type plants. GIR1 was expressed in all tissues, but at substantially different levels. Consistent with its effect on root hair and with the lack thereof on trichomes, GIR1 expression levels were the highest in the root, and they were the lowest, by ca. 10-fold relative to the root, in both rosette and cauline leaves. GIR1 expression in stems and siliques was also low, by ca. 5-fold in comparison to roots, whereas its expression in flowers and in seedlings occurred at intermediate levels (Fig. S2A). We then analyzed GIR1 expression within different zones of the root using a β-glucuronidase (GUS) reporter under the control of the GIR1 native promoter. In the whole Arabidopsis seedlings, the reporter GUS activity was observed predominantly in the shoot and root meristems (Fig. S2B, arrows). Within the root, the GUS reporter was expressed mainly in the meristematic zone and in the elongation zone of the root (Fig. S2C), where root hairs initiate [18,19]. As the root matures and elongates, root hairs emerge and also mature, and we observed virtually no GUS reporter activity in this mature zone of the root (Fig. S2C), suggesting that GIR1 is expressed during the onset of root hair development. Interestingly, this expression pattern resembles the known pattern of expression of GL2 in the Arabidopsis root [20].
4. Discussion
GIR1 and GIR2 comprise a new plant-specific two-member family of closely related GL2-interacting proteins that may function as adaptor proteins that link to transcriptional repressor(s) and suppress root hair development. Furthermore, they are specific for higher, vascular plants, and are not found in non-tracheophytes, such as mosses or algae. GIR1 and GIR2 exhibit significant degree of functional redundancy as inactivation of one of them produces no discernable phenotypes, but abolishing expression of both genes results in substantial and specific phenotypic changes. On the other hand, these changes can be reversed to the wild-type-like phenotype by genetic complementation with only one of the GIR proteins, i.e., GIR1. This redundancy may explain the recalcitrance of the GIR1 and GIR2 loci to functional annotation which, in Arabidopsis, is often done by forward genetics of T-DNA insertional mutant populations [21]. GIR1 is expressed in the meristematic and elongation zones of the root, and GIR1/2 loss-of-function mutant gir1-1 gir2-1 develops increased root hair density whereas gain-of-function of GIR1 represses root hair development.
Interestingly, although both GIR1 and GIR2 interact, and most likely function, with GL2, this functionality is more narrowly tissue-specific than that of GL2 itself. In particular, GL2 regulates development of two very diverse organs, root hair and trichomes, by two opposing mechanisms. In the root epidermis, GL2 suppresses formation of root hair in N cells by repressing its target genes [2,20,22,23] whereas, in the leaf epidermis, GL2 induces formation of trichomes by activating its target genes [3]. Potentially, this functional dichotomy may be due to the nature of the specific chromatin-remodeling factors with which GL2 associates in the target cells; this is by analogy to the lysine-specific demethylase 1 (LSD1/KDM1) [24,25], the activity of which promotes gene silencing or activation, depending on its interaction partners [26]. GIR1 and GIR2 may represent such GL2-associated factors because they participate in GL2-mediated control only of the root hair development and do not appear to be involved in development of trichomes. This notion is consistent with very low levels of expression of GIR1 in the leaf tissues.
Supplementary Material
Acknowledgments
The work in the V.C. laboratory is supported by grants from NIH (GM50224), NSF (MCB 1118491), USDA/NIFA (2013-02918), and BARD (IS-4605-13C) to V.C.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2017.05.084.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2017.05.084.
References
- [1].Ishida T, Kurata T, Okada K, Wada T, A genetic regulatory network in the development of trichomes and root hairs, Annu. Rev. Plant Biol 59 (2008) 365–386. [DOI] [PubMed] [Google Scholar]
- [2].Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC, The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein, Plant Cell 11 (1999) 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khosla A, Paper JM, Boehler AP, Bradley AM, Neumann TR, Schrick K, HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23, Plant Cell 26 (2014) 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qing L, Aoyama T, Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator, J. Integr. Plant Biol 54 (2012) 729–737. [DOI] [PubMed] [Google Scholar]
- [5].Taheri A, Gao P, Yu M, Cui D, Regan S, Parkin I, Gruber M, A landscape of hairy and twisted: hunting for new trichome mutants in the Saskatoon Arabidopsis T-DNA population, Plant Biol. 17 (2015) 384–394. [DOI] [PubMed] [Google Scholar]
- [6].Murashige T, Skoog F, A revised medium for rapid growth and bio assays with tobacco tissue cultures, Physiol. Plant 15 (1962) 473–497. [Google Scholar]
- [7].Zaltsman A, Krichevsky A, Loyter A, Citovsky V, Agrobacterium induces expression of a plant host F-box protein required for tumorigenicity, Cell Host Microbe 7 (2010) 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method, Methods 25 (2001) 402–408. [DOI] [PubMed] [Google Scholar]
- [9].Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V, pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants, Plant Mol. Biol 57 (2005) 503–516. [DOI] [PubMed] [Google Scholar]
- [10].Bent A, Arabidopsis thaliana floral dip transformation method, Methods Mol. Biol 343 (2006) 87–104. [DOI] [PubMed] [Google Scholar]
- [11].Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R, A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex, Mol. Cell. Biol 21 (2001) 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H, Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system, Mol. Cell. Biol 15 (1995) 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ballas N, Citovsky V, Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein, Proc. Natl. Acad. Sci. U. S. A 94 (1997) 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T, Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta, J. Mol. Biol 362 (2006) 1120–1131. [DOI] [PubMed] [Google Scholar]
- [15].Yang C, Li H, Zhang J, Luo Z, Gong P, Zhang C, Li J, Wang T, Zhang Y, Lu Y, Ye Z, A regulatory gene induces trichome formation and embryo lethality in tomato, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hu CD, Chinenov Y, Kerppola TK, Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation, Mol. Cell 9 (2002) 789–798. [DOI] [PubMed] [Google Scholar]
- [17].Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, Rhee S, Raikhel NV, Citovsky V, High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta, Plant Physiol. 135 (2004) 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J, Root Hairs, vol. 12, The Arabidopsis book/American Society of Plant Biologists, 2014, p. e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW, The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root, Dev. Biol 166 (1994) 740–754. [DOI] [PubMed] [Google Scholar]
- [20].Masucci JD, Schiefelbein JW, Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root, Plant Cell 8 (1996) 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR, Genome-wide insertional mutagenesis of Arabidopsis thaliana, Science 301 (2003) 653–657. [DOI] [PubMed] [Google Scholar]
- [22].Lee MM, Schiefelbein J, WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning, Cell 99 (1999) 473–483. [DOI] [PubMed] [Google Scholar]
- [23].Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J, The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root, Dev. Camb. Engl 130 (2003) 6431–6439. [DOI] [PubMed] [Google Scholar]
- [24].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y, Histone demethylation mediated by the nuclear amine oxidase homolog LSD1, Cell 119 (2004) 941–953. [DOI] [PubMed] [Google Scholar]
- [25].Zhou X, Ma H, Evolutionary history of histone demethylase families: distinct evolutionary patterns suggest functional divergence, BMC Evol. Biol 8 (2008) 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R, LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription, Nature 437 (2005) 436–439. [DOI] [PubMed] [Google Scholar]
- [27].Saitou N, Nei M, The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol 4 (1987) 406–425. [DOI] [PubMed] [Google Scholar]
- [28].Felsenstein J, Confidence limits on phylogenies: an approach using the bootstrap, Evol. Int. J. Org. Evol 39 (1985) 783–791. [DOI] [PubMed] [Google Scholar]
- [29].Zuckerkandl E, Pauling L, Evolutionary divergence and convergence in proteins, in: Bryson V, Vogel HJ (Eds.), Evolving Genes and Proteins, Academic Press, New York, 1965, pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


