Abstract
Biosynthesis of extracellular cellulases in the cellulose-degrading actinomycete Thermobifida fusca is controlled by a transcriptional regulator, CelR, and cellobiose, which acts as an inducer interfering with the CelR-DNA interaction. We report the identification and characterization of a mutation in the celR gene that changes Ala55 in the hinge helix of CelR to Thr. The wild-type and mutant celR genes were cloned in Escherichia coli, and their protein products were characterized. The CelR mutant protein bound DNA more weakly than the wild-type protein and formed a less stable complex with DNA in the presence of cellobiose. The results of Western analysis and gel retardation experiments suggest that CelR is produced constitutively and its DNA-binding activity is regulated through posttranslational modification.
Many soil actinomycetes degrade cellulose in plant residues by using secreted enzymes. Biosynthesis of extracellular cellulases in Cellulomonas uda, C. fimi, Thermomonospora curvata, T. fusca, Acidothermus cellulolyticus, Streptomyces reticuli, S. halstedii, and other members of the family Actinomycetaceae is regulated by induction and repression (1–3, 9, 17, 20, 23). In several of these species, regulation occurs at the transcriptional level (2, 3, 11, 23). The results of these studies suggest a complex control of cellulase genes in actinomycetes.
Cellulase synthesis in T. fusca (recently reclassified as Thermobifida fusca [26]) is induced by cellulose and cellobiose and is subject to sugar catabolite repression (9). The cellulolytic system of T. fusca is encoded by at least six unlinked genes, designated celA through celF, that form a regulon (6, 8, 25; D. Irwin, unpublished data). The six extracellular enzymes produced by this species (endocellulases E1, E2, and E5; exocellulases E3 and E6; and the processive endocellulase E4) act synergistically, degrading cellulose to cellobiose and other soluble sugars (5).
The close correlation between the levels of the celE transcript and the level of the protein product of the celE gene, endoglucanase E5, produced on different carbon sources provided evidence that cellulase biosynthesis is regulated through the control of transcription (11). A 14-bp inverted repeat (TGGGAGCGCTCCCA) located in the 5′-upstream regions of all cel genes was identified as a cis-acting regulatory element (10), and a transcription regulator, CelR, that specifically binds to the 14-bp inverted repeat was isolated (19). In vitro experiments showed that cellobiose at physiological concentrations acts as an effector causing dissociation of the CelR-DNA complex. It was suggested that transcription of the cel genes is controlled by CelR and cellobiose.
A partially constitutive mutant strain, CC-2, with enhanced carboxymethyl cellulose (CMC)-hydrolyzing activity was isolated from T. fusca (9). In this paper, we show that the strain has a mutation in the celR gene. We used biochemical approaches to characterize the properties of the wild-type and mutant CelR proteins. Our results are consistent with the role of CelR as a repressor controlled by cytoplasmic cellobiose levels and posttranscriptional modification.
Bacterial strains, plasmids, and culture conditions.
The following T. fusca strains were used in this study: YX (wild type); CC-2, a partially cellulase constitutive strain (9); and ER1, an extracellular protease-negative strain (24). Strains CC-2 and ER1 were derived from T. fusca YX. Escherichia coli DH5α was used for cloning and plasmid isolation; E. coli BL21(DE3) was used for protein production. Plasmid pNS1 was described earlier (19), and plasmids pNS2 and pNS3 were constructed as described below.
T. fusca was grown on Hagerdahl medium (4) supplemented with either 0.5% cellobiose, glucose, or xylose (Sigma Chemical Co.) or 1% Solka Floc (microcrystalline cellulose; James River Corporation) at 52 to 55°C. E. coli strains containing recombinant plasmids were grown in Luria broth or plated on Luria agar plates containing ampicillin at 0.1 mg/ml (pNS1) or kanamycin at 30 μg/ml (pNS2 and pNS3).
Enzyme activity and protein assays.
T. fusca culture samples were centrifuged at 5,000 × g for 5 min, and the supernatants were assayed for cellulase activity using filter paper (FP) or 1% low-viscosity CMC (Sigma Chemical Co.) as the substrate. The amount of reducing sugar (primarily cellobiose) produced was measured spectrophotometrically at 600 nm after reaction with dinitrosalicylic acid as previously described (18).
Cell density was estimated from cytoplasmic protein. The cell pellet was washed two times in lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride and 0.1 mM dithiothreitol. Cells were resuspended in lysis buffer and lysed with a French press at 4°C, and the cell lysate was centrifuged at 10,000 × g for 20 min. Culture supernatants and cell lysates were stored at −20°C. Cytoplasmic protein in the cell lysate and total secreted (extracellular) protein in the culture supernatant were measured by the method of Lowry et al. (12).
Subcloning, expression, and purification of wild-type and mutant celR in E. coli.
Genomic DNA was isolated from T. fusca CC-2 as previously described (19). The celR gene was amplified from genomic DNA with PCR using the Expand high-fidelity DNA polymerase system (Boehringer Mannheim) and the primers GCCGCGCACGCTGCCATTGAG and TCCGCCTGCCTCCCGTTGTCCTC. A DNA fragment containing the celR gene was isolated by gel electrophoresis and purified with the QIAquick gel extraction kit (QIAGEN). The wild-type celR gene (from pNS1) and PCR products containing the celR mutant gene (from strain CC-2) were used for subcloning. An NdeI restriction site, CATATG, was introduced by PCR immediately upstream from the start codon of the genes by using primers GGGTTGGGGGAACACATATGGAGCGTC and CTTTGCGCGGGCCCCTCATCC (pNS1) or primers GGGTTGGGGGAACACATATGGAGCGTC and TCCGCCTGCCTCCCGTTGTCCTC (CC-2). PCR products were cut with NdeI and BamHI, gel purified, and ligated to pET26b(+) vector DNA (Novagen) that had been cut with the same enzymes. The resulting plasmids, pNS2 (containing wild-type celR) and pNS3 (containing the mutant celR gene from strain CC-2), were electroporated into E. coli DH5α. DNA was isolated from transformants and checked for undesired PCR mutations by sequencing. Specific primers for sequencing were synthesized, and DNA sequencing was performed by the dideoxy-chain termination method (14) at the BioResource Center, Cornell University.
Electrophoretically pure CelR and mutant CelR were isolated from E. coli BL21(DE3) transformed with pNS2 or pNS3. Cells were grown in 0.5 liter of M9 medium containing kanamycin at 30 μg/ml and 0.8% glucose at 37°C, induced with isopropyl-β-d-thiogalactopyranoside after 6 h, and cultured overnight. Cells were harvested and lysed, and CelR protein was isolated and purified by chromatography on phenyl Sepharose CL-4B and heparin Sepharose CL-4B columns as previously described (19). Precipitation with streptomycin sulfate was omitted. The protein concentration was determined with the bicinchoninic acid reagent (Pierce Chemical Co.) using bovine serum albumin (Sigma Chemical Co.) as the standard.
celE promoter-binding assay and quantitation of the CelR protein in T. fusca.
The celE promoter-binding activities of the wild-type and mutant CelR proteins were measured in vitro with a gel retardation assay by determining the alteration of the electrophoretic mobility of the 32P-labeled celE promoter region after formation of a complex with the DNA-binding protein as previously described (19).
Accurate quantitation of the CelR protein in the wild type and strain CC-2 was not possible because of the high proteolytic activity. Protease-negative strain ER1 was used for CelR quantitation by Western blotting and gel retardation assay. Cells were grown on 0.5% cellobiose. Stationary-phase cultures were used to inoculate the medium, supplemented with either 0.5% cellobiose, glucose, or xylose or 1% Solka Floc. Cells were cultured at 55°C and 200 rpm in triplicate on each carbon source. T. fusca cytoplasmic proteins were isolated as described above, separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (7), and electrophoretically transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore). Rabbit polyclonal antiserum raised against pure CelR protein and goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate were used for CelR detection. Dilutions of pure CelR protein separated on the same gels were used as standards. The CelR bands were detected with a Vistra ECL Western blotting kit and scanned with a STORM 840 scanner, and the images were quantitated with the ImageQuant program (Molecular Dynamics). The abundance of the CelR protein was also calculated from celE promoter-binding activity in T. fusca extracts measured with a gel retardation assay. Electrophoretically pure CelR protein used as a standard in gel retardation and Western blotting experiments was purified from T. fusca grown on Solka Floc as previously described (19).
Cellulase activity in T. fusca wild-type and CC-2 mutant strains.
Wild-type T. fusca YX and partially constitutive strain CC-2 were grown on minimal medium with microcrystalline cellulose (Solka Floc), cellobiose, or glucose as the carbon source. Cellulase activity was measured with soluble CMC, which is a preferred substrate for endocellulases E1 and E5, and with microcrystalline cellulose (FP), which is degraded by the synergistic action of all six enzymes, after 24 and 72 h of cultivation (Table 1). There were characteristic differences in the growth of T. fusca on different carbon sources (data not shown). Cellobiose induced fast growth of both strains. Glucose was a poorer carbon source, as evidenced by delayed growth and decreased production of intracellular protein. Both strains showed continuous accumulation of extracellular protein in the medium, and the level was higher in cultures grown on inducing carbon sources.
TABLE 1.
Cellulase activity and extracellular protein levels in T. fusca cultures grown on microcrystalline cellulose, cellobiose, and glucose
| Strain and carbon source(s)a | Avg extracellular proteinb ± SD at:
|
Avg FP-hydrolyzing activityc ± SD at:
|
Avg CMC-hydrolyzing activityc ± SD at:
|
|||
|---|---|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | |
| YX | ||||||
| SF | 147 ± 16 | 530 ± 55 | 109 ± 11 | 131 ± 7 | 8,650 ± 1,680 | 12,550 ± 2,670 |
| CB | 123 ± 2 | 228 ± 12 | 58 ± 3 | 36 ± 4 | 1,980 ± 80 | 2,080 ± 450 |
| GL | 61 ± 2 | 187 ± 14 | NRd | NR | 970 ± 10 | 442 ± 23 |
| SF + GL | 149 ± 5 | 413 ± 46 | NR | 68 ± 10 | 1,743 ± 27 | 7,576 ± 215 |
| CC-2 | ||||||
| SF | 43 ± 2 | 579 ± 40 | 84 ± 6 | 169 ± 24 | 4,190 ± 435 | 11,780 ± 2,370 |
| CB | 113 ± 1 | 227 ± 17 | 86 ± 6 | 79 ± 19 | 3,210 ± 290 | 5,350 ± 90 |
| GL | 36 ± 2 | 208 ± 26 | NR | 49 ± 2 | 2,310 ± 697 | 6,390 ± 1,570 |
| SF + GL | 56 ± 2 | 575 ± 45 | NR | 96 ± 7 | 3,236 ± 165 | 7,595 ± 449 |
Abbreviations: SF, microcrystalline cellulose; CB, cellobiose; GL, glucose.
Micrograms per milliliter.
Nanomoles of glucose per milligram of protein per minute.
NR, target digestion was not reached.
Wild-type cultures reached saturating cell density by 24 h on every carbon source, but accumulation of extracellular protein and cellulase activity in the supernatant continued until the end of cultivation. There was significant repression of cellulase synthesis in cultures grown on glucose, as clearly shown by both the CMC and filter paper assays. When both cellulose and glucose were present in the medium, cells preferentially metabolized glucose and the level of cellulase synthesis remained low during the first 24 h of cultivation. The glucose in the medium was then exhausted and cellulase activity increased, reaching 60% of the fully induced level by 72 h of cultivation.
Strain CC-2 was characterized by enhanced cellulase production, slower growth, and slower production of extracellular protein. It displayed higher FP-hydrolyzing activity on every carbon source and higher CMC-hydrolyzing activity in cultures grown on sugars. The final cellulase levels in cultures grown on glucose and cellobiose were 54 and 45%, respectively, of the level reached on microcrystalline cellulose. Strain CC-2 grown on glucose showed enhanced CMCase activity both after 24 h of cultivation, when the glucose in the medium was not completely exhausted, and 48 h later. Strain CC-2 grown on microcrystalline cellulose in the presence of glucose showed a lowered glucose repression of CMCase activity after 24 h of cultivation (23% instead of the 80% of the wild type) that increased by the end of cultivation (34% instead of the 54% of the wild type). We could not measure glucose repression of FP-hydrolyzing activity after 24 h because the high level of glucose in the medium interfered with the assay.
Partially constitutive stain CC-2 carries a mutation in the celR gene.
The celR gene in partially constitutive strain CC-2 carries a G-to-A substitution at the first position of the 55 triplet which changed the alanine residue to threonine in the mutant protein. Residue 55 lies within the hinge helix connecting the DNA-binding domain to the corepressor-binding domain. Crystallographic studies of another member of the GalR-LacI family, PurR, showed that the repressor-operator complex consists of two PurR molecules bound to the palindromic operator sequence. The hinge helix participates in DNA-protein complex formation by binding to the DNA minor groove and by forming a series of contacts with the DNA-binding domain, the corepressor-binding domain, and the hinge helix of the other PurR monomer (16). Ala55 in CelR is a homologue of Val50 in PurR, which forms van der Waals contacts with the side chain of Val50 in the other PurR monomer. Mutagenesis studies of the LacI repressor from E. coli showed that this region is particularly sensitive to mutations. Amino acid replacements in the hinge helix resulted in defective repressors with altered operator binding, inducer binding, or allosteric transition (13).
celE promoter-binding activities of wild-type and mutant CelR proteins.
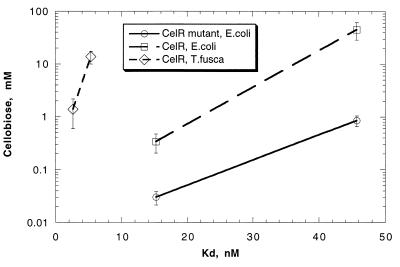
The wild-type and mutant CelR proteins were purified to homogeneity from the overproducing E. coli strains, and their properties were compared to those of wild-type CelR purified from T. fusca. Both the wild-type and mutant CelR proteins produced in E. coli had an apparent molecular mass of 41.5 kDa, identical to that of CelR from T. fusca. The dissociation constant for the CelR-celE promoter complex was calculated as the concentration of CelR that caused 50% of the DNA to bind under the conditions of the assay. CelR expressed in E. coli had a slightly lower celE promoter-binding affinity (Kd = 1.9 × 10−9 M) than CelR isolated from T. fusca (Kd = 1 × 10−9 M). The CelR mutant protein had even weaker DNA binding (Kd = 4.1 × 10−9 M).
The effect of cellobiose on DNA-protein complex formation was measured with the gel retardation assay using CelR and the mutant CelR protein expressed in E. coli. It was shown that the mutant protein formed a less stable intermolecular complex with the celE promoter than did wild-type CelR (see Fig. 2). A weaker dissociation constant and lower stability of the CelR mutant-DNA complex in the presence of cellobiose may be responsible for the partial constitutive cellulase synthesis in the CC-2 mutant. The CelR protein purified from T. fusca formed a significantly more stable CelR-celE promoter complex in the presence of cellobiose than did CelR expressed in E. coli.
FIG. 2.
Effect of cellobiose on the dissociation constant of the CelR-celE promoter complex. The values are averages ± the standard deviations. The celE promoter-binding activities of electrophoretically pure CelR and mutant CelR expressed in E. coli and CelR isolated from T. fusca were measured with the gel retardation assay.
celE promoter-binding activity and abundance of the CelR protein in T. fusca.
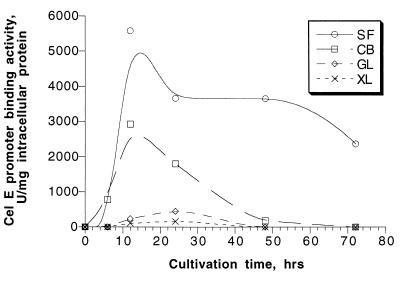
The highest celE promoter-binding activity was found in T. fusca cultures grown on microcrystalline cellulose (Fig. 1). The maximum level of activity was observed after 12 h of cultivation and remained high during 72 h of growth. Significant binding activity was present in cells grown on cellobiose. In this case, activity decreased rapidly and was almost undetectable after 48 h of cultivation. Cells grown on glucose and xylose showed very low activity after 12 and 24 h of cultivation.
FIG. 1.
celE promoter-binding activity in T. fusca ER1 grown on Solka Floc (SF), cellobiose (CB), glucose (GL), and xylose (XL) measured with the gel retardation assay. One unit of celE promoter-binding activity is the amount of DNA-binding protein that converts 50% of the DNA fragment to a DNA-protein complex under the assay conditions used.
The abundance of the CelR protein in crude T. fusca extracts was measured after 20 h of cultivation, the peak of celE promoter-binding activity. Dilutions of cell extracts were analyzed by Western blot analysis and by the gel retardation assay. Both Western blotting and the gel retardation assay showed similar CelR levels in cells grown on cellulose and cellobiose (Table 2). However, the gel retardation assay showed very low CelR levels in cells grown on glucose and xylose, while Western analysis revealed significant levels of the protein in these cells. These results indicate that most of the CelR produced under noninducing conditions cannot bind its DNA target.
TABLE 2.
Levels of CelR protein in T. fusca ER1 cultures grown for 20 h on different carbon sources as determined by Western blotting and gel retardation assay
| Carbon source | Avg CelR protein level (% of intracellular protein) ± SD
|
|
|---|---|---|
| Western blotting | Gel retardation assay | |
| Solka Floc | 0.31 ± 0.05 | 0.37 ± 0.04 |
| Cellobiose | 0.19 ± 0.01 | 0.19 ± 0.02 |
| Glucose | 0.16 ± 0.03 | 0.031 ± 0.003 |
| Xylose | 0.16 ± 0.02 | 0.032 ± 0.008 |
The high levels of CelR in T. fusca grown under both inducing and noninducing conditions from Western blotting analysis is evidence for its constitutive synthesis. The fact that only a minor fraction of the CelR in cells grown under noninducing conditions can bind its target DNA indicates that the activity of CelR may be regulated through posttranslational modification. Another indication of posttranslational modification is the different stabilities of the CelR-celE promoter complexes formed by the CelR proteins expressed in T. fusca and E. coli in the presence of cellobiose (Fig. 2). There is no CelR-binding site in the upstream region of the celR gene that would allow its negative autoregulation. Based on these results, we suggest that CelR is produced constitutively but its DNA-binding activity is regulated by a protein kinase or some other mechanism.
In many respects, regulation of the cel genes in T. fusca is similar to transcriptional regulation of polysaccharide degradation genes in phylogenetically related actinomycetes. In Streptomyces reticuli, transcription of the cel1 cellulase gene is controlled by a CebR repressor that shows high homology to CelR and binds to the same target sequence (15). α-Amylase (aml) genes in S. coelicolor are under the negative control of MalR, another member of the GalR-LacI family (21, 22). Similar to the cel genes in T. fusca that are induced by cellobiose (the end product of cellulase), aml genes in S. coelicolor are induced by maltose and maltotriose derived from starch by the enzymatic action of products of the aml genes. Both the CelR and MalR regulatory proteins are expressed constitutively. Similar to celR, which is located immediately downstream of the bglABC operon that encodes components of an energy-dependent sugar (probably cellobiose) transport system, cebR and malR are located in the vicinity of gene clusters encoding components of similar systems for cellobiose-cellotriose and maltose utilization. These common features indicate a close evolutionary relationship among the polysaccharide catabolic pathways of the three species.
Acknowledgments
We gratefully thank Diana Irwin for advice and helpful suggestions and Joseph Calvo for discussion and critical reading of the manuscript.
This work was supported by grant DE-FG02-84ER13233 from the Department of Energy Basic Energy Research Program.
REFERENCES
- 1.Fennington G, Neubauer D, Stutzenberger F. Cellulase biosynthesis in a catabolite repression-resistant mutant of Thermomonospora fusca. Appl Environ Microbiol. 1984;47:201–204. doi: 10.1128/aem.47.1.201-204.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Abalos J M, Ruiz-Arribas A, Garda A L, Santamaria R I. Effect of carbon source on the expression of celA-1, a cellulase-encoding gene from Streptomyces halstedii JM8. FEMS Microbiol Lett. 1997;153:97–103. doi: 10.1111/j.1574-6968.1997.tb10469.x. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg N M, Warren R A J, Kilburn D G, Miller R C., Jr Regulation, initiation and termination of the cenA and cex transcripts of Cellulomonas fimi. J Bacteriol. 1987;169:646–653. doi: 10.1128/jb.169.2.646-653.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagerdahl B G R, Ferchak J D, Pye E K. Cellulolytic enzyme system of Thermomonospora sp. grown on microcrystalline cellulose. Appl Environ Microbiol. 1978;36:606–612. doi: 10.1128/aem.36.4.606-612.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwin D, Walker L, Spezio M, Wilson D. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotech Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 6.Jung E D, Lao G, Irwin D, Barr B K, Benjamin A, Wilson D B. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl Environ Microbiol. 1993;59:3032–3043. doi: 10.1128/aem.59.9.3032-3043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Lao G, Ghangas G S, Jung E D, Wilson D B. DNA sequences of three β-1,4-endoglucanase genes from Thermomonospora fusca. J Bacteriol. 1991;173:3397–3407. doi: 10.1128/jb.173.11.3397-3407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin E, Wilson D B. Regulation of β-1,4-endoglucanase synthesis in Thermomonospora fusca. J Bacteriol. 1987;53:1352–1357. doi: 10.1128/aem.53.6.1352-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin E, Wilson D B. Identification of a celE binding protein and its potential role in induction of the celE gene in Thermomonospora fusca. J Bacteriol. 1988;170:3843–3846. doi: 10.1128/jb.170.9.3843-3846.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin E, Wilson D B. Transcription of the celE gene in Thermomonospora fusca. J Bacteriol. 1988;170:3838–3842. doi: 10.1128/jb.170.9.3838-3842.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 13.Markiewicz P, Kleina L G, Cruz C, Ehret S, Miller J H. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and nonessential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlösser A, Jantos J, Hackmann K, Schrempf H. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl Environ Microbiol. 1999;65:2636–2643. doi: 10.1128/aem.65.6.2636-2643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher M A, Choi K Y, Zalkin H, Brennan R G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 17.Shiang M, Linden J C, Mohagheghi A, Grohmann K, Himmel M E. Regulation of cellulase synthesis in Acidothermus cellulolyticus. Biotechnol Prog. 1991;7:315–322. [Google Scholar]
- 18.Spiridonov N A, Wilson D B. Regulation of biosynthesis of individual cellulases in Thermomonospora fusca. J Bacteriol. 1998;180:3529–3532. doi: 10.1128/jb.180.14.3529-3532.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiridonov N A, Wilson D B. Characterization and cloning of CelR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J Biol Chem. 1999;274:13127–13132. doi: 10.1074/jbc.274.19.13127. [DOI] [PubMed] [Google Scholar]
- 20.Stoppok W, Rapp P, Wagner F. Formation, location, and regulation of endo-1,4-β-glucanases and β-glucosidases from Cellulomonas uda. Appl Environ Microbiol. 1982;44:44–53. doi: 10.1128/aem.44.1.44-53.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Wezel G P, White J, Bibb M J, Postma P W. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol Gen Genet. 1997;254:604–608. doi: 10.1007/s004380050458. [DOI] [PubMed] [Google Scholar]
- 22.van Wezel G P, White J, Young P, Postma P W, Bibb M J. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol Microbiol. 1997;23:537–549. doi: 10.1046/j.1365-2958.1997.d01-1878.x. [DOI] [PubMed] [Google Scholar]
- 23.Walter S, Schrempf H. The synthesis of the Streptomyces reticuli cellulase (Avicelase) is regulated by both activation and repression mechanisms. Mol Gen Genet. 1996;251:186–195. doi: 10.1007/BF02172917. [DOI] [PubMed] [Google Scholar]
- 24.Wilson D B. Biochemistry and genetics of actinomycete cellulases. Crit Rev Biotechnol. 1992;12:45–63. doi: 10.3109/07388559209069187. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Lao G, Wilson D B. Characterization of a Thermomonospora fusca exocellulase. Biochemistry. 1995;34:3386–3395. doi: 10.1021/bi00010a030. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Wang Y, Ruan J. Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol. 1998;48:411–422. doi: 10.1099/00207713-48-2-411. [DOI] [PubMed] [Google Scholar]