Abstract
Programmable nucleases are the most important tool for manipulating the genes and genomes of both prokaryotes and eukaryotes. Since the end of the 20th century, many approaches were developed for specific modification of the genome. The review briefly considers the advantages and disadvantages of the main genetic editors known to date. The main attention is paid to programmable nucleases from the family of prokaryotic Argonaute proteins. Argonaute proteins can recognize and cleave DNA sequences using small complementary guide molecules and play an important role in protecting prokaryotic cells from invading DNA. Argonaute proteins have already found applications in biotechnology for targeted cleavage and detection of nucleic acids and can potentially be used for genome editing.
Keywords: programmable nucleases, Argonaute proteins, guide DNA, biosensors, genome editing
MAIN GROUPS OF PROGRAMMABLE NUCLEASES
Targeted changes in the DNA sequence at the level of specific genes and regulatory regions of the genome are highly important for fundamental studies of the mechanisms of genome functioning and the role of individual proteins, as well as for solving applied problems, from the obtaining of producer strains to the treatment of genetic diseases. The most important tool in these studies are programmable nucleases, enzymes that can be directed to a specific region in the genome to make changes in the DNA sequence. Catalytically inactive variants of programmable nucleases can be used to edit epigenetic marks and regulate gene expression by attracting various functional domains to certain regions in the genome and to visualize the target regions in cells. Programmable nucleases are also used to identify target sequences in biological samples and to cleave nucleic acids in vitro.
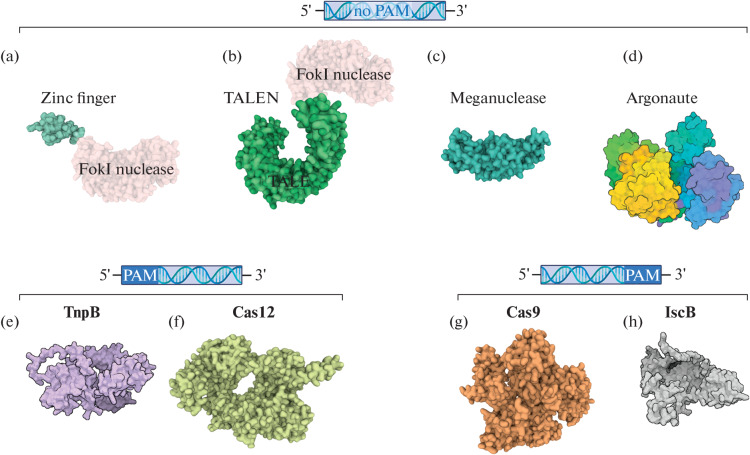
To date, several groups of programmable nucleases of natural and synthetic origin are known, differing in the mechanism of DNA recognition and cleavage and the potential for practical application. In all cases, the introduction of breaks in the target DNA region can then lead to the loss of the functionality of this gene, or to the required change in its sequence during repair. Two large groups of such enzymes can be distinguished: nucleases that recognize the cleavage site using DNA-protein interactions, and nucleases that recognize DNA through an associated short complementary RNA or DNA. Although historically the enzymes of the first group were found and used earlier, the enzymes of the second group have much greater flexibility in recognizing target DNA regions and have been used as the main tools for genome editing during last years. A comparison of different classes of programmable nucleases is shown in Fig. 1.
Fig. 1.
Variety of programmable nucleases. (a–d) Endonucleases that do not require a PAM sequence in the target for catalytic activity; (e, f) endonucleases that require the presence of PAM (protospacer adjacent motif) in the 5'-region from the recognition site; (g, h) endonucleases that require the presence of PAM in the 3'-region from the recognition site. (a) Chimeric endonuclease based on the zinc finger DNA-binding domain (PDB ID: 7ZNF) and FokI (PDB ID: 2FOK) (protein structures are combined for illustration); (b) chimeric endonuclease based on the TALE DNA-binding domain (PDB ID: 4HPZ) and FokI (PDB ID: 2FOK) (protein structures are combined for illustration); (c) meganuclease I-AabMI (PDB ID: 4YIT); (d) Argonaute protein can be an RNA- or DNA-guided endonuclease and cleave RNA or DNA (TtAgo, PDB ID: 4NCB); (e) RNA-dependent DNA-endonuclease TnpB (TnpB ISDra2 structure was predicted using the AlphaFold2 algorithm [1]); (f) RNA-dependent DNA-endonuclease Cas12 (PDB ID: 7EU9) [2]; (g) RNA-dependent DNA-endonuclease Cas9 (PDB ID: 6M0X) [3]; and (h) RNA-dependent DNA-endonuclease IscB (the structure was predicted using the AlphaFold2 algorithm [1]).
Meganucleases
Meganucleases recognize long (from 12 to 44 bp) DNA regions [4, 5]. Naturally occurring meganucleases belong to the group of homing endonucleases that promote mobility of transposable elements. Meganucleases cleave DNA strands in the area of four central nucleotides in the recognition site with the formation of a double-strand break with 3'-terminal 4-nucleotide overhang [6, 7]. The small size of these proteins with a long DNA recognition site has drawn attention to it as tools for genome manipulation. The limitation is that the recognition site must be artificially introduced into the genome. By the early 2000s, several hundred endonucleases with different recognition sites had been discovered and described [5], and enzymes with altered specificity were created [8–10].
Chimeric Nucleases
The idea of creating chimeric nucleases by combining the nuclease domain of one protein and the DNA-recognition domain of another protein in one molecule was proposed in 1994 by Kim & Chandrasegaran [11]. The researchers combined the nuclease domain of the Flavobacterium okeanokoites FokI restriction endonuclease, which does not have specificity for a certain DNA sequence, and the homeodomain of the Drosophila melanogaster Ultrabithorax regulatory protein, which recognizes a 9 nucleotide long DNA sequence. The authors also proposed variants of DNA-recognition domains from other proteins, which were indeed later used to create new chimeric nucleases.
SGN
SGN (structure-guided endonuclease) is a structure-specific endonuclease. Nucleases can be targeted to a specific location in the genome by recognizing specific structures in DNA. An example is the nuclease consisting of the flap endonuclease (flap endonuclease-1) FEN1 of the archaea Archaeoglobus fulgidus and the FokI nuclease domain [12, 13]. To introduce a break at the desired location in the DNA, DNA guides are used that are complementary to the desired site, with the exception of one 3'-terminal nucleotide. This nucleotide is recognized by FEN1 within the SGN, then FokI introduces a DNA break at a distance of 9‒10 nucleotides from the guide 3' end. However, SGN can also recognize 3'-protruding single-stranded fragments that arise during repair and replication. This leads to significant non-target rearrangements of the genome [12, 13]. Along with low efficiency, this has become a significant limitation for the development of SGN technology.
Zinc Fingers
The zinc finger domain (ZF) was first described by J. Miller et al. in 1985 [14] while studying the transcription factor TFIIIA from frog oocytes. TFIIIA consists of 9 repeats each containing 30 amino acid residues (a.a.), including two conserved histidines and two cysteines; polar and basic amino acids are concentrated at the top of each “finger” responsible for interactions with nucleic acids [14, 15]. One zinc finger is responsible for recognition of a 3-nucleotide sequence [16, 17]. The first site-specific nuclease containing zinc fingers as a DNA recognition domain and FokI nuclease domain (ZFN, zinc finger nuclease) was obtained and described by Y. Kim et al. in 1996 [18]. The recognition of individual nucleotides by individual amino acids within the zinc finger and the ability of the ZF to work as independent modules in recognizing extended DNA sequences make it possible to use these domains to program artificial protein tools designed to work with nucleic acids. Studies of chimeric ZFN molecules in Drosophila and mammalian cells revealed their high cytotoxicity due to nonspecific DNA recognition and cutting (off-target effect) [16]. An obvious way to increase the target recognition specificity with ZFN is to increase the number of zinc fingers. However, Y. Shimizu et al. [19] found that the activity of nucleases with five or six zinc fingers is much lower compared to proteins with one such domain. Thus, the creation of functional nucleases with multiple zinc fingers is a difficult task.
TALEN
TALEN (transcription activator-like effectors nuclease) is an effector nuclease similar to transcription activators. TALE is a large family of transcriptional activators that belong to the virulence factors of phytopathogenic bacteria of the genus Xanthomonas. In 1989, U. Bonas et al. [20] showed that these proteins contain repeating elements. Repeats have the same sequence, except for amino acids at positions 12 and 13 (repeat variable di-residues, RVDs). This pair of amino acids is responsible for the specific recognition of a single nucleotide in DNA [21–23]. The modular structure and specificity to individual nucleotides of TALE DNA-recognition domains opens up the possibility of their application as a component of chimeric nucleases. The first TALE-based chimeric nucleases (TALEN–TALE nuclease) were constructed on the basis of a repeat-containing fragment of the TALE protein and the FokI nuclease domain [24].
Cas Nucleases
In recent years, Cas nucleases, which are part of the CRISPR–Cas systems of prokaryotic adaptive immunity (clustered regularly interspaced short palindromic repeats), have been most widely used in genomic editing in recent years. CRISPR–Cas systems have a modular organization. The two main components are the module responsible for adaptation (inserting new spacers into the CRISPR cassette) and the effector module responsible for interference. The modules are represented by complexes of Cas proteins, the genes of which are located in the same operon. Based on the structure of the effector module, two classes of CRISPR–Cas systems are distinguished. In Class 1 systems, the effector module is represented by a multisubunit complex, which consists of several Cas proteins. In Class 2 systems, effector module function is performed by one large protein with several domains. Class 1 includes CRISPR–Cas systems of types I, III, IV, and Class 2 includes II, V and VI [25]. Class 2 effector modules are the most interesting for practical application, since they are represented by a single protein molecule with nuclease activity.
Cas9 nuclease from the bacterium Streptococcus pyogenes (SpCas9), a representative of type II CRISPR–Cas systems, has been most thoroughly studied. Two RNAs are required for Cas9 activity: crRNA (CRISPR RNA) containing the spacer sequence and tracrRNA (trans-activating crRNA). Cas9 loaded with crRNA-tracrRNA introduces a double-strand break in the target DNA sequence corresponding to the spacer. For practical applications of Cas9, crRNA and tracrRNA can be combined into one RNA molecule, single guide RNA (sgRNA). Cas9 is a large DNA endonuclease (SpCas9 consists of 1368 a.a.), introducing a double-strand break with the formation of a blunt end within the region recognized by sgRNA, at a distance of 3 nucleotides from its right end. DNA cleavage is carried out by two nuclease domains. The HNH-like domain cuts the DNA strand complementary to the guide RNA (target strand). The RuvC-like domain has an RNase H-like fold and cuts the opposite strand (non-target strand) [26]. In early 2013, three studies were published that showed the possibility of using Cas9 as a programmable DNase to introduce changes in the genome in human cell cultures [27–29]. This marked the beginning of the Cas9 nuclease use in science and biotechnology.
sgRNA allows directing Cas9 to any DNA sequence containing a PAM motif (protospacer adjacent motif) to the right of the target sequence. PAM recognized by SpCas9 is a trinucleotide 5'-NGG-3', where N is any of the four DNA nucleotides [30], the length and sequence of this motif may vary for different proteins of the Cas family. To expand the range of sequences available for Cas9 recognition, variants of the enzyme with altered PAM specificity were constructed [31, 32], and Cas9 proteins from other bacteria were also used [33–40].
The second problem with the use of Cas9 is the off-target cleavage of partially complementary DNA. Errors in DNA recognition may result from inaccurate pairing of guide and target outside of the primary binding region (seed) [40, 41]. There are two fundamental approaches to reducing off-target effects: increasing the specificity of Cas9 [42] and limiting the time of its action on DNA [40, 43–47].
Other representatives of class II CRISPR–Cas systems have also found practical application. In 2015, the effector nuclease Cas12a (also known as Cpf1) from the type V system of bacteria Acidaminococcus and Lachnospiraceae bacterium was described [48]. Cas12a cleaves both strands of double-stranded DNA with the help of one RuvC nuclease domain, which leads to the formation of a double-strand break with 4‒5-nucleotide 5' sticky ends [48, 49]. As in the case of Cas9, the presence of a PAM motif in Cas12a is necessary for recognition of the target, in this case, to the left of the recognized sequence. After cutting the target DNA and releasing the products, Cas12a can perform non-specific exonuclease cleavage of single-stranded DNA. This phenomenon has been called “collateral activity.” In the cell, such cleavage can disrupt various steps of DNA metabolism. Based on the collateral activity of Cas12a, systems for detecting target DNA sequences in biological samples HOLMES (an one-HOur Low-cost Multipurpose highly Efficient System) [50] and DETECTR (DNA endonuclease-targeted CRISPR trans porter) [51] have been developed.
The type VI effector nuclease of the CRISPR–Cas system, Cas13 (also known as C2c2), has two HEPN (higher eukaryotes and prokaryotes nucleotide binding) RNase domains. This type of nuclease was first isolated in 2016 from the bacterium Leptotrichia shahii [52]. Cas13 recognizes and cleaves single-stranded RNA targets using crRNA, also showing nonspecific collateral activity. Cas13 can be directed to target transcripts in bacterial, plant, and mammalian cells [53]. The efficiency of target RNA cleavage is comparable to the activity of RNA interference using short RNAs (see below). However, Cas13 showed significantly fewer off-target effects. Based on the collateral activity of Cas13, a highly sensitive method for the nucleic acids detection SHERLOCK (Specific High-Sensitivity Enzymatic Reporter Unlocking) was developed [54, 55]. Currently, detection technologies using Cas nucleases continue to be developed and improved. Thus, in 2020-2021, many systems were developed on this platform for the detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of COVID-19 [56].
At the end of 2021, two groups of programmable prokaryotic transposon nucleases were described. Transposons of the IS200/IS605 group encode the IcsB proteins, the ancestral forms of Cas9, and TnpB, from which the Cas12 nucleases originated. They can be guided to the target DNA sequence by guide RNA: ωRNA for IscB and reRNA (right end RNA) for TnpB. As in the case of Cas nucleases, the interaction of these transposon nucleases with target DNA requires the presence of a specific short sequence TAM (target-adjacent motif, or transposon-associated motif) to the right or to the left of the recognized sequence for IcsB and TnpB, respectively. The possibility of introducing targeted changes into the cell genome using these nucleases was shown [57, 58].
ARGONAUTE PROTEINS—ANOTHER GROUP OF PROGRAMMABLE NUCLEASES
Eukaryotic Argonautes—Key RNA Interference Proteins
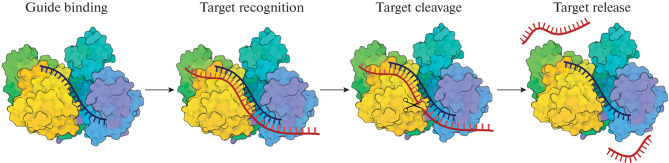
Argonaute (Ago) proteins, like Cas proteins, recognize and cut the target when loaded with a complementary guide nucleic acid (Fig. 2). Unlike Cas nucleases, no additional sequences in the target (such as a PAM motif) are required for this. Argonaute proteins are found in all domains of life: in many bacteria, archaea, and in almost all eukaryotes, suggesting their ancient origin and functional importance [59–61]. In eukaryotes, Argonaute proteins are involved in the regulation of gene expression by small RNAs, suppression of transposon activity, and protection of cells from viral infections [62–68]. They bind short interfering RNAs (siRNAs), microRNAs (miRNAs), or RNAs associated with the PIWI proteins (piRNAs) and coordinate subsequent gene silencing events usually by interacting with other protein factors [68–71]. The resulting effector complexes recognize complementary target RNAs and can cleave it due to the endonuclease activity of Argonautes. Also, Argonaute-mediated cleavage-independent RNA destabilization and repression of transcription and translation through interactions with other proteins have been described [72, 73].
Fig. 2.
Cleavage of nucleic acids by Argonaute proteins. Argonaute is loaded with a guide single-stranded nucleic acid molecule (guide binding), followed by a target search based on complementary interactions (target recognition). If the target is complementary to the guide, the conformation of the enzyme changes, catalysis occurs (target cleavage) and the cleaved target is released (target release).
There are three groups of eukaryotic Argonaute proteins [74–76]. The first includes AGO-like proteins that are involved in cytoplasmic post-transcriptional gene silencing. The second group includes PIWI-like proteins, which are found predominantly in animal germ cells; they suppress the expression of transposable genetic elements and thus contribute to maintaining the integrity of the genome [69, 77–79]. The third group, WAGO, is represented exclusively by worm proteins and was studied in the Caenorhabditis elegans model. Different types of organisms have different numbers of Argonaute proteins of different groups. For example, four proteins belonging to the AGO group and four to the PIWI group are known in humans [68, 80]. Different RNA interference pathways can operate in the same organism and even in the same cell [81].
RNA interference involving proteins of the AGO group occurs via two alternative pathways, at the level of RNA or chromatin [80, 82–85]. An RNA–protein complex, RISC (RNA induced silencing complex) is responsible for the suppression of gene expression. siRNAs are double-stranded RNAs 20–30 nucleotides long and are formed in the cytoplasm as a result of the processing of exogenous and endogenous double-stranded RNAs by the Dicer enzyme. miRNAs are formed in the nucleus by the Drosha endonuclease from RNA precursors containing a hairpin structure [70, 77, 86–90]. After loading of the Argonaute protein with short double-stranded RNAs, the dissociation of one strand of the RNA duplex occurs, after which such complex with single stranded guide RNA can recognize a fully or partially complementary target mRNA. This results in translation inhibition, endonuclease cleavage of the target by Argonaute, or exonuclease degradation of mRNA by other nucleases [59, 91–93]. RNA interference can also occur at the chromatin level via histone modification or DNA methylation, which also makes it possible to suppress gene expression or, in rare cases, leads to transcription activation [80, 94, 95]. PIWI proteins also translocate into the nucleus upon piRNA loading and suppress transposon expression via transcriptional gene silencing [69, 84, 96, 97]. In general, RNA interference pathways are involved in a wide range of cellular functions, including growth, development, apoptosis, as well as in pathophysiological processes such as carcinogenesis [98].
All eukaryotic Argonautes have four domains: N‑terminal, PAZ (Piwi-Argonaute-Zwille), MID (middle) and PIWI (P-element Induced Wimpy Testis), forming a channel in which nucleic acids are located [83, 99–101].
The PIWI domain of Argonaute proteins contains a conserved tetrad of amino acid residues DEDX (where X is an aspartic acid, histidine, or lysine residue), which coordinates two divalent cations required for catalysis [59]. Many Argonaute proteins contain substitutions of negatively charged amino acid residues of the catalytic tetrad and are not capable of target cleavage [59–61].
The MID domain contains a pocket for the 5' end guide binding. In some cases, the 5'-terminal nucleotide of the guide binds specifically. Thus, human hAgo2, KpAgo from the yeast Kluyveromyces polysporus, and SIWI from the silkworm Bombyx mori bind guides containing an uridine residue at the 5' end [99, 102, 103]. The corresponding unpaired nucleotide of the target strand can be specifically recognized in a separate pocket of the PIWI domain [104].
PAZ is a small domain (~140 a.a.) that is involved in the binding of the 3' end of the guide molecule, but is not absolutely necessary for this. Interestingly, PAZ is present not only in Argonautes, but also in the Dicer protein. Probably, the PAZ domain of the ancestor of modern eukaryotic Argonautes underwent a duplication and appeared in Dicer [105].
The N-terminal domain is the least conserved and, possibly, promotes separation of the RNA duplex [99, 106–108].
Prokaryotic Argonautes: Diversity of Structures
The existence of Argonaute genes in bacteria and archaea has been known for a long time, and AfAgo from the archaea Archaeoglobus fulgidus [109–112] and AaAgo from the bacterium Aquifex aeolicus [113–115] were even used as models for structural studies of RNA interference. The first description of the diversity of prokaryotic Argonautes was made in 2009 [59]. K. Makarova et al. found 85 homologues of eukaryotic Argonautes encoded in 80 prokaryotic genomes in the non-redundant protein database RefSeq. The rapid growth in the number of sequenced genomes of bacteria and archaea in recent years has made it possible to find many more Argonautes: 487 in 2014 [61], 1010 in 2018 [60], and 1711 in 2020 [116]. In general, about 10–20% of bacterial genomes and about a quarter of archaeal genomes encode at least one Argonaute, and in rare cases, several Argonaute genes at once.
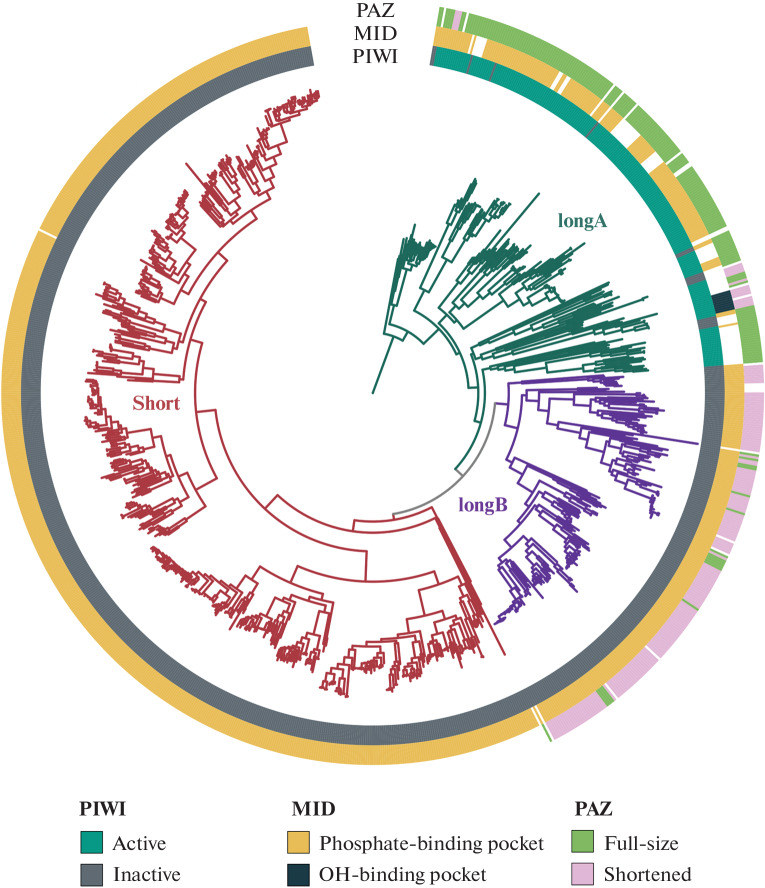
Two large groups can be distinguished on phylogenetic tree of Argonaute proteins (Fig. 3): long Argonautes, containing the same 4 domains as eukaryotic proteins, and short, in which the N-terminal and PAZ domains are absent [59–61]. All short Argonautes have substitutions in the active site in the PIWI domain, which indicates their lack of catalytic activity. Long Argonautes are divided into two groups, called “longA” and “longB” [60]. All representatives of group B are inactive and often have a truncated PAZ domain, while most of the longA Argonautes have a catalytic tetrad and a full-length PAZ. Nevertheless, some representatives of different branches of group A also contain substitutions in the catalytic tetrad, which indicates the multiple appearance of inactive forms in the course of evolution [59–61]. Phylogenetic analysis has shown that eukaryotic Argonautes are a monophyletic group and originate from the longA prokaryotic Argonautes [59–61]. The phylogenetic tree of prokaryotic Argonautes does not correspond to the taxonomic tree of living organisms. From this, it can be concluded that horizontal gene transfer significantly contributes to the distribution of Argonaute genes among bacteria and archaea [59–61].
Fig. 3.
Phylogenetic tree of prokaryotic Argonaute proteins. The tree was built according to the data published in [59]. Multiple alignment of the MID and PIWI domains was performed using the MAFFT program (v7.487) [117]. The tree was constructed using the IQ-TREE (v2.1.4-beta) program [118–120]. The colors of the branches correspond to the groups of Argonautes: longA (green), longB (blue), short (burgundy). In circles, color indicates the features of the structure of PIWI domains (inner circle; active proteins—green, inactive—gray), MID (second circle; phosphate-binding domain—yellow, OH-binding domain—dark blue) and PAZ (outer circle; full-length PAZ—green, shortened—pink); the PAZ domain is missing in “short” Argonautes.
To date, several dozen of prokaryotic Argonautes from different branches of the phylogenetic tree have been characterized. Table 1 describes the properties of most prokaryotic Argonaute proteins studied to date. Attention was mostly focused on in vitro studies of active Argonautes from bacteria and archaea, which have nuclease activity. Unlike eukaryotic Argonautes, which work in the cell with RNA guides and RNA targets, prokaryotic Argonautes preferentially recognize DNA targets. Some of them can also cleave target RNA, although less efficiently; the functional significance of this phenomenon has not yet been elucidated [100, 126, 148]. Most prokaryotic Argonautes bind DNA guides, but some (MpAgo from the bacterium Marinitoga piezophila and its closest homologues) prefer RNA guides [130]. It is known that some eukaryotic Argonautes can also use DNA guides for in vitro target recognition (for example, hAgo2) [107, 156] and even participate in DNA repair (AGO1 from Arabidopsis thaliana) [157], but whether they are capable to interact with DNA in vivo remains unknown.
Table 1. .
Characteristics of catalytically active prokaryotic and eukaryotic Argonaute proteins
| Host | Argonaute, protein ID |
Guide, lengthа | 5' end of the guideb |
Target | Reaction temperatureс | Ion | Mismatches in a duplexd |
Catalytic activity | References |
|---|---|---|---|---|---|---|---|---|---|
| Prokaryotic Argonaute proteins | |||||||||
| Aquifex aeolicus |
AaAgo, |
DNA 18‒24 |
5′-P; ? |
RNA, (DNA ?) |
opt. 55°C; up to 75°C |
Mg2+, Mn2+ |
? | Guide-dependent | [113–115] |
|
Clostridium butyricum (strains TK520, CWBI 1009) |
CbAgo, CbcAgo, |
DNAe (RNA weakly) 16‒18 |
5′-P; none |
DNA* (RNA weakly) |
opt. 37°C; 30‒60°C (CbcAgo opt. 37‒42°C; 25‒55°C) |
Mg2+, Mn2+, Co2+ |
↓5, 6, 12‒15 nt | Guide-dependent; guide-independent |
[116, |
| Clostridium perfringens | CpAgo, WP_003477422.1 |
DNA 15‒30 (≥12) |
5′-P; none | DNA (RNA) |
opt. 37°C; 4–70°C |
Mg2+, Mn2+ |
↓12‒15 nt | Guide-dependent | [124] |
| Ferroglobus placidus | FpAgo, WP_012966655.1 |
DNA 15‒16 |
5′-P; G1 |
DNA | 75–99°C |
Mg2+, Mn2+, Co2+, Ni2+ |
↑7 nt; ↓3,8,11‒15 nt |
Guide-dependent | [125] |
| Intestinibacter bartlettii | IbAgo, WP_007287731.1 |
DNA 15‒30 (≥14) |
5′-P; none | DNA |
opt. 37°C; 4–60°C |
Mg2+, Mn2+ |
↓14‒16 nt | Guide-dependent | [124] |
|
Kurthia massiliensis |
KmAgo, WP_010289662.1 |
DNA (RNA) 16‒20 (≥12) |
5′-P; none |
DNA (RNA) |
opt. 37‒60°С; from 25°С |
Mn2+, Mg2+, weakly Co2+ |
↓4, 5, 10‒15 nt (DNA‒DNA); ↓4, 8‒11 nt (DNA‒RNA); ↓3‒11 nt (RNA‒DNA) |
Guide-dependent; guide-independent (37°С) | [126, 127] |
| Limnothrix rosea | LrAgo, WP_075892274.1 |
DNAe 16‒18 (≥10) |
5′-P; none | DNA |
opt. 37°C; 30–54°C |
Mn2+, Mg2+, weakly Co2+ |
↑4‒8 nt; ↓10‒15 nt |
Guide-dependent; guide-independent | [122] |
| Methanocaldococcus fervens | MfAgo, WP_015791216.1 |
DNA, RNA 16 |
5′-P DNA and RNA; 5′-OH DNA |
DNA |
opt. 80‒90°C; from 54°C |
Mn2+, Mg2+, Co2+ |
? | Guide-dependent | [128] |
| Marinitoga piezophila | MpAgo, WP_014295921.1 |
RNA 16‒40 |
5′-OH; none | DNA (RNA) | 37, 55, 60°C |
Mn2+, Mg2+, weakly Ni2+ |
↓5, 7, 8 nt; ↓5‒15 nt dinucleotide mismatches |
Guide-dependent | [129–131] |
|
Methanocaldococcus jannaschii |
MjAgo, WP_010870838.1 |
DNA 15‒41 |
5′-P; purines |
DNA |
opt. 75‒95°C; from 37°C |
Mg2+ |
? ↓10‒11 dinucleotide mismatches |
Guide-dependent; guide-independent | [101, 132, 133] |
| Natronobacterium gregoryi | NgAgo, WP_005580376.1 |
DNA 22‒24 |
5′-P; ? |
DNA ? (RNA ?) |
37°C | Mg2+ | ? | Guide-dependent | [134–138] |
|
Pyrococcus furiosus |
PfAgo, |
DNA 15‒31 |
5′-P; none | DNA |
opt. 87‒99.9°C; from 37°C |
Mn2+, Co2+ |
? | Guide-dependent; guide-independent | [139–141] |
| Synechococcus elongatus | SeAgo, WP_011244830.1 |
DNAe 14‒24 |
5′-P; none | DNAe | opt. 37°C | Mn2+, weakly Mg2+ |
↑5, 6, 10 nt; ↓7, 12‒15, 18 nt |
Guide-dependent; guide-independent | [142] |
| Thermotoga profunda | TpAgo, WP_041081268.1 |
RNA 21 |
5′-OH; ? | DNA | 60°C |
Mg2+, Mn2+ |
? | Guide-dependent | [130] |
|
Thermus thermophilus |
TtAgo, WP_011174533.1 |
DNAe 13‒25 |
5′-P; C1 G1' |
DNAe (RNA) |
opt. 73‒75°C; ⩾20 °C ssDNA; ⩾ 65°C plasmids |
Mn2+, Mg2+ |
↓12‒15 nt | Guide-dependent; guide-independent | [100, 106, 143–149] |
| Eukaryotic Argonaute proteins | |||||||||
| Homo sapiens | hAgo2, NP_036286.2 |
RNAe (DNA) 20‒21 |
5′-P; U1 A1' |
RNAe | 37°C | Mg2+ |
↓2‒8 nt (target binding); ↓8‒11 triple mismatch |
Guide-dependent | [99, 139, 150–152] |
| Kluyveromyces polysporus (Vanderwaltozyma polyspora) | KpAgo, XP_001644461.1 |
RNAe (DNA) 12‒17 (11‒25) |
5′-P; U1 |
RNAe | 25‒30°C | Mg2 |
↓7‒14 nt (↓↓9‒13 nt) |
Guide-dependent | [102, 153] |
| Bombyx mori | SIWI, NP_001098066.2 |
RNAe 28‒30 |
5′-P; U1 |
RNAe | 26, 37°C |
Mg2+, Mn2+ |
↓2‒7 nt (target binding) | Guide-dependent | [103, 154, 155] |
aThe type of guide nucleic acid and its length (number of nucleotides). bCharacteristics of the 5'-end of the guide: P—phosphate group, OH—hydroxyl group; the presence of specificity to the nitrogenous base is indicated: none—no specificity, A/U/G/C— types of nitrogenous bases, 1—the first nucleotide of the guide chain; 1'—the first nucleotide of the target chain. Here and further: ?—unknown. cThe temperature at which Argonaute protein could cleave target; opt.—the temperature optimum, the temperature range at which the Argonaute cleaves targets is also given. Discrete temperature values are given for publications where measurements were carried out only under these conditions. dThe effect of mismatches (unpaired nucleotides of the duplex guide–target) on the efficiency of target cleavage; the positions of the nucleotides (nt) are numbered relative to the 5'-end of the guide. ↑—increase in cleavage efficiency, ↓—decrease in cleavage efficiency. eBoth in vitro and in vivo.
Unlike eukaryotic Argonautes, prokaryotic Argonautes do not require accessory proteins to be loaded with guide nucleic acids. For TtAgo from Thermus thermophilus, MjAgo from archaea Methanocaldococcus jannaschii, SeAgo from Synechococcus elongatus, CbAgo from Clostridium butyricum, and LrAgo from Limnothrix rosea, guide-independent processing of plasmid DNA in vitro and loading of plasmid fragments as guides without additional protein factors were shown [122, 132, 142, 145].
The MID and PIWI domains are the most conserved and are present in all Argonaute proteins. The MID domain plays a key role in the guide 5' end binding. In prokaryotic Argonautes and eukaryotic PIWI proteins, the amino acid residues of the MID pocket coordinate a divalent cation (Mg2+ or Mn2+) involved in the binding of the 5'-terminal phosphate of guide molecule [100, 103, 110, 111, 148, 158]. However, among the active prokaryotic Argonautes, there is a small group of proteins with a hydrophobic pocket in the MID domain. Structural analysis of one of the representatives of this group, MpAgo from Marinitoga piezophila, showed that such Argonautes do not bind the divalent cation in the MID pocket and prefer guides that are not phosphorylated at the 5' end [60, 130].
Some Argonautes of bacteria and archaea discri-minate guides by the nitrogenous base of the 5'-terminal nucleotide: RsAgo from the bacterium Rhodobacter sphaeroides binds guides containing uracil at the 5' end, TtAgo from the bacterium Thermus thermophilus‒cytosine, FpAgo from the archaea Ferroglobus placidus‒guanine, MjAgo from archaea Methanocaldococcus jannaschii–purines. At the same time, other studied prokaryotic Argonautes do not show specificity for the 5'-terminal nucleotide (Table 1) [101–103, 122, 124, 125, 130, 141, 142, 146, 159]. The corresponding unpaired nucleotide of the target chain can be specifically recognized in a separate pocket of the PIWI domain (G for TtAgo, A for RsAgo) [145, 158].
Being bound in the MID pocket, the 5'-terminal nucleotide of the guide molecule does not participate in the formation of hydrogen bonds with the complementary nucleotide in target and is called an anchor. There are several regions that can be distinguished in the guide molecule: the primary binding region (“seed”) – 2–8 guide nucleotides, the cleavage site – 10–11 nucleotides, the 3'-supplementary site – 12–16 nucleotides, the 3'-terminal region, and the last nucleotide which usually binds in the pocket of the PAZ domain [151, 160–164]. Most active Argonautes cleave the target between positions complementary to 10 and 11 nucleotides of the guide [100, 101, 132, 133, 148]. Some prokaryotic Argonautes, which normally bind 5'-phosphorylated guides, can also use 5'-OH guides, for example, CbAgo, LrAgo, KmAgo, SeAgo, but in this case the cleavage site is shifted by one nucleotide [122, 123, 126, 142].
Binding of a complementary target by a guide-loaded Argonaute is accompanied by structural rearrangements, which include rotation of the PAZ domain and changes in the position of several loops in the PIWI domain [100, 129, 144, 148]. The guide 3' end is released from PAZ and a catalytically active ternary Argonaute complex with the guide and target is formed [165, 166].
For eukaryotic Argonautes, guide-target pairing at positions 2–8 of the duplex in the seed region play a crucial role. Non-complementary nucleotides and insertions in the guide opposite the target cleavage site also significantly reduce the efficiency of target cleavage, while mismatches in the 3' terminal region do not have a significant effect [151, 163, 167]. Such a general pattern cannot be revealed for prokaryotic Argonautes, probably due to their diversity. Interactions of guides with not fully complementary targets may affect the activity of prokaryotic Argonautes in different ways. It has been shown that insertions in the seed region do not affect guide-target duplex binding by TtAgo and RsAgo [158, 168, 169]. In LrAgo and SeAgo, non-complementary interactions in the seed region even stimulate target cleavage [122, 142]. It is interesting to note that the presence of additional or non-complementary nucleotides in the seed region stimulates the release of the guide-target hybrid from hAgo2 and prokaryotic RsAgo, which may be the mechanism for the replacement of Argonaute -associated guide molecules [150, 158]. In contrast to eukaryotic Argonautes, CbAgo, LrAgo, SeAgo, and TtAgo are sensitive to the presence of mismatches in the 3' supplementary region of the DNA guide [122, 142, 143]. In general, such experiments are necessary to understand the specificity of target binding and cleavage by new prokaryotic Argonautes from different groups.
It is still unknown which structural features of prokaryotic Argonautes are responsible for the discrimination of RNA/DNA guides and targets, as well as for sensitivity to base-pairing in the guide–target duplex. Probably, these features are primarily determined by variations in the structures of the MID and PIWI domains. To answer this question, further studies of the activity and structures of prokaryotic Argonaute proteins are required.
Argonaute Proteins Protect Prokaryotic Cells from Foreign Nucleic Acids
A possible role of prokaryotic Argonautes in protecting cells from viruses was suggested in 2009 [59]. At the same time, experimental data confirming this hypothesis have appeared only recently.
First, it has been shown that Argonaute genes are often located in the so-called “defense islands,” regions of the genome with increased mobility, in which various genes are encoded that protect bacteria and archaea from foreign nucleic acids. The best known prokaryotic defense systems are restriction-modification systems and CRISPR–Cas. To date, more than a hundred such systems are known, and this number continues to grow [170–173]. Genes with an unknown function, often encoded in defense islands, are also likely to be involved in defense systems [59, 174–176]. Interestingly, some groups of prokaryotic Argonautes are associated with certain defense systems. For example, MpAgo, which uses RNA guides, and its closest homologues are encoded inside CRISPR-Cas systems [130]. A more extensive analysis showed that active prokaryotic Argonautes in most cases are encoded in genomes that also contain CRISPR-Cas systems, while genomes without Argonautes or with inactive Argonautes contain CRISPR-Cas systems much less often [116]. A more detailed analysis of the association of various groups of Argonautes with other defense systems of prokaryotes is the subject of further research.
Secondly, some prokaryotic Argonautes are associated with additional domains that perform protective functions. Thus, many short Argonautes contain the SIR2 (silent information regulator 2) or TIR (Toll/interleukin-1 receptor) domains or their coding sequences are located in the neighboring gene [59, 60]. These domains are also part of the Thoeris defense system found in bacteria: in response to infection of a cell with a virus, they cleave NAD+, which leads to cell death and stops the spread of the virus in the population [175, 177, 178]. It has recently been shown that the SIR2/Ago and TIR/Ago systems protect bacterial cells from phages and plasmids: short Argonautes with MID-PIWI domains recognize foreign DNA, while SIR2 or TIR cleaves NAD+ [178, 179]. The TIR/Ago system is also involved in antiplasmid defense. Some Argonautes are associated in operons with nucleases from different families, which are presumably involved in the production of guides or act as protein effectors of defense systems [59, 60].
Finally, direct experimental confirmation of the protective role of long Argonautes in bacteria has been obtained in recent years. Thus, the CbAgo protein protects cells from phages [116], the CbAgo [116], PfAgo (from the archaea Pyrococcus furiosus) [141], and RsAgo [159] proteins reduce the efficiency of transformation and promote plasmid degradation in bacterial cells.
In addition to protecting cells from foreign DNA, Argonautes can perform other functions. To date, one such example is known: it has been shown that, in the absence of gyrase, TtAgo is involved in cell division, resolving chromosome catenanes after replication in Thermus thermophilus cells [149]. The diversity of the structures and activities of prokaryotic Argonautes may indicate that their functional role in bacterial and archaeal cells is not limited to currently known examples.
PROSPECTS FOR THE PRACTICAL APPLICATION OF PROKARYOTIC ARGONAUTE PROTEINS
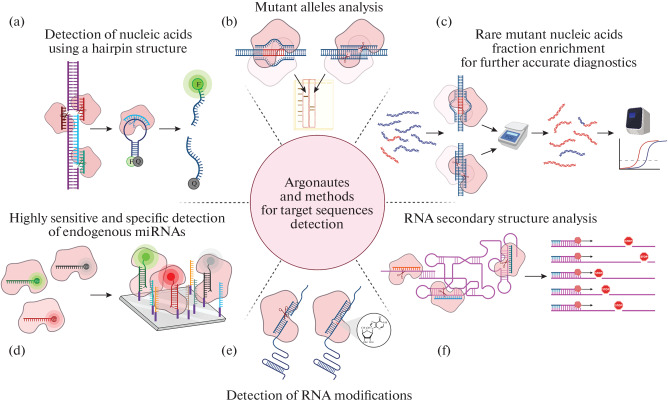
Argonaute proteins are considered promising candidates for directed nucleic acid manipulation and genome editing. The ability of Argonaute proteins to act as site-specific programmable nucleases opens up wide opportunities for their use in vitro. The currently known applications of bacterial Argonaute proteins are summarized in Fig. 4.
Fig. 4.
Detection of target sequences in biological samples using Argonaute proteins. (a) Detection of nucleic acids using Argonautes. The Argonaute is programmed to cut out the target sequence from the DNA; in addition, an oligonucleotide probe complementary to this sequence is added to the sample; the probe has a hairpin structure, to the ends of which a fluorescent label (F) and a quencher (Q) are attached. During the first round of catalysis, the Argonaute cuts out the target sequence in the sample, after which the Argonaute binds this sequence as a guide and performs the second round of catalysis, as a result of which the fluorescent probe is cleaved. The presence of the target sequence is assessed by increasing the fluorescence intensity [180, 181]. (b) Mutant alleles analysis in biological samples. To detect the mutant allele, DNA is first amplified, and then PCR products are incubated at 98°C with Argonaute loaded with two DNA guides corresponding to one of the variants of the target sequence; then the products are separated in an agarose gel [182]. (c) Detection of rare DNA variants. The mutant DNA sample is incubated with Argonaute loaded with guides corresponding to the wild-type sequence; only mutant DNA remains intact after catalysis; the amount of such DNA can be estimated using Real-time PCR or other methods [183]. (d) For miRNA detection, catalytically inactive Argonaute is loaded with a guide corresponding to the target sequence and containing a fluorescent label; after incubation with miRNAs immobilized on a chip, the fluorescence signal of guides immobilized in a complex with Argonautes and the target miRNA is detected [184]. (e) Detection of RNA modifications [131]. (f) RNA secondary structure analysis. Structured RNAs are incubated with Argonautes loaded with guides to different sequence sites; cleavage sites can be detected directly or by reverse transcription [126, 127, 153].
Unlike restriction endonucleases, Argonaute proteins can be used to make a cut in potentially any DNA sequence and obtain fragments with “sticky” ends of the required length and nucleotide composition. Since Argonautes have a single active site and can cleave only one strand of DNA, double-stranded DNA cleavage requires the loading of Argonautes with a pair of guides that are complementary to opposite strands at the cutting site. For example, PfAgo from a thermophilic archaea was successfully used to assemble genetic constructs and to analyze them [185]. In a large-scale study using the guide panel for the TtAgo, patterns of preference for guide sequences were determined. Based on the results obtained in the course of this work, it is possible to optimize the using of TtAgo as a programmable endonuclease [143, 186].
An important area of application of Argonaute proteins is the detection of target sequences and nucleotide modifications in biological samples. A system for detecting the desired sequence in the sample was developed on the basis of PfAgo called PAND (PfAgo-mediated Nucleic acid Detection). The technique uses reporter DNA, upon cleavage of which by Argonaute, the fluorescent label and quencher are uncoupled and a fluorescent signal appears (Fig. 4). The guide for the reporter cleavage is produced from the target DNA sequence and is cut from the target DNA of the sample by PfAgo using a combination of three synthetic guides. When using reporters with different fluorescent beacons, it is possible to detect several sequences in one tube [180]. This method has been successfully adapted to identify SARS-CoV-2 RNA and its mutant variant (D614G in the spike protein) in patient samples [187]. Combining two methods, PAND and ligase chain reaction made it possible to simplify the detection of nucleic acids. The combined protocol was called PLCR (PfAgo coupled with modified Ligase Chain Reaction for nucleic acid detection). A thermostable ligase joins two halves of the guide using the target DNA as a template. Then, in the course of several cycles, guide is amplified. The guide is loaded into the Argonaute, and the reporter DNA is cut, which leads to appearance of fluorescent signal [181].
Other proposed methods are based on the decreased activity of Argonautes in the presence of mismatches or modifications in the target, compared to the situation when the guide and the target are completely complementary (Fig. 4). A quick analysis of the presence of insertions or deletions in one or two alleles of the target gene that occurs during genome editing is possible if guides are selected to the editing nuclease cleavage site. In the case of the wild-type allele, the DNA will be cut by the Argonaute. The reaction products can be detected in a gel [182]. The NAVIGATER method (Nucleic Acids of clinical interest Via DNA-Guided Argonaute from Thermus thermophilus) has been developed to detect rare single nucleotide substitutions that occur, for example, in cancer cells. To detect DNA or RNA with a single nucleotide substitution, it is necessary to choose a guide that is complementary to the predominant wild-type sequences in the sample, and a rare variant has a substitution at the 10th or 11th position of the target, counting from the 5' end of the guide. The presence of the substitution dramatically reduces the nuclease activity of TtAgo. In this case, the wild-type DNA will be cleaved by the Argonaute, and the relative amount of the rare variant will increase. Such enrichment makes it possible to detect the rare variant by standard methods (various PCR techniques or sequencing) [183].
To detect substitutions and modifications in RNA, it is necessary to use Argonaute with the appropriate specificity. MpAgo uses RNA as guides and can bind complementary target RNA. For target recognition, nucleotides 6 and 7 are most important. By determining the dissociation constant of the guide RNA with the target, it is possible to identify mismatches, as well as modified nucleotides such as inosine (Fig. 4) [131]. The difference in the cleavage efficiency of single- and double-stranded RNA can be used to study the structure of highly structured RNA. The ability to guide the Argonaute to specific RNA sequences makes it possible to study the structure of specific RNA regions under various conditions [126, 127, 153].
Argonautes also found application in super-resolution microscopy to visualize structures beyond the diffraction limit of microscopy. The DNA-PAINT (DNA Point Accumulation In Nanoscale Topology) method is based on the detection of association and dissociation of a fluorescent DNA probe with a fixed DNA target. If labelled DNA is loaded into CbAgo, the rate of the process increases due to the fact that Argonaute organizes guide DNA for optimal target binding [188]. The ability of Argonaute to accelerate the binding of a fluorescently labeled DNA guide to target RNA was also used for specific visualization of miRNA. The authors, S. Shin et al. [184], called this technology Ago-FISH (Argonaute-based Fluorescence In Situ Hybridization) (Fig. 4).
The ability of Argonautes to introduce site-specific cuts in nucleic acids allows us to consider them as potential tools for editing the genome and transcriptome. As such, Argonautes may have some advantages over Cas nucleases. First, Argonautes do not require the presence of a PAM sequence in the target DNA. Second, the guides for most prokaryotic Argonautes are short DNA molecules while Cas nucleases require long RNA guides. DNA synthesis is much cheaper, which can facilitate development of the Argonaute-based genome editing system. Third, the size of the Argonaute molecule is smaller than that of Cas9, so it is easier to deliver them inside cells [189, 190].
In 2016, a work was published on editing the human cell genome using Argonaute from the halophilic archaea Natronobacterium gregoryi (NgAgo) [189]. However, the authors soon retracted the article, since other groups of researchers did not confirm the reliability of the results presented in it when trying to introduce changes using NgAgo in the genome of human cells, mouse embryos and Danio rerio embryos, as well as the hepatitis B virus [135, 137, 138, 191–194]. An attempt to edit the genome of human cells using Argonaute TtAgo also failed [135]. Thus, the question of the possibility of using Argonautes as a tool for editing the genome remains open. It should be noted that the choice of the Argonaute protein can significantly affect the results of such experiments. In published works, Argonautes from thermophilic microorganisms were used, which have an optimum efficiency at high temperatures. In addition, NgAgo is a protein from the halophilic archaea, and it is known that the expression and folding of such proteins occurs inefficiently under normal salt conditions [195]. In some studies, which did not give a positive result, a nuclear localization signal (NLS) was added to the C‑terminus of the Argonaute [135, 137, 193]. This could affect the activity of the protein, since the C-terminus of Argonaute is important for catalysis [144]. Guides to sequences located on opposite DNA strands at a great distance from each other were also used [194]. Several studies using NgAgo have noticed a decrease in the mRNA level of the target gene without making changes to the corresponding genome region, possibly due to Argonaute interactions with target RNAs [137, 138]. Thus, for further experiments on genome editing, it is worth to choose Argonautes highly specific for DNA targets, whose activity is optimal under physiological conditions.
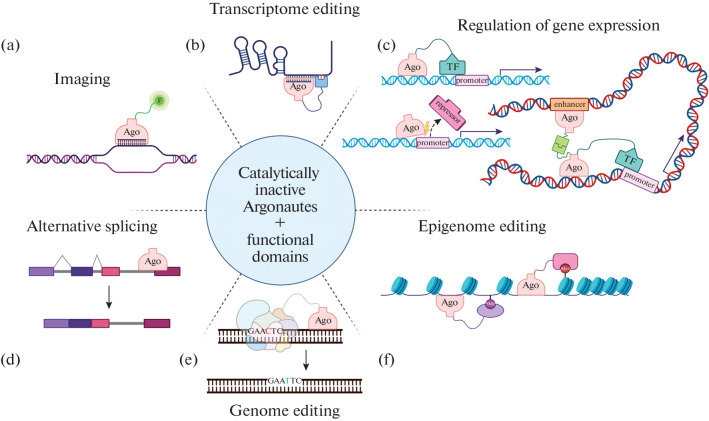
Argonautes with a non-functional catalytic center, inactive Argonautes or mutant forms of active Argonautes, also have great potential for practical applications. Such Argonautes recognize target DNA or RNA sequences using guides and can potentially be used as a DNA recognition domain in the construction of chimeric proteins with various functions. Previously, similar approaches were successfully used in the case of Cas nucleases. Possible applications of catalytically inactive bacterial Argonaute proteins are summarized in Fig. 5. For example, the combination of Argonautes with enzymes that modify nitrogenous bases may be used to make changes in the DNA or RNA sequence without creating a double-strand break. Cytidine deaminase converts cytidine to uridine, which leads to the replacement of the C:G pair with T:A during DNA replication [196]. A reverse substitution of the T:A pair with C:G can be carried out by a chimeric protein containing adenosine deaminase as a functional domain, which converts adenine to inosine, which is recognized by polymerases as guanine [197].
Fig. 5.
Promising areas of applications of catalytically inactive Argonautes in biotechnology. (a) Argonaute (Ago) can be linked to a fluorescent label (F) and loaded with a guide complementary to the sequence of interest. (b) Argonaute may be combined with enzymes that replace or modify nucleotides in RNA. (c) Combining Argonaute with a transcription factor (TF) may enhance the transcription of a particular gene; a similar effect may be achieved if Argonaute competes with a transcriptional repressor protein; Argonaute with additional domains may enhance the interactions of the enhancer with the promoter of the gene of interest and thereby activate transcription. (d) Pre-mRNA splicing may be regulated by targeting Argonaute to its specific region. (e) Argonaute may be linked to various enzymes of other DNA editing systems to modify specific regions of the genome. (f) Argonaute may be combined with histone and DNA modifying enzymes for epigenome editing (orange circle indicates histone modification, lilac circle indicates cytosine methylation in DNA).
Argonautes may be used as a DNA recognition domain to recruit factors that change epigenetic marks and thus activate or inactivate gene expression (Fig. 5). For this, approaches previously proposed for catalytically inactive variants of Cas nucleases can be used. Activation or suppression of gene expression may be achieved by combining Argonaute with transcriptional activators or repressors, respectively. For example, directing cytosine-DNA-methyltransferase DNMT3a to the promoter of the target gene can reduce its expression. The use of the KRAB domain (Krüppel-associated box), which forms a complex with two histone DNA methyltransferases, leads to the repression of target genes. Fusion with p300 acetyltransferase leads to acetylation of lysine at position 27 of histone H3 and, as a result, transcriptional activation of target genes [40, 198]. Changes in the arrangement of DNA regions in the nucleus can lead to changes in gene expression, for example, when promoters and enhancers become closer to each other [199]. For this, inactive Argonautes with heterodimerizing protein domains can be used. Argonautes may be also used as a DNA recognition domain for visualizing various structures and processes in the nucleus when fused with fluorescent proteins, as was shown for TALEN nucleases [200]. Also Argonautes could be used as an RNA recognition domain for visualizing the location of target RNA in the cell (Fig. 5). Inactive Argonautes interacting with RNA targets could be used to attract additional functional domains to RNA to regulate gene expression at the post-transcriptional level. For example, the effector domains of the YTHDF1 and YTHDF2 proteins (YT521-B Homology Domain Family) in eukaryotic cells recognize N6-methyladenosine in RNA. YTHDF1 activates translation of the target RNA, while YTHDF2, on the contrary, leads to its degradation. Replacement of the N6-methyladenosine-recognizing domain with an RNA-recognizing domain of guide loaded Argonaut makes it possible to direct these functions to any RNA [201]. It is also possible to perform directed RNA editing using the ADAR1 or human ADAR2 deaminase domain (Adenosine Deaminase Acting on RNA). This protein converts adenosine to inosine, mainly in cases where adenosine is opposite cytidine in the RNA duplex [202]. Similar to published experiments using dCas9 (a catalytically inactive variant of Cas-9), RNA-recognizing Argonautes could also be used to regulate alternative splicing by changing the efficiency of incorporation of certain exons into mature mRNA (Fig. 5) [203].
The tools for genome editing has evolved from the search and creation of proteins capable of recognizing specific DNA sequences (meganucleases, TALEN, zinc finger nucleases) to the discovery of universal programmable nucleases, the specificity of which is determined by guide nucleic acids. These targeted nucleases include enzymes based on Cas proteins, which are currently actively used for gene editing, as well as Argonaute proteins. Although no success was yet achieved in the use of Argonautes for genome editing, their diversity, both structural and functional, allows us to hope that they will soon provide complimentary tools fro genome manitpulations and biotechnplogy along with Cas nucleases.
At present, studies of the prokaryotic Argonaute family proteins already led to their practical applications in new highly sensitive methods for detecting nucleic acids, including SARS-CoV-2 RNA. In the near future, we could expect the appearance of works using Argonautes as effective tools for manipulations with nucleic acids in vitro, and in vivo.
AUTHOR CONTRIBUTIONS
E.V. Kropocheva, L.A. Lisitskaya, A.A. Agapov, and D.M. Esyunina contributed equally to this work.
FUNDING
This work was supported by the Russian Science Foundation, grant 19-14-00359.
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
REFERENCES
- 1.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B., Luo D., Li Y., Perčulija V., Chen J., Lin J., Ye Y., Ouyang S. Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1. Nat. Commun. 2021;12:3476. doi: 10.1038/s41467-021-23876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Zhang H., Xu X., Wang Y., Chen W., Wang Y., Wu Z., Tang N., Wang Y., Zhao S., Gan J., Ji Q. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat. Catal. 2020;3:813–823. doi: 10.1038/s41929-020-00506-9. [DOI] [Google Scholar]
- 4.Hafez M., Hausner G. Homing endonucleases: DNA scissors on a mission. Genome. 2012;55:553–569. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- 5.Paques F., Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 6.Jurica M.S., Monnat R.J., Stoddard B.L. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-Crel. Mol. Cell. 1998;2:469–476. doi: 10.1016/S1097-2765(00)80146-X. [DOI] [PubMed] [Google Scholar]
- 7.McMurrough T.A., Brown C.M., Zhang K., Hausner G., Junop M.S., Gloor G.B., Edgell D.R. Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases. Nucleic Acids Res. 2018;46:11990–12007. doi: 10.1093/nar/gky976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith J., Grizot S., Arnould S., Duclert A., Epinat J.C., Chames P., Prieto J., Redondo P., Blanco F.J., Bravo J., Montoya G., Pâques F., Duchateau P. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier B.S., Kortemme T., Chadsey M.S., Baker D., Monnat R.J., Stoddard B.L. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/S1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 10.Li H., Pellenz S., Ulge U., Stoddard B.L., Monnat R.J. Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y.G., Chandrasegaran S. Chimeric restriction endonuclease. Proc. Natl. Acad. Sci. U. S. A. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshney G.K., Burgess S.M. DNA-guided genome editing using structure-guided endonucleases. Genome Biol. 2016;17:187. doi: 10.1186/s13059-016-1055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S., Cao S., Zou B., Yue Y., Gu C., Chen X., Wang P., Dong X., Xiang Z., Li K., Zhu M., Zhao Q., Zhou G. An alternative novel tool for DNA editing without target sequence limitation: the structure-guided nuclease. Genome Biol. 2016;17:186. doi: 10.1186/s13059-016-1038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J., McLachlan A.D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasegaran S., Carroll D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavletich N.P., Pabo C.O. Zinc finger–DNA recognition: crystal structure of a Zif268–DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu Y., Şöllü C., Meckler J.F., Adriaenssens A., Zykovich A., Cathomen T., Segal D.J. Adding fingers to an engineered zinc finger nuclease can reduce activity. Biochemistry. 2011;50:5033–5041. doi: 10.1021/bi200393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonas U., Stall R.E., Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 21.Kay S., Hahn S., Marois E., Hause G., Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 22.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 23.Moscou M.J., Bogdanove A.J. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 24.Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin E.V., Makarova K.S. 2019. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc., B.374 (1772), 20180087. [DOI] [PMC free article] [PubMed]
- 26.Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 27.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee P., Lee J., Nip L., Koseki S.R.T., Tysinger E., Sontheimer E.J., Jacobson J.M., Jakimo N. A Cas9 with PAM recognition for adenine dinucleotides. Nat. Commun. 2020;11:2474. doi: 10.1038/s41467-020-16117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B, Shalem O., Wu X., Makarova K.S., Koonin E.V., Sharp P.A., Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z., Wang S., Zhang C., Gao N., Li M., Wang D., Wang D., Liu D., Liu H., Ong S.G., Wang H., Wang Y. A compact Cas9 ortholog from Staphylococcus auricularis (Sauricas9) expands the DNA targeting scope. PLoS Biol. 2020;18:e3000686. doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karvelis T., Gasiunas G., Young J, Bigelyte G., Silanskas A., Cigan M., Siksnys V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.R.J., Aryee M.J., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heler R., Wright A.V., Vucelja M., Bikard D., Doudna J.A., Marraffini L.A. Mutations in Cas9 enhance the rate of acquisition of viral spacer sequences during the CRISPR-Cas immune response. Mol. Cell. 2017;65:168–175. doi: 10.1016/j.molcel.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakade S., Yamamoto T., Sakuma T. Cas9, Cpf1 and C2c1/2/3—what’s next? Bioengineered. 2017;8:265–273. doi: 10.1080/21655979.2017.1282018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribeiro L.F., Ribeiro L.F.C., Barreto M.Q., Ward R.J. Protein engineering strategies to expand CRISPR-Cas9 applications. Int. J. Genomics. 2018;2018:1652567. doi: 10.1155/2018/1652567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai S.Q., Joung J.K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maji B., Moore C.L., Zetsche B., Volz S.E., Zhang F., Shoulders M.D., Choudhary A. Multidimensional chemical control of CRISPR-Cas9. Nat. Chem. Biol. 2017;13:9–11. doi: 10.1038/nchembio.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zetsche B., Volz S.E., Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nihongaki Y., Kawano F., Nakajima T., Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 48.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul B., Montoya G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020;43:8–17. doi: 10.1016/j.bj.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 51.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:557–558. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., Lander E.S., Voytas D.F., Ting A.Y., Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021;29:689–703. doi: 10.1016/j.chom.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altae-Tran H., Kannan S., Demircioglu F.E., Oshiro R., Nety S.P., McKay L.J., Dlakić M., Inskeep W.P., Makarova K.S., Macrae R.K., Koonin E.V., Zhang F. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374:57–65. doi: 10.1126/science.abj6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karvelis T., Druteika G., Bigelyte G., Budre K., Zedaveinyte R., Silanskas A., Kazlauskas D., Venclovas Č., Siksnys V. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. 2021;599:692–696. doi: 10.1038/s41586-021-04058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makarova K.S., Wolf Y.I., van der Oost J., Koonin E. V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryazansky S., Kulbachinskiy A., Aravin A.A. The expanded universe of prokaryotic Argonaute proteins. MBio. 2018;9:e01935–18. doi: 10.1128/mBio.01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 63.Miesen P., Joosten J., van Rij R.P. PIWIs go viral: arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016;12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miesen P., Ivens A., Buck A.H., van Rij R.P. Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl. Trop. Dis. 2016;10:e0004452. doi: 10.1371/journal.pntd.0004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mussabekova A., Daeffler L., Imler J.L. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol. Life Sci. 2017;74:2039–2054. doi: 10.1007/s00018-017-2453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pumplin N., Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 67.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 68.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 69.Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Chien M., Russo J.J., Ju J., Sheridan R., Sander C., Zavolan M., Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 70.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 71.Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/S0092-8674(02)00908-X. [DOI] [PubMed] [Google Scholar]
- 72.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pratt A.J., MacRae I.J. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 75.Tolia N.H., Joshua-Tor L. Slicer and the Argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 76.Vaucheret H. Plant Argonautes. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 77.Aravin A.A., Naumova N.M., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/S0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 78.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 79.Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 80.Ma X., Zuo Z., Shao W., Jin Y., Meng Y. The expanding roles of Argonautes: RNA interference, splicing and beyond. Brief Funct. Genomics. 2018;17:191–197. doi: 10.1093/bfgp/elx045. [DOI] [PubMed] [Google Scholar]
- 81.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu W., Duttke S.H., Hetzel J., Groth M., Feng S., Gallego-Bartolome J., Zhong Z., Kuo H.Y., Wang Z., Zhai J., Chory J., Jacobsen S.E. RNA-directed DNA methylation involves co-transcriptional small RNA-guided slicing of Pol V transcripts in Arabidopsis. Nat. Plants. 2018;4:181–188. doi: 10.1038/s41477-017-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olina A.V., Kulbachinskiy A.V., Aravin A.A., Esyunina D.M. Argonaute proteins and mechanisms of RNA interference in eukaryotes and prokaryotes. Biochemistry. 2018;83:483–497. doi: 10.1134/S0006297918050024. [DOI] [PubMed] [Google Scholar]
- 84.Pezic D., Manakov S.A., Sachidanandam R., Aravin A.A. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014;28:1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I.S., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ameres S.L., Martinez J., Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 87.Aravin A.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 89.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 90.Vagin V V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 91.Förstemann K., Horwich M.D., Wee L.M., Tomari Y., Zamore P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 93.Tomari Y., Du T., Zamore P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le Thomas A., Rogers A.K., Webster A., Marinov G.K., Liao S.E., Perkins E.M., Hur J.K., Aravin A.A., Tóth K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sienski G., Dönertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Czech B., Munafò M., Ciabrelli F., Eastwood E.L., Fabry M.H., Kneuss E., Hannon G.J. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Webster A., Li S., Hur J.K., Wachsmuth M., Bois J.S., Perkins E.M., Patel D.J., Aravin A.A. Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol. Cell. 2015;59:564–575. doi: 10.1016/j.molcel.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhattacharjee S., Roche B., Martienssen R.A. RNA-induced initiation of transcriptional silencing (RITS) complex structure and function. RNA Biol. 2019;16:1133–1146. doi: 10.1080/15476286.2019.1621624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human Argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017;2:17035. doi: 10.1038/nmicrobiol.2017.35. [DOI] [PubMed] [Google Scholar]
- 102.Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsumoto N., Nishimasu H., Sakakibara K., Nishida K.M., Hirano T., Ishitani R., Siomi H., Siomi M.C., Nureki O. Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell. 2016;167:484–497.e9. doi: 10.1016/j.cell.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 104.Schirle N.T., Sheu-Gruttadauria J., Chandradoss S.D., Joo C., MacRae I.J. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. eLife. 2015;4:e07646. doi: 10.7554/eLife.07646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koonin E. V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol. Direct. 2017;12:5. doi: 10.1186/s13062-017-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Willkomm S., Zander A., Grohmann D., Restle T. Mechanistic insights into archaeal and human Argonaute substrate binding and cleavage properties. PLoS One. 2016;11:e0164695. doi: 10.1371/journal.pone.0164695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kwak P.B., Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 109.Parker J.S., Roe S.M., Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′ -end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parker J.S., Roe S.M., Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan Y.R., Pei Y., Chen H.Y., Tuschl T., Patel D.J. A potential protein–RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rashid U.J., Paterok D., Koglin A., Gohlke H., Piehler J., Chen J.C.H. Structure of Aquifex aeolicus Argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007;282:13824–13832. doi: 10.1074/jbc.M608619200. [DOI] [PubMed] [Google Scholar]
- 116.Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A., Maslova O., Ninova M., Ryazansky S., Leach D., Aravin A.A., Kulbachinskiy A. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020;587:632–637. doi: 10.1038/s41586-020-2605-1. [DOI] [PubMed] [Google Scholar]
- 117.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019;47:5809–5821. doi: 10.1093/nar/gkz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019;47:5822–5836. doi: 10.1093/nar/gkz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.García-Quintans N., Bowden L., Berenguer J., Mencía M. DNA interference by a mesophilic Argonaute protein, CbcAgo. F1000Res. 2019;8:321. doi: 10.12688/f1000research.18445.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T., Shi W., Li C., Wang Y., Zhao F., Wang H. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37°C. Cell Discov. 2019;5:38. doi: 10.1038/s41421-019-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo X., Sun Y., Chen L., Huang F., Liu Q., Feng Y. A hyperthermophilic Argonaute from Ferroglobus placidus with specificity on guide binding pattern. Front. Microbiol. 2021;12:654345. doi: 10.3389/fmicb.2021.654345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kropocheva E., Kuzmenko A., Aravin A.A., Esyunina D., Kulbachinskiy A. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 2021;49:4054–4065. doi: 10.1093/nar/gkab182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Y., Li W., Jiang X., Wang Y., Zhang Z., Liu Q., He R., Chen Q., Yang J., Wang L., Wang F., Ma L. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021;49:1597–1608. doi: 10.1093/nar/gkaa1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chong Y., Liu Q., Huang F., Song D., Feng Y. Characterization of a recombinant thermotolerant Argonaute protein as an endonuclease by broad guide utilization. Bioresour. Bioprocess. 2019;6:21. doi: 10.1186/s40643-019-0254-8. [DOI] [Google Scholar]
- 129.Doxzen K.W., Doudna J.A. DNA recognition by an RNA-guided bacterial Argonaute. PLoS One. 2017;12:e0177097. doi: 10.1371/journal.pone.0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4057–4062. doi: 10.1073/pnas.1524385113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lapinaite A., Doudna J.A., Cate J.H.D. Programmable RNA recognition using a CRISPR-associated Argonaute. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3368–3373. doi: 10.1073/pnas.1717725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stöckl S., Tinnefeld P., Schneider S., Klingl A., Albers S.V., Werner F., Grohmann D. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017;2:17034. doi: 10.1038/nmicrobiol.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014;11:45–56. doi: 10.4161/rna.27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fu L., Xie C., Jin Z., Tu Z., Han L., Jin M., Xiang Y., Zhang A. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019;47:3568–3579. doi: 10.1093/nar/gkz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.O’Geen H., Ren C., Coggins N.B., Bates S.L., Segal D.J. Unexpected binding behaviors of bacterial Argonautes in human cells cast doubts on their use as targetable gene regulators. PLoS One. 2018;13:e0193818. doi: 10.1371/journal.pone.0193818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lao Y.H., Li M., Gao M.A., Shao D., Chi C.W., Huang D., Chakraborty S., Ho T.C., Jiang W., Wang H.X., Wang S., Leong K.W. HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute. Adv. Sci. 2018;5:1700540. doi: 10.1002/advs.201700540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qi J., Dong Z., Shi Y., Wang X., Qin Y., Wang Y., Liu D. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 2016;26:1349–1352. doi: 10.1038/cr.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu Z., Tan S., Xu L., Gao L., Zhu H., Ma C., Liang X. NgAgo–gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017;145:20–23. doi: 10.1016/j.antiviral.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 139.Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 140.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 141.Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015;43:5120–5129. doi: 10.1093/nar/gkv415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olina A., Kuzmenko A., Ninova M., Aravin A.A., Kulbachinskiy A., Esyunina D. Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 2020;17:677–688. doi: 10.1080/15476286.2020.1724716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hunt E.A., Evans T.C., Tanner N.A. Single-stranded binding proteins and helicase enhance the activity of prokaryotic Argonautes in vitro. PLoS One. 2018;13:e0203073. doi: 10.1371/journal.pone.0203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U. S. A. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M., Zhang Y., Zhao H., Lou J., Wang Y., Joo C., van der Oost J. Auto-nomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell. 2017;65:985–998. doi: 10.1016/j.molcel.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J., van der Oost J. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Swarts D.C., Koehorst J.J., Westra E.R., Schaap P.J., van der Oost J. Effects of Argonaute on gene expression in Thermus thermophilus. PLoS One. 2015;10:e0124880. doi: 10.1371/journal.pone.0124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D. Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell. 2020;182:1545–1559. doi: 10.1016/j.cell.2020.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Park J.H., Shin S.Y., Shin C. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 2017;45:1569–1583. doi: 10.1093/nar/gkx029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schirle N.T., Sheu-Gruttadauria J., MacRae I.J. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dayeh D.M., Cantara W.A., Kitzrow J.P., Musier-Forsyth K., Nakanishi K. Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA. Nucleic Acids Res. 2018;46:e98. doi: 10.1093/nar/gky496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anzelon T.A., Chowdhury S., Hughes S.M., Xiao Y., Lander G.C., MacRae I.J. Structural basis for piRNA targeting. Nature. 2021;597:285–289. doi: 10.1038/s41586-021-03856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nishida K.M., Iwasaki Y.W., Murota Y., Nagao A., Mannen T., Kato Y., Siomi H., Siomi M.C. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 2015;10:193–203. doi: 10.1016/j.celrep.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 156.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 157.Schalk C., Cognat V., Graindorge S., Vincent T., Voinnet O., Molinier J. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2965–2975. doi: 10.1073/pnas.1618834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu Y., Esyunina D., Olovnikov I., Teplova M., Kulbachinskiy A., Aravin A.A., Patel D.J. Accommodation of helical imperfections in Rhodobacter sphaeroides Argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 2018;24:453–462. doi: 10.1016/j.celrep.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A. Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Globyte V., Kim S.H., Joo C. Single-molecule view of small RNA-guided target search and recognition. Annu. Rev. Biophys. 2018;47:569–593. doi: 10.1146/annurev-biophys-070317-032923. [DOI] [PubMed] [Google Scholar]
- 161.Klum S.M., Chandradoss S.D., Schirle N.T., Joo C., MacRae I.J. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018;37:75–88. doi: 10.15252/embj.201796474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sheu-Gruttadauria J., MacRae I.J. Structural foundations of RNA silencing by Argonaute. J. Mol. Biol. 2017;429:2619–2639. doi: 10.1016/j.jmb.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Willkomm S., Makarova K.S., Grohmann D. DNA silencing by prokaryotic Argonaute proteins adds a new layer of defense against invading nucleic acids. FEMS Microbiol. Rev. 2018;42:376–387. doi: 10.1093/femsre/fuy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hur J.K., Zinchenko M.K., Djuranovic S., Green R. Regulation of Argonaute slicer activity by guide RNA 3' end interactions with the N-terminal lobe. J. Biol. Chem. 2013;288:7829–7840. doi: 10.1074/jbc.M112.441030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jung S.R., Kim E., Hwang W., Shin S., Song J.J., Hohng S. Dynamic anchoring of the 3′-end of the guide strand controls the target dissociation of argonaute-guide complex. J. Am. Chem. Soc. 2013;135:16865–16871. doi: 10.1021/ja403138d. [DOI] [PubMed] [Google Scholar]
- 167.Dahlgren C., Zhang H.Y., Du Q., Grahn M., Norstedt G., Wahlestedt C., Liang Z. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 2008;36:e53. doi: 10.1093/nar/gkn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lisitskaya L., Aravin A.A., Kulbachinskiy A. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018;9:5165. doi: 10.1038/s41467-018-07449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sheng G., Gogakos T., Wang J., Zhao H., Serganov A., Juranek S., Tuschl T., Patel D.J., Wang Y. Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes. Nucleic Acids Res. 2017;45:9149–9163. doi: 10.1093/nar/gkx547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tal N., Sorek R. SnapShot: bacterial immunity. Cell. 2022;185:578. doi: 10.1016/j.cell.2021.12.029. [DOI] [PubMed] [Google Scholar]
- 171.Payne L.J., Todeschini T.C., Wu Y., Perry B.J., Ronson C.W., Fineran P.C., Nobrega F.L., Jackson S.A. Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types. Nucleic Acids Res. 2021;49:10868–10878. doi: 10.1093/nar/gkab883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tesson F., Hervé A., Mordret E., Touchon M., D’Humières C., Cury J., Bernheim A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022;13:2561. doi: 10.1038/s41467-022-30269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rocha E.P.C., Bikard D. Microbial defenses against mobile genetic elements and viruses: who defends whom from what? PLoS Biol. 2022;20:e3001514. doi: 10.1371/journal.pbio.3001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Noto M.J., Kreiswirth B.N., Monk A.B., Archer G.L. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 2008;190:1276–1283. doi: 10.1128/JB.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Doron S., Melamed S., Ofir G., Leavitt A., Lopatina A., Keren M., Amitai G., Sorek R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359:1008–1009. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rousset F., Depardieu F., Miele S., Dowding J., Laval A.-L., Lieberman E., Garry D., Rocha E.P.C., Bernheim A., Bikard D. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe. 2022;30:740–753.e5. doi: 10.1016/j.chom.2022.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ka D., Oh H., Park E., Kim J.H., Bae E. Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+ degradation. Nat. Commun. 2020;11:28–16. doi: 10.1038/s41467-020-16703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ofir G., Herbst E., Baroz M., Cohen D., Millman A., Doron S., Tal N., Malheiro D.B.A., Malitsky S., Amitai G., Sorek R. Antiviral activity of bacterial TIR domains via signaling molecules that trigger cell death. Nature. 2021;600:116–120. doi: 10.1038/s41586-021-04098-7. [DOI] [PubMed] [Google Scholar]
- 179.Koopal B., Potocnik A., Mutte S.K., Aparicio-Maldonado C., Lindhoud S., Vervoort J.J.M., Brouns S.J.J., Swarts D.C. Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA. Cell. 2022;185:1471–1486. doi: 10.1016/j.cell.2022.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.He R., Wang L., Wang F., Li W., Liu Y., Li A., Wang Y., Mao W., Zhai C., Ma L. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. (Camb.) 2019;55:13219–13222. doi: 10.1039/C9CC07339F. [DOI] [PubMed] [Google Scholar]
- 181.Wang L., He R., Lv B., Yu X., Liu Y., Yang J., Li W., Wang Y., Zhang H., Yan G., Mao W., Liu L., Wang F., Ma L. Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction for detection of SARS-CoV-2 and HPV. Talanta. 2021;227:122154. doi: 10.1016/j.talanta.2021.122154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xiao G., Fu X., Zhang J., Liu S., Wang Z., Ye T., Zhang G. Rapid and cost-effective screening of CRISPR/Cas9-induced mutants by DNA-guided Argonaute nuclease. Biotechnol. Lett. 2021;43:2105–2110. doi: 10.1007/s10529-021-03177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J., Carpenter E.L., van der Oost J., Bau H.H. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus Argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2020;48:e19. doi: 10.1093/nar/gkz1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shin S., Jung Y., Uhm H., Song M., Son S., Goo J., Jeong C., Song J.-J., Kim V.N., Hohng S. Quantification of purified endogenous miRNAs with high sensitivity and specificity. Nat. Commun. 2020;11:6033. doi: 10.1038/s41467-020-19865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Enghiad B., Zhao H. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017;6:752–757. doi: 10.1021/acssynbio.6b00324. [DOI] [PubMed] [Google Scholar]
- 186.Hunt E.A., Tamanaha E., Bonanno K., Cantor E.J., Tanner N.A. Profiling Thermus thermophilus Argonaute guide DNA sequence preferences by functional screening. Front. Mol. Biosci. 2021;8:670940. doi: 10.3389/fmolb.2021.670940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang F., Yang J., He R., Yu X., Chen S., Liu Y., Wang L., Li A., Liu L., Zhai C., Ma L. PfAgo-based detection of SARS-CoV-2. Biosens. Bioelectron. 2021;177:112932. doi: 10.1016/j.bios.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Filius M., Cui T.J., Ananth A.N., Docter M.W., Hegge J.W., van der Oost J., Joo C. High-speed super-resolution imaging using protein-assisted DNA-PAINT. Nano Lett. 2020;20:2264–2270. doi: 10.1021/acs.nanolett.9b04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gao F., Shen X.Z., Jiang F., Wu Y., Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2016;34:768–773. doi: 10.1038/nbt.3547. [DOI] [PubMed] [Google Scholar]
- 190.Hegge J.W., Swarts D.C., van der Oost J. Prokaryotic argonaute proteins: novel genome-editing tools? Nat. Rev. Microbiol. 2018;16:5–11. doi: 10.1038/nrmicro.2017.73. [DOI] [PubMed] [Google Scholar]
- 191.Burgess S., Cheng L, Gu F., Huang J., Huang Z., Lin S., Li J., Li W., Qin W., Sun Y., Songyang Z., Wei W., Wu Q., Wang H., Wang X., Xiong J.W., Xi J., Yang H., Zhou B., Zhang B. Questions about NgAgo. Protein Cell. 2016;7:913–915. doi: 10.1007/s13238-016-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lee S., Turchiano G., Ata H., Nowsheen S., Romito M., Lou Z., Ryu S.M., Ekker S.C., Cathomen T., Kim J.S. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2016;35:17–18. doi: 10.1038/nbt.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cai M., Si Y., Zhang J., Tian Z., Du S. Zebrafish embryonic slow muscle is a rapid system for genetic analysis of sarcomere organization by CRISPR/Cas9, but not NgAgo. Mar. Biotechnol. 2018;20:168–181. doi: 10.1007/s10126-018-9794-8. [DOI] [PubMed] [Google Scholar]
- 194.Khin N.C., Lowe J.L., Jensen L.M., Burgio G. No evidence for genome editing in mouse zygotes and HEK293T human cell line using the DNA-guided Natronobacterium gregoryi Argonaute (NgAgo) PLoS One. 2017;12:e0178768. doi: 10.1371/journal.pone.0178768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lee K.Z., Mechikoff M.A., Kikla A., Liu A., Pandolfi P., Fitzgerald K., Gimble F.S., Solomon K.V. NgAgo possesses guided DNA nicking activity. Nucleic Acids Res. 2021;49:9926–9937. doi: 10.1093/nar/gkab757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Waryah C.B., Moses C., Arooj M., Blancafort P. Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing. Methods Mol. Biol. 2018;1767:19–63. doi: 10.1007/978-1-4939-7774-1_2. [DOI] [PubMed] [Google Scholar]
- 199.Morgan S.L., Mariano N.C., Bermudez A., Arruda N.L., Wu F., Luo Y., Shankar G., Jia L., Chen H., Hu J.F., Hoffman A.R., Huang C.C., Pitteri S.J., Wang K.C. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 2017;8:15993. doi: 10.1038/ncomms15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pederson T. Repeated TALEs: Visualizing DNA sequence localization and chromosome dynamics in live cells. Nucleus. 2014;5:28–31. doi: 10.4161/nucl.28143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rauch S., He C., Dickinson B.C. Targeted m6A reader proteins to study epitranscriptomic regulation of single RNAs. J. Am. Chem. Soc. 2018;140:11974–11981. doi: 10.1021/jacs.8b05012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]