Abstract
Background
Most hospitals use traditional infection prevention (IP) methods for outbreak detection. We developed the Enhanced Detection System for Healthcare-Associated Transmission (EDS-HAT), which combines whole-genome sequencing (WGS) surveillance and machine learning (ML) of the electronic health record (EHR) to identify undetected outbreaks and the responsible transmission routes, respectively.
Methods
We performed WGS surveillance of healthcare-associated bacterial pathogens from November 2016 to November 2018. EHR ML was used to identify the transmission routes for WGS-detected outbreaks, which were investigated by an IP expert. Potential infections prevented were estimated and compared with traditional IP practice during the same period.
Results
Of 3165 isolates, there were 2752 unique patient isolates in 99 clusters involving 297 (10.8%) patient isolates identified by WGS; clusters ranged from 2–14 patients. At least 1 transmission route was detected for 65.7% of clusters. During the same time, traditional IP investigation prompted WGS for 15 suspected outbreaks involving 133 patients, for which transmission events were identified for 5 (3.8%). If EDS-HAT had been running in real time, 25–63 transmissions could have been prevented. EDS-HAT was found to be cost-saving and more effective than traditional IP practice, with overall savings of $192 408–$692 532.
Conclusions
EDS-HAT detected multiple outbreaks not identified using traditional IP methods, correctly identified the transmission routes for most outbreaks, and would save the hospital substantial costs. Traditional IP practice misidentified outbreaks for which transmission did not occur. WGS surveillance combined with EHR ML has the potential to save costs and enhance patient safety.
Keywords: whole-genome sequencing, surveillance, machine learning, hospital-associated infections, outbreaks
Whole-genome sequencing surveillance of bacterial pathogens and machine learning of the electronic health record finds previously undetected outbreaks and their transmission routes, which can increase patient safety and save costs.
Approaches for healthcare outbreak detection have remained essentially unchanged for decades [1]. When an outbreak is suspected, a method to establish genetic relatedness such as whole-genome sequencing (WGS) may be performed. This approach can miss outbreaks and falsely identify suspected outbreaks that are refuted by WGS.
Although WGS surveillance has been useful for identifying otherwise undetected transmission events, identifying the responsible transmission route has had limited success. This is because investigations have focused primarily on geotemporal clustering, which can miss complex transmission routes [2, 3].
In late 2016 we began development of the Enhanced Detection System for Healthcare-Associated Transmission (EDS-HAT), which combines WGS surveillance with machine learning (ML) of the electronic health record (EHR) to detect outbreaks and identify their routes of transmission [4–8]. We have found EHR ML useful for transmission routes that cannot be identified by traditional means [4, 5, 7].
The EDS-HAT was run with an at least 6-month lag between infection and WGS so that its performance could be compared with our practice of using WGS in reaction to suspected outbreaks. We conducted a detailed analysis of EDS-HAT compared with traditional infection prevention (IP) practice.
METHODS
Study Setting
This study was performed at the University of Pittsburgh Medical Center–Presbyterian Hospital (UPMC), an adult tertiary care hospital with 758 total beds, 134 critical care beds, and over 400 annual solid-organ transplants. An independent chronic care facility with 32 beds is physically embedded within the UPMC. Transfer of patients between this facility and UPMC is common. Ethics approval was obtained from the University of Pittsburgh Institutional Review Board.
Isolate Collection
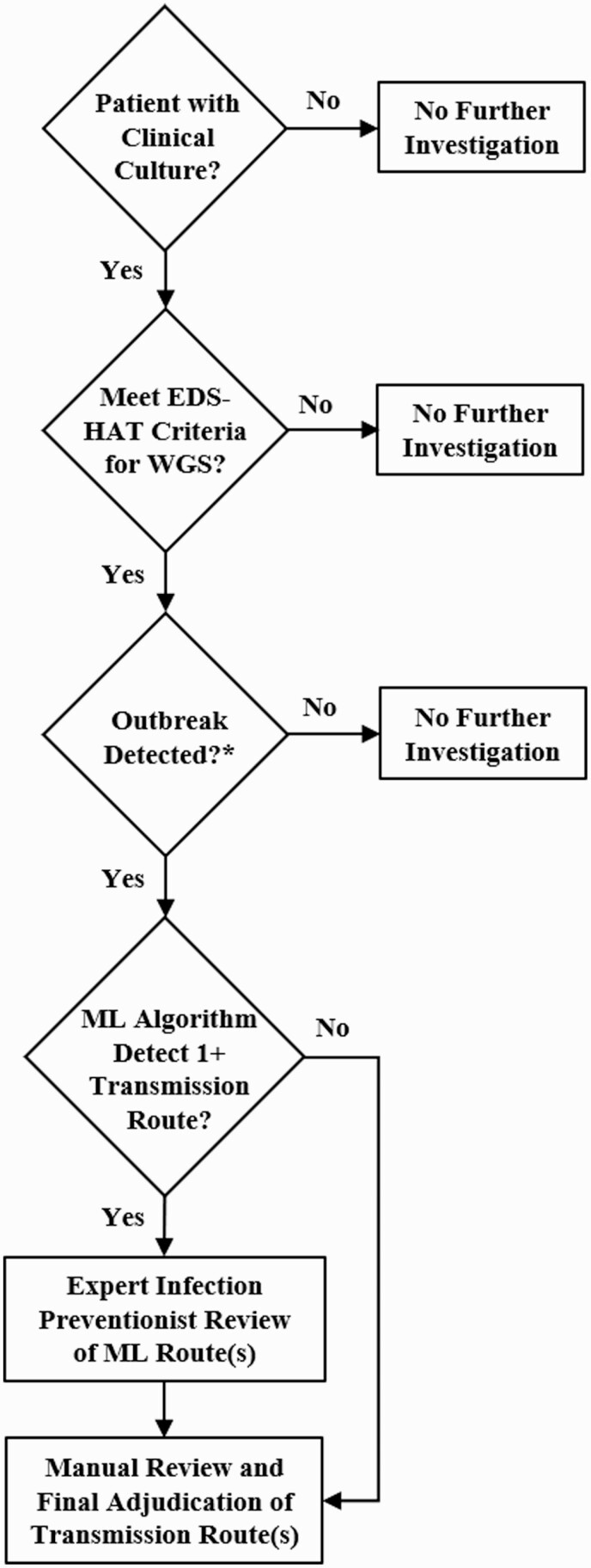
A description of the outbreak detection process is shown in Figure 1. For WGS surveillance, we collected select bacterial pathogens isolated from clinical specimens between November 2016 and November 2018: Acinetobacter species, Pseudomonas species, extended-spectrum B-lactamase-producing (ESBL) Escherichia coli, Klebsiella species, Clostridioides difficile, ESBL Enterobacter species, vancomycin-resistant Enterococcus (VRE), methicillin-resistant Staphylococcus aureus (MRSA), Stenotrophomonas species, Serratia species, Burkholderia species, Legionella species, Providencia species, Proteus species, and Citrobacter species. These pathogens were selected because they cause serious infections and healthcare-associated outbreaks. For Clostridioides difficile, we performed culture of stool specimens that were culture-independent diagnostic test–positive for C. difficile. Inclusion criteria were hospital admission or observation 3 or more days before the culture date and/or a recent inpatient or outpatient encounter in the 30 days before the culture date.
Figure 1.
Flow diagram of the EDS-HAT outbreak detection process, from clinical culture through adjudication of transmission route(s). ∗As described in Methods. Abbreviations: EDS-HAT, Enhanced Detection System for Healthcare-Associated Transmission; ML, machine learning; WGS, whole-genome sequencing.
Whole-Genome Sequencing
Whole-genome sequencing was performed on the NextSeq 500 platform (Illumina, San Diego, CA). Reads were assembled with SPAdes v3.13 [9], annotated with Prokka v1.14 [10], and multilocus sequence types (STs) were assigned using PubMLST typing schemes (https://github.com/tseemann/mlst) [11].
Pairwise core genome single nucleotide polymorphism (cgSNP) differences were calculated using Snippy v4.3.0 (https://github.com/tseemann/snippy) within species STs having 2 or more isolates. Genetically related clusters were assigned using initial SNP cutoffs using hierarchical clustering with single linkage [5, 6]. Based on our experience and the literature [3, 5, 6, 12–19], clusters were defined as isolates from more than 1 patient having 15 or fewer pairwise cgSNPs for all species except for C. difficile, for which 5 or fewer pairwise cgSNPs were used to identify clusters. For this organism, we defined clusters as all isolates that were within 0–2 cgSNPs, regardless of whether a transmission route was identified, and included cases that were within 3–5 cgSNPs of one another only if we could identify a statistically significant transmission route detected at 0–2 cgSNPs.
Extraction and Processing of Electronic Health Record Data
All patient encounters including inpatient, emergency room, and same-day surgery were mined for charge transaction codes, clinical microbiologic data, admission data, discharge data, and length of stay [4]. Charge transaction codes were included because they reflect many types of exposures associated with transmission, such as medical procedures, medical services, and medications. Data were assigned a unique identification number using De-ID software (De-ID Data, Philadelphia, PA). The names of healthcare workers who signed clinical notes were also extracted and de-identified. Procedures with multiple charge codes were aggregated into groups for transmission route analysis.
Machine Learning Algorithm
An ML algorithm based on point estimates for model parameters and incorporating case-control methodology was used [4, 7]. Case patients were defined as those with clinical isolates that clustered by WGS as defined above and control patients were all patients who were hospitalized in the 30 days prior to a case patient’s culture date and did not have a positive result for the genetically related strain. Only route exposures on or prior to a case patient’s culture date were considered.
The ML algorithm scores each outbreak by the maximum log-likelihood ratio of observing the case infections, given that exposure to the principal transmission route probabilistically causes infection over the likelihood of a nontransmission explanation. A constant patient-to-patient transmission likelihood is added for each case infection not exposed to the principal transmission route. Empirical P values are computed by estimating the likelihood of a higher outbreak score, given that no relationship exists between the case patients. This is done by sampling random sets of patients of equal size and computing their outbreak score maximized over routes. Importance sampling is used to improve efficiency of this process. Model parameters were fit using 9 historical outbreaks between 2012 and 2016, which are separate from the analysis presented in this manuscript (Supplementary Table 1). Parameter estimation was accomplished by transforming the outbreak detection problem into logistic regression, as previously described [4, 7].
Transmission routes for clustered isolates with statistically significant odds ratios (ORs) (P < .05) from the algorithm for category types (eg, procedures, locations, and providers) underwent manual EHR review for accuracy and biological plausibility. The manual EHR review was performed by an experienced infection preventionist (A. J. S.), who subsequently reviewed the findings with 2 senior investigators (L. H. H. and G. M. S.), all who have experience in hospital epidemiology and outbreak investigation. The purpose of the manual EHR review was to determine the most likely transmission route predicted by the ML algorithm or to investigate routes of transmission that were not identified by the algorithm. For some clusters, more than 1 transmission route was considered plausible (eg, transmission from a medical device with subsequent hospital unit–based transmission).
Clinical and Economic Modeling
Clinical and economic impact analysis was conducted from a hospital’s perspective. The analysis utilized the transmission network of outbreaks, effectiveness of IP interventions by transmission route, and time needed to implement IP interventions to estimate the expected number of transmissions under EDS-HAT, based on the method we previously described [8]. Since the effectiveness of IP interventions can decrease with time, we estimated lower and upper impact boundaries, with the true value likely between these estimates. For the lower boundary, we assumed that effectiveness would decline linearly and measured effectiveness from the time when the IP team first intervened. The effect of subsequent IP interventions that would have been implemented whenever an additional patient was infected through the same route was ignored. For the upper boundary, intervention effectiveness was assumed to remain constant. For outbreaks with more than 1 plausible transmission route, we weighted routes by the OR generated by the ML algorithm. If any route was missed by ML but detected by manual EHR review, we conservatively assigned the lowest OR score. Additionally, we performed a downstream cluster analysis to calculate the number of preventable infections if an intervention based on 1 outbreak could potentially prevent another outbreak using the same IP effectiveness parameters. For example, if EDS-HAT detected an outbreak in a hospital unit and an intervention was implemented, theoretically that intervention could prevent a subsequent outbreak.
Outcomes were incremental costs per transmission averted, number of readmissions averted, and lives saved. Probabilistic sensitivity analysis was conducted to assess the impact of uncertainty in parameter values of EDS-HAT. Data sources are described in Supplementary Table 2. All costs were adjusted to 2020 using the medical component of the Consumer Price Index [20]. Costs and benefits were discounted at 3%. Readmissions at 7 and 30 days postdischarge were recorded. An EHR review was performed to ascertain if readmissions were attributable to the infection; attributable readmissions were incorporated into the economic impact analysis.
Traditional Infection Prevention Practice
Whole-genome sequencing was performed in reaction to IP requests (reactive WGS) for suspected outbreaks. For the 2-year study period, the number of outbreaks detected by EDS-HAT versus traditional IP practice was determined.
RESULTS
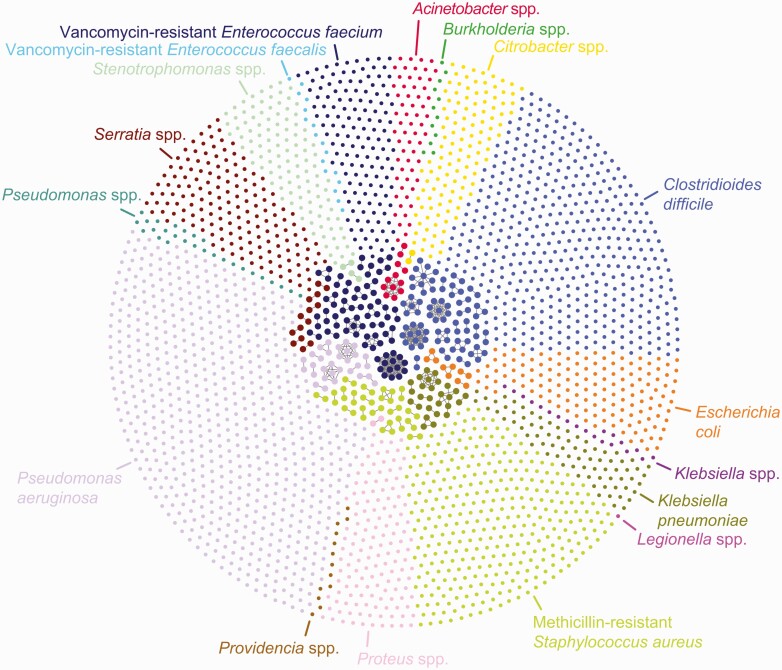
Of 3165 clinical isolates that underwent WGS, 2752 unique patient isolates were clustered by ST. A total of 297 (10.8%) isolates representing 99 distinct, genetically related clusters ranging in size between 2 and 14 isolates were identified (Figure 2, Table 1). A total of 269 (90.6%) isolates were from inpatient cultures, 27 (9.1%) were from the emergency room, and 1 (0.3%) was from an outpatient visit. EDS-HAT detected potential transmission routes for 65 (65.7%) clusters containing 221 (74.4%) of the related isolates (Supplementary Table 3). No significant transmission routes were detected by the EDS-HAT ML algorithm or manual review in the remaining 34 clusters, which ranged in size from 2 to 5 patients and contained 76 isolates. A brief description of high-impact or notable outbreaks and transmission routes detected by EDS-HAT ML is provided in Table 2, while Supplementary Table 3 describes all outbreaks.
Figure 2.
Cluster network of EDS-HAT isolates sequenced, grouped by bacterial species. The outer circle shows patient isolates that are not genetically related. The inner circle shows outbreaks of genetically related isolates as defined by cgSNP cutoffs described in Methods. The network plot was visualized with Gephi. Abbreviations: cgSNP, core genome single nucleotide polymorphism; EDS-HAT, Enhanced Detection System for Healthcare-Associated Transmission.
Table 1.
EDS-HAT Isolates Sequenced and Attributable Readmissions
| Sequenced | Attributable Readmissions | |||||
|---|---|---|---|---|---|---|
| Species | Collected | Unique Patient Isolates | No. Relateda (%) | Clusters | 7-Day | 30-Day |
| Acinetobacter species | 83 | 72 | 12 (16.7) | 3 | 1 | 1 |
| Burkholderia species | 12 | 12 | 0 (0) | 0 | 0 | 0 |
| Citrobacter species | 126 | 118 | 2 (1.7) | 1 | 0 | 0 |
| Clostridioides difficile | 558 | 524 | 80 (15.3) | 21 | 2 | 10 |
| Escherichia coli (ESBL) | 170 | 149 | 10 (6.7) | 4 | 0 | 1 |
| Klebsiella species (ESBL, not pneumoniae) | 25 | 20 | 0 (0) | 0 | 0 | 0 |
| Klebsiella pneumoniae (ESBL) | 111 | 102 | 27 (26.5) | 8 | 0 | 1 |
| Legionella species | 1 | 1 | 0 (0) | 0 | 0 | 0 |
| Methicillin-resistantStaphylococcus aureus | 425 | 365 | 39 (10.7) | 18 | 1 | 5 |
| Proteus species | 151 | 140 | 2 (1.4) | 1 | 0 | 0 |
| Providencia species | 14 | 13 | 0 (0) | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 881 | 693 | 31 (4.5) | 10 | 2 | 3 |
| Pseudomonas species (not aeruginosa) | 28 | 27 | 0 (0) | 0 | 0 | 0 |
| Serratia species | 181 | 173 | 14 (8.1) | 7 | 1 | 3 |
| Stenotrophomonas species | 127 | 114 | 4 (3.5) | 2 | 0 | 0 |
| Vancomycin-resistantEnterococcus faecalis | 17 | 17 | 0 (0) | 0 | 0 | 0 |
| Vancomycin-resistantEnterococcus faecium | 247 | 212 | 76 (35.8) | 24 | 5 | 16 |
| Total | 3165 | 2752 | 297 (10.8) | 99 | 12 | 40 |
Abbreviations: EDS-HAT, Enhanced Detection System for Healthcare-Associated Transmission; ESBL, extended-spectrum B-lactamase; SNP, single nucleotide polymorphism.
Fifteen or fewer pairwise SNPs for all organisms except for C. difficile (≤2 SNPs) (see Methods).
Table 2.
High-Impact or Notable Outbreaks Detected by EDS-HAT
| Outbreak | Details |
|---|---|
| Vancomycin-resistant Enterococcus faecium outbreak associated with IR and injection of sterile contrast [6] | This outbreak involved 10 initial patients and was ongoing when it was discovered. The EDS-HAT ML algorithm identified IR as a significant transmission route (OR: 43.8; P < .01; 95% CI: 5.6 to 346). Nine patients, including 3 with bacteremia, were identified as having IR procedures involving unsterile practices in the preparation of contrast. Safe practices and enhanced environmental cleaning were implemented and no additional IR-associated infections occurred. Subsequently, transmission of the outbreak strain occurred among 4 patients on shared hospital units. |
| Pseudomonas aeruginosa outbreak associated with gastroscopy [5] | This outbreak comprised 6 patients housed on different units over 7 months. Two patients had bacteremia, 3 had pneumonia, and 1 had a urinary tract infection. The EDS-HAT ML algorithm detected gastroscopy as a significant route for 4 patients (OR: 300.6; P < .01; 95% CI: 15.8 to 5690.5) with a fifth patient who did not have a charge code that reflected the gastroscopy procedure but who had a clinical note reflecting the procedure that was identified on manual EHR review. A post-disinfection gastroscope culture performed as part of routine IP practice was positive for P. aeruginosa; the isolate was sequenced and belonged to the outbreak, confirming gastroscopy as the responsible transmission route. |
| Outbreaks of multiple pathogens at the embedded chronic care facility | EDS-HAT ML identified 11 clusters involving 38 patients over 22 months, with a range of 2–9 total patients per cluster; 25 (65.8%) patients had this facility as a plausible transmission route. Pathogens included C. difficile (6 clusters), Klebsiella pneumoniae (1 cluster), MRSA (1 cluster), P. aeruginosa (2 clusters), and VRE (1 cluster). Three patients with C. difficile in 3 clusters were subsequently transferred to our institution and had unit-based commonalities with 3 additional patients who later developed C. difficile infection suggesting continuing transmission. |
| Outbreaks of multiple pathogens on an ICU | There were 12 clusters with 57 patients (range: 2–14), of whom 28 (49.1%) had a single ICU stay identified by EDS-HAT ML as the potential transmission route. Organisms included C. difficile (3 clusters involving 10 patients), K. pneumoniae (3 clusters involving 16 patients), P. aeruginosa (1 cluster involving 3 patients), Serratia marcescens (1 cluster involving 2 patients), and VRE (4 clusters involving 26 patients). |
| C. difficile outbreaks associated with wound care | There were 9 C. difficile clusters, ranging in size from 2 to 12 patients. Of 52 patients, 29 (55.8%) had wound care service identified as a potential transmission route, with exposures occurring 1–92 days (mean: 16 days; median: 9 days) before the positive test for C. difficile. This consult service involved nurses providing management of sacral pressure ulcer wounds. |
| MRSA infections associated with EEG | This cluster consisted of 2 patients with culture dates separated by 8 days. The EDS-HAT ML algorithm identified EEG as a transmission route. Manual EHR review determined that both patients had a bedside EEG performed on the same day on separate units by the same physician and technician, 2 and 10 days before positive culture dates. |
Abbreviations: CI, confidence interval; EDS-HAT, Enhanced Detection System for Healthcare-Associated Transmission; EEG, electroencephalography; EHR, electronic health record; ICU, intensive care unit; IP, infection prevention; IR, interventional radiology; ML, machine learning; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio; VRE, vancomycin-resistant Enterococcus faecium.
Outbreaks Detected by Traditional Infection Prevention Practice
During the study period, our IP department requested reactive WGS for 15 suspected and potentially actionable outbreaks while EDS-HAT was running in parallel (2 Acinetobacter baumannii, 1 Burkholderia cepacia, 6 C. difficile, 1 Klebsiella pneumoniae, 3 Serratia marcescens, 2 Serratia maltophilia) involving 133 patients. Of these 15 suspected clusters, 5 (3.8%) patient isolates from 2 clusters (A. baumannii and Stenotrophomonas maltophilia) were found to be genetically related. Of these 5 patients with related isolates, 2 of the transmissions involving A. baumannii were also detected by EDS-HAT.
Clinical and Economic Impact Analysis
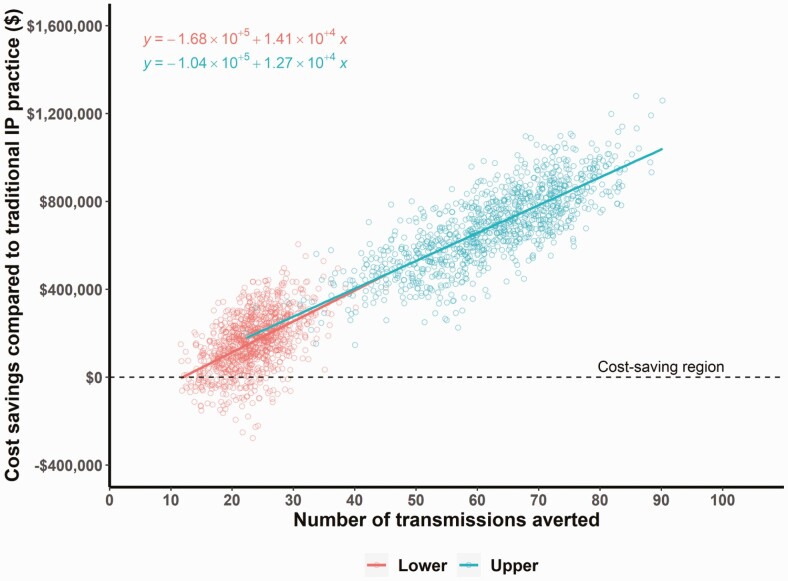
EDS-HAT could have prevented 25 (lower bound) to 63 (upper bound) transmissions. Moreover, 3.1–8.0 fewer 30-day attributable readmissions and 1.6–3.3 fewer deaths would have occurred had EDS-HAT been running in real time. Under EDS-HAT, the increase in cost of sequencing would be offset by savings in costs of treating infections, resulting in overall cost-savings of $192 408 to $692 532 over the study period. EDS-HAT was found to be a more-effective and cost-saving program than traditional IP practice by providing savings of $7745–$10 939 for each transmission averted. Based on the lower bound estimates, EDS-HAT remained cost-saving and more effective in various independent scenarios: when the time needed for effective intervention was increased to 21 days, the proportion of time spent towards outbreak detection under EDS-HAT was doubled (20%), effectiveness against procedures and healthcare workers was reduced to 30% (relative risk = 0.7), the duration after which the IP intervention’s effectiveness would become zero was reduced to 13 weeks for all transmission routes except for instruments, or the proportion of untreated cases was increased to 70% for respiratory, 50% for urine, 25% for wound, or 10% for stool. In a probabilistic sensitivity analysis, EDS-HAT was cost-saving and more effective than traditional IP practice alone in more than 88% of simulations in lower and 99% in upper bound scenarios (Figure 3, Supplementary Table 4).
Figure 3.
EDS-HAT cost-savings and effectiveness plot for estimated lower and upper bound boundaries (see Methods). Cost-savings of EDS-HAT were examined by estimated costs associated with number of transmissions averted, using 1000 simulations in probabilistic sensitivity analysis comparing EDS-HAT with traditional infection prevention practice. Each point represents 1 simulation of the economic model. The best-fit linear model is shown as a straight line. Abbreviation: EDS-HAT, Enhanced Detection System for Healthcare-Associated Transmission.
DISCUSSION
In this study, we demonstrate the value of combining WGS surveillance with ML of the EHR for enhanced hospital outbreak detection. EDS-HAT detected consequential outbreaks and transmission routes that were undetected by traditional IP practice, whereas the latter mostly identified suspected outbreaks that were not confirmed by reactive WGS. Both components of EDS-HAT are essential: WGS surveillance is used to “connect the dots” between seemingly unrelated patients to signal an outbreak and ML, in combination with review by an IP expert, then identifies the responsible transmission route. In our study, we found that 10.8% of sequenced isolates were related, which is in line with other studies of WGS surveillance [12, 16, 21–24].
The results of our clinical and economic impact analysis suggest that, had it been running in real time, EDS-HAT would be highly cost-saving. The cost of sequencing 1 bacterial isolate is low ($70) relative to the high costs of treating a single, potentially preventable infection (eg, >$24 000 for Pseudomonas pneumonia). Recent budget and clinical impact analyses of WGS surveillance of multidrug-resistant pathogens in Australia also demonstrated that this approach is cost-saving [25, 26]. Our analysis showed cost-savings despite our conservative modeling assumptions, which included the effectiveness of various types of interventions and the fact that we did not consider the cost of personal protective equipment and other costs associated with isolation precautions of patients. By using this conservative approach, we likely underestimated the true impact and cost-savings of EDS-HAT.
The inability to demonstrate transmission routes that do not involve geotemporal clustering is a serious limitation of previous studies of WGS surveillance for outbreak detection in hospitals [2, 3]. EDS-HAT overcomes this limitation by incorporating EHR ML [27–29]. Outbreaks that were detected exclusively by EDS-HAT tended to involve common hospital pathogens that lacked geographic clustering and had transmission routes that were not readily apparent on manual EHR review. For example, the interventional radiology VRE outbreak identified a newly discovered procedural vulnerability, the outbreak of Pseudomonas aeruginosa affirmed known risks of endoscopy, outbreaks in the chronic care facility highlighted the problem of high-risk transmission in this vulnerable patient population, the outbreak associated with wound care highlighted operational susceptibilities in the nature of care provided, and the cluster of MRSA associated with electroencephalography and specific providers shows how EDS-HAT can detect unusual and specific routes.
Implementation of real-time WGS surveillance and ML of the EHR will require investment in healthcare infrastructure; the results of our economic analysis provide evidence that implementation can be cost-saving for hospitals that perform reactive WGS. Parcell et al [30] highlight barriers to implementation and methods for integration into IP practice. We view EDS-HAT as complementary to IP practice because it alerts to possible outbreaks, which prompts additional investigating and intervention. EDS-HAT requires input from infection preventionists to evaluate the transmission routes that are generated and determine what interventions are needed.
There are several limitations to our study. First, it is unlikely that all outbreaks and outbreak patients were captured in this study, because, for example, some infected patients may not have cultures taken or cultures may have been negative because of recent antibiotic administration. In addition, our exclusion of cultures during the first 3 days of hospitalization likely led us to miss transmission events. Second, we did not include surveillance swabs, meaning that we likely missed transmission events for, for example, VRE. Third, the retrospective nature of the study did not allow us to investigate and confirm potential transmission routes for some of our outbreaks; this limitation can be alleviated and the potential impact will likely increase when EDS-HAT is run in real time. Fourth, during this 2-year evaluation, we had fewer transmissions identified by traditional IP practice at our institution than usual [4, 31, 32]. However, EDS-HAT would likely have detected any IP-identified outbreak more quickly. Fifth, our economic modeling of real-time interventions may not reflect true intervention effectiveness and timeliness. However, we adjusted for both conservative and loose parameters to estimate the true effectiveness in between those bounds. Sixth, we did not account for potential asymptomatic carriage of urinary and wound cultures in our model. However, infection preventionists would intervene regardless of clinical presentation given that it would aid in interrupting future transmission. In addition, many of these positive cultures are treated and, therefore, incur costs, whether the treatment is appropriate or not. Finally, we included only a limited number of pathogens in WGS surveillance because of feasibility and cost and therefore likely missed outbreaks caused by other pathogens.
Advances in microbial genomics and bioinformatics, digitalization of healthcare data, and ML technology have made enhanced outbreak detection in hospitals feasible. Taken together, our results suggest that EDS-HAT represents a potential paradigm shift in how outbreaks are detected in hospitals. If instituted in real time, this approach can reduce healthcare-related costs and significantly improve patient safety.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the UPMC Presbyterian Infection Prevention team.
Disclaimer. The National Institutes of Health played no role in data collection, analysis, or interpretation; study design; writing of the manuscript; or decision to submit for publication.
Financial support. This work was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant numbers R21Al109459 and R01AI127472). D. V. T. reports support from the University of Pittsburgh Department of Medicine (funding to support D. V. T.’s contributions).
Contributor Information
Alexander J Sundermann, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA; Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Jieshi Chen, Auton Lab, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA.
Praveen Kumar, Department of Health Policy and Management, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Ashley M Ayres, Department of Infection Control and Hospital Epidemiology, UPMC Presbyterian, Pittsburgh, Pennsylvania, USA.
Shu Ting Cho, Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Chinelo Ezeonwuka, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Marissa P Griffith, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
James K Miller, Auton Lab, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA.
Mustapha M Mustapha, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
A William Pasculle, Department of Pathology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Melissa I Saul, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USAand.
Kathleen A Shutt, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Vatsala Srinivasa, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Kady Waggle, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Daniel J Snyder, Department of Microbiology and Molecular Genetics and Center for Evolutionary Biology and Medicine, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Vaughn S Cooper, Department of Microbiology and Molecular Genetics and Center for Evolutionary Biology and Medicine, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Daria Van Tyne, Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Graham M Snyder, Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA; Department of Infection Control and Hospital Epidemiology, UPMC Presbyterian, Pittsburgh, Pennsylvania, USA.
Jane W Marsh, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Artur Dubrawski, Auton Lab, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA.
Mark S Roberts, Department of Health Policy and Management, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USAand.
Lee H Harrison, Microbial Genomic Epidemiology Laboratory, Center for Genomic Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA; Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
References
- 1. Peacock SJ, Parkhill J, Brown NM.. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology 2018; 164:1213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sherry NL, Lee RS, Gorrie CL, et al. . Pilot study of a combined genomic and epidemiologic surveillance program for hospital-acquired multidrug-resistant pathogens across multiple hospital networks in Australia. Infect Control Hosp Epidemiol 2020; 1–9. doi: 10.1017/ice.2020.1253. [DOI] [PubMed] [Google Scholar]
- 3. Ward DV, Hoss AG, Kolde R, et al. . Integration of genomic and clinical data augments surveillance of healthcare-acquired infections. Infect Control Hosp Epidemiol 2019; 40:649–55. [DOI] [PubMed] [Google Scholar]
- 4. Sundermann AJ, Miller JK, Marsh JW, et al. . Automated data mining of the electronic health record for investigation of healthcare-associated outbreaks. Infect Control Hosp Epidemiol 2019; 40:314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sundermann AJ, Chen J, Miller JK, et al. . Outbreak of Pseudomonas aeruginosa infections from a contaminated gastroscope detected by whole genome sequencing surveillance. Clin Infect Dis 2020; 73:e638–e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sundermann AJ, Babiker A, Marsh JW, et al. . Outbreak of vancomycin-resistant Enterococcus faecium in interventional radiology: detection through whole-genome sequencing-based surveillance. Clin Infect Dis 2020; 70:2336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller JK, Chen J, Sundermann A, et al. . Statistical outbreak detection by joining medical records and pathogen similarity. J Biomed Inform 2019; 91:103126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar P, Sundermann AJ, Martin EM, et al. . Method for economic evaluation of bacterial whole genome sequencing surveillance compared to standard of care in detecting hospital outbreaks. Clin Infect Dis 2020; 73:e9–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30:2068–9. [DOI] [PubMed] [Google Scholar]
- 11. Jolley KA, Maiden MCJ.. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berbel Caban A, Pak TR, Obla A, et al. . PathoSPOT genomic epidemiology reveals under-the-radar nosocomial outbreaks. Genome Med 2020; 12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jakharia KK, Ilaiwy G, Moose SS, et al. . Use of whole-genome sequencing to guide a Clostridioides difficile diagnostic stewardship program. Infect Control Hosp Epidemiol 2019; 40:804–6. [DOI] [PubMed] [Google Scholar]
- 14. Gona F, Comandatore F, Battaglia S, et al. . Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb Genom 2020; 6. doi: 10.1099/mgen.0.000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose R, Nolan DJ, Moot S, et al. . Molecular surveillance of methicillin-resistant Staphylococcus aureus genomes in hospital unexpectedly reveals discordance between temporal and genetic clustering. Am J Infect Control 2021; 49:59–64. [DOI] [PubMed] [Google Scholar]
- 16. Marmor A, Daveson K, Harley D, Coatsworth N, Kennedy K.. Two carbapenemase-producing Enterobacteriaceae outbreaks detected retrospectively by whole-genome sequencing at an Australian tertiary hospital. Infect Dis Health 2020; 25:30–3. [DOI] [PubMed] [Google Scholar]
- 17. Sherry NL, Lane CR, Kwong JC, et al. . Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012 to 2016. J Clin Microbiol 2019; 57:e00573–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwong JC, Lane CR, Romanes F, et al. . Translating genomics into practice for real-time surveillance and response to carbapenemase-producing Enterobacteriaceae: evidence from a complex multi-institutional KPC outbreak. PeerJ 2018; 6:e4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raven KE, Gouliouris T, Brodrick H, et al. . Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin Infect Dis 2017; 64:886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bureau of Labor Statistics. Medical care in US city average, all urban consumers, not seasonally adjusted. Washington, DC: Bureau of Labor Statistics, 2020. [Google Scholar]
- 21. Leong KWC, Cooley LA, Anderson TL, et al. . Emergence of vancomycin-resistant Enterococcus faecium at an Australian hospital: a whole genome sequencing analysis. Sci Rep 2018; 8:6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. García-Fernández S, Frentrup M, Steglich M, et al. . Whole-genome sequencing reveals nosocomial Clostridioides difficile transmission and a previously unsuspected epidemic scenario. Sci Rep 2019; 9:6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eyre DW, Shaw R, Adams H, et al. . WGS to determine the extent of Clostridioides difficile transmission in a high incidence setting in North Wales in 2015. J Antimicrob Chemother 2019; 74:1092–100. [DOI] [PubMed] [Google Scholar]
- 24. Miles-Jay A, Weissman SJ, Adler AL, Baseman JG, Zerr DM.. Whole genome sequencing detects minimal clustering among Escherichia coli sequence type 131-H30 isolates collected from United States children’s hospitals. J Pediatric Infect Dis Soc 2021; 10:183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon LG, Elliott TM, Forde B, et al. . Budget impact analysis of routinely using whole-genomic sequencing of six multidrug-resistant bacterial pathogens in Queensland, Australia. BMJ Open 2021; 11:e041968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. National Center for Biotechnology Information. Genomic surveillance, characterization and intervention of a polymicrobial multidrug-resistant outbreak in critical care—PubMed. Available at: https://pubmed.ncbi.nlm.nih.gov/33599607/. Accessed 20 February 2021. [DOI] [PMC free article] [PubMed]
- 27. Bartels MD, Larner-Svensson H, Meiniche H, et al. . Monitoring meticillin resistant Staphylococcus aureus and its spread in Copenhagen, Denmark, 2013, through routine whole genome sequencing. Euro Surveill 2015; 20. doi: 10.2807/1560-7917.es2015.20.17.21112. [DOI] [PubMed] [Google Scholar]
- 28. Mellmann A, Bletz S, Böking T, et al. . Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 2016; 54:2874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price JR, Cole K, Bexley A, et al. . Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: a longitudinal cohort study based on whole-genome sequencing. Lancet Infect Dis 2017; 17:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parcell BJ, Gillespie SH, Pettigrew KA, Holden MTG.. Clinical perspectives in integrating whole-genome sequencing into the investigation of healthcare and public health outbreaks—hype or help? J Hosp Infect 2021; 109:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marsh JW, Mustapha MM, Griffith MP, et al. . Evolution of outbreak-causing carbapenem-resistant Klebsiella pneumoniae ST258 at a tertiary care hospital over 8 years. mBio 2019; 10. doi: 10.1128/mBio.01945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galdys AL, Marsh JW, Delgado E, et al. . Bronchoscope-associated clusters of multidrug-resistant Pseudomonas aeruginosa and carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol 2019; 40:40–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.