Abstract
The role of salicylic acid in iron metabolism was examined in two wild-type strains (mc2155 and NCIMB 8548) and three mutant strains (mc21292 [lacking exochelin], SM3 [lacking iron-dependent repressor protein IdeR] and S99 [a salicylate-requiring auxotroph derived in this study]) of Mycobacterium smegmatis. Synthesis of salicylate in SM3 was derepressed even in the presence of iron, as was synthesis of the siderophores exochelin, mycobactin, and carboxymycobactin. S99 was dependent on salicylate for growth and failed to grow with the three ferrisiderophores, suggesting that salicylate fulfills an additional function(s) other than being a precursor of mycobactin and carboxymycobactin. Salicylic acid at 100 μg/ml repressed the formation of a 29-kDa cell envelope protein (putative exochelin receptor protein) in S99 grown both iron deficiently and iron sufficiently. In contrast, synthesis of this protein was affected only under iron-limited conditions in the parent strain, mc2155, and remained unaltered in SM3, suggesting an interaction between the IdeR protein and salicylate. Thus, salicylate may also function as a signal molecule for recognition of cellular iron status. Growth of all strains and mutants with p-aminosalicylate (PAS) at 100 μg/ml increased salicylate accumulation between three- and eightfold under both iron-limited and iron-sufficient growth conditions and decreased mycobactin accumulation by 40 to 80% but increased carboxymycobactin accumulation by 50 to 55%. Thus, although PAS inhibited salicylate conversion to mycobactin, presumptively by blocking salicylate AMP kinase, PAS also interferes with the additional functions of salicylate, as its effect was heightened in S99 when the salicylate concentration was minimal.
Mycobacterium smegmatis, a saprophytic mycobacterium, secretes two extracellular siderophores (exochelin and carboxymycobactin) and has an intracellular siderophore termed mycobactin (28, 30). De Voss et al. (8) demonstrated the importance of the siderophores mycobactin and carboxymycobactin in M. tuberculosis by showing that a mutant incapable of producing these two siderophores failed to grow within macrophages. Carboxymycobactin is structurally related to mycobactin but is probably not derived from it (2). While carboxymycobactin is the sole extracellular siderophore of pathogenic mycobacteria (30), it is only a minor one in saprophytic mycobacteria, which use exochelin as the major siderophore for iron acquisition (28). Salicylic acid is produced extracellularly by most mycobacteria in greatly increased quantities (up to 17 μg/ml) when grown under iron-deficient conditions (28, 33) as opposed to iron-sufficient conditions. Although salicylate is a precursor of mycobactin (31), mycobactin could not spare the requirement for salicylate in a salicylate-requiring auxotrophic mutant of M. smegmatis (32), suggesting that salicylate fulfills a second but unknown role in this bacterium.
Salicylate and its derivatives are important as analgesics, antipyretics, anticoagulants, and anti-inflammatory drugs (15, 41), and in addition, salicylate is also important in plants, where it induces flowering and is involved in resistance to systemic diseases through multiple signal transduction pathways (14). The role of salicylate in iron metabolism in bacteria is much less clearly defined. Salicylate appears to be synthesized by the members of only four genera: Pseudomonas, Azospirillum, Yersinia, and Mycobacterium. For the former two genera, it has been suggested that salicylate acts as a siderophore in its own right (21, 35, 37, 40), although this can only happen in the absence of competing ions such as phosphate, which quickly precipitate iron from ferrisalicylate and render the iron inaccessible for uptake into cells (34). Thus, a role for salicylate as a siderophore must be very limited and could not, in any case, apply to pathogenic bacteria, which would encounter high concentrations of phosphate in host tissues (1).
Salicylate also induces a multiple antibiotic resistance (mar) operon in Escherichia coli (5): it binds to the MarR (repressor) protein and thereby decreases its affinity for operator sites containing the MarR recognition sequence in vitro (18). Salicylate also inactivates EmrR, a MarR-like repressor of an unrelated promoter involved in efflux of several hydrophobic antibacterials in E. coli (38). The elucidation of the sequence of the M. tuberculosis genome has identified two proteins similar to MarA of E. coli (6), suggesting the operation of a mar system in mycobacteria. Salicylate therefore acts as a regulator of metabolism in a number of diverse systems, although its function in mycobacteria remains unclear.
In the present study, the function of salicylic acid has been examined in wild-type M. smegmatis mc2155 and mutant strains SM3, mc21292, and S99. Some studies were also performed with a second wild-type strain, NCIMB 8548. The results confirm the central role of salicylate as the precursor of mycobactin and carboxymycobactin and, in addition, point to an as yet unidentified role(s) for it in the assimilation of iron.
MATERIALS AND METHODS
Strains and growth conditions.
The phenotypic characteristics and sources of all of the strains used in the present study are given in Table 1. mc2155 was used as the principal wild-type strain. An additional wild-type strain, NCIMB 8548, was used in some experiments. The mutant SM3 was kindly supplied by Issar Smith, Public Health Research Institute, New York, N.Y., and strain mc21292 was kindly provided by William R. Jacobs, Jr., Howard Hughes Medical Institute, Albert Einstein College of Medicine, Bronx, N.Y. All strains were routinely grown in minimal medium treated for iron removal (33) and containing the following (grams per liter): KH2PO4, 5; glycerol, 10; asparagine, 5 (pH 7.6). Prior to inoculation, the medium was supplemented with the following (micrograms per milliliter): Zn2+, 0.45; Mn2+, 0.1; Mg2+, 40; Fe2+, 0.05 (for iron-deficient growth); Fe2+, 2.0 (for iron-sufficient growth). Cultures (100 ml) were grown for 5 days at 37°C with shaking; cell dry weights and exochelin, mycobactin, and carboxymycobactin were estimated as previously described (30). Salicylic acid and p-aminosalicylic acid (PAS) were obtained from Sigma. The salicylic acid-requiring mutant S99 was isolated from wild-type strain mc2155 by N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis (13). Transposon mutagenesis was performed by using a temperature-sensitive system (T. Parish and N. G. Stoker, personal communication).
TABLE 1.
Phenotypic characteristics of the various strains of M. smegmatis used in the present study
| Strain | Phenotype | Source or reference |
|---|---|---|
| NCIMB 8548 | Wild type | National Collection of Industrial and Marine Bacteria |
| mc2155 | Efficient plasmid transformation (Ept) mutant (wild-type parent of mutants listed below) | 36 |
| SM3 | Iron-dependent repressor (IdeR)-deficient mutant | 10 |
| mc21292 | Exochelin-deficient mutant | 11 |
| S99 | Salicylic acid-requiring mutant | This study |
Assay of salicylic acid.
Cells were harvested by centrifugation. Culture supernatants were acidified with 10 M H2SO4 to pH 1.5 to 2.0. The medium was filtered and extracted three times with 0.5 volume of chloroform; the extract was then washed with double-distilled water, and the solvent was allowed to evaporate under reduced pressure. The residue was dissolved in methanol-water (70:30, vol/vol)–3% (vol/vol) acetic acid and analyzed by high-pressure liquid chromatography (HPLC) using a Lichrosorb RP-18 reverse-phase column (250 by 3.2 mm) at room temperature. Salicylic acid was eluted with a linear gradient of methanol-acetic acid-water (50:0.1:50, by volume) to methanol-water (70:30, vol/vol) running at 0.5 ml/min over 20 min and then holding at the final concentration for a further 20 min. All solvents were continuously degassed with helium. The eluate was continuously monitored at 296 nm. The efficiency of extraction for all concentrations of salicylic acid was 50 to 55%. Salicylic acid concentration was determined from a standard curve generated by using 2.5 to 100 μg of salicylic acid; the detector response is linear within these limits, and correction was applied for the extraction efficiency.
Separation of salicylic acid and PAS.
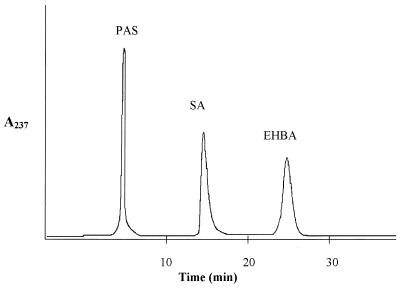
Salicylic acid and PAS were extracted as described above from the supernatants of the cultures grown in the presence of PAS and separated by HPLC at room temperature using KH2PO4-H3PO4 (0.01 mol/liter, pH 2.3)-acetonitrile-methanol (70:25:5, by volume). The elution rate was 0.5 ml/min, and eluates were monitored by measuring A237 (see Fig. 2). Salicylic acid and PAS concentrations were determined from the standard graphs generated by measuring A237 and using 2.5 to 100 μg of salicylic acid and PAS; the detector response is linear within these limits for both compounds.
FIG. 2.
Separation of salicylic acid (SA) and PAS by HPLC with ethyl 4-hydroxybenzoic acid (EHBA) as the internal standard. Eluants were monitored by measuring A237.
SDS-PAGE of membrane proteins of M. smegmatis.
Cell envelope proteins were extracted from cultures grown under conditions of iron deficiency or iron sufficiency after ultrasonication for 10 30-s periods with 15-s cooling intervals using a Dawe Soniprobe, type 7533A, as previously described (9). The proteins were analyzed by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) (16).
RESULTS AND DISCUSSION
Isolation and characterization of a salicylic acid-requiring mutant.
An attempt was made to isolate a salicylic acid-requiring mutant by transposon mutagenesis using a temperature-sensitive system (T. Parish and N. G. Stoker, personal communication), but for unknown reasons, no such mutant was isolated, although around 25,000 colonies were screened and the system did produce other auxotrophs at a frequency of 2.3 × 10−4. Thus, N-methyl-N′-nitro-N-nitrosoguanidine was used to mutagenize M. smegmatis mc2155 as previously described (13). Over 20,000 colonies were screened by replica plating on solid medium containing 25 μg of salicylic acid per ml and a single salicylic acid-requiring mutant, designated S99, was found among 16 putative mutants.
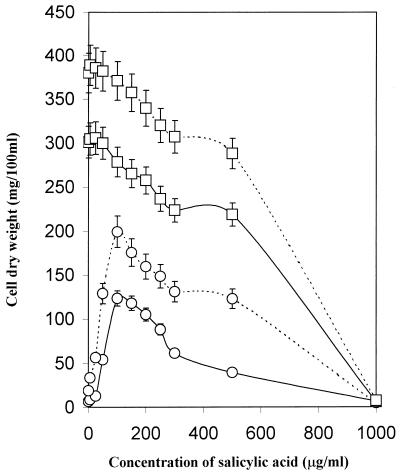
Growth of S99 was directly proportional to the concentration of salicylic acid added to the growth medium, and optimal growth was achieved with 100 μg of salicylic acid/ml (Fig. 1). A decline in growth was noted with higher concentrations of salicylic acid under both low- and high-iron conditions. However, the growth of this mutant was only 60% of that observed in the parent, even in the presence of salicylic acid. The growth of parent strain mc2155 was not affected by up to 100 μg of salicylic acid/ml under iron-deficient or iron-sufficient growth conditions. Salicylic acid at 1,000 μg/ml, however, was lethal to both of the strains, irrespective of the iron status (Fig. 1). This mutant produced all of the siderophores (exochelin, mycobactin, and carboxymycobactin) when grown with salicylic acid (see Table 3), suggesting that the mutation had not affected the conversion of salicylic acid to mycobactin and carboxymycobactin.
FIG. 1.
Responses of wild-type M. smegmatis mc2155 (□) and salicylate-requiring mutant S99 (○) to salicylate under iron-insufficient (———) and iron-sufficient (–––) conditions.
TABLE 3.
Production of siderophores and salicylic acid by different strains of M. smegmatis in the presence and absence of PAS at 100 μg/mla
| Strain | Iron in medium | Cell dry wt (mg/100 ml)
|
Exochelin (mg/100 ml)
|
Salicylic acidb (μg/100 ml)
|
Mycobactin (mg/g of cell dry wt)
|
Carboxymycobactinb (μg/100 ml)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MMc | MM + PAS | MM | MM + PAS | MM | MM + PAS | MM | MM + PAS | MM | MM + PAS | ||
| NCIMB 8548 (wild type) | Low | 326 | 297 | 33 | 30 | 154 | 808 | 40 | 24 | 558 | 862 |
| High | 417 | 302 | 0.7 | 0.6 | 69 | 370 | 0.4 | 0.4 | 65 | 69 | |
| mc2155 (wild type) | Low | 295 | 265 | 30 | 27 | 126 | 772 | 35 | 15 | 496 | 754 |
| High | 436 | 307 | 0.7 | 0.6 | 72 | 290 | 0.4 | 0.4 | 72 | 77 | |
| SM3 (IdeR− mutant) | Low | 309 | 275 | 18 | 17 | 694 | 4,980 | 10 | 2 | 684 | 1,070 |
| High | 410 | 297 | 4 | 4 | 1,520 | 4,706 | 3 | 0.7 | 424 | 682 | |
| mc21292 (Exo− mutant) | Low | 297 | 203 | 0 | 0 | 494 | 7,762 | 17.7 | 3 | 774 | 1,170 |
| High | 423 | 265 | 0 | 0 | 157 | 2,436 | 0.5 | 0.2 | 154 | 245 | |
| S99 (salicylate-requiring auxotroph) | Low | 119 | NGd | 8 | NG | NQe | NG | 16 | NG | 124 | NG |
| High | 240 | NG | 0.4 | NG | NQ | NG | 0.2 | NG | 35 | NG | |
Cultures were grown for 5 days in glycerol-asparagine medium (100 ml), pH 7.6, under low-iron (0.05 μg of Fe/ml) and high-iron (2 μg of Fe/ml) conditions and analyzed for the characteristics shown.
The efficiency of extraction for salicylic acid and carboxymycobactin was 50 to 55%, and the equivalent correction factor has been applied for these values. S99 was grown in medium containing 100 μg of salicylic acid/ml. Thus, salicylic acid from this strain could not be quantified.
MM, minimal medium.
NG, no growth.
NQ, not quantified.
To characterize the auxotrophic nature of salicylate-requiring mutant strain S99, the growth medium was supplemented at 5 μg/ml with various siderophores that normally occur in wild-type M. smegmatis. While exochelin and mycobactin failed to restore the growth of S99 under both low- and high-iron conditions, there was partial (50%) restoration of growth with carboxymycobactin or citrate, but only under iron-sufficient conditions and not under iron-deficient growth conditions (data not shown). A previous study (32) prompted us to assess the influence of 2,3- and 2,4-dihydroxybenzoic acids, which at 100 μg/ml restored the growth of this present mutant, but only under high-iron conditions (data not shown).
Mutant strain S99 failed to grow with any of the siderophores, added as ferric complexes, in the absence of salicylic acid and grew only very slightly when combinations of the siderophores were used (Table 2). The growth of the mutant with ferrisiderophores as the sole source of iron was significantly enhanced only when salicylic acid was added. A combination of 2,3- or 2,4-dihydroxybenzoic acid or citrate and ferrimycobactin (sole source of iron) failed to support the growth of S99 (data not shown). Parent strain mc2155 grew better with the individual ferrisiderophores than it did in iron-deficient medium, indicating that the additional iron in the form of ferrisiderophores contributed to the extra growth of the cells (Table 2).
TABLE 2.
Effects of mycobacterial ferrisiderophores as the sole sources of iron on the growth of mutant strain S99 and parent strain mc2155 in the presence and absence of salicylic acida
| Source(s) of iron | Cell dry wt (mg/100 ml)
|
|||
|---|---|---|---|---|
| S99
|
mc2155
|
|||
| No SA | 100 SAb | No SA | 100 SA | |
| MM (no iron) | 6 | 12 | 39 | 43 |
| MM + A | 8 | 119 | 240 | 252 |
| MM + B | 11 | 207 | 434 | 443 |
| MM + C | 14 | 113 | 301 | 324 |
| MM + D | 12 | 106 | 412 | 432 |
| MM + E | 10 | 117 | 393 | 414 |
| MM + C + D | 23 | 135 | 425 | 437 |
| MM + C + E | 20 | 143 | 413 | 421 |
| MM + D + E | 25 | 139 | 419 | 429 |
| MM + C + D + E | 40 | 183 | 446 | 463 |
Cultures were grown for 5 days in glycerol-asparagine medium (100 ml), pH 7.6, in the presence of various iron sources. MM, minimal medium; SA, salicylic acid; A, 0.05 μg of Fe/ml (iron-deficient medium); B, 2 μg of Fe/ml (iron-sufficient medium); C, 5 μg of ferriexochelin/ml (equivalent to 0.32 μg of iron/ml); D, 5 μg of ferrimycobactin/ml (equivalent to 0.32 μg of iron/ml); E, 5 μg of ferricarboxymycobactin/ml (equivalent to 0.32 μg of iron/ml).
100 SA, salicylic acid at 100 μg/ml.
The results in Table 2 confirm the earlier finding that mycobactin could not replace or spare the salicylate requirement of a salicylate-requiring mutant (32). In addition, the present work shows that neither mycobactin nor carboxymycobactin, nor exochelin, can substitute for salicylate, either individually or in combination. If salicylate acted simply as an extracellular siderophore, as had been suggested with Pseudomonas species and Azospirillum lipoferum (21, 35, 37, 40), then one of the other mycobacterial siderophores should have been able to substitute for it, and if salicylate was needed only for mycobactin and/or carboxymycobactin synthesis, then either or both of these should have been adequate substitutes. As this did not occur, we conclude that salicylate must fulfill a second function that does not involve its further metabolism, as no compound containing a salicylate moiety, other than mycobactin or carboxymycobactin, has been recognized in mycobacteria (28, 31). The evidence that salicylate acts as a siderophore for the solubilization and uptake of Fe(III) is, however, extremely tenuous: in the presence of phosphate ions, any ferric salicylate is rapidly converted to the insoluble ferric phosphate, which is not transportable (34). However, synthesis of salicylate is considerably increased during iron-deficient growth, as opposed to iron-sufficient growth. Moreover, this increased synthesis occurs as an early response to the onset of iron deprivation (26), thus firmly implying that its function, besides that of a mycobactin precursor, is connected with iron metabolism.
Iron regulation in mycobacteria.
The production of salicylic acid and the siderophores is negatively regulated by iron in the two wild-type strains (Table 3). Dusserget et al. (10) have shown that the synthesis of mycobactin and exochelin is regulated by the IdeR protein when activated by iron and that the synthesis of these siderophores is derepressed in mutant strain SM3. We have extended these observations by showing that the synthesis of salicylic acid and carboxymycobactin is also derepressed in the IdeR-deficient mutant, suggesting that IdeR also mediates the regulation of their respective biosyntheses by iron. However, the pattern of regulation of salicylate synthesis differed from that of the siderophores since the production of salicylate in this mutant under high-iron conditions, unlike that of its parent strain (mc2155), was twice as high as under low-iron conditions (Table 3). In fact, the level of salicylic acid in the IdeR mutant was 10 times the concentration seen in the wild-type cultures grown under iron-deficient conditions. These results suggest that repression by IdeR is probably not the sole control mechanism for the synthesis of salicylic acid. In contrast, the siderophore levels under high-iron conditions were significantly lower than those found in iron-deficient cultures, suggesting that the derepression was only partial. In addition, the concentrations of exochelin and mycobactin were lower in SM3 than in the wild-type strain grown under low-iron conditions, in agreement with the findings of Dusserget et al. (10). The concentration of carboxymycobactin was similar to that found in the parent strain.
These results suggest that IdeR does not account for all adaptive responses to iron starvation and that additional mechanisms control the expression of the various components of the mycobacterial iron uptake machinery. In E. coli, expression of fepA, encoding the receptor for colicin B, which is necessary for ferric enterochelin uptake, has been shown to be partially derepressed in fur mutants while other ferric uptake genes are constitutively expressed (7), suggesting that fepA is under the control of an additional regulator. It is now clear that there are complex regulatory mechanisms for iron acquisition in some bacteria; for instance, multiple regulatory elements, in addition to Fur, have been described in Campylobacter jejuni (39), Vibrio anguillarum, P. putida, and P. aeruginosa (7). This view is further supported by the identification of multiple iron-dependent regulators in the M. tuberculosis genome (6, 44).
Interestingly, salicylate synthesis was also considerably increased in the exochelin-negative mutant mc21292 under both iron-sufficient and iron-deficient conditions (Table 3), although salicylate was at its highest concentration in cells grown under iron-deficient conditions. This mutant also produced 55% more carboxymycobactin than its parent, allowing it to compensate for the lack of exochelin, and this, in turn, led to a decrease in the mycobactin levels in the cells. The increased production of carboxymycobactin by mc21292 also explains how it is able to grow in the absence of exochelin (see also reference 11). Mycobactin and carboxymycobactin are derived from the same, as yet unidentified, precursor. The high concentration of salicylate present in the mc21292 mutant suggests that the cells now have to rely entirely on the secondary routes of iron uptake in the absence of exochelin. Higher concentrations of salicylate may therefore be necessary to ensure maximum synthesis of carboxymycobactin, as well as optimal assimilation of iron into the cells.
PAS toxicity and resistance mechanism.
PAS is a potent antitubercular drug (3, 43). While M. tuberculosis is much more sensitive to inhibition by PAS than is M. smegmatis, which can tolerate high concentrations of PAS, its site of action remains unknown. We have now investigated the effect of PAS on the synthesis of siderophores and salicylic acid in M. smegmatis. All strains were grown with 100 μg of PAS/ml and without PAS, and the cultures were then analyzed for formation of the siderophores. Salicylic acid was separable from PAS by HPLC (Fig. 2) and could therefore be quantified in its presence.
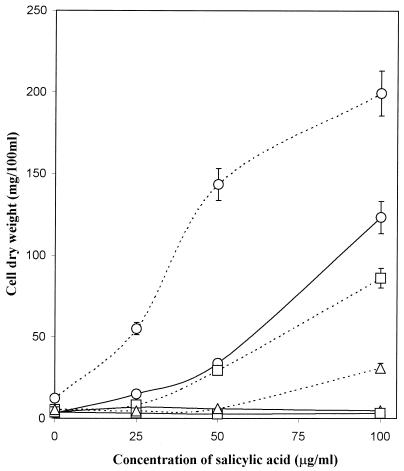
PAS at 100 μg/ml prevented the growth of mutant S99 on salicylic acid under both low- and high-iron conditions (Table 3). In contrast, the growth of all of the other strains was only slightly affected (25 to 35% inhibition) and a 3- to 10-fold salicylic acid increase occurred in all strains that were able to grow in the presence of PAS, irrespective of the iron status of the medium. The production of carboxymycobactin increased by 50 to 55% in the two wild-type strains, as well as in the mutants SM3 and mc21292 in the presence of PAS, whereas the mycobactin levels decreased (40 to 80%) in all of these cells under iron-deficient growth conditions. PAS (1 μg/ml) completely inhibited the growth of mutant strain S99 under iron-deficient growth conditions (Fig. 3). An increase in the concentration of salicylic acid in the medium counteracted the toxic effects of PAS partially, but only under high-iron conditions. The present results support and extend the initial proposals that PAS is an antimetabolite of salicylic acid (3, 29). The overproduction of salicylate in M. smegmatis in the presence of PAS (Table 3) indicates that PAS probably acts as an analog of salicylate and blocks the action of salicylate kinase, which forms salicyloyl-AMP from salicylate as the first step in mycobactin and carboxymycobactin synthesis (25). However, this inhibition was not complete, as indicated by the continued synthesis of mycobactin and carboxymycobactin under iron-restricted conditions (Table 3), and although mycobactin formation was decreased by this action of PAS, the synthesis of carboxymycobactin was increased. Overall, however, there was a diminished flux of salicylate along this pathway to the common precursor of mycobactin and carboxymycobactin. Extremely high concentration of salicylate accumulated in both SM3 (IdeR mutant) and mc21292 (exochelin-deficient mutant) after their growth in the presence of PAS, reaching between about 5 and 8 mg/100 ml, respectively (Table 3). This increase could be attributed to the dual effect of the lack of repression of salicylate synthesis in both SM3 and exochelin-deficient strains and the probable inhibition of salicylate kinase by PAS.
FIG. 3.
Influence of exogenous salicylate on the growth of S99 in the absence of PAS (○) and in the presence of 1 (□) or 5 (▵) μg of PAS per ml under iron-deficient (———) and iron-sufficient (–––) conditions.
The increased sensitivity of the salicylate auxotroph S99 to PAS also suggests that PAS is an antagonist of salicylic acid. With little salicylate being available in this mutant and with PAS gaining access to the cell via an uptake system separate from that of salicylate (3), PAS was clearly able to exert its inhibitory action without competition from salicylate. Even when an equal amount of salicylate was added with PAS in the medium, no growth took place (Table 3). However, when the concentration of PAS was decreased to 1 μg/ml, growth could be restored partially by adding salicylate at 100 μg/ml, but only under iron-sufficient conditions (Fig. 3). These results are explainable by salicylate and PAS having separate uptake systems and PAS being able to outcompete salicylate in whatever function salicylate normally fulfills. The addition of mycobactin to a previous salicylate auxotroph treated with PAS did not restore its growth (3), again indicating that salicylate has a role beyond mycobactin synthesis. As a mycobactin-requiring auxotroph of M. smegmatis also retained this high sensitivity to PAS, even in the presence of mycobactin (3), PAS could also act at additional sites other than the conversion of salicylate to mycobactin.
The much higher reported PAS sensitivity of pathogenic mycobacteria, where 1.0 μg/ml is typically sufficient to cause inhibition (43), might possibly be due to the lower concentrations of salicylate synthesized by them. M. bovis BCG grown under iron-deficient conditions accumulated only about 20% of the salicylate seen with M. smegmatis (33), but this could also have been due to increased uptake of PAS via the p-aminobenzoate permease (3). Either case would lead to a higher intracellular concentration of PAS and a higher PAS-salicylate ratio in the cells, which is clearly crucial for effective inhibition.
Expression of the 29-kDa envelope protein.
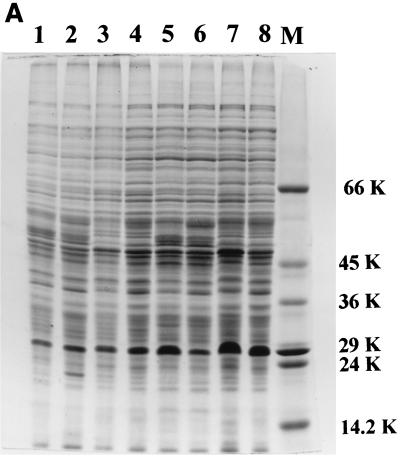
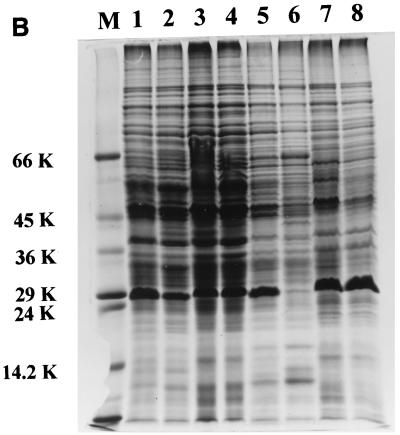
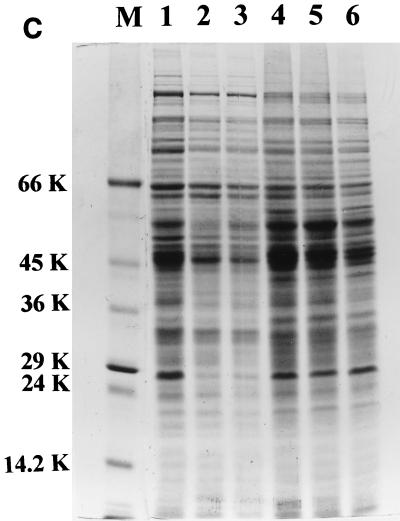
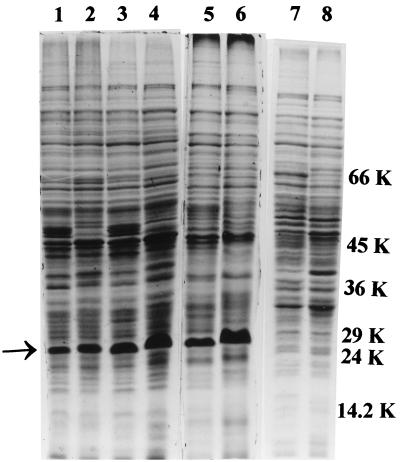
The uptake of ferrisiderophore complexes is mediated by specific proteins that occur on the outer membrane surface of microbial cells (23). In order to study the consequence of mutation on the expression of these proteins, the envelope protein profile was analyzed in all of the strains. The 29-kDa iron-regulated envelope protein described by Hall et al. (12) and considered by Dover and Ratledge (9) to be a possible exochelin-binding protein was found to be constitutively expressed in the NCIMB wild-type strain, irrespective of the iron status of the medium (see lanes 1 and 4 in Fig. 5C), in accordance with Dover and Ratledge (9). The N-terminal sequence of this protein shows 75% homology to that of a 29-kDa protein of M. tuberculosis which has been reported to show increased expression under iron-sufficient cells (4). Surprisingly, all of the strains derived from mc2155 showed increased expression of this 29-kDa protein under high-iron conditions, with the single exception of the S99 mutant strain, in which diminished synthesis was evident, irrespective of the iron status of the growth medium (lanes 7 and 8 of Fig. 4).
FIG. 5.
SDS-PAGE profile of cell envelope proteins of M. smegmatis. The effect of exogenous salicylic acid on the 29-kDa protein in the SM3 mutant (lanes 1 to 4) and the parent strain mc2155 (lanes 5 to 8) is shown. Lanes: 1 and 5, low iron; 2 and 6, low iron plus salicylic acid; 3 and 7, high iron; 4 and 8, high iron plus salicylic acid. Salicylic acid was added at 25 (A) and 100 (B) μg/ml. (C) Wild-type M. smegmatis NCIMB 8548 grown with different concentrations of salicylic acid under iron-deficient (lanes 1 to 3) and iron-sufficient (lanes 4 to 6) conditions. Lanes: 1 and 4, no salicylic acid; 2 and 5, 50 μg of salicylic acid/ml; 3 and 6, 100 μg of salicylic acid/ml. Lanes M contained marker proteins whose molecular masses (kilodaltons) are shown beside the panels.
FIG. 4.
SDS-PAGE profile of cell envelope proteins of M. smegmatis mutant SM3 (lanes 1 and 2), parent mc2155 (lanes 3 and 4), mutant mc21292 (lanes 5 and 6), and mutant S99 (lanes 7 and 8) under iron-deficient and -sufficient conditions, respectively. The arrow indicates the 29-kDa iron-regulated envelope protein. Molecular masses (kilodaltons) are shown on the right.
To investigate whether the decrease in the 29-kDa protein was the result of a pleiotropic effect of the mutation in strain S99, or the consequence of the high concentration (100 μg/ml) of exogenous salicylic acid required for its growth, strain SM3 and parental strain mc2155 were each grown in the presence of 25 and 100 μg of salicylic acid per ml under low- and high-iron conditions (Fig. 5A and B). There was a marked decrease of the 29-kDa protein in mc2155 grown with 25 μg of salicylic acid/ml and complete repression of the protein with 100 μg of salicylic acid/ml. However, the effect of salicylic acid was seen only under iron-deficient conditions, leaving the 29-kDa protein unaltered under iron-sufficient conditions. There was no significant change in this protein in SM3, the IdeR-deficient mutant. The results obtained with wild-type strain NCIMB 8548 (Fig. 5C) were similar to those obtained with mc2155.
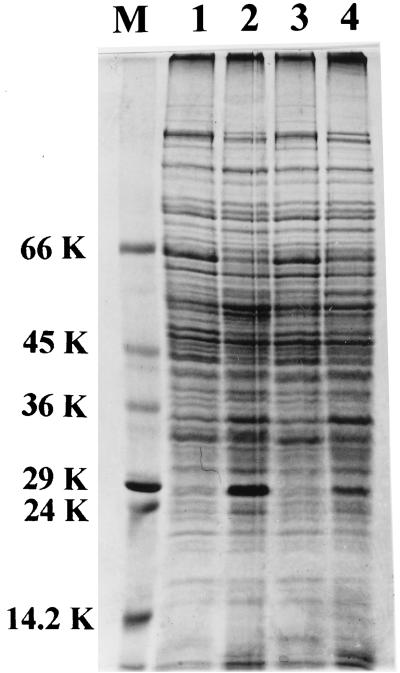
These results suggest a possible interaction between salicylic acid and the regulatory protein IdeR to bring about repression of the synthesis of the 29-kDa protein. Surprisingly, PAS, like salicylic acid, also repressed the formation of this protein in the wild-type mc2155 strain, and again, this effect was only seen under iron-deficient conditions (Fig. 6). The combination of PAS and salicylic acid led to a decrease in the 29-kDa protein even under iron-sufficient conditions.
FIG. 6.
SDS-PAGE profile of cell envelope proteins of parent strain mc2155 grown with 100 μg of PAS ml−1 under low- and high-iron conditions in the absence of salicylic acid (lanes 1 and 2) and in the presence of salicylic acid (lanes 3 and 4). Molecular mass marker (lane M) sizes are shown on the left in kilodaltons.
Conclusions.
Our results lead us to suggest that salicylate fulfills three distinct roles. First, salicylate acts as the precursor of mycobactin and carboxymycobactin, and PAS clearly inhibits the first reaction of this pathway, causing additional accumulation of salicylate. Second, salicylate is probably involved in the intracellular transfer of iron from the siderophores exochelin, carboxymycobactin, or mycobactin following the reduction of Fe(III) to Fe(II), which allows the iron to be desequestered (19). Ferrosalicylate could then transfer the iron into bacterioferritin, which acts as the cytoplasmic store of iron (28), and also into apoproteins and porphyrins. If this second role were proved to be correct, then this could explain the dependency of the salicylate auxotroph S99 on salicylate for utilization of the iron from ferrisiderophores (Table 2). Interestingly, when M. smegmatis was grown under iron-sufficient conditions, which is when the concentration of bacterioferritin would be optimal, either 2,3- or 2,4-dihydroxybenzoic acid could restore the growth of the S99 auxotroph. Thus, as these two phenolic acids would serve to complex Fe(II) similarly to salicylate, the loading of iron into bacterioferritin might be accomplished by alternatives to salicylate. Citrate, which also supported limited growth of the auxotroph under iron-sufficient conditions, may also be capable of acting as an Fe(II) transfer agent. This role of salicylate, as a means of moving and transferring Fe(II) from molecule to molecule, may not be entirely stringent, but as under iron-deficient conditions, the dihydroxybenzoic acids and citrate do not substitute for salicylate, this suggests that salicylate then fulfills a further and separate role under such conditions.
The third role for salicylate could be as a signal molecule to recognize the iron status within the cell. Salicylate would then be expected to act together with the IdeR protein that is involved in the regulation of a number of genes associated with iron metabolism (10). Support for this hypothesis comes from the expression of the 29-kDa envelope protein. Antibodies against this protein prevented iron uptake into whole cells in the presence of ferriexochelin (12), suggesting that it is involved in iron uptake as a putative exochelin receptor (9). The N terminus of this protein is distinctive (24) and shows 75 to 90% homology to several proteins: N termini of a DNA-binding protein (HupB) (6), 28- and 29-kDa proteins from M. tuberculosis (4), a histone-like DNA-binding protein from M. smegmatis (17), and a 40-kDa outer membrane protein of M. smegmatis (22). The repression of the 29-kDa protein by salicylate suggests a commonality with salicylate binding to the repressors MarR and EmrR of E. coli and inhibition of their function (18, 38). As there are reports of mar-like systems in mycobacteria (20), salicylic acid could also modulate gene expression in mycobacteria, perhaps by interacting with IdeR, and thereby serve as an iron signal molecule.
Cloning and characterization of the putative salicylate synthetase gene(s) from M. tuberculosis are under way, and preliminary studies indicate that entD (6) could be involved in the formation of salicylate in mycobacteria. However, it is possible that both entD and trpE2 (mbtI) (6, 25) are involved in the formation of salicylate.
ACKNOWLEDGMENTS
We thank the University of Hull for a Sir Brynmor Jones Research studentship to T.A.
We thank Maureen Ewing for technical assistance and Neil Stoker and Tanya Parish for generous hospitality in the Stoker laboratory and assistance with the transposon mutagenesis system.
REFERENCES
- 1.Barclay R, Wheeler P R. Metabolism of mycobacteria in tissues. In: Ratledge C, Stanford J L, Grange J M, editors. The biology of the mycobacteria. Vol. 3. London, United Kingdom: Academic Press Ltd.; 1989. pp. 137–196. [Google Scholar]
- 2.Barclay R, Ratledge C. Iron-binding compounds of Mycobacterium avium, M. intracellulare, M. scrofulaceum, and mycobactin-dependent M. paratuberculosis and M. avium. J Bacteriol. 1983;153:1138–1146. doi: 10.1128/jb.153.3.1138-1146.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown K A, Ratledge C. The effect of p-aminosalicylic acid on iron transport and assimilation in mycobacteria. Biochim Biophys Acta. 1975;385:207–220. doi: 10.1016/0304-4165(75)90349-9. [DOI] [PubMed] [Google Scholar]
- 4.Calder K M, Horwitz M A. Identification of iron-regulated proteins of Mycobacterium tuberculosis and cloning of tandem genes encoding a low iron-induced protein and a metal transporting ATPase with similarities to two-component metal transport systems. Microb Pathog. 1998;24:133–143. doi: 10.1006/mpat.1997.9999. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S T, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Crossa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Voss J J, Rutter K, Schroeder B G, Barry C E., III Iron acquisition and metabolism by mycobacteria. J Bacteriol. 1999;181:4443–4451. doi: 10.1128/jb.181.15.4443-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dover L G, Ratledge C. Identification of 29 kDa protein in the envelope of Mycobacterium smegmatis as a putative ferri-exochelin receptor. Microbiology. 1996;142:1521–1530. doi: 10.1099/13500872-142-6-1521. [DOI] [PubMed] [Google Scholar]
- 10.Dussurget O, Rodriguez M, Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- 11.Fiss E H, Yu S, Jacobs W R., Jr Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol Microbiol. 1994;14:557–569. doi: 10.1111/j.1365-2958.1994.tb02189.x. [DOI] [PubMed] [Google Scholar]
- 12.Hall R M, Sritharan M, Messenger A J M, Ratledge C. Iron transport in Mycobacterium smegmatis: occurrence of iron-regulated envelope proteins as potential receptors for iron uptake. J Gen Microbiol. 1987;133:2107–2114. doi: 10.1099/00221287-133-8-2107. [DOI] [PubMed] [Google Scholar]
- 13.Holland K T, Ratledge C. A procedure for selecting and isolating specific auxotrophic mutants of Mycobacterium smegmatis. J Gen Microbiol. 1971;66:115–118. doi: 10.1099/00221287-66-1-115. [DOI] [PubMed] [Google Scholar]
- 14.Klessig D F, Malamy J. The salicylic acid signal in plants. Plant Mol Biol. 1994;26:1439–1458. doi: 10.1007/BF00016484. [DOI] [PubMed] [Google Scholar]
- 15.Kopp E, Ghosh S. Inhibition of NF-κB by sodium salicylate and aspirin. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lee B H, Murugasu-Oei B, Dick T. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol Gen Genet. 1998;260:475–479. doi: 10.1007/s004380050919. [DOI] [PubMed] [Google Scholar]
- 18.Martin R G, Rosner J L. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCready K A, Ratledge C. Ferrimycobactin reductase activity from Mycobacterium smegmatis. J Gen Microbiol. 1979;113:67–72. [Google Scholar]
- 20.McDermott P F, White D G, Podglajen I, Lekshun M N A, Levy S B. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J Bacteriol. 1998;180:2995–2998. doi: 10.1128/jb.180.11.2995-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer J M, Azelvandre P, Georges C. Iron metabolism in Pseudomonas: salicylic acid, a siderophore of Pseudomonas fluorescens CHA0. Biofactors. 1992;4:23–27. [PubMed] [Google Scholar]
- 22.Mukhopadhyay S, Basu D, Chakrabarti P. Characterization of a porin from Mycobacterium smegmatis. J Bacteriol. 1997;179:6205–6207. doi: 10.1128/jb.179.19.6205-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neilands J B. Microbial envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- 24.Nixon G. Studies in the iron metabolism of mycobacteria. Ph.D. thesis. Hull, United Kingdom: University of Hull; 1999. [Google Scholar]
- 25.Quadri L E, Sello J, Keating T A, Weinreb P H, Walsh C T. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 26.Ratledge C. The biosynthesis of salicylic acid in Mycobacterium smegmatis via the shikimic acid pathway. Biochim Biophys Acta. 1969;192:148–150. doi: 10.1016/0304-4165(69)90023-3. [DOI] [PubMed] [Google Scholar]
- 27.Ratledge C. Nutrition, growth and metabolism. In: Ratledge C, Stanford J L, editors. Biology of the mycobacteria. Vol. 1. London, United Kingdom: Academic Press Ltd.; 1982. pp. 186–212. [Google Scholar]
- 28.Ratledge C. Iron metabolism. In: Ratledge C, Dale J, editors. Mycobacteria: molecular biology and virulence. Oxford, United Kingdom: Blackwell Science Publishers Ltd.; 1999. pp. 260–286. [Google Scholar]
- 29.Ratledge C, Brown K A. Inhibition of mycobactin formation in Mycobacterium smegmatis by p-aminosalicylate. A new proposal for the mode of action of p-aminosalicylate. Am Rev Respir Dis. 1972;106:774–776. doi: 10.1164/arrd.1972.106.5.774. [DOI] [PubMed] [Google Scholar]
- 30.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 31.Ratledge C, Hall M J. Uptake of salicylic acid into mycobactin S by growing cells of Mycobacterium smegmatis. FEBS Lett. 1970;10:309–312. doi: 10.1016/0014-5793(70)80460-4. [DOI] [PubMed] [Google Scholar]
- 32.Ratledge C, Hall M J. Isolation and properties of auxotrophic mutants of Mycobacterium smegmatis requiring either salicylic acid or mycobactin. J Gen Microbiol. 1972;72:143–150. doi: 10.1099/00221287-72-1-143. [DOI] [PubMed] [Google Scholar]
- 33.Ratledge C, Winder F G. The accumulation of salicylic acid by mycobacteria during growth on iron-deficient medium. Biochem J. 1962;84:501–506. doi: 10.1042/bj0840501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratledge C, Macham L P, Brown K A, Marshall B J. Iron transport in Mycobacterium smegmatis: a restricted role for salicylic acid in the extracellular environment. Biochim Biophys Acta. 1974;372:39–51. doi: 10.1016/0304-4165(74)90071-3. [DOI] [PubMed] [Google Scholar]
- 35.Saxena B, Modi M, Modi V V. Isolation and characterization of siderophores from Azospirillum lipoferum D-2. J Gen Microbiol. 1986;132:2219–2224. [Google Scholar]
- 36.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterisation of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 37.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 38.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: proto-typic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 39.Van Vliet A H M, Wooldridge K G, Ketley J M. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visca P, Ciervo A, Sanfilippo V, Orsi N. Iron-regulated salicylate synthesis by Pseudomonas spp. J Gen Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]
- 41.Weismann G. Aspirin. Sci Am. 1991;264:58–64. [Google Scholar]
- 42.Wheeler P R, Ratledge C. Metabolism of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: American Society for Microbiology; 1994. pp. 353–385. [Google Scholar]
- 43.Winder F G. The antibacterial action of streptomycin, isoniazid and PAS. In: Barry V C, editor. Chemotherapy of tuberculosis. London, United Kingdom: Butterworth & Co. Ltd.; 1964. pp. 111–149. [Google Scholar]
- 44.Wong D K, Lee B-Y, Horwitz M A, Gibson B W. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect Immun. 1999;67:327–336. doi: 10.1128/iai.67.1.327-336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]