Abstract
Background
Microsporidia are rarely reported to cause outbreaks of diarrhea. We describe a foodborne outbreak of microsporidiosis from a workplace canteen in November 2020 in Denmark.
Methods
A probable case was defined as any person using the canteen between 4 November and 13 December 2020, reporting at least one gastrointestinal symptom, whereas a confirmed case also had an Enterocytozoon bieneusi positive stool sample. A web-based questionnaire was used to collect clinical, epidemiological, and food exposure data. We performed a retrospective cohort study and tested stool samples from affected individuals for bacterial, viral, and parasitic pathogens, including E. bieneusi.
Results
Altogether, 195 individuals completed the questionnaire. We identified 52 cases (65% male; median age 45 years [range 25–65]). Diarrhea (90%), fatigue (83%), and abdominal pain (79%) were the most commonly reported symptoms. Eight cases were laboratory-confirmed and had E. bieneusi genotype C. The incubation period was between 5 and 12 days, and polymerase chain reaction (PCR)-detectable spore shedding occurred up to 43 days after symptom onset. Disease was associated with consuming food from the workplace canteen on 4 November 2020 (relative risk [RR[, 2.8 [95% confidence interval [CI]: 1.4 – 5.4]) and lunchboxes containing open sandwiches (RR, 3.2 [95% CI: 1.4 – 7.2]) served that day.
Conclusions
This is the second documented foodborne outbreak of E. bieneusi genotype C-associated diarrhea worldwide. Epidemiological findings advocated an open sandwiches lunchbox from 4 November 2020, as a likely source. E. bieneusi may be an under-reported cause of outbreaks of diarrhea, and testing for it might be useful in foodborne outbreak investigations.
Keywords: Enterocytozoon, foodborne disease, microsporidia, microsporidiosis, outbreak
Enterocytozoon bieneusiwas identified as the cause of a foodborne outbreak of diarrhea affecting >70 individuals. The incubation period was 5–12 days. Spores were shed in stool for up to 43 days after symptom onset.
Microsporidia are single-celled, spore-forming obligate intracellular organisms comprising more than 1500 species belonging to more than 200 genera [1–4]. Although microsporidia readily infect a wide range of vertebrate and invertebrate hosts worldwide, human infections are rarely reported [2, 5–8]. Only 17 species, including Enterocytozoon bieneusi (E. bieneusi), are considered human pathogens [9]. Infection may lead to clinical symptoms and disease, which is referred to as microsporidiosis.
E. bieneusi is an important cause of opportunistic infections, particularly among immunocompromised individuals, such as patients with AIDS or cancer patients, as well as patients receiving organ transplants [10–13]. However, documented cases remain overall rare [14]. The incubation period of microsporidiosis remains unclear, but a previous investigation of an outbreak of diarrhea due to E. bieneusi in a hotel in Sweden suggested that it is between 0 and 21 days [8]. Existing literature indicates that diarrhea is the most common symptom caused by E. bieneusi, typically accompanied by other gastrointestinal symptoms [9, 15].
The infection can result from fecal-oral transmission of spores from infected humans or animals through contaminated food or water [14]. Fresh produce (berries, vegetables, lettuce, parsley, and sprouts) [7, 16] and dairy products [17] can contain E. bieneusi and potentially cause foodborne infections in humans. Microsporidia can also be present in drinking water, wastewater, and surface and recreational waters [2]. Two reports from France showed that E. bieneusi can be waterborne [13, 18]. The conclusions from the foodborne outbreak of E. bieneusi in Sweden in 2009 highlighted the potential of this pathogen to cause gastrointestinal disease in humans after ingestion of contaminated food items [11].
On 23 November 2020, Statens Serum Institut (SSI) was informed by the Danish Veterinary and Food Administration (DVFA) about an outbreak of diarrhea in 77 individuals of a private company in Denmark. A foodborne outbreak originating from the canteen of the company was suspected. Altogether, 49 stool samples from 24 affected individuals tested negative for common gastrointestinal bacterial and viral pathogens at the regional clinical microbiology department. Additional stool samples were collected, and these were analyzed at SSI. On 1 December 2020, E. bieneusi, genotype C was identified in some of the stool samples.
We investigated the outbreak and aimed to provide a comprehensive description of its epidemiological and clinical aspects. Furthermore, we aimed to assess pathogen-related aspects, such as incubation period and duration of polymerase chain reaction (PCR)-detectable spore shedding.
METHODS
Case Definition
A probable case was defined as any person who worked at the company reporting at least 1 of the following symptoms: diarrhea, abdominal pain, nausea, or vomiting, during the period of 4 November to 13 December 2020, and who consumed lunch from the workplace canteen. A confirmed case was defined as any person who worked at the company with a positive test for E. bieneusi in at least one stool sample obtained during the period of 4 November to 13 December 2020.
Study Population and Epidemiological Investigation
In order to identify the potential source of the outbreak, we conducted a retrospective cohort study. Between 4 December 2020 and 5 January 2021, all individuals at the affected company were asked to complete a web-based questionnaire concerning clinical manifestations and their food and water consumption at lunch of the company’s canteen, as well as secondary exposure to a person with diarrhea or other symptoms, during the period of 2 November to 13 November 2020. We sent reminder emails to enhance participation as well as reminders for stool sample submission. Individuals who only submitted stool samples and who reported to be sick but did not fill in the questionnaire were excluded from the analyses. This means that only confirmed and probable cases who answered the questionnaire survey were included. Due to increased hygiene measures during the COVID-19 pandemic, only pre-packed lunchboxes instead of buffet-style food were offered for purchase at the canteen of the affected company. All exposure items were assessed by self-reported consumption using a dichotomous variable.
The χ2 test or Fisher exact test, and the Kruskal-Wallis rank-sum test were used to compare proportions and means between groups, respectively. Exposure-specific attack rates (AR), relative risks (RR), and 95% confidence intervals (CI) were calculated for canteen lunch day and for food and water items on a given canteen lunch day. Confounding was assessed by comparing crude and Mantel–Haenszel (MH)-adjusted RR. Stratified analysis was conducted by comparing strata-specific RR according to a relevant exposure item. Woolf test of homogeneity was used to assess statistically significant associations on effect modification. Data analyses were performed using R© base version 4.03 [19], tidyverse metapackage version 1.3.0 [20], and mStats package 3.4.0.
Laboratory Analyses
Individuals of the affected company who developed symptoms during the above-mentioned period were encouraged to submit stool samples for microbiology testing. Samples were tested by PCR for parasites and viruses, and by culture for bacteria. Specifically, samples were screened for the following pathogens: Cryptosporidium spp., Giardia duodenalis, rotavirus, adenovirus, astrovirus, sapovirus, norovirus, Salmonella spp., Shigella spp., Yersinia spp., Campylobacter spp., Clostridium difficile, Aeromonas spp., Plesiomonas shigelloides, Vibrio spp., Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus. In addition, culture for Escherichia coli was conducted with screening for specific virulence genes for Shiga toxin-producing E. coli, enteropathogenic E. coli, enterotoxigenic E. coli, enteroinvasive E. coli, and attaching and effacing E. coli.
After testing negative for all the above-mentioned pathogens, samples were then tested for E. bieneusi by real-time PCR [21] and genotyped using 2 sets of primers described by Buckholt et al [22]. targeting the internal transcribed spacer (ITS) region of E. bieneusi. For these purposes, DNA was directly extracted from stool samples using an eMAG® (BioMérieux) after initial disruption of the spores with a TissueLyser II (Qiagen, Germany). To identify the genotype of E. bieneusi, PCR products obtained after nested PCR from the previous step were column-purified and subject to bidirectional Sanger sequencing as described by Buckholt et al [22]. The Basic Local Alignment Search Tool (National Center for Biotechnology Information) was used to compare the nucleotide sequences with sequences in the GenBank database. Individuals were encouraged to submit stool samples for testing at Statens Serum Institut from 23 November 2020, the beginning of the outbreak investigation, until 5 January 2021 when the cohort study ended. However, once testing positive for E. bieneusi, the affected individuals were encouraged to keep submitting stool samples until they tested negative in order to identify the duration of spore shedding. The last stool sample was submitted on 19 January 2021.
Environmental Investigations
The DVFA performed environmental investigations on the company’s premises and canteen during November and December 2020. These covered structural conditions on food storage, food handling, and cooked-food security at the canteen and interviews of food handlers. Due to the closure of the canteen for four days, at the time of visit by the DVFA, food and water items from the period of the outbreak were no longer available and could not be collected for testing for E. bieneusi.
RESULTS
Descriptive Epidemiology
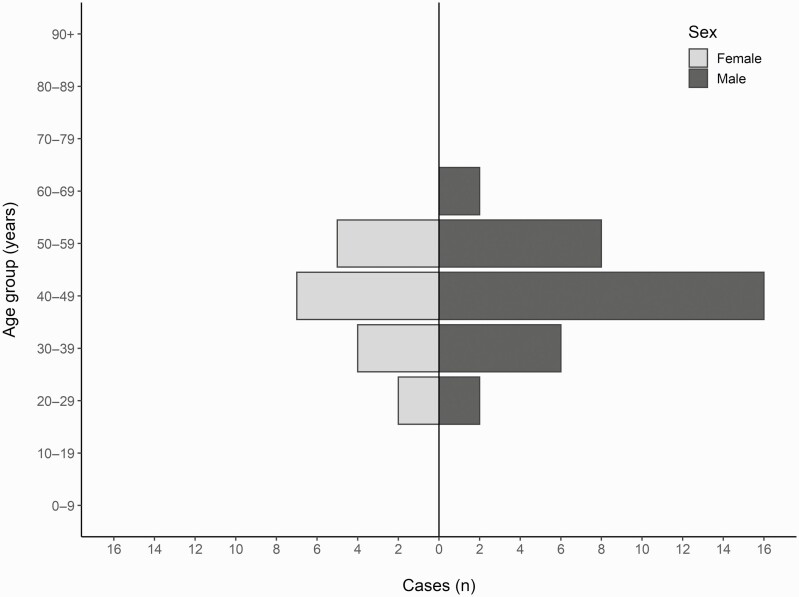
Overall, 195 out of approximately 300 individuals from the company completed the questionnaire. The cohort comprised 44 probable cases and 8 confirmed cases (attack rate [AR] 27%), and 143 non-cases. The cohort comprised 80 women and 115 men (Table 1), and approximately two thirds of all cases were males (Table 2). The median age was 47 years for all participants (range 24–66) (Table 1) and 45 years for all cases (range 25–65) (Table 2). There was no statistically significant difference between those who reported symptoms and those who did not among all respondents, by sex, age (Table 1), or department of work (data not shown). No cases were reported among the staff of the canteen.
Table 1.
General Demographic Characteristics of Non-cases and Cases During an Enterocytozoon bieneusi Outbreak, Denmark, 2020 Identified Through a Questionnaire Investigation (N = 195)
| Non-cases (N = 143) | Cases (N = 52) | Total (N = 195) | P-value | |
|---|---|---|---|---|
| Sex, n (%) | .32a | |||
| Female | 62 (43) | 18 (35) | 80 (41) | |
| Male | 81 (57) | 34 (65) | 115 (59) | |
| Age, years | .43b | |||
| Median (range) | 47.0 (24.0–66.0) | 45.0 (25.0–65.0) | 46.0 (24.0–66.0) | |
| Missing (n) | 9 | 4 | 13 |
Fisher exact test with simulated P-value (based on 1000 replicates).
Kruskal-Wallis rank sum test.
Table 2.
Summary of General Demographic Characteristics, Clinical Presentation, and Duration of Illness Reported by Probable and Confirmed Cases Identified During an Enterocytozoon bieneusi Outbreak, Denmark, 2020 (N = 52)
| Probable Cases (N = 44) | Confirmed Cases (N = 8) | Total (N = 52) | P-value | |
|---|---|---|---|---|
| Sex, n (%) | .99a | |||
| Female | 15 (34) | 3 (38) | 18 (35) | |
| Male | 29 (66) | 5 (62) | 34 (65) | |
| Age, years | .68b | |||
| Median (range) | 45.5 (25.0–65.0) | 42.5 (30.0–57.0) | 45.0 (25.0–65.0) | |
| Missing, n | 4 | 0 | 4 | |
| Clinical manifestations | ||||
| Any gastrointestinal symptom(s), n (%) | .99a | |||
| No | 0 (0) | 0 (0) | 0 (0) | |
| Yes | 43 (98) | 8 (100) | 51 (98) | |
| Do not remember | 1 (2) | 0 (0) | 1 (2) | |
| Diarrhea, n (%) | .99a | |||
| No | 5 (11) | 0 (0) | 5 (10) | |
| Yes | 39 (89) | 8 (100) | 47 (90) | |
| Do not remember | 0 (0) | 0 (0) | 0(0) | |
| Abdominal pain, n (%) | .99c | |||
| No | 9 (20) | 1 (12) | 10 (19) | |
| Yes | 34 (77) | 7 (88) | 41 (79) | |
| Do not remember | 1 (2) | 0 (0) | 1 (2) | |
| Nausea, n (%) | .09a | |||
| No | 15 (34) | 0 (0) | 15 (29) | |
| Yes | 29 (66) | 8 (100) | 37 (71) | |
| Do not remember | 0 (0) | 0 (0) | 0(0) | |
| Vomiting, n (%) | .6a | |||
| No | 33 (75) | 7 (88) | 40 (77) | |
| Yes | 11 (25) | 1 (12) | 12 (23) | |
| Do not remember | 0 (0) | 0 (0) | 0(0) | |
| Fever, n (%) | .05c | |||
| No | 30 (68) | 2 (25) | 32 (62) | |
| Yes | 12 (27) | 6 (75) | 18 (35) | |
| Do not remember | 2 (5) | 0 (0) | 2 (4) | |
| Headache, n (%) | .06a | |||
| No | 23 (52) | 1 (12) | 24 (46) | |
| Yes | 21 (48) | 7 (88) | 28 (54) | |
| Do not remember | 0 (0) | 0 (0) | 0(0) | |
| Muscle pain, n (%) | .01c | |||
| No | 28 (64) | 1 (12) | 29 (56) | |
| Yes | 15 (34) | 7 (88) | 22 (42) | |
| Do not remember | 1 (2) | 0 (0) | 1 (2) | |
| Fatigue, n (%) | .44c | |||
| No | 8 (18) | 0 (0) | 8 (15) | |
| Yes | 35 (80) | 8 (100) | 43 (83) | |
| Do not remember | 1 (2) | 0 (0) | 1 (2) | |
| Incubation period, days | .02b | |||
| Median (range) | 11.5 (1–38) | 9 (5–12) | 11 (1–38) | |
| Duration of illness, days, – n (%) | .01c | |||
| 1–7 | 27 (61) | 1 (13) | 28 (54) | |
| 8–14 | 11 (25) | 3 (38) | 14 (27) | |
| 15–21 | 2 (5) | 0 (0) | 2 (4) | |
| 22 | 4 (9) | 4 (50) | 8 (15) | |
| Hospital admission, n (%) | … | |||
| No | 44 (100) | 8 (100) | 52 (100) | |
| Yes | 0 (0) | 0 (0) | 0(0) | |
| Contact with symptomatic person (last 2 weeks), n (%) | .99c | |||
| No | 40 (91) | 8 (100) | 48 (92) | |
| Yes | 3 (7) | 0 (0) | 3 (6) | |
| Do not know | 1 (2) | 0 (0) | 1 (2) | |
Fisher exact test.
Kruskal-Wallis rank sum test.
Fisher exact test with simulated P-value (based on 1000 replicates).
Outbreak Signal and Clinical Manifestations
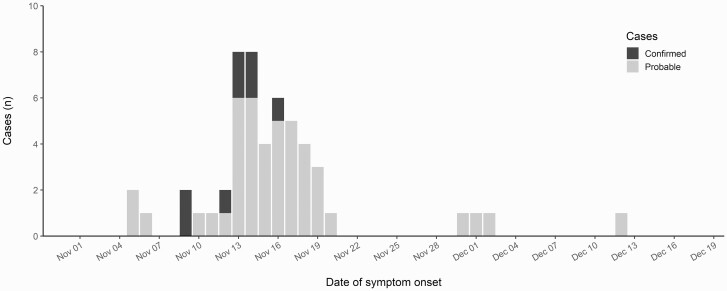
Dates of symptom onset reported by the 52 cases ranged from 5 November to 12 December 2020, with a peak on 14 and 15 November 2020 (Figures 1 and 2). Four probable cases with symptom onset between 30 November and 12 December 2020 could be either secondary cases or ill from other causes.
Figure 1.
Distribution of cases (N = 48) in the Enterocytozoon bieneusi-associated outbreak in Denmark 2020, by sex and 10-year age groups.
Figure 2.
Distribution of cases (N = 52) of the Enterocytozoon bieneusi-associated outbreak in Denmark 2020, by date of symptom onset and case definition.
Diarrhea (90%), fatigue (83%), abdominal pain (78%), and nausea (71%) were the most commonly reported symptoms among all cases. The duration of illness ranged between 14 and 22 days, with 81% (N = 42) of all cases reporting a duration of illness of up to 14 days (Table 2). Furthermore, some cases reported time-varying symptoms, presenting with diarrhea in the first days followed by unspecific influenza-like symptoms, including fatigue, pain, and fever, lasting for 2–3 weeks. No cases were hospitalized, and no treatment was given to any of the cases as symptoms resolved spontaneously (Table 2).
Laboratory Tests
Altogether, 49 stool samples from 24 individuals were tested. Dates of stool sampling ranged from 24 November 2020 to 19 January 2021, with most samples (84%) submitted before 31 December 2020. The samples were negative for all tested pathogens, except for 27 samples from 14 individuals that were positive for E. bieneusi genotype C. Of these, 8 individuals responded to the web-based questionnaire. Six individuals with E. bieneusi-positive samples submitted 2 or more samples (median 4; range 2–7) over separate days. The time from symptom onset until the last positive PCR test, indicative of duration of spore shedding, among the three individuals who submitted multiple stool samples and answered the web-based questionnaire was 25, 31, and 43 days, respectively. Samples from all of them tested negative by 7 January 2021, and the very last stool sample, also negative, was submitted on 19 January 2021.
Analysis of Food and Water Exposures
Individuals who consumed lunch from the company’s canteen on 4 November 2020 had a 2.8 times higher risk of being a case (RR 2.8 [95% CI: 1.4–5.4] P = .001) compared with those who did not. Having had lunch in the canteen on 5 November 2020 was associated with a 2.0 times higher risk of being a case (RR 2.0 [95% CI: 1.1–3.6] P = .02) (Table 3). There were 32 cases and 56 non-cases who ate in the canteen on both days (RR 3.3 [95% CI: 1.5–7.5] P = .0008). The risk of being a case was highest for those who had consumed any of the three offered open sandwich combinations (RR 3.2 [95% CI: 1.4–7.2] P = .002), especially the egg/shrimp and corned beef sandwiches (RR 3.6 [95% CI: 2.1–6.4] P = .00001) served on 4 November (Table 4 and Supplementary Table 1). Overall, 52% (N = 27) of the cases had consumed any of the three open sandwich combinations, whereas 37% (N = 19) of the cases had consumed the egg/shrimp and corned beef sandwiches (Table 4). Mantel–Haenszel and stratified risk estimates by consumption of egg/shrimp and corned beef sandwiches for food items with statistically significant crude risk estimates are shown in Table 5 and Supplementary Table 2. MH-adjusted RR estimates for food and water exposures were statistically nonsignificant and smaller by at least 15% than the respective crude estimates. Effect modification by consumption of egg/shrimp and corned beef sandwiches was not observed for the analyzed food items, namely, the baked pumpkin salad (Woolf P = .40) and the chocolate cake (Woolf P = .58).
Table 3.
Univariate Analysis of Data On Exposures, by Date of Lunch at the Canteen of the Company Affected by the Enterocytozoon bieneusi outbreak in Denmark, 2020
| Exposure | Exposed | Unexposed | 95% CI | % Cases | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (Lunch) | Total | Cases | AR (%) | Total | Cases | AR (%) | RR | Low | Upper | P-value | Exposed |
| Between 2 and 15 November | 160 | 46 | 29 | 32 | 4 | 13 | 2.3 | .9 | 5.9 | .08 | 88 |
| 2 November | 121 | 38 | 31 | 58 | 11 | 19 | 1.7 | .9 | 3.0 | .11 | 73 |
| 3 November | 117 | 34 | 29 | 61 | 13 | 21 | 1.4 | .8 | 2.4 | .29 | 65 |
| 4 November | 104 | 37 | 36 | 70 | 9 | 13 | 2.8 | 1.4 | 5.4 | .00 | 71 |
| 5 November | 106 | 35 | 33 | 72 | 12 | 17 | 2.0 | 1.1 | 3.6 | .02 | 67 |
| 6 November | 79 | 23 | 29 | 97 | 23 | 24 | 1.2 | .7 | 2.0 | .49 | 44 |
| 9 November | 109 | 34 | 31 | 67 | 13 | 19 | 1.6 | .9 | 2.8 | .11 | 65 |
| 10 November | 110 | 33 | 30 | 66 | 15 | 23 | 1.3 | .8 | 2.2 | .38 | 63 |
| 11 November | 98 | 29 | 30 | 76 | 18 | 24 | 1.2 | .8 | 2.1 | .40 | 56 |
| 12 November | 105 | 29 | 28 | 67 | 17 | 25 | 1.1 | .7 | 1.8 | .86 | 56 |
| 13 November | 81 | 27 | 33 | 89 | 19 | 21 | 1.6 | .9 | 2.6 | .09 | 52 |
Abbreviations: AR, attack rate; CI, confidence interval; RR, relative risk.
Table 4.
Univariate Analysis and Risk Estimates by Specific Food Items Exposures, by Date of Lunch at the Canteen of the Company Affected by the Enterocytozoon bieneusi Outbreak in Denmark, 2020
| Exposure | Exposed | Unexposed | 95% CI | % Cases | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (Lunch menu) | Total | Cases | AR (%) | Total | Cases | AR (%) | RR | Low | Upper | P-value | Exposed |
| November 4 | |||||||||||
| Fish and rolled sausage sandwiches | 28 | 10 | 36 | 97 | 23 | 24 | 1.5 | .8 | 2.8 | .23 | 19 |
| Egg/shrimp and corned beef sandwiches | 32 | 19 | 59 | 86 | 14 | 16 | 3.6 | 2.1 | 6.4 | .00 | 37 |
| Fish and roast beef sandwiches | 34 | 9 | 27 | 84 | 21 | 25 | 1.1 | .5 | 2.1 | 1.00 | 17 |
| Potato and roast beef sandwiches | 14 | 4 | 29 | 106 | 27 | 26 | 1.1 | .5 | 2.7 | .75 | 8 |
| Avocado/tomato salad | 3 | 1 | 33 | 126 | 34 | 27 | 1.2 | .2 | 6.3 | 1.00 | 2 |
| Iceberg salad | 10 | 4 | 40 | 112 | 31 | 28 | 1.4 | .6 | 3.3 | .47 | 8 |
| Noodle salad | 26 | 7 | 27 | 97 | 28 | 29 | 0.9 | .5 | 1.9 | 1.00 | 13 |
| Carrot and raisin salad | 28 | 10 | 36 | 88 | 22 | 25 | 1.4 | .8 | 2.6 | .33 | 19 |
| Any (2) fish sandwiches | 59 | 18 | 31 | 66 | 15 | 23 | 1.3 | .7 | 2.4 | .42 | 35 |
| Any (2) fish or egg/shrimp sandwiches | 72 | 27 | 38 | 51 | 6 | 12 | 3.2 | 1.4 | 7.2 | .00 | 52 |
| November 5 | |||||||||||
| Cod roe sandwich | 13 | 3 | 23 | 106 | 29 | 27 | 0.8 | .3 | 2.4 | 1.00 | 6 |
| Ham and cheese sandwich | 43 | 16 | 37 | 77 | 15 | 20 | 1.9 | 1.1 | 3.5 | .05 | 31 |
| Smoked turkey sandwich | 19 | 5 | 26 | 94 | 24 | 26 | 1.0 | 5 | 2.4 | 1.00 | 10 |
| Pastrami sandwich | 15 | 6 | 40 | 103 | 23 | 22 | 1.8 | .9 | 3.7 | .20 | 12 |
| Broccoli salad | 24 | 7 | 29 | 87 | 21 | 24 | 1.2 | .6 | 2.5 | .60 | 13 |
| Baked vegetables salad | 9 | 3 | 33 | 107 | 25 | 23 | 1.4 | .5 | 3.8 | .45 | 6 |
| Pumpkin hummus | 8 | 4 | 50 | 108 | 26 | 24 | 2.1 | 1.0 | 4.5 | .20 | 8 |
| Green salad | 10 | 3 | 30 | 100 | 25 | 25 | 1.2 | .4 | 3.3 | .71 | 6 |
| November 6 | |||||||||||
| Pâté tapas | 56 | 16 | 29 | 46 | 8 | 17 | 1.6 | .8 | 3.5 | .24 | 44 |
| Pasta salad | 14 | 4 | 29 | 81 | 17 | 21 | 1.4 | .5 | 3.5 | .50 | 8 |
| Baked vegetables salad | 8 | 4 | 50 | 87 | 17 | 20 | 2.6 | 1.1 | 5.8 | .07 | 8 |
| Pineapple salad | 20 | 7 | 35 | 75 | 15 | 20 | 1.8 | .8 | 3.7 | .23 | 13 |
| Chocolate cake | 43 | 16 | 37 | 59 | 9 | 15 | 2.4 | 1.2 | 5.0 | .02 | 31 |
| Tap water (during the 2 weeks) | 138 | 44 | 32 | 43 | 5 | 12 | 2.7 | 1.2 | 6.5 | .01 | 85 |
| Contact with symptomatic person (last 2 weeks) | 10 | 3 | 30 | 182 | 48 | 26 | 1.1 | .43 | 3.02 | .73 | 6 |
Abbreviations: AR, attack rate; CI, confidence interval; RR, relative risk.
Table 5.
Mantel-Haenszel and Stratified Analysis, and Risk Estimates for Specific Food and Water Exposures, by Consumption of Egg/Shrimp and Corned Beef Sandwiches for Lunch at the Canteen of the Company Affected by the Enterocytozoon bieneusi Outbreak, Denmark, 2020
| Did Not Eat Egg/Shrimp and Corned Beef Sandwich | Ate Egg/Shrimp and Corned Beef Sandwich | P-value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exposure | RRc | 95% CI | RRMH | 95% CI | RR0 | 95% CI | RR1 | 95% CI | |||||
| Baked pumpkin salad | 2.3 | 1.2 | 4.4 | 1.2 | .8 | 1.9 | 1.0 | .7 | 1.5 | 2.2 | .4 | 13.0 | .40 |
| Chocolate cake | 2.4 | 1.2 | 5.0 | 1.3 | 1.0 | 1.8 | 1.2 | .9 | 1.7 | 1.7 | .6 | 5.3 | .58 |
| Baked vegetables salad | 2.6 | 1.1 | 5.8 | 1.6 | .8 | 3.2 | 1.3 | .7 | 2.3 | Inf | … | … | … |
| Tap water (during the 2 weeks) | 2.7 | 1.2 | 6.5 | 1.2 | .9 | 1.6 | 1.0 | .8 | 1.4 | 2.2 | 1.1 | 4.4 | .04 |
Woolf test of homogeneity was used to assess statistically significant associations on effect modification.
Abbreviations: CI, confidence interval; RRc, relative risk (crude); RRMH, relative risk (Mantel-Haenszel adjusted); RR0, relative risk for stratum 0 (not exposed); RR1, relative risk for stratum 1 (exposed).
Consumption of tap water between 2 and 15 November 2020 (Table 4) presented a positive crude association to being a case (RR 2.7 [95% CI: 1.2–6.5] P = .01). However, those who had consumed tap water but had not eaten the egg/shrimp and corned beef sandwiches did not present a higher risk of being a case (RR 1.0 [95% CI: .8–1.4]). On the contrary, those who consumed both tap water and these sandwiches had a 2.2 times higher risk of being a case (RR 2.2 [95% CI: 1.1–4.4] Woolf P = .04). Additionally, the association between frequency of tap water consumption and being a case was not statistically significant (Cochran-Armitage trend test, Z = –1.72, P = .09).
Environmental Investigation
DVFA did not identify structural deficiencies in terms of food storage, food handling, or cooked-food security at the canteen. There were no cases among food handlers working at the canteen. No food samples were tested for E. bieneusi, as the DVFA did not have the chance to collect any leftover food items due to closure of the canteen. Trace-back investigation from food suppliers did not reveal contamination by microsporidia.
DISCUSSION
We describe a foodborne outbreak of E. bieneusi genotype C, the first reported in Denmark and to our knowledge the second reported in the literature. Our epidemiological investigation revealed a point-source outbreak, and stool samples from 14 individuals testing positive for E. bieneusi genotype C and negative for other pathogens strongly indicated that E. bieneusi was the likely cause of the outbreak. Hence, the evidence pointed toward a single common source of infection. The only other foodborne outbreak with E. bieneusi genotype C documented to date was likely caused by contaminated cucumbers [8]. Our results add to the knowledge on characteristic features of E. bieneusi infections with regard to symptoms, incubation period, and spore shedding duration. The main symptom of E. bieneusi genotype C infection in this outbreak was diarrhea accompanied by other gastrointestinal symptoms. Some cases reported that symptoms changed over time, beginning with diarrhea in the first days and then changing to influenza-like illness, which lasted for 2–3 weeks.
The incubation period for microsporidiosis in immunocompetent individuals is currently not well understood. The data from the E. bieneusi genotype C outbreak in Sweden suggested an incubation period of up to 21 days and a median incubation period of 9 days [8], which is in line with our data, and which is longer than the incubation period for foodborne bacteria and viruses causing gastrointestinal disease. Noteworthy in this outbreak was the high attack rate (AR) in apparently healthy individuals, which was similar to the Swedish study [10]. This is remarkable, considering that microsporidiosis has been often described in immunocompromised individuals [11, 12, 14, 23].
Univariate analysis revealed that the egg/shrimp and corned beef sandwiches served on 4 November were the most suspected food items for this outbreak based on the point-source epidemic curve, timing of the initial reported cases and incubation period. Consumption of chocolate cake on 6 November and consumption of tap water during the 2 weeks were also initially considered. However, the chocolate cake from November 6 was unlikely to be the source of infection, as probable cases of E. bieneusi had already been recorded by this date. Tap water was an unlikely source as stratified and frequency-response associations were not statistically significant. Because the lunchboxes contained shared ingredients and more than 1 type of sandwich, it was not possible to identify the exact food item causing this outbreak.
One limitation of our investigation was that there were no food items available that could have been tested for the presence of E. bieneusi. Furthermore, not all cases provided stool samples for laboratory analysis, and therefore most cases could only be categorized as probable. Another limitation was that the symptomology of all cases was broad, which could lead to a misclassification of some of the cases and potentially widen the range of incubation period and symptoms. Because our results were based on a questionnaire that asked individuals from the company to report on symptoms, food consumption, and dates, there is the possibility of a recall bias since cases may remember what they ate differently from others who did not fall ill.
In summary, we report here a foodborne outbreak of E. bieneusi genotype C where the organism was confirmed in several stool samples from cases, providing new information on symptoms and incubation period. Because testing for microsporidia is rarely performed worldwide, microsporidiosis may be underreported, and outbreaks might go undetected. We recommend considering including E. bieneusi in the laboratory test panels used for investigating potential causes of foodborne outbreaks. Testing for E. bieneusi can be useful even weeks after exposure due to long PCR-detectable spore shedding.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors would like to thank the laboratory technicians for their technical assistance with sample screening. They would also like to thank the affected company for their commitment and assistance for this outbreak investigation and to the affected individuals for providing stool samples.
Authors’ contributions.
D. A.: Conceptualization, data collection, formal analysis, writing—original draft preparation, writing—review and editing.
L. A. S.: Conceptualization, data collection, formal analysis, writing—original draft preparation, writing—review and editing.
L. M.: Conceptualization, data collection, writing—review and editing.
P. J.: Formal analysis, writing—review and editing.
S. E.: Writing—review and editing.
L. S. V.: Data collection, writing—review and editing.
S. S.: Data collection, formal analysis, writing—review and editing.
S. M.: Data collection, writing—review and editing.
C. W. J.: Data collection, writing—review and editing.
L. D. R.: Writing—review and editing.
C. R. S.: Data collection, formal analysis, writing—review and editing.
Financial support. This work was supported by the European Programme for Intervention Epidemiology Training (EPIET), European Centre for Disease Prevention and Control, (ECDC), Stockholm, Sweden, the European Programme for Public Health Microbiology (EUPHEM), European Centre for Disease Prevention and Control, (ECDC); and the European Union’s Horizon 2020 Research and Innovation Programme (grant number 773830: One Health European Joint Programme, PARADISE-project).
Contributor Information
Daniela Michlmayr, Department of Bacteria, Parasites and Fungi, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark; European Public Health Microbiology Training Programme (EUPHEM), European Centre for Disease Prevention and Control (ECDC), Solna, Sweden.
Luís Alves de Sousa, Department of Infectious Disease Epidemiology and Prevention, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark; European Programme for Intervention Epidemiology Training (EPIET), European Centre for Disease Prevention and Control (ECDC), Solna, Sweden.
Luise Müller, Department of Infectious Disease Epidemiology and Prevention, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
Pikka Jokelainen, Department of Bacteria, Parasites and Fungi, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
Steen Ethelberg, Department of Infectious Disease Epidemiology and Prevention, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark; Department of Public Health, Global Health Section, University of Copenhagen, Copenhagen, Denmark.
Lasse Skafte Vestergaard, Department of Bacteria, Parasites and Fungi, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark; Department of Infectious Disease Epidemiology and Prevention, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
Susanne Schjørring, Department of Bacteria, Parasites and Fungi, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
Sarah Mikkelsen, Danish Veterinary Food Administration (DVFA), Copenhagen, Denmarkand.
Carl Widstrup Jensen, Danish Veterinary Food Administration (DVFA), Copenhagen, Denmarkand.
Lasse Dam Rasmussen, Department of Virus & Microbiological Special Diagnostics, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
Christen Rune Stensvold, Department of Bacteria, Parasites and Fungi, Infectious Disease Preparedness, Statens Serum Institut, Copenhagen, Denmark.
References
- 1. Matos O, Lobo ML, Xiao L.. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res 2012; 2012:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stentiford GD, Becnel JJ, Weiss LM, et al. Microsporidia: emergent pathogens in the global food chain. Trends Parasitol 2016; 32:657. [DOI] [PubMed] [Google Scholar]
- 3. Keeling P. Five questions about microsporidia. PLoS Pathog 2009; 5:e1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corradi N, Keeling PJ.. Microsporidia: a journey through radical taxonomical revisions. Fung Biol Rev 2009; 23(1):1–8. [Google Scholar]
- 5. Vávra J, Lukeš J.. Microsporidia and “the art of living together.” Adv Parasitol 2013; 82:253–319. [DOI] [PubMed] [Google Scholar]
- 6. Lobo ML, Augusto J, Antunes F, et al. Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi and other intestinal parasites in young children in Lobata province, Democratic Republic of São Tomé and Príncipe. PLoS One 2014; 9:e97708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J, Shi K, Sun F, et al. Identification of human pathogenic Enterocytozoon bieneusi, Cyclospora cayetanensis, and Cryptosporidium parvum on the surfaces of vegetables and fruits in Henan, China. Int J Food Microbiol 2019; 307:108292. [DOI] [PubMed] [Google Scholar]
- 8. Decraene V, Lebbad M, Botero-Kleiven S, Gustavsson AM, Lofdahl M.. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol Infect 2012; 140:519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathis A, Weber R, Deplazes P.. Zoonotic potential of the microsporidia. Clin Microbiol Rev 2005; 18:423–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop 2005; 94:61–76. [DOI] [PubMed] [Google Scholar]
- 11. Lores B, López-Miragaya I, Arias C, Fenoy S, Torres J, del Aguila C.. Intestinal microsporidiosis due to Enterocytozoon bieneusi in elderly human immunodeficiency virus-negative patients from Vigo, Spain. Clin Infect Dis 2002; 34:918–21. [DOI] [PubMed] [Google Scholar]
- 12. Wichro E, Hoelzl D, Krause R, Bertha G, Reinthaler F, Wenisch C.. Microsporidiosis in travel-associated chronic diarrhea in immune-competent patients. Am J Trop Med Hyg 2005; 73:285–7. [PubMed] [Google Scholar]
- 13. Cotte L, Rabodonirina M, Chapuis F, et al. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J Infect Dis 1999; 180:2003–8. [DOI] [PubMed] [Google Scholar]
- 14. Didier ES, Stovall ME, Green LC, Brindley PJ, Sestak K, Didier PJ.. Epidemiology of microsporidiosis: sources and modes of transmission. Vet Parasitol 2004; 126:145–66. [DOI] [PubMed] [Google Scholar]
- 15. Li W, Feng Y, Santin M.. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol 2019; 35:436–51. [DOI] [PubMed] [Google Scholar]
- 16. Jedrzejewski S, Graczyk TK, Slodkowicz-Kowalska A, Tamang L, Majewska AC.. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl Environ Microbiol 2007; 73:4071–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee JH. Molecular detection of Enterocytozoon bieneusi and identification of a potentially human-pathogenic genotype in milk. Appl Environ Microbiol 2008; 74:1664–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desoubeaux G, Nourrisson C, Moniot M, et al. Genotyping approach for potential common source of Enterocytozoon bieneusi infection in hematology unit. Emerg Infect Dis 2019; 25:1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 20. Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. JOpen Source Softw 2019; 4:1686. [Google Scholar]
- 21. Verweij JJ, Ten Hove R, Brienen EA, van Lieshout L.. Multiplex detection of Enterocytozoon bieneusi and Encephalitozoon spp. in fecal samples using real-time PCR. Diagn Microbiol Infect Dis 2007; 57:163–7. [DOI] [PubMed] [Google Scholar]
- 22. Buckholt MA, Lee JH, Tzipori S.. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol 2002; 68:2595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fournier S, Liguory O, Garrait V, et al. Microsporidiosis due to Enterocytozoon bieneusi infection as a possible cause of traveller’s diarrhea. Eur J Clin Microbiol Infect Dis 1998; 17:743–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.