Abstract
Background
Observational studies have reported the association between tea consumption and the risk of lower respiratory tract infections (LRTIs). However, a consensus has yet to be reached, and whether the observed association is driven by confounding factors or reverse causality remains unclear.
Method
A two-sample Mendelian randomization (MR) analysis was conducted to determine whether genetically predicted tea intake is causally associated with the risk of common LRTI subtypes. Genome-wide association study (GWAS) from UK Biobank was used to identify single-nucleotide polymorphisms (SNPs) associated with an extra cup of tea intake each day. The summary statistics for acute bronchitis, acute bronchiolitis, bronchiectasis, pneumonia, and influenza and pneumonia were derived from the FinnGen project.
Results
We found that genetically predicted an extra daily cup of tea intake was causally associated with the decreased risk of bronchiectasis [odds ratio (OR) = 0.61, 95% confidence interval (CI) = 0.47–0.78, P < 0.001], pneumonia (OR = 0.90, 95% CI = 0.85–0.96, P = 0.002), influenza and pneumonia (OR = 0.91, 95% CI = 0.85–0.97, P = 0.002), but not with acute bronchitis (OR = 0.91, 95% CI = 0.82–1.01, P = 0.067) and acute bronchiolitis (OR = 0.79, 95% CI = 0.60–1.05, P = 0.100). Sensitivity analyses showed that no heterogeneity and pleiotropy could bias the results.
Conclusions
Our findings provided new evidence that genetically predicted an extra daily cup of tea intake may causally associated with a decreased risk of bronchiectasis, pneumonia, and influenza and pneumonia.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00394-022-02994-w.
Keywords: Lower respiratory tract infections, Tea, Mendelian randomization
Introduction
Lower respiratory tract infections (LRTIs), which include acute bronchitis, acute bronchiolitis, bronchiectasis, pneumonia and others, remain to be the leading cause of morbidity and mortality around the world, particularly affecting adults over 70 years and children under 5 years old [1, 2]. LRTIs are caused by various pathogens, such as bacteria (Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, etc.), viruses (respiratory syncytial virus [RSV], influenza virus, human coronavirus [hCoV], etc.), mycoplasma, chlamydia, and legionella [2]. The systematic analysis for the Global Burden of Disease Study 2016 estimated that there were 2,377,697 deaths due to LRTIs globally in 2016 [3]. Over the past decades, many countries have taken a range of actions to prevent and treat LRTIs, including the improvement of nutrition and hygiene, distribution and usage of influenza antibiotics and antivirals, and global promotion of Haemophilus influenzae type b and pneumococcal conjugate vaccines [4, 5]. Despite remarkable progress has been made with these measures, LRTIs still bring huge burden to individuals and society. As a result, sustained efforts and preventive strategies remain significant for managing LRTIs.
For thousands of years, plants have exerted an important role in maintaining health and improving the quality of life for humans [6]. Tea is the most popular beverages after water, consumed by more than two-thirds of the general population worldwide [7]. Two polyphenols, catechins and theaflavins, were found in abundance in green and black tea, and were reported to have multiple physiological activities, such as antibacterial, antiviral, antioxidative, and anticancer effects, etc. [8, 9]. Accumulating evidence has linked tea intake to various health benefits, including the prevention of tumors, cardiovascular diseases, and kidney stones [10, 11]. The association between tea consumption and LRTIs risk has also been investigated in many epidemiologic researches. However, these studies failed to reach a consensus. Several studies observed that tea consumption has preventive effects against LRTIs [12, 13], but others did not find similar link [14, 15]. Traditional epidemiologic studies are susceptible to underlying confounding factors and reverse causality, which can overestimate or underestimate the causal association between determinants and outcomes. Thus, it is uncertain whether the observed relationship between tea intake and LRTIs risk is causal.
Mendelian randomization (MR), which integrates summary data from genome-wide association studies (GWAS) to infer the causality in the putative exposure–outcome pathway, is an emerging genetic epidemiologic method [16, 17]. Since the genetic variations are randomly arranged in meiosis and fixed after fertilization, MR approach enables to overcome the shortcomings of traditional epidemiologic studies, including confounding factors, reverse causation and selection biases [18]. The method has never been used to explore the association between tea intake and LRTIs risk. Hence, we conducted a two-sample MR analysis to determinate whether tea consumption is causally correlated with the risk of the common subtypes of LRTIs.
Materials and methods
All analyses performed in the current study were based on publicly available GWAS summary statistics. No additional ethical statements or informed consent were required.
Data source
Single-nucleotide polymorphisms (SNPs) were selected as instrumental variables (IVs) for exposure (tea intake) and outcome (LRTIs) from the GWAS databases. SNPs associated with tea intake were derived from a large GWAS conducted by Neale Lab, which involves 349,376 samples of European ancestry. The GWAS adjusted for sex, age, age2, sex × age, sex × age2, and first twenty ancestry principal components. Information about tea intake were acquired by a dietary questionnaire. Participants were asked the following question at baseline: “How many cups of tea do you drink every day? (Include black and green tea)”. Details regarding the study design, data analyses, and ethical approval were described at https://github.com/Nealelab/UK_Biobank_GWAS.
SNPs associated with common LRTI subtypes were extracted from the data of FinnGen Release 5 (released to public on May 11, 2021), including acute bronchitis, acute bronchiolitis, bronchiectasis, pneumonia, and influenza and pneumonia. We limited LRTI data to samples of European ancestry to avoid potential bias aroused by population stratification. The dataset consists of 7338 acute bronchitis cases (208,689 controls), 976 acute bronchiolitis cases (208,689 controls), 1107 bronchiectasis cases (186,723 controls), 27,370 pneumonia cases (191,422 controls), and 29,924 influenza and pneumonia cases (188,868 controls). FinnGen project is a public–private partnership that integrates disease endpoint genetic data from the Finnish biobanks and the Finnish health registries. Detailed documentations were recorded at FinnGen website (https://finngen.gitbook.io/documentation/).
Instrumental SNPs selection
To ensure the correctness and robustness of the conclusion on the causal link between tea intake and the risk of LRTIs, a series of quality control steps were adopted to select valid IVs associated with tea consumption. First, SNPs had a genome-wide significant association (P < 5E-08) with tea intake were selected as IVs. Second, the minor allele frequency (MAF) threshold of selected SNPs should more than 0.01. Third, due to the presence of strong linkage disequilibrium (LD) among the selected SNPs may bias the results, the clumping process (r2 < 0.001, clumping distance = 10,000 kb) were carried out to eliminate the LD between the included IVs. Fourth, to guarantee that the impact of SNPs on exposure corresponds to the same allele as the impact on the outcome, palindromic SNPs with intermediate allele frequencies were excluded. Fifth, when the exposure-related SNPs were not available in the outcome dataset, the proxy SNPs significantly correlated with the variants of interest were used (r2 > 0.8).
MR assumptions
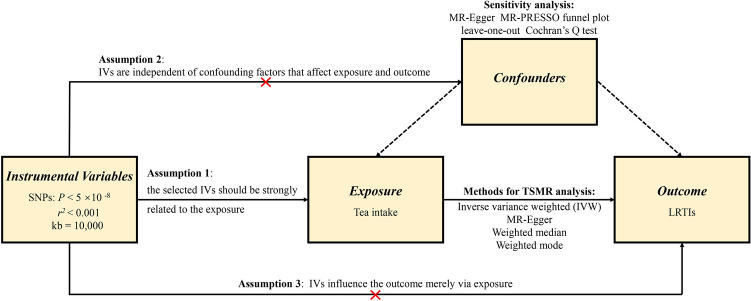
To minimize the bias on the results, the MR approach must obey three key assumptions. First, the selected IVs must be closely associated with the exposure. Herein, the strength of the correlation between IVs and exposure was evaluated by F statistic, which is presented as R2(n−k−1)/k(1−R2). In this formula, R2 represents the cumulative interpreted variance of selected SNPs on exposure, k refers to the number of selected SNPs and n is the sample size. If F value is more than 10, the correlation is considered strong enough to avoid bias caused by weak IVs. Second, IVs are independent of confounders that influence exposure and outcome. Third, IVs affect the outcome merely via exposure, that is, there is no horizontal pleiotropy effect between IVs and outcome. Figure 1 presents an overview of the design for the current study.
Fig. 1.
An overview of this Mendelian randomization study design
Effect size estimate
A two-sample MR analysis was conducted to evaluate the causal relationship between exposure (tea intake) and outcome (LRTIs). The inverse variance weighted (IVW) method was perceived as the primary analysis. IVW is essentially a meta-analysis approach, which translates into a weighted regression of outcome effects of IVs on exposure effects to acquire an overall estimate of causal effect, with a cut-off limit of zero. In the absence of horizontal pleiotropic effect, IVW can avoid the influence of confounding factors and acquire unbiased estimation [19]. In addition, three complementary methods were adopted to correct horizontal pleiotropy, despite at the expense of reduced statistical power. First, the MR-Egger regression was conducted. MR-Egger may be strongly affected by outlier genetic instruments, resulting in inaccurate estimates. However, even though all the selected IVs are invalid, MR-Egger method can still calculate unbiased estimates [20]. Second, the weighted median method was conducted, which combines data from multiple genetic instruments for consistency analysis by calculating a single weighted median estimator [21]. Third, the weighted mode method was performed, which is flexible for genetic variables that violate the pleiotropy hypothesis [22].
Sensitivity analysis
To ensure the reliability of the conclusion, six sensitivity analyses were adopted to verify whether heterogeneity and pleiotropy within the genetic variables could bias the MR results. First, the MR-Egger regression was applied to detect and adjust for the underlying horizontal pleiotropic effects among the selected IVs through the assessment of the intercept. Second, MR pleiotropy residual sum and outlier (MR-PRESSO) test was used to detect potential horizontal pleiotropy, and MR-PRESSO global test of heterogeneity was conducted to identify the underlying horizontal pleiotropy. Third, the asymmetry of funnel plots was also used as an indicator of horizontal pleiotropy. Fourth, the Cochran’s Q statistic was applied to quantify the heterogeneity across the selected genetic instruments. Fifth, the leave-one-out sensitivity analysis was implemented by deleting each SNP in turn, which can ensure that the MR estimates are not driven by certain strong SNPs. Finally, the PhenoScanner database was searched to check whether any of the SNPs were significantly associated (P < 5E-08) with other phenotypes at risk of affecting the five LRTIs subtypes independent of the tea consumption [23]. We further assessed the effect of removing these SNPs from the MR estimates to exclude potential pleiotropic effects.
A Bonferroni corrected P-value less than 0.01 (correcting for five outcomes) was defined as statistically significant. All data analyses were carried out using the “TwoSampleMR” and “MRPRESSO” packages of the R 4.0.3 software (www.r-project.org).
Results
Instrumental variables selection
A total of 45 SNPs (all P < 5E-08, r2 < 0.001) associated with tea consumption were identified, explaining 0.76% of the variance for tea intake. Supplementary Table S1 provided the detailed information of the 45 SNPs, including effect allele, other allele, beta, SE, P-value, F statistic, etc. The F statistic of these SNPs ranged from 30 to 408 with the median value of 40, which was greater than the conventional threshold of 10, indicating that the instrument bias was weak and cannot significantly affect the estimation of causal effects. Three SNPs (rs12901092, rs140775622, rs28676340) were excluded because they were not available in the outcome datasets, and four SNPs (rs11487328, rs2315024, rs397074, rs79413667) were excluded due to palindrome. Ultimately, a total of 38 SNPs with two proxy SNPs were selected as IVs for the following MR analysis.
MR analysis and sensitivity analysis
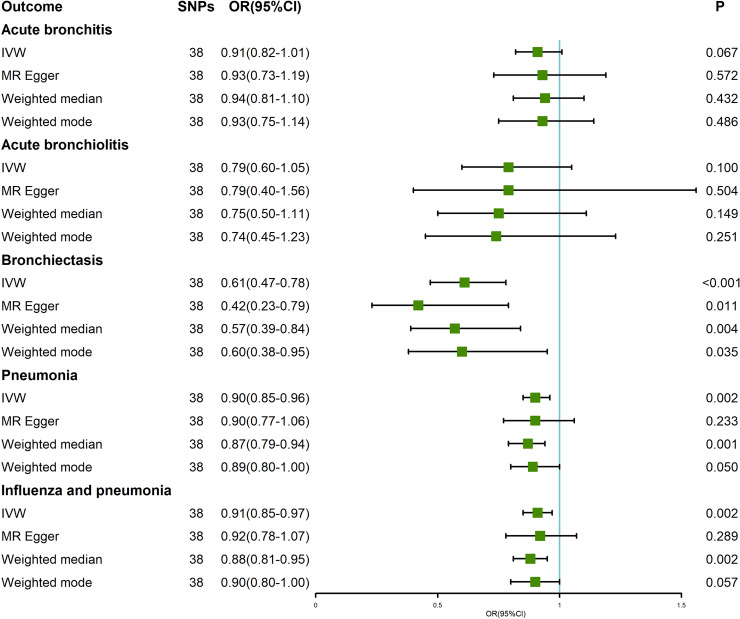
The results of IVW analyses revealed that genetically predicted an extra daily cup of tea intake was causally associated with the decreased risk of bronchiectasis (odds ratio (OR) = 0.61, 95% confidence interval (CI) = 0.47–0.78, P < 0.001), pneumonia (OR = 0.90, 95% CI = 0.85–0.96, P = 0.002), influenza and pneumonia (OR = 0.91, 95% CI = 0.85–0.97, P = 0.002) (Table 1, Fig. 2). Moreover, MR Egger, weighted median and weighted mode also obtained similar effect estimates, although wider CIs led to lower statistical power (Figs. 2, 3). The horizontal pleiotropy between IVs and outcomes was evaluated by MR-Egger regression, and the results indicated that there was weak evidence of horizontal pleiotropy (Table 1). Similarly, the MR-PRESSO test also identified no outlier SNPs or a horizontal pleiotropic effect of tea intake on the risk of bronchiectasis (P = 0.095), pneumonia (P = 0.121), and influenza and pneumonia (P = 0.117). The funnel plots showed symmetric results, by which the directional and horizontal pleiotropy was not significant (Supplementary Figure S1). The Cochran’s Q test suggested that there was no significant heterogeneity across the selected genetic instruments (P > 0.05; Table 1). The leave-one-out and forest plots showed that there was no single SNP driving the MR estimates, indicating that the results of the current MR analysis were robust (Supplementary Figure S2 and S3). Using the PhenoScanner tool, we found that two SNPs (rs199621380 and rs79217743) were significantly associated hematological traits (eg., monocyte count, eosinophil count and neutrophil percentage of white cells). It is known that changes in hematological parameters in patients with LRTIs are very common [24]. To avoid horizontal pleiotropy due to possible causal mechanistic associations between hematological traits and LRTIs subtypes, we removed these SNPs. Notably, the significant associations between tea intake and bronchiectasis, pneumonia and influenza and pneumonia did not change (Supplementary Figure S4).
Table 1.
MR estimates from each method of assessing the causal effects of tea intake on LRTIs risk
| Outcome | Nsnp | Methods | Beta | SE | OR (95% CI) | P-value | Horizontal pleiotropy | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Egger intercept | SE | P-value | Cochran’s Q | P-value | |||||||
| Bronchitis | 38 | MR Egger | − 0.07 | 0.13 | 0.93 (0.73–1.19) | 0.572 | − 0.0016 | 0.01 | 0.852 | 32.57 | 0.633 |
| IVW | − 0.09 | 0.05 | 0.91 (0.82–1.01) | 0.067 | |||||||
| Weighted median | − 0.06 | 0.08 | 0.94 (0.81–1.10) | 0.432 | |||||||
| Weighted mode | − 0.08 | 0.11 | 0.93 (0.75–1.14) | 0.486 | |||||||
| Bronchiolitis | 38 | MR Egger | − 0.24 | 0.348 | 0.79 (0.40–1.56) | 0.504 | 0.0002 | 0.02 | 0.994 | 22.16 | 0.966 |
| IVW | − 0.23 | 0.141 | 0.79 (0.60–1.05) | 0.100 | |||||||
| Weighted median | − 0.29 | 0.203 | 0.75 (0.50–1.11) | 0.149 | |||||||
| Weighted mode | − 0.30 | 0.256 | 0.74 (0.45–1.23) | 0.251 | |||||||
| Bronchiectasis | 38 | MR Egger | − 0.86 | 0.32 | 0.42 (0.23–0.79) | 0.011 | 0.0259 | 0.02 | 0.229 | 35.87 | 0.475 |
| IVW | − 0.50 | 0.13 | 0.61 (0.47–0.78) | < 0.001 | |||||||
| Weighted median | − 0.56 | 0.19 | 0.57 (0.39–0.84) | 0.004 | |||||||
| Weighted mode | − 0.51 | 0.23 | 0.60 (0.38–0.95) | 0.035 | |||||||
| Pneumonia | 38 | MR Egger | − 0.10 | 0.08 | 0.90 (0.77–1.06) | 0.233 | − 0.0001 | 0.01 | 0.984 | 48.12 | 0.085 |
| IVW | − 0.10 | 0.03 | 0.90 (0.85–0.96) | 0.002 | |||||||
| Weighted median | − 0.14 | 0.04 | 0.87 (0.79–0.94) | 0.001 | |||||||
| Weighted mode | − 0.11 | 0.05 | 0.89 (0.80–1.00) | 0.050 | |||||||
| Influenza and pneumonia | 38 | MR Egger | − 0.09 | 0.08 | 0.92 (0.78–1.07) | 0.289 | − 0.0009 | 0.01 | 0.868 | 48.30 | 0.083 |
| IVW | − 0.10 | 0.03 | 0.91 (0.85–0.97) | 0.002 | |||||||
| Weighted median | − 0.13 | 0.04 | 0.88 (0.81–0.95) | 0.002 | |||||||
| Weighted mode | − 0.11 | 0.06 | 0.90 (0.80–1.00) | 0.057 | |||||||
P < 0.01 was considered statistically significant
CI confidence interval, IVW inverse variance weighted, LRTI lower respiratory tract infection, MR Mendelian randomization, OR odds ratio, SNP single nucleotide polymorphism, SE standard error
Fig. 2.
Mendelian randomization estimates of tea consumption with lower respiratory tract infections risk
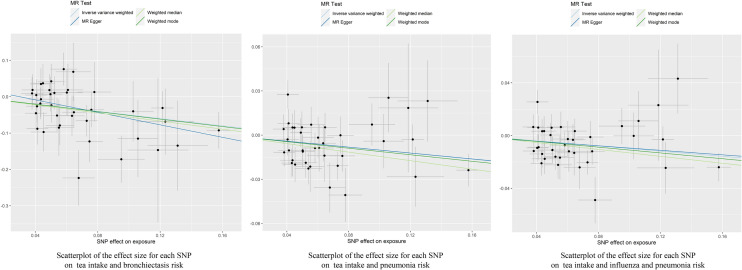
Fig. 3.
Scatterplot of the effect size for each SNP on tea consumption and the risk of bronchiectasis, pneumonia, and influenza and pneumonia
On the contrary, the results of IVW method revealed that genetically predicted tea intake was not causally associated with the risk of acute bronchitis (OR = 0.91, 95% CI = 0.82–1.01, P = 0.067) and acute bronchiolitis (OR = 0.79, 95% CI = 0.60–1.05, P = 0.100) (Table 1, Fig. 2). The methods of MR Egger, weighted median and weighted mode also provided consistent results (Table 1, Figs. 2, 4). The MR-Egger regression analyses indicated that there was weak evidence of horizontal pleiotropy (Table 1). Additionally, no outlier SNPs or a horizontal pleiotropic effect of tea intake on the risk of acute bronchitis (P = 0.629) and acute bronchiolitis (P = 0.986) were identified by the MR-PRESSO approaches. The funnel plots were also visually symmetric (Supplementary Figure S5). The Cochran’s Q test found no significant heterogeneity across the selected IVs (P > 0.05; Table 1). The leave-one-out and forest plots further proved the robustness of the MR results (Supplementary Figure S6 and S7).
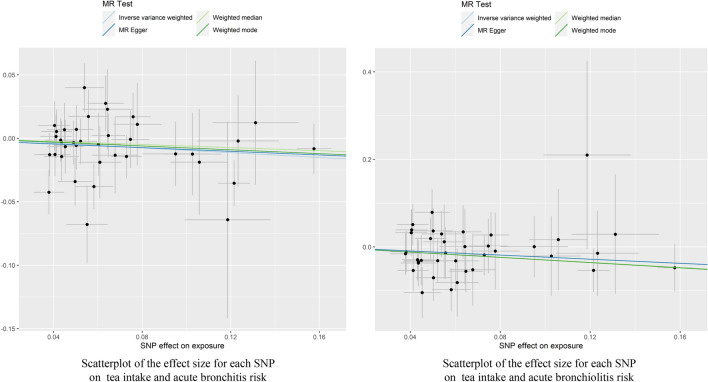
Fig. 4.
Scatterplot of the effect size for each SNP on tea consumption and the risk of acute bronchitis and acute bronchiolitis
Discussion
To the best of our knowledge, this is the first study to determinate the causal relationship between tea consumption and LRTIs risk. The main finding was that genetically predicted higher tea intake was causally associated with the decreased risk of bronchiectasis, pneumonia, and influenza and pneumonia. However, we did not find the association between tea intake and the risk of acute bronchitis and acute bronchiolitis.
The association between tea consumption and LRTIs risk remains controversial in previous epidemiologic studies. In a large cohort of 19,079 males and 21,493 males, Watanabe et al. found that tea consumption was associated with a lower risk of death from pneumonia in Japanese women [13]. In the CoPanFlu-France cohort of 1121 participants, tea consumption at least twice a week was a protective factor for influenza infection [25]. In an observational study, Park et al. reported that the consumption of 1–5 cups per day of tea may prevent influenza infection in children [12]. However, a case–control study conducted by Kondo et al. observed no association between pneumonia and tea consumption [15]. The discrepancy between these studies may be caused by the residual confounders, such as selection bias, recall bias, and detection bias.
In the current MR analysis, genetically predicted intake of one extra cup of tea per day was associated with 39% lower risk of bronchiectasis, 10% lower risk of pneumonia, and 9% lower risk of influenza and pneumonia. Several potential biological mechanisms may account for the inverse association between tea intake and these LRTIs. LRTIs are usually caused by various pathogenic microorganisms. Experimental studies have indicated that tea may be effective against many Gram-positive and gram-negative microorganisms as well as some viruses, fungi and parasites [26–28]. Polyphenols are the most important components in tea, which exert antimicrobial effects by damaging the bacterial cell membrane, inhibiting fatty acid synthesis, enzyme activity and inflammation [26]. Moreover, polyphenols can also bind to virus’s hemagglutinin molecule, thereby inhibiting the virus’s adsorption to host cells and preventing its assembly or maturation lysis [29]. Epigallocatechin gallate (EGCG), a major and highly bioactive catechin, was verified to reduce the infectivity of influenza A virus and influenza B virus in Madin–Darby canine kidney cells [27, 30]. ECGG can not only inhibit the migration of neutrophils through endothelial monolayer, thus decreasing vascular permeability [31], but also reduce neutrophil elastase, which is a proteolytic molecule involved in the body’s inflammatory response that has been associated with increased alveolar epithelial permeability [32]. The above provides a theoretical basis for the role of tea in preventing bronchiectasis, pneumonia and influenza, but the specific mechanisms remain to be further elucidated.
The strengths of this study are that it has large sample size, and it is the first MR analysis to evaluate the causal association between tea consumption and LRTIs risk. The application of MR approach reduces the interference of confounding factors and reverse causality on the results and is more convincing than observational studies. However, several shortcomings worth further consideration. First, the data of this study were derived from large-scale GWAS analyses, and the detailed information of the subjects and tea type (black or green tea) was not available, which makes it impossible to conduct stratification analyses or analyses adjusted for other covariables. Second, all the included subjects were of European ancestry, so the extrapolation of the findings to other ethnic groups may be limited. Third, there might be overlapping participants in the exposure and outcome studies, but the degree of sample overlap was difficult to estimate. Fortunately, the strong IVs (F statistic much larger than 10) used in this study minimizes the bias due to sample overlap. Finally, standard MR method assumes a linear relationship between exposure and outcome, so the non-linear association and threshold effect between tea intake and LRTIs cannot be detected.
In summary, our study found that genetically predicted an extra daily cup of tea intake may causally associated with the reduced risk of bronchiectasis, pneumonia and influenza and pneumonia. The finding of this study provided new evidence for the prevention and treatment of these LRTIs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the UK Biobank for providing summary statistics. We also want to acknowledge the participants and investigators of FinnGen study.
Author contributions
YC, JS: conceptualization, methodology, software; YC, JS: data curation, writing-original draft preparation; YW: visualization, investigation; MN, YD, XS, XW: supervision; TZ: software; FP: validation; ZT: writing—reviewing and editing.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81773514, 82073655) and the funds for academic and technical leaders in Anhui province (2017D140) and Clinical Medicine Discipline construction project of Anhui Medical University (2021lcxk043).
Declarations
Conflict of interest
The authors have no conflicts of interest or sources of funding to declare.
Footnotes
Yuting Chen and Jiran Shen contributed equally to this work and should be considered co-first author.
Contributor Information
Faming Pan, Email: famingpan@ahmu.edu.cn.
Zhiru Tang, Email: tang_zhiru2004668@sina.com.
References
- 1.Langelier C, Kalantar KL, Moazed F, Wilson MR, Crawford ED, Deiss T, Belzer A, Bolourchi S, Caldera S, Fung M, Jauregui A, Malcolm K, Lyden A, Khan L, Vessel K, Quan J, Zinter M, Chiu CY, Chow ED, Wilson J, Miller S, Matthay MA, Pollard KS, Christenson S, Calfee CS, DeRisi JL. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA. 2018;115(52):E12353–e12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll KC, Adams LL. Lower respiratory tract infections. Microbioly Spectr. 2016 doi: 10.1128/microbiolspec.DMIH2-0029-2016. [DOI] [PubMed] [Google Scholar]
- 3.(2018) Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18(11):1191–1210. 10.1016/s1473-3099(18)30310-4 [DOI] [PMC free article] [PubMed]
- 4.Murdoch DR, Howie SRC. The global burden of lower respiratory infections: making progress, but we need to do better. Lancet Infect Dis. 2018;18(11):1162–1163. doi: 10.1016/s1473-3099(18)30407-9. [DOI] [PubMed] [Google Scholar]
- 5.Kamata K, Thein KN, Di Ja L, Win NC, Win SMK, Suzuki Y, Ito A, Osada H, Chon I, Phyu WW, Aizawa Y, Ikuse T, Ota T, Kyaw Y, Tin HH, Shobugawa Y, Watanabe H, Saito R, Saitoh A. Clinical manifestations and outcome of viral acute lower respiratory infection in hospitalised children in Myanmar. BMC Infect Dis. 2022;22(1):350. doi: 10.1186/s12879-022-07342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemler EC, Hu FB. Plant-based diets for personal, population, and planetary health. Adv Nutr (Bethesda, Md) 2019;10(Suppl_4):S275–S283. doi: 10.1093/advances/nmy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat Rev Cancer. 2009;9(6):429–439. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mhatre S, Srivastava T, Naik S, Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomed Int J Phytother Phytopharmacol. 2021;85:153286. doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neill EJ, Termini D, Albano A, Tsiani E. Anti-cancer properties of Theaflavins. Molecules (Basel, Switzerland) 2021 doi: 10.3390/molecules26040987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trevisanato SI, Kim YI. Tea and health. Nutr Rev. 2000;58(1):1–10. doi: 10.1111/j.1753-4887.2000.tb01818.x. [DOI] [PubMed] [Google Scholar]
- 11.Khan N, Mukhtar H. Tea polyphenols in promotion of human health. Nutrients. 2018 doi: 10.3390/nu11010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park M, Yamada H, Matsushita K, Kaji S, Goto T, Okada Y, Kosuge K, Kitagawa T. Green tea consumption is inversely associated with the incidence of influenza infection among schoolchildren in a tea plantation area of Japan. J Nutr. 2011;141(10):1862–1870. doi: 10.3945/jn.110.137547. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe I, Kuriyama S, Kakizaki M, Sone T, Ohmori-Matsuda K, Nakaya N, Hozawa A, Tsuji I. Green tea and death from pneumonia in Japan: the Ohsaki cohort study. Am J Clin Nutr. 2009;90(3):672–679. doi: 10.3945/ajcn.2009.27599. [DOI] [PubMed] [Google Scholar]
- 14.Ide K, Yamada H, Matsushita K, Ito M, Nojiri K, Toyoizumi K, Matsumoto K, Sameshima Y. Effects of green tea gargling on the prevention of influenza infection in high school students: a randomized controlled study. PLoS One. 2014;9(5):e96373. doi: 10.1371/journal.pone.0096373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Suzuki K, Washio M, Ohfuji S, Adachi S, Kan S, Imai S, Yoshimura K, Miyashita N, Fujisawa N, Maeda A, Fukushima W, Hirota Y. Association between coffee and green tea intake and pneumonia among the Japanese elderly: a case-control study. Sci Rep. 2021;11(1):5570. doi: 10.1038/s41598-021-84348-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 19.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14(10):577–590. doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45(6):1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics (Oxford, England) 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Q, Wen X, Wang L, Han X, Shen Y, Cao J, Peng Q, Xu J, Zhao L, He J, Yuan H. Role of hematological parameters in the diagnosis of influenza virus infection in patients with respiratory tract infection symptoms. J Clin Lab Anal. 2020;34(5):e23191. doi: 10.1002/jcla.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delabre RM, Lapidus N, Salez N, Mansiaux Y, de Lamballerie X, Carrat F. Risk factors of pandemic influenza A/H1N1 in a prospective household cohort in the general population: results from the CoPanFlu-France cohort. Influenza Other Respir Viruses. 2015;9(1):43–50. doi: 10.1111/irv.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reygaert WC. The antimicrobial possibilities of green tea. Front Microbiol. 2014;5:434. doi: 10.3389/fmicb.2014.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmann J, Buer J, Pietschmann T, Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br J Pharmacol. 2013;168(5):1059–1073. doi: 10.1111/bph.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falcinelli SD, Shi MC, Friedlander AM, Chua J. Green tea and epigallocatechin-3-gallate are bactericidal against Bacillus anthracis. FEMS Microbiol Lett. 2017 doi: 10.1093/femsle/fnx127. [DOI] [PubMed] [Google Scholar]
- 29.Imanishi N, Tuji Y, Katada Y, Maruhashi M, Konosu S, Mantani N, Terasawa K, Ochiai H. Additional inhibitory effect of tea extract on the growth of influenza A and B viruses in MDCK cells. Microbiol Immunol. 2002;46(7):491–494. doi: 10.1111/j.1348-0421.2002.tb02724.x. [DOI] [PubMed] [Google Scholar]
- 30.Song JM, Park KD, Lee KH, Byun YH, Park JH, Kim SH, Kim JH, Seong BL. Biological evaluation of anti-influenza viral activity of semi-synthetic catechin derivatives. Antiviral Res. 2007;76(2):178–185. doi: 10.1016/j.antiviral.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Donà M, Dell'Aica I, Calabrese F, Benelli R, Morini M, Albini A, Garbisa S. Neutrophil restraint by green tea: inhibition of inflammation, associated angiogenesis, and pulmonary fibrosis. J Immunol (Baltimore, Md: 1950) 2003;170(8):4335–4341. doi: 10.4049/jimmunol.170.8.4335. [DOI] [PubMed] [Google Scholar]
- 32.Xiaokaiti Y, Wu H, Chen Y, Yang H, Duan J, Li X, Pan Y, Tie L, Zhang L, Li X. EGCG reverses human neutrophil elastase-induced migration in A549 cells by directly binding to HNE and by regulating α1-AT. Sci Rep. 2015;5:11494. doi: 10.1038/srep11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.